Abstract
Localization of uranium within cells is mandatory for the comprehension of its cellular mechanism of toxicity. Secondary Ion Mass Spectrometry (SIMS) has recently shown its interest to detect and localize uranium at very low levels within the cells. This technique requires a specific sample preparation similar to the one used for Transmission Electronic Microscopy, achieved by implementing different chemical treatments to preserve as much as possible the living configuration uranium distribution into the observed sample. This study aims to compare the bioaccumulation sites of uranium within liver or kidney cells after chemical fixation and cryomethods preparations of the samples: SIMS analysis of theses samples show the localization of uranium soluble forms in the cell cytoplasm and nucleus with a more homogenous distribution when using cryopreparation probably due to the diffusible portion of uranium inside the cytoplasm.
Keywords: cell culture, ion imaging, sample preparations, uranium
1. INTRODUCTION
For the last few decades microscopy techniques have been increasingly used in life sciences to study biological samples. Secondary Ion Mass Spectrometry (SIMS) technique allows investigating the elemental and isotopic distributions at cell level. This technique requires a specific preparation of the biological samples. The morphological and chemical preservations of the sample are of major importance as distribution of elements observed by SIMS has to be the closest to the native state.
Several reviews have recently presented the state of the art of preparation techniques of biological samples essentially for studies by Transmission Electron Microscope (TEM) (Bell & Safiejko‐Mroczka, 1997; Hurbain & Sachse, 2011; Lešer, Drobne, Pipan, Milani, & Tatti, 2009; McDonald, 2014; Mielanczyk, Matysiak, Michalski, Buldak, & Wojnicz, 2014; Weston, Armer, & Collinson, 2010).The chemical preparation procedure is the most currently used. Aldehydes at room temperature are used to stabilize the sample macromolecular structures, and then the sample is dehydrated with organic solvents prior to infiltration and embedded in a plastic resin. Osmium tetroxide and uranyl acetate can be used as a second fixation step for lipids and some proteins. They also act as an electron stain. This conventional sample preparation can introduce different artifacts modifying the cell structure and the chemical composition: structural reorganization, aggregation proteins, loss of lipids, light ions and small molecules, and also chemical modifications. Cryopreparation methods have also been developed to preserve biological fine structures. This is a quick freezing technic to vitrify the cells water content (Dubochet, 2007) that immobilizes the sample chemical composition without changing the cell morphology. Other freezing techniques are used in connection with the volume of the sample (Hurbain & Sachse, 2011; McDonald, 2014; Mielanczyk et al., 2014; Vanhecke, Graber, & Studer, 2008; Weston et al., 2010). Then the sample is dehydrated at low temperature and embedded in a resin at room temperature (Bell & Safiejko‐Mroczka, 1997; Lešer et al., 2009; Weston et al., 2010). Another method consists of keeping the sample frozen and hydrated, and cutting it into thin sections before being observed under the cryomicroscope (CEMOVIS) (Al‐Amoudi et al., 2004; Dubochet, 2007; Dubochet, Al‐Amoudi, Bouchet‐Marquis, Eltsov, & Zuber, 2009; Dubochet & Sartori Blanc, 2001; Hurbain & Sachse, 2011; Mielanczyk et al., 2014; Milne et al., 2013; Vanhecke et al., 2008). To our knowledge, very few studies on SIMS imaging presents a comparative study of cell structure using two types of samples preparations (Clerc, Fourré, & Fragu, 1997; Guerquin‐Kern, Wu, Quintana, & Croisy, 2005). It is then poorly known if the sample preparation techniques can influence the detection and micro‐localization of uranium a known cytotoxic substance.
This radioelement is also a heavy metal that humans can be exposed to as a result of its natural presence or human activities such as mill tailings, nuclear industry or military use. It can thus be found in quantities that vary between areas by a factor of more than a thousand (Wrenn et al., 1985). Kidneys and more precisely the proximal tubular cells are one of the main accumulation sites and toxicity targets of soluble uranium compounds (Guéguen et al., 2017; Vicente‐Vicente et al., 2010). It has been previously shown by us and others that at level below 1 µg/g of kidney for in vivo experiments and below 100 µM for in vitro experiments, close to realistic exposure levels, uranium did not induced direct deleterious toxic effects but induced biological pathways related to cell and tissue defense (Guéguen et al., 2015; Leggett et al., 1989; Prat et al., 2010). Recent in vivo experimental studies have confirmed that uranium distribution is highly heterogeneous within the kidney and can reach concentration tenfold higher than the mean renal tissue level (Homma‐Takeda et al., 2015).
The uranium penetration into different cell types, as well as its cytotoxicity, have been first studied at precipitate form by TEM in vitro (Carrière et al., 2005, 2008; Ghadially, Lalonde, & Yang‐Steppuhn, 1982; Mirto et al., 1999). Recently we have shown, at lower noncytotoxic concentration, by SIMS that soluble or precipitate forms of depleted uranium inside the cells play a role in biological cell response (Guéguen et al., 2015; Rouas et al., 2010). Moreover, we have displayed for the first time the presence of soluble uranium within the nuclei using the SIMS technique in different cell cultures (kidney, hepatocyte and neuron) after 15 min to 24 hr of exposure to concentrations lower than 100 µM. Until now chemical sample preparation has been used for SIMS analyses.
To continue our study of the distribution of soluble uranium in cells cryo or chemical fixation preparation processes have been compared. Two cell models were used, Human Hepatoma cells and Human Kidney cells (HepG2 and HK‐2), two target organs for uranium (Alexandra, Miller, Smith, & Page, 2004; Bao et al., 2013; Dedieu et al., 2009; Jalal Pourahmad, Tanbakosazan, Ghalandari, Ettehadi, & Dahaghin, 2010; Prat et al., 2010) after, respectively, 30 min and 2 hr exposure to a 50 µM uranium solution and SIMS analyses were performed. The concentration and duration of exposure were chosen according to our previous studies showing soluble uranium inside the cells without uranium precipitates (Guéguen et al., 2015).
2. MATERIALS AND METHODS
2.1. Materials
A solution of uranyl nitrate hexahydrate (UO2(NO3)2.6H2O) (AREVA‐COGEMA, France), certified for its isotopic composition (238 U = 99.74%, 235 U = 0.255%, and 234 U = 0.0055% (AREVA‐COGEMA), was prepared to obtain depleted uranium (DU) concentration of 10 mM by dissolving the powder in 100 mM sodium bicarconate (Na+ ).
Human Hepatome cell line (HepG2) and Human kidney line (HK‐2) cells were obtained from ATCC (Molsheim, France) Roswell Park Memorial Institute medium (RPMI 1640, ref. 21875‐034); DMEM F12 (ref 12634‐010), penicillin/streptomycin 10,000 U/ml (PS, ref. 15140), fetal bovine serum (FBS, ref. 10270‐106), and L‐glutamine (ref. 25030‐024) were purchased from Life Technologies (Cergy‐Pontoise, France).
For the cryopreparation, the LEICA CPC (Cryo‐Plonge Chamber) and LEICA AFS (Automate Freezing Substitution) have been used. The samples are cut using a Leica UC6 ultra microtome mounted with a DIATOME histo diamond knife (DITAOME diamond knives, LFG Distribution, France) to obtain 500 nm thickness sections.
The embedded sample is prepared with an epoxy resin, consisting of a mixture; of EMbed‐812, DDS (Dodecenyl Succinic Anhydride), NMA (Methyl‐5‐Norbornene‐2,3‐Dicarboxylic Anhydride), DMP‐30 (2,4,6‐Tri(dimethylaminomethyl) phenol) (Kit EMbed 812 réf: RT14120, EMS, Hatfield). In ours preparations, DMP‐30 were replaced by BDMA (N‐Benzyl‐N, N‐Dimethylamine), BDMA proves to be a much better choice for an accelerator than DMP‐30. It is much less viscous, has a longer shelf life, and offers better penetration of the tissue (technical tip EMS, Hatfield).
To reach the optimal experimental conditions, several supports have been tested (sapphire, termanox, aclar®). By comparing adhesion, proliferation, and cell morphology, we have selected the use of Aclar® (embedding film 7.8 mil thickness, ref 50425 EMS, Hatfield) discs, allowing better performance. These discs were made using a cutting mat and a calibrated punch (punch 5 mm diameter) allowing insertion into a silicone mold Pelco 107 (Tedd Pella, P.O. Box 492477, Redding, CA 96049‐2477) for final embedding.
2.2. Cell cultures
HepG2 cells were grown in a monolayer culture in RPMI supplemented with 10% FBS, 1% PS in an incubator with humidified atmosphere (i.e., 37 °C, 5% CO2) to a confluence of 80%. HK‐2 cells were grown in a monolayer culture in DMEM F12 supplemented with 10% SVF, 1% antibio, and L‐Glutamine 2 mM. These cells were grown on Aclar® discs, cut out of an Aclar sheet with a calibrated circular punch. First these discs have been degreased in absolute ethanol following by sterilization (Kingsley & Cole, 1988). Then the discs were laid into 96‐well culture plates containing the culture medium.
2.3. Uranium exposure
The DU stock solution (10 mM, PH 2.8) was prepared immediately before use by dissolving 0.5% (w/v) of uranyl nitrate in Na+ solution (100 mM). Its exact concentration was checked by inductively coupled plasma‐mass spectrometry. DU solutions used for experiments were prepared by diluting stock solution in cell culture media. HepG2 and HK‐2 cells were incubated with 50 µM DU for 30 min and 2 hr, respectively.
Control cells were treated similarly without addition of DU in the culture medium.
2.4. Preparations of the biological samples for SIMS analysis
2.4.1. Conventional preparation
Following the different DU‐exposure phases (30 min or 2 hr), the culture media was removed and the cells underwent a standard chemical fixation procedure. Cells on Aclar® discs were fixed with a solution containing 2.5% of glutaraldehyde for 1 hr at room temperature, then dehydrated in ethanol baths, and infiltration with an ethanol/Epon mixture, and finally embedded in pure Epon‐type resin 48 hr at 60 °C. Osmium tetroxide has not been used to avoid the possible interferences at mass 238; moreover, it increases the peak noise of a decade. Uranium being our element of interest, uranyl acetate has also been excluded.
2.4.2. Cryopreparations
2.4.2.1 Plunge freezing
The Aclar® disc (embedding film 7.8 mil thickness, ref 50425 EMS, Hatfield) on which the monolayer of cells has grown underwent a very rapid immersion into cryogen with liquid propane at 77 K in the Leica CPC. The cooling rate was equal to at least 104 K/s. This sample was embedded in a thin layer of vitrified water. When diving, the disc transits from CPC to AFS in a liquid nitrogen environment so as not to break the thermal effect. Once in AFS, the temperature rise for the impregnation is controlled by a temperature probe and slow.
2.4.2.2. Freeze substitution and embedding
After vitrification the sample was transferred without breaking cold and dehydrated at 183 K using an organic solvent such acetone, in the Leica AFS. The sample was then held at 183 K for 10 hr, then ramp back up to 128 K for 2 hr, and finally reached at room temperature. Between the steps, the temperature was raised by 1 K/min. Then the sample was embedded into Epon resin (Matsko & Mueller, 2005).
2.4.3. Resin section
The embedded sample were cut in serial thin sections (0.5 µm) and laid on polished ultrapure silicon holders for SIMS analysis (to avoid relief effects and minimize charge effects) or glass slides for histological controls using an optical microscope.
2.5. SIMS microscopy
SIMS microscopy analyzes the elemental and isotopic composition of a solid surface through an ion beam coupled with a mass spectrometer. The principle of this technique has been previously described by Rouas et al. (2010) and Tessier et al. (2012). The SIMS analyses were performed with a CAMECA IMS 4F E7 instrument. For this study, beam bombardment was used to enhance the ionization field composed of electropositive species such as uranium. The primary beam is focused into a small shaft (around 0.5 µm), which scans the sample surface. The secondary ions collected after mass filtering can be measured with an electron multiplier and also sequentially converted into an image. Mass resolution can reach M/ΔM = 10,000, where M is a molecular mass of the detected ion. This apparatus can discriminate the molecular mass of two very close elements, separated by a distance of ΔM = 10−4 M. Lateral resolution is essentially dependent on the size of the probe and the number of image points (pixels). With O+ primary ions, the probe has routinely a 0.5 µm diameter. As a result, any particle that is below this size would not be exploitable in imaging. For each area analyzed, mass spectra at around the 238 uranium isotope mass and ionic images were acquired. 40Ca+, 23Na+, and 39K+ images show the histological structure of the cells and 238U+ images the uranium micro‐distribution within cellular compartments.
3. RESULTS
3.1. Control condition for uranium detection by SIMS
Under these SIMS experimental conditions, the mass spectra of control cells (HK‐2 and HepG2) and for both types of preparations, which were not exposed to uranium, have been recorded around on 238U+ mass at low mass resolution (M/ΔM = 300). The orange zone do not show the presence of a significant peak at mass 238. The blue peaks represent the intensity pierced by the device, the dashed blue line indicates the background noise of the analysis (about 1 cps/s), and it is voluntarily placed in this zone because below we are in limit of detection for one element (Figure 1).
Figure 1.
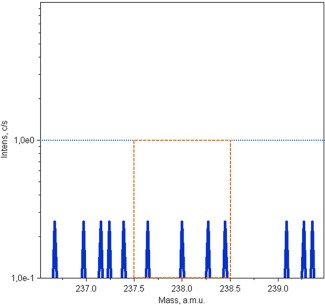
238U+ mass spectrum of control cells recorded at low mass resolution (M/ΔM = 300). The area delimited by the orange dotted line zone show the absence of a significant peak at mass 238. The blue peaks represent the intensity pierced by the device. The dashed blue line delimitates the background noise of the analysis (around 1 cps/s). Then the peaks are under the limit of detection for one element and as expected no uranium is detect on these control samples [Color figure can be viewed at http://wileyonlinelibrary.com]
These results confirm that naturally present uranium is not detected, and no polyatomic ions are superimposed on the element of interest at a low mass resolution. In this case, working at low resolution is necessary because at high mass resolution, secondary ion transmission is improved and, therefore, the detection limits are lower.
3.2. 238U+ localization in HepG2 and HK‐2 cells
The SIMS images (40Ca+, 23Na+,39K+, 238U+ and superposed 238U+/23Na+) and mass spectra of HepG2 and HK‐2 cells, which have been exposed, respectively, for 30 min and 2 hr to a low concentration of uranium (50 µM), and histological view are presented in Figures 2 and 3 for chemical preparation, and in Figures 4 and 5 for cryopreparation. In SIMS ionic images, hot colors show the higher concentrations of the element except for the superposed images where uranium is in red and sodium in green.
Figure 2.
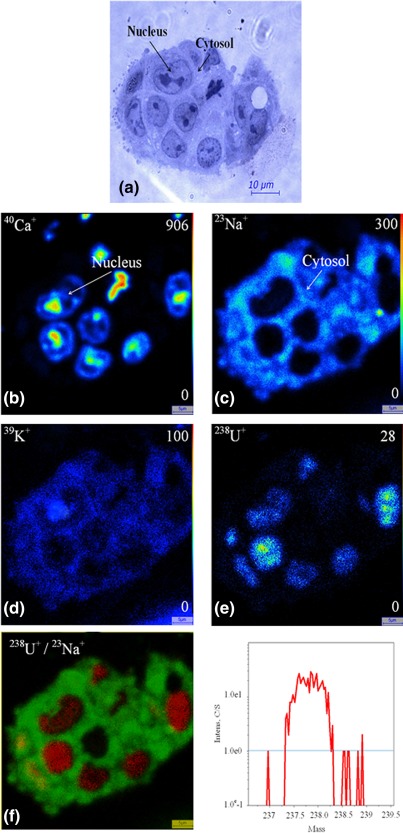
Uranium distribution inside the HepG2 cells form after exposure to 50 μM of uranyl nitrate for 30 min, chemical preparation. (a) Histological view, SIMS ionic images (b) 40Ca+, (c) 23Na+, (d) 39K+, (e) 238U+, (f) superposition of 238U+(red)/23Na+(green), (g) 238U+ mass spectrum. Field 50 × 50 μm2 [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
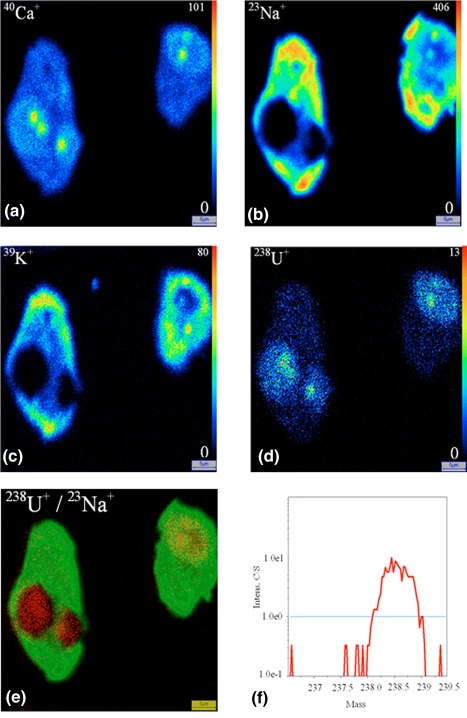
Uranium distribution inside the HK‐2 cells after exposure to 50 μM of uranyl nitrate for 2 hr, chemical preparation. SIMS ionic images (a) 40Ca+, (b) 23Na+, (c) 39K+, (d) 238U+, (e) superposition of 238U+(red)/23Na+(green), (f) 238U+ mass spectrum. Field 50 × 50 μm2 [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 4.
Uranium distribution inside the HepG2 cells exposure to 50 μM of uranyl nitrate for 30 min, cryopreparation, SIMS ionic images (a) 40Ca+, (b) 23Na+, (c) 39K+, (d) 238U+, (e) superposition of 238U+(red)/23Na+(green), (f) 238U+ mass spectrum. Field 50 × 50 μm2 [Color figure can be viewed at http://wileyonlinelibrary.com]
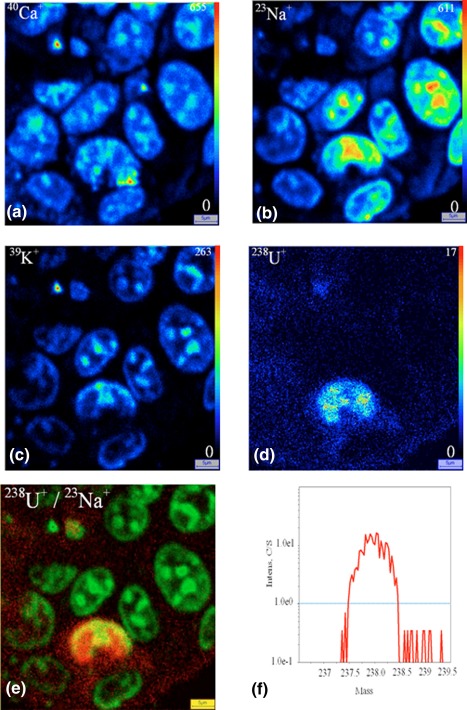
Figure 5.
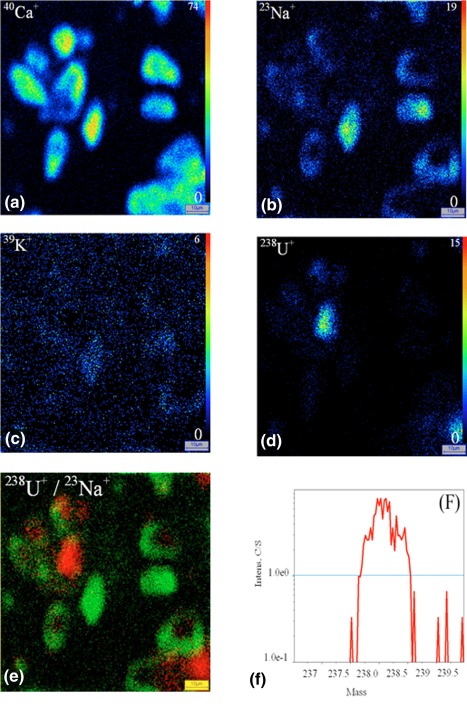
Uranium distribution inside the HK‐2 cells after exposure to 50 μM of uranyl nitrate for 2 hours. cryopreparation. SIMS ionic images (a) 40Ca+, (b) 23Na+, (c) 39K+, (d) 238U+, (e) superposition of 238U+(red)/23Na+(green), (f) 238U+ mass spectrum. Field 100 × 100 μm2 [Color figure can be viewed at http://wileyonlinelibrary.com]
The 40Ca+, 23Na+, and 39K+ images allow showing the different cellular compartments (cytoplasm and nucleus) (Figure 2). 40Ca+ is present in all the cells with a higher accumulation in the nucleus, especially within the chromatin for chemical preparation; this element is relatively homogeneously distributed in the cytoplasm and nucleus for cryopreparation. The 23Na+ and 39K+ elements are mainly localized in the cytoplasm for chemical preparation and in the nucleus for cryopreparation.
For the two cellular types, the 238U+ images display a radionuclide accumulation in the cell, whatever the sample preparation technique: cytoplasm and nucleus. The distribution of uranium inside the cells appears more homogenous when using cryomethods compared with chemical preparation method. The conventional chemical preparation shows a higher localization of soluble uranium inside the nuclei both for HepG2 cells and HK‐2 cells.
4. DISCUSSION
In the literature, speciation studies have shown that actinides, especially uranium, could bind to several biological ligands: proteins, lipids or amino acids and DNA (Ansoborlo et al., 2006; Bresson, Ansoborlo, & Vidaud, 2011; Carrière et al., 2004; Dedieu et al., 2009; Hartsock, Cohen, & Segal, 2007; Huynh, Bourgeois, Basset, Vidaud, & Hagège, 2015; Safi et al., 2013). Moreover, previous studies have displayed, in cytotoxic uranium conditions (U > 300 µM), uranyl phosphate precipitates in the cytoplasm of cells (Carrière et al., 2005, 2008; Ghadially et al., 1982; Mirto et al., 1999). These precipitated forms have been detected by TEM and more recently by our group using the SIMS technique (Rouas et al., 2010) at 100 µM after 24 hr of exposure. By contrast, with noncytotoxic uranium concentrations from 10 to 100 µM, we have shown for the first time the presence of the soluble form of uranium inside cells mainly in the nuclei (Guéguen et al., 2015; Rouas et al., 2010). These previous in vitro experiments have been achieved using samples chemically prepared for SIMS or MET.
The use of cryofixation methods have been discussed for ultrastructure analysis or detection of other toxic element (Ortega et al., 2009; Weston et al., 2010) but not yet for uranium detection. In this work we have compared for the first time the uranium detection with SIMS using chemical and cryopreparations on two human cells lines (HepG2 and HK‐2) exposed to uranium (50 µM) for 30 min and 2 hr, respectively. At these concentrations and duration of exposure, we showed that uranium precipitates are not detected with SIMS. The chemical preparation consists of stabilizing highly hydrated biological structures by creating new chemical bonds. Long insoluble chains are produce by bridging organic molecules and chemical agent to permit dehydration. The cryopreparation permits the vitrification of the aqueous phase lowering very strongly and quickly the temperature of the sample. It is a physical action which essentially involves a temperature variation.
SIMS analyses show the presence of soluble uranium inside the cell nuclei no matter the sample preparation technique (Figure 2, 3, 4, 5) and confirms the previous data related to the uranium distribution within the nuclei (Guéguen et al., 2015; Rouas et al., 2010). Nevertheless uranium content in the cytoplasm seems to be lower when using the chemical preparation (Figures 2 and 3) compared with the cryopreparation (Figures 4 and 5). This radioelement is considered as a heavy metal which is probably strongly chemically fixed in the nucleus in contrast with highly diffusible element (Na, Ca, K…). The soluble form of uranium may remain diffusible in cell but can also interact with cell resulting either in precipitation or in binding with biological element, and finally fixed inside a particular cell compartment such as the nuclei. In the same human kidney cell model (HK‐2), previous work has shown that uranium can be bond to 64 proteins displaying varied function (Dedieu et al., 2009). Grovenor et al. (2006) have shown that the distribution of the sequestered metallic species is poorly disturbed. It can be hypothesized a decrease of uranium content from the cytoplasm during the different steps of the chemical preparation. This observation could be documented by a lesser binding to the macromolecules within the cytoplasm (Bresson et al., 2011; Frelon, Mounicou, Lobinski, Gilbin, & Simon, 2013; Ortega et al., 2009; Perrin, Carmona, Roudeau, & Ortega, 2015). In addition, we have previously shown from in vivo experiments i using SIMS microscopy that uranium is heterogeneously distributed in the nephron and localized mainly in cell nuclei of proximal convoluted tubules (Poisson et al., 2014; Tessier et al., 2012). Similar uranium distribution has been observed using different imaging technic in case of acute renal contamination in rats (Homma‐Takeda et al., 2015). The lower resolution of particle induced X‐ray emission (PIXE) images in this study, compared to the SIMS microscopy did not allowed observing n heterogeneous subcellular distribution of uranium. Nevertheless, the combination of PIXE, scanning transmission ion microscopy, and backscattering spectrometry on an in vitro model exposed to cobalt allowed the detection of subcellular heterogeneous distribution and quantification of this element with a higher content in the nucleus than in the cytosol (Ortega et al., 2009).
In our work, the 40Ca+, 23Na+, and 39K+ SIMS images have been used to trace the main cellular compartments, cytoplasm and nucleus. The biodistribution of light elements and highly diffusible ions can be changed at the sample preparation step (Figures 2, 3, 4, 5).
In our study, we observed that the sodium and potassium microdistributions are different depending on the preparation process. In chemical preparation, 23Na+ and 39K+ images show the same localization mostly in the cytoplasm (Figures 2 and 3). In cryoprepared sample, these elements are mainly localized in the nucleus (Figures 4 and 5). The 40Ca+ distribution is comparable for the two sample preparations; the Figures 2, 3, 4, 5 display localization both in the nucleus and the cytoplasm. In previous studies, some authors have analyzed the redistribution of these elements within the cell to carry out the 39K+/40Ca+ or 39K+/23Na+ abundance ratios (Arlinghaus et al., 2006; Chandra, 2008; Grignon, Halpern, Jeusset, Briançon, & Fragu, 1997). The authors have shown in the case of cryopreparation that the 39K+/23Na+ cellular ratio matches with that of the living cell; the intracellular concentration of 39K+ is greater than that 23Na+, and inversely in the culture medium. The SIMS technique allows semi‐quantitative analysis for our biological matrices. In addition, being unable to perfectly control the sputtering efficiency of the sample and its ionization rate in the matrix, quantification is not possible. With the chemical method the cellular ratio 39K+/23Na+ is closer to the cell culture medium. Both diffusible elements and molecules are prone to artifactinduced‐relocation at subcellular scale if the sample preparation is compromised. Therefore, it is recognized that sample preparation technic is critical for the analysis of the distribution of elements by SIMS and needs specific and devoted experiments especially for diffusible elements like Ca, Na and K.
The direct comparison of chemical or cryogenic preparation using two different cell types prior to SIMS analysis confirmed the presence of uranium within the cell nuclei and showed that this distribution is poorly altered with the sample preparation process. The cryogenic method is more tricky and longer to implement successfully. Whatever the cell type (renal or hepatic) and preparation used (chemical or cryogenic), exposure to low concentration of uranium (U < 100 µM) leads to a distribution of soluble form in the nuclei, and also in the cytoplasm. The choice of preparation will depend on the analyzed element. Other recent cryogenic fixation methods, such as high pressure freezing or slam‐freezing (Mielanczyk et al., 2014) would be even closer to the native state. This is the technique of choice for the study of very labile elements. Only the complexity and sometimes the cost, might limit its use. Finally, the authors reach to the conclusion that the study of the cellular distribution of toxic element such as uranium with SIMS technic is nicely helpful for the understanding of their biological effects particularly in the low‐dose range (Guéguen et al., 2015).
Acknowledgments
This work was supported by the Institute for Radioprotection and Nuclear Safety (IRSN). This study was part of the ENVIRHOM research program of the IRSN.
Suhard D, Tessier C, Manens L, et al. Intracellular uranium distribution: Comparison of cryogenic fixation versus chemical fixation methods for SIMS analysis. Microsc Res Tech. 2018;81:855–864. 10.1002/jemt.23047
Funding information Institute for Radioprotection and Nuclear Safety (IRSN)
References
- Al‐Amoudi, A. , Chang, J. J. , Leforestier, A. , McDowall, A. , Salamin, L. M. , Norlen, L. P. , … Dubochet, J. (2004). Cryo‐electron microscopy of vitreous sections. The Embo Journal, 23(18), 3583–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandra, C. , Miller, K. B. , Smith, J. , & Page, N. (2004). Effect of the militarily‐relevant heavy metals, depleted uranium and heavy metal tungsten‐alloyon gene expression in human liver carcinoma cells(HepG2). Molecular and Cellular Biochemistry, 255(1/2), 247–256. [DOI] [PubMed] [Google Scholar]
- Ansoborlo, E. , Prat, O. , Moisy, P. , Den Auwer, C. , Guilbaud, P. , Carriere, M. , … Moulin, V. (2006). Actinide speciation in relation to biological processes. Biochimie, 88(11), 1605–1618. [DOI] [PubMed] [Google Scholar]
- Arlinghaus, H. F. , Kriegeskotte, C. , Fartmann, M. , Wittig, A. , Sauerwein, W. , & Lipinsky, D. (2006). Mass spectrometric characterization of elements and molecules in cell cultures and tissues. Applied Surface Science, 252(19), 6941–6948. [Google Scholar]
- Bao, Y. , Wang, D. , Li, Z. , Hu, Y. , Xu, A. , Wang, Q. , … Chen, H. (2013). Efficacy of a novel chelator BPCBG for removing uranium and protecting against uranium‐induced renal cell damage in rats and HK‐2 cells. Toxicology and Applied Pharmacology, 269(1), 17–24. [DOI] [PubMed] [Google Scholar]
- Bell, P. B., Jr. , & Safiejko‐Mroczka, B. (1997). Preparing whole mounts of biological specimens for imaging macromolecular structures by light and electron microscopy. International Journal of Imaging Systems and Technology, 8(3), 225–239. [Google Scholar]
- Bresson, C. , Ansoborlo, E. , & Vidaud, C. (2011). Radionuclide speciation: A key point in the field of nuclear toxicology studies. Journal of Analytical Atomic Spectrometry, 26(3), 593–601. [Google Scholar]
- Carrière, M. , Avoscan, L. , Collins, R. , Carrot, F. , Khodja, H. , Ansoborlo, E. , & Gouget, B. (2004). Influence of uranium speciation on Normal Rat Kidney (NRK‐52E) proximal cell cytotoxicity. Chemical Research in Toxicology, 17(3), 446–452. [DOI] [PubMed] [Google Scholar]
- Carrière, M. , Gouget, B. , Gallien, J. P. , Avoscan, L. , Gobin, R. , Verbavatz, J. M. , & Khodja, H. (2005). Cellular distribution of uranium after acute exposure of renal epithelial cells: SEM, TEM and nuclear microscopy analysis. Nuclear Instruments and Methods in Physics Research, Section B: Beam Interactions with Materials and Atoms, 231(1–4), 268–273. [Google Scholar]
- Carrière, M. , Proux, O. , Milgram, S. , Thiebault, C. , Avoscan, L. , Barre, N. , … Gouget, B. (2008). Transmission electron microscopic and X‐ray absorption fine structure spectroscopic investigation of U repartition and speciation after accumulation in renal cells. Journal of Biological Inorganic Chemistry, 13(5), 655–662. [DOI] [PubMed] [Google Scholar]
- Chandra, S. (2008). Challenges of biological sample preparation for SIMS imaging of elements and molecules at subcellular resolution. Applied Surface Science, 255(4), 1273–1284. [Google Scholar]
- Clerc, J. , Fourré, C. , & Fragu, P. (1997). Sims microscopy: Methodology, problems and perspectives in mapping drugs and nuclear medicine compounds. Cell Biology International, 21(10), 619–633. [DOI] [PubMed] [Google Scholar]
- Dedieu, A. , Berenguer, F. , Basset, C. , Prat, O. , Quemeneur, E. , Pible, O. , & Vidaud, C. (2009). Identification of uranyl binding proteins from human kidney‐2 cell extracts by immobilized uranyl affinity chromatography and mass spectrometry. Journal of Chromatography A, 1216(28), 5365–5376. [DOI] [PubMed] [Google Scholar]
- Dubochet, J. (2007). The physics of rapid cooling and its implications for cryoimmobilization of cells. Methods in Cell Biology, 79, 7–21. [DOI] [PubMed] [Google Scholar]
- Dubochet, J. , Al‐Amoudi, A. , Bouchet‐Marquis, C. , Eltsov, M. , & Zuber, B. (2009). CEMOVIS: Cryo‐electron microscopy of vitreous sections (pp. 259–289). Boca Raton, FL: CRC Press.
- Dubochet, J. , & Sartori Blanc, N. (2001). The cell in absence of aggregation artifacts. Micron (Oxford, England: 1993), 32(1), 91–99. [DOI] [PubMed] [Google Scholar]
- Frelon, S. , Mounicou, S. , Lobinski, R. , Gilbin, R. , & Simon, O. (2013). Subcellular fractionation and chemical speciation of uranium to elucidate its fate in gills and hepatopancreas of crayfish Procambarus clarkii. Chemosphere, 91(4), 481–490. [DOI] [PubMed] [Google Scholar]
- Ghadially, F. N. , Lalonde, J. M. A. , & Yang‐Steppuhn, S. (1982). Uraniosomes produced in cultured rabbit kidney cells by uranyl acetate. Virchows Archiv B Cell Pathology Including Molecular Pathology, 39(1), 21–30. [DOI] [PubMed] [Google Scholar]
- Grignon, N. , Halpern, S. , Jeusset, J. , Briançon, C. , & Fragu, P. (1997). Localization of chemical elements and isotopes in the leaf of soybean (Glycine max) by secondary ion mass spectrometry microscopy: Critical choice of sample preparation procedure. Journal of Microscopy, 186(1), 51–66. [Google Scholar]
- Grovenor, C. R. M. , Smart, K. E. , Kilburn, M. R. , Shore, B. , Dilworth, J. R. , Martin, B. , … Rickaby, R. E. M. (2006). Specimen preparation for NanoSIMS analysis of biological materials. Applied Surface Science, 252(19), 6917–6924. [Google Scholar]
- Guéguen, Y. , Suhard, D. , Poisson, C. , Manens, L. , Elie, C. , Landon, G. , … Tessier, C. (2015). Low‐concentration uranium enters the HepG2 cell nucleus rapidly and induces cell stress response. Toxicology In Vitro, 30(1), 552–560. [DOI] [PubMed] [Google Scholar]
- Guéguen, Y. , Roy, L. , Hornhardt, S. , Badie, C. , Hall, J. , Baatout, S. , Pernot, E. , Tomasek, L. , Laurent, O. , Ebrahimian, T. , Ibanez, C. , Grison, S. , Kabacik, S. , Laurier, D. , Gomolka, M. (2017). Biomarkers for Uranium Risk Assessment for the Development of the CURE (Concerted Uranium Research in Europe) Molecular Epidemiological Protocol. Radiat Res, 187(1), 107–127. [DOI] [PubMed] [Google Scholar]
- Guerquin‐Kern, J.‐L. , Wu, T.‐D. , Quintana, C. , & Croisy, A. (2005). Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry (SIMS microscopy). Biochimica et Biophysica Acta (BBA)‐General Subjects, 1724(3), 228–238. [DOI] [PubMed] [Google Scholar]
- Hartsock, W. J. , Cohen, J. D. , & Segal, D. J. (2007). Uranyl acetate as a direct inhibitor of DNA‐binding proteins. Chemical Research in Toxicology, 20(5), 784–789. [DOI] [PubMed] [Google Scholar]
- Homma‐Takeda, S. , Kitahara, K. , Suzuki, K. , Blyth, B. J. , Suya, N. , Konishi, T. , … Shimada, Y. (2015). Cellular localization of uranium in the renal proximal tubules during acute renal uranium toxicity. Journal of Applied Toxicology, 35(12), 1594–1600. [DOI] [PubMed] [Google Scholar]
- Hurbain, I. , & Sachse, M. (2011). The future is cold: Cryo‐preparation methods for transmission electron microscopy of cells. Biology of the Cell, 103(9), 405–420. [DOI] [PubMed] [Google Scholar]
- Huynh, T. N. S. , Bourgeois, D. , Basset, C. , Vidaud, C. , & Hagège, A. (2015). Assessment of CE‐ICP/MS hyphenation for the study of uranyl/protein interactions. Electrophoresis, 36(11–12), 1374–1382. [DOI] [PubMed] [Google Scholar]
- Jalal Pourahmad, F. S. , Tanbakosazan, F. , Ghalandari, R. , Ettehadi, H. A. , & Dahaghin, E. (2010). Protective effects of fungal stress cytotoxicity induced by depleted uranium in isolated rat hepatocytes. Human and Experimental Toxicology, 30(3), 173–181. [DOI] [PubMed] [Google Scholar]
- Kingsley, R. E. , & Cole, N. L. (1988). Preparation of cultured mammalian cells for transmission and scanning electron microscopy using Aclar film. Journal of Electron Microscopy Technique, 10(1), 77–85. [DOI] [PubMed] [Google Scholar]
- Lešer, V. , Drobne, D. , Pipan, Ž. , Milani, M. , & Tatti, F. (2009). Comparison of different preparation methods of biological samples for FIB milling and SEM investigation. Journal of Microscopy, 233(2), 309–319. [DOI] [PubMed] [Google Scholar]
- Matsko, N. , & Mueller, M. (2005). Epoxy resin as fixative during freeze‐substitution. Journal of Structural Biology, 152(2), 92–103. [DOI] [PubMed] [Google Scholar]
- McDonald, K. L. (2014). Out with the old and in with the new: Rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma, 251(2), 429–448. [DOI] [PubMed] [Google Scholar]
- Mielanczyk, L. , Matysiak, N. , Michalski, M. , Buldak, R. , & Wojnicz, R. (2014). Closer to the native state. Critical evaluation of cryo‐techniques for Transmission Electron Microscopy: Preparation of biological samples. Folia Histochemica et Cytobiologica, 52(1), 1–17. [DOI] [PubMed] [Google Scholar]
- Milne, J. L. S. , Borgnia, M. J. , Bartesaghi, A. , Tran, E. E. H. , Earl, L. A. , Schauder, D. M. , … Subramaniam, S. (2013). Cryo‐electron microscopy ‐ A primer for the non‐microscopist. The Febs Journal, 280(1), 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirto, H. , Hengé‐Napoli, M. H. , Gibert, R. , Ansoborlo, E. , Fournier, M. , & Cambar, J. (1999). Intracellular behaviour of uranium(VI) on renal epithelial cell in culture (LLC‐PK1): Influence of uranium speciation. Toxicology Letters, 104(3), 249–256. [DOI] [PubMed] [Google Scholar]
- Ortega, R. , Bresson, C. , Fraysse, A. , Sandre, C. , Devès, G. , Gombert, C. , … Moulin, C. (2009). Cobalt distribution in keratinocyte cells indicates nuclear and perinuclear accumulation and interaction with magnesium and zinc homeostasis. Toxicology Letters, 188(1), 26–32. [DOI] [PubMed] [Google Scholar]
- Perrin, L. , Carmona, A. , Roudeau, S. , & Ortega, R. (2015). Evaluation of sample preparation methods for single cell quantitative elemental imaging using proton or synchrotron radiation focused beams. Journal of Analytical Atomic Spectrometry, 30(12), 2525–2532. [Google Scholar]
- Poisson, C. , Stefani, J. , Manens, L. , Delissen, O. , Suhard, D. , Tessier, C. , … Guéguen, Y. (2014). Chronic uranium exposure dose‐dependently induces glutathione in rats without any nephrotoxicity. Free Radical Research, 48(10), 1218–1231. [DOI] [PubMed] [Google Scholar]
- Prat, O. , Bérenguer, F. , Steinmetz, G. , Ruat, S. , Sage, N. , & Quéméneur, E. (2010). Alterations in gene expression in cultured human cells after acute exposure to uranium salt: Involvement of a mineralization regulator. Toxicology In Vitro, 24(1), 160–168. [DOI] [PubMed] [Google Scholar]
- Rouas, C. , Bensoussan, H. , Suhard, D. , Tessier, C. , Grandcolas, L. , Rebiere, F. , … Gueguen, Y. (2010). Distribution of soluble uranium in the nuclear cell compartment at subtoxic concentrations. Chemical Research in Toxicology, 23(12), 1883–1889. [DOI] [PubMed] [Google Scholar]
- Safi, S. , Creff, G. , Jeanson, A. , Qi, L. , Basset, C. , Roques, J. , … Den Auwer, C. (2013). Osteopontin: A uranium phosphorylated binding‐site characterization. Chemistry ‐ A European Journal, 19(34), 11261–11269. [DOI] [PubMed] [Google Scholar]
- Tessier, C. , Suhard, D. , Rebière, F. , Souidi, M. , Dublineau, I. , & Agarande, M. (2012). Uranium microdistribution in renal cortex of rats after chronic exposure: A study by secondary ion mass spectrometry microscopy. Microscopy and Microanalysis, 18(1), 123–133. [DOI] [PubMed] [Google Scholar]
- Vanhecke, D. , Graber, W. , & Studer, D. (2008). Chapter 9 Close‐to‐Native Ultrastructural Preservation by High Pressure Freezing. Methods in Cell Biology, 88, 151–164. [DOI] [PubMed] [Google Scholar]
- Vicente‐Vicente, L. , Quiros, Y. , Pérez‐Barriocanal, F. , López‐Novoa, J. M. , López‐Hernández, F. J. , & Morales, A. I. (2010). Nephrotoxicity of uranium: Pathophysiological, diagnostic and therapeutic perspectives. Toxicological Sciences, 118(2), 324–347. [DOI] [PubMed] [Google Scholar]
- Weston, A. E. , Armer, H. E. J. , & Collinson, L. M. (2010). Towards native‐state imaging in biological context in the electron microscope. Journal of Chemical Biology, 3(3), 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn, M. E. , Durbin, P. W. , Howard, B. , Lipsztein, J. , Rundo, J. , Still, E. T. , & Willis, D. L. (1985). Metabolism of ingested U and RA. Health Physics, 48(5), 601–633. [DOI] [PubMed] [Google Scholar]


