Abstract
The cyclic-nucleotide modulated ion channel family includes cyclic nucleotide-gated (CNG) and hyperpolarization-activated and cyclic nucleotide-modulated (HCN) channels, which play essential roles in visual and olfactory signaling and the heart pacemaking activity. Functionally, these channels have been extensively characterized by electrophysiological techniques from protein heterologously expressed in Xenopus oocytes and mammalian cells. On the other hand, expression and purification of these proteins for biophysical and structural analyses in vitro is problematic and expensive and, accordingly, only limited information on the purified channels is available in the literature. Here we describe a protocol for binding studies of fluorescently labeled cyclic nucleotides to a homologue of eukaryotic CNG channels. Furthermore, we describe how to directly probe binding of unlabeled cyclic nucleotides in a competition assay. The use of fluorescence as a sensitive probe for ligand binding reduces the amount of protein needed and enables fast and easy measurements using standard laboratory equipment.
Keywords: Fluorescence titration, Ligand binding, Ligand discrimination, Fluorescent nucleotides, Competition assay
Background
Understanding the function of a protein in molecular detail requires extensive microscopic characterization. For ligand-gated ion channels different assays are needed to gain information about the specific interaction of the protein with the ligand, the communication between the ligand binding site and the pore, as well as channel-specific characteristics such as ionic throughput and inactivation or desensitization properties. Together with structural data for channel conformations corresponding to various functional states, this allows the development of a complete mechanistic description of channel function and regulation. Cyclic nucleotide-gated (CNG) ion channels are tetrameric potassium channels that are of particular interest due to their function in olfactory and visual signaling cascades (Kaupp and Seifert, 2002; Craven and Zagotta, 2006). However, there is only very limited data available on purified CNG channels under defined conditions, mostly from single molecule force spectroscopy ( Higgins et al., 2002 ; Maity et al., 2015 ; Goldschen- Ohm et al., 2016 ; Mazzolini et al., 2018 ). Electrophysiological characterization of CNG channels expressed in Xenopus oocytes or mammalian cells ( Biel et al., 1993 and 1994; Baumann et al., 1994 ; Weyand et al., 1994 ; Yu et al., 1996 ; Zagotta and Siegelbaum, 1996), shows that they are activated by micromolar concentrations of cyclic nucleotides (cNMP), but cAMP and cGMP can act differentially on different channel subtypes. High resolution structures of a few cyclic nucleotide-modulated channels have recently been solved ( James et al., 2017 ; Lee and MacKinnon, 2017; Li et al., 2017 ; Rheinberger et al., 2018 ), highlighting the need for biophysical assays to characterize these channels in order to gain a better understanding of the molecular interactions during gating and regulation.
Recently, we introduced SthK, a bacterial homologue of eukaryotic CNG channels, as a model to analyze the allosteric regulation of these channels in vitro ( Schmidpeter et al., 2018 ). One interesting observation was that cAMP and cGMP activate SthK with considerably different efficacies but interact with the protein with very similar affinities. We used fluorescently labeled cyclic nucleotides (Figure 1) to measure the binding affinity of these ligands to full-length SthK channels. The fluorescence of the NBD moieties increases upon interaction with a binding partner and thus directly reports the complex formation with SthK. To probe direct binding of unlabeled nucleotides to the channel we employed fluorescence-based competition assays. In the following we will provide detailed protocols for both experiments which can be performed in any protein-biochemistry laboratory using standard equipment and are applicable to protein-ligand interactions in general. Furthermore, this assay can be adapted to a plate-reader format which might increase the throughput and at the same time account for possible pipetting errors.
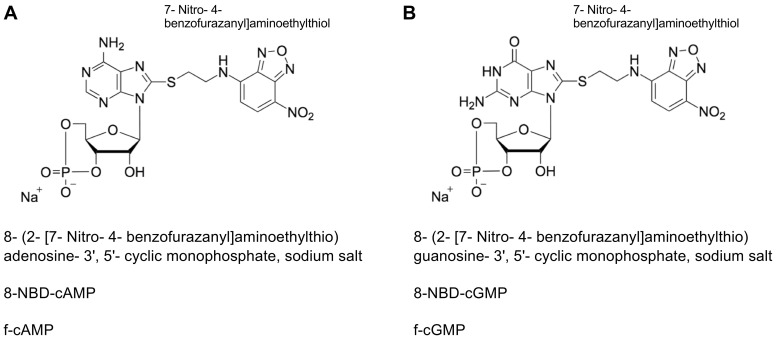
Figure 1. Structural formula of fluorescently labeled cyclic nucleotides.
A. 8-NBD-cAMP. B. 8-NBD-cGMP. For both compounds the fluorescent group is labelled separately, the exact chemical name, the abbreviation and the trivial name are given. The representations were adapted from Biolog (Bremen, Germany, www.biolog.de).
Materials and Reagents
Amicon Ultra-15 Centrifugal Filter MWCO 100 kDa (Merck, catalog number: UFC910024)
Amicon Ultra-4 Centrifugal Filter MWCO 100 kDa (Merck, catalog number: UFC810024)
BenchMarkTM Pre-stained Protein Ladder (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10748010)
Detergent Removal Column (Thermo Fisher Scientific, PierceTM, catalog number: 87779)
Ethanol ≥ 95% (Decon Labs, catalog number: V1116)
Amphipol A8-35 (Anatrace, catalog number: A835)
3',5'-cyclic AMP, cAMP (Alfa Aesar, catalog number: J62174-03)
3',5'-cyclic GMP, cGMP (Sigma-Aldrich, catalog number: G6129)
8-NBD-cAMP, f-cAMP (BIOLOG, catalog number: N 002)
8-NBD-cGMP, f-cGMP (BIOLOG, catalog number: N 001)
Glutaraldehyde (Sigma-Aldrich, catalog number: G6257)
HEPES (Simga-Aldrich, catalog number: H3375)
KCl (Simga-Aldrich, catalog number: P9333)
Superdex 200 10/300 GL (GE Healthcare, catalog number: 17517501)
Amphipol stock solution (see Recipes)
200 mM cAMP stock solution (see Recipes)
Assay buffer (see Recipes)
Equipment
Stir bar (suitable for the cuvette)
Set of single channel pipettes, ranging from 0.1-1,000 μl (VWR, catalog number: 75788-460)
Äkta Chromatography System (GE Healthcare)
NanoDropTM (Thermo Fisher Scientific)
Fluorescence-spectrophotometer (HORIBA, PTI QuantaMasterTM, model: 800)
UV cuvette (suitable for wavelengths between 230 and 800 nm, volume according to the specifications of the spectrometer between 1 and 4 ml)
Refrigerated Microcentrifuge (Eppendorf, model: 5418 R)
Rotator (Fixed Speed Rotator, Cole-Parmer, Stuart, model: SB2)
Software
Data can be analyzed using any software capable of programmable, non-linear least-squares fitting (We analyzed the data using GraFit 5)
GraFit 5.0.13 (Erithacus Software Limited, http://www.erithacus.com/grafit/)
Procedure
-
Sample preparation and cAMP removal
The expression and purification details of SthK are specified in Schmidpeter et al. (2018) and in the following we will only describe the details for the sample preparation in order to perform fluorescence titrations starting after eluting the sample from the immobilized metal affinity column. The ligand-removal step is only applicable for protein samples that are not stable in detergent in the absence of ligand and thus require purification in the presence of ligand.
-
Concentrate the protein solution to ~15 mg/ml (using Amicon Ultra-15 Centrifugal Filter MWCO 100 kDa) and add additional cAMP to ~400 μM.
Note: Make sure to saturate SthK with cAMP at this stage as this helps the stability of the protein. However, add additional cAMP only after you determined the protein concentration as the absorbance of nucleotides at 260 nm will influence the reading at 280 nm. By adding cAMP from a 200 mM stock solution the dilution will not significantly affect protein concentration (1:1,000 dilution).
-
Briefly spin down aggregates (15,000 × g, 4 °C, 10 min) and add amphipol A8-35 in a ratio of A8-35:SthK 3:1 (weight/weight).
Note: Amphipol should be hydrated in ddH2O overnight at 4 °C before use. Aliquots of 100 mg/ml A8-35 can be stored at -20 °C but avoid repeated freezing and thawing.
Incubate protein amphipol mixture for 1 h at 4 °C under constant agitation (such as slowly rotating).
Wash detergent removal columns (PierceTM Detergent Removal Spin Column, 4 ml) by gravity flow with 3 column volumes of ddH2O and equilibrate with 3 column volumes of buffer without detergent and reduced cAMP concentrations (20 mM HEPES, 100 mM KCl, 100 μM cAMP, pH 7.4).
Load 1 ml of the SthK:amphipol mixture onto the detergent removal column and let solution enter the column bed completely.
Add 4 ml of buffer to the column and immediately collect fractions of 500 μl.
-
Check all fractions for their protein content by measuring absorbance spectra.
Note: By measuring spectra from 350 nm to 220 nm, it is possible to quickly distinguish between protein-containing fractions (at 280 nm) and cAMP-containing fractions (at 260 nm) that will elute towards the end of the detergent removal column.
-
Pool the protein-containing fractions and concentrate to ~10 mg/ml using an Amicon® Ultra-4 concentrator with 100 kDa cutoff.
Note: SthK is stable in amphipol even in the absence of cAMP. At this point the concentration of cAMP can be reduced by diluting the protein in buffer without cAMP and concentrating it again. Reducing the cAMP concentration before the gel filtration helps to achieve complete removal of the ligand.
-
Load SthK in amphipol on a Superdex200 (10/300) column in assay buffer and perform the gel filtration (Figure 2A) using an Äkta system (GE Healthcare).
Note: For details about size exclusion chromatography as well as practical aspects, the handbook from GE Healthcare Life Sciences is a good resource.
Collect the peak containing tetrameric SthK (Figures 2A and 2B) and concentrate to ~400 μM using an Amicon® Ultra-4 concentrator with 100 kDa cutoff.
-
Determine the final protein concentration from the absorbance spectrum using the extinction coefficient of your protein (for SthK, the extinction coefficient is ε280 = 55,900 M-1 cm-1).
Note: The absorbance ratio A260/A280 can be used to evaluate the efficiency of cAMP removal. Typically, we reach a value of 0.5 for SthK without cAMP.
-
-
Titration of fluorescent cyclic nucleotides with SthK
-
Prepare SthK stock solution for the titration by mixing SthK in amphipol with f-cAMP or f-cGMP and assay buffer to reach the desired concentrations (concentrations can be easily adjusted according to Table 1).
Note: As for every fluorescence titration the molecule that generates the signal should be kept at a constant concentration throughout the experiment to avoid signal changes due to changes in the concentration. Thus, f-cAMP or f-cGMP should be added to the titrand stock in the final concentration (see Step B6).
-
Turn on the fluorescence spectrometer and wait for at least 20 min in order for the lamp to reach a stable temperature and set the excitation wavelength to 463 nm, the emission wavelength to 536 nm each with 5 nm bandwidth.
Note: The bandwidth settings depend strongly on the fluorometer. If fluorescence bleaching is observed during the experiment the excitation bandwidth can be reduced. If the recorded signal intensity is low the emission bandwidth can be increased.
Wash a UV-compatible cuvette 2 x with ddH2O, 2 x 70% Ethanol and again 2 x ddH2O, quickly dry it using pressured air, put a clean stir bar inside the cuvette and place the cuvette in the fluorometer.
-
Add 1,999.8 μl of assay buffer to the cuvette and adjust the speed of the stir bar to medium.
Note: Constant stirring of the solution throughout the entire experiment ascertains homogeneous equilibration of the components and furthermore increases signal stability by reducing effects of local fluorescence bleaching.
-
Equilibrate the buffer for 2 min and measure the fluorescence at 536 nm after excitation at 463 nm.
Note: There are two main modes of measuring the fluorescence. Some instruments read a set number of points and average the fluorescence. Make sure to set the value to at least 5 measurements. The other possibility is to monitor the fluorescence over time and average manually over at least 1 min.
-
Add 0.2 μl of f-cAMP or f-cGMP from a 1 mM stock solution to the cuvette and equilibrate for 2 min before measuring the fluorescence (0 μM SthK value).
Note: Using 0.1 μM of fluorescent cNMP turned out to give a good signal in our case. The concentration can be adjusted according to the sensitivity of the spectrometer and the KD value of the interaction that is being analyzed.
Add 2 μl of the SthK/f-cAMP or SthK/f-cGMP stock solution to the cuvette (according to Table 1), equilibrate for 2 min and measure the fluorescence.
-
Repeat Step B7 with the volumes defined by your pipetting scheme (see example in Table 1).
Note: Equilibrate for 2 min every time you add SthK from the stock solution to make sure the complex formation reaches a stable equilibrium state.
Table 1 provides an example of how a spreadsheet can be set up to easily adjust volumes and concentrations for the titration experiment. The volume and concentrations of the interacting molecules in the stock solution are given in the first column. The final volume in the cuvette (column 3) and the final SthK concentration (column 4) should be calculated as functions of the stock concentration and the added volume (column 1 and 2, respectively) to enable easy adjustments of the experiment. By varying the titrand concentration in the stock solution and the volumes that will be added to the cuvette in every step it is possible to span a wide range of concentrations throughout the titration.
-
-
Competition assay between labeled cAMP and unlabeled cNMP
-
Prepare stock solution of SthK in amphipol, f-cAMP and cNMP according to your pipetting scheme (Table 2).
Note: The concentrations for f-cAMP and SthK are fixed and cNMP (the titrand/competitor) is added at higher concentration. This ensures that the concentrations of the components that provide the read-out (complex between SthK and f-cAMP) are constant throughout the experiment.
-
Turn on the fluorescence spectrometer and wait for at least 20 min in order for the lamp to reach a stable temperature and set the excitation wavelength to 463 nm, the emission wavelength to 536 nm each with 5 nm bandwidth.
Note: The bandwidth settings depend strongly on the fluorometer. If fluorescence bleaching is observed during the experiment, the excitation bandwidth can be reduced. If the recorded signal intensity is low, the emission bandwidth can be increased.
Wash a UV-compatible cuvette 2 x with ddH2O, 2 x 70% Ethanol and again 2 x ddH2O, quickly dry it using pressured air, put a clean stir bar inside the cuvette and place the cuvette in the fluorometer.
Add assay buffer to the cuvette and adjust the speed of the stir bar to medium. Omit volume for f-cAMP and SthK from the buffer volume in order to reach 2,000 μl for the first data point (see Steps C5 and C6).
Add 0.3 μM f-cAMP to the cuvette, equilibrate for 2 min and measure the fluorescence (this serves as control measurement).
Add 0.3 μM SthK to the cuvette, equilibrate for 2 min and measure the fluorescence (this is the 0 μM cNMP value).
Add 1 μl of the stock solution (defined by your pipetting scheme in Table 2) to the cuvette and equilibrate for 2 min before measuring the fluorescence.
-
Repeat Step C7 with the volumes defined by your pipetting scheme (Table 2, column 2).
Note: Equilibrate for 2 min every time you add SthK from the stock solution to make sure the complex formation reaches a stable equilibrium state.
Table 2 provides an example of how a spreadsheet can be set up to easily adjust volumes and concentrations for the competition experiment. The volume and concentrations of the interacting molecules in the stock solution are given in the first column. The final volume in the cuvette (column 3) and the final concentration of cNMP (column 4) should be calculated as functions of the stock concentration and the added volume (column 1 and 2, respectively) to enable easy adjustments of the experiment. By varying the competitor concentration (cNMP) in the stock solution and the volumes that will be added to the cuvette in every step, it is possible to span a wide range of concentrations throughout the competition experiment.
-
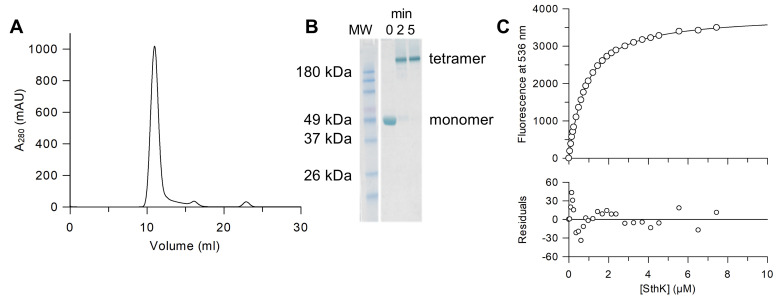
Figure 2. Sample preparation and titration of 8-NBD-cAMP with SthK in amphipol.
A. Gel filtration profile of SthK after amphipol exchange (Superdex200 10/300 in 20 mM HEPES, 100 mM KCl, pH 7.4) to remove cAMP. B. SDS-PAGE of the crosslinking of SthK in amphipol A8-35 using 0.13% glutaraldehyde (Kapoor, 2015) (at 37 °C for 5 min) shows that the main peak from the gel filtration is tetrameric SthK. All samples were heated to 98 °C for 10 min before SDS-PAGE was performed according to the manufacturer’s instructions. Selected bands of the molecular weight standard (MW, BenchMarkTM Pre-stained Protein Ladder, Thermo Fisher Scientific) are labeled. C. Example of a single titration experiment between 8-NBD-cAMP and SthK (upper panel) with averaged fluorescence values (circles) and the fit according to equation 6 (line) corrected by the fluorescence of 8-NBD-cAMP in the absence of SthK (KD = 0.72 μM). The residuals between the fit and the data points (lower panel) demonstrate the goodness of the fit.
Table 1. Example of a pipetting scheme for 0.1 μM f-cAMP and increasing concentrations of SthK.
| Stock solution | Volume of stock solution to be added (μl) | Final volume in cuvette (μl) | [SthK] (μM) in cuvette |
|
400 μl volume 0.1 μM f-cAMP 50 μM SthK |
0 | 2,000 | 0.00 |
| 2 | 2,002 | 0.05 | |
| 2 | 2,004 | 0.10 | |
| 2 | 2,006 | 0.15 | |
| 2 | 2,008 | 0.20 | |
| 2 | 2,010 | 0.25 | |
| 5 | 2,015 | 0.37 | |
| 5 | 2,020 | 0.50 | |
| 5 | 2,025 | 0.62 | |
| 5 | 2,030 | 0.74 | |
| 5 | 2,035 | 0.86 | |
| 5 | 2,040 | 0.98 | |
| 10 | 2,050 | 1.22 | |
| 10 | 2,060 | 1.46 | |
| 10 | 2,070 | 1.69 | |
| 10 | 2,080 | 1.92 | |
| 10 | 2,090 | 2.15 | |
| 10 | 2,100 | 2.38 | |
| 20 | 2,120 | 2.83 | |
| 20 | 2,140 | 3.27 | |
| 20 | 2,160 | 3.70 | |
| 20 | 2,180 | 4.13 | |
| 20 | 2,200 | 4.55 | |
| 50 | 2,250 | 5.56 | |
| 50 | 2,300 | 6.52 | |
| 50 | 2,350 | 7.45 |
Table 2. Example of a pipetting scheme for the competition of cNMP with the complex formed between 0.3 μM f-cAMP and 0.3 μM SthK.
| Stock solution | Volume of stock solution to be added (μl) | Final volume in cuvette (μl) | [cNMP] (μM) in cuvette |
|
200 μl volume 0.3 μM f-cAMP 0.3 μM SthK 30 mM cNMP |
0 | 2,000 | 0.00 |
| 1 | 2,001 | 14.99 | |
| 1 | 2,002 | 29.97 | |
| 1 | 2,003 | 44.93 | |
| 2 | 2,005 | 74.81 | |
| 2 | 2,007 | 104.63 | |
| 3 | 2,010 | 149.25 | |
| 5 | 2,015 | 223.33 | |
| 5 | 2,020 | 297.03 | |
| 10 | 2,030 | 443.35 | |
| 10 | 2,040 | 588.24 | |
| 20 | 2,060 | 873.79 | |
| 20 | 2,080 | 1,153.85 | |
| 20 | 2,100 | 1,428.57 | |
| 20 | 2,120 | 1,698.11 | |
| 20 | 2,140 | 1,962.62 | |
| 20 | 2,160 | 2,222.22 |
Data analysis
In our case, we recorded the fluorescence for every SthK concentration for 1 min with 1 sec intervals and exported the signal as text files. The data were imported into Excel, averaged manually, and the averaged values were then plotted as function of the ligand concentration (SthK for the titration with fluorescent cyclic nucleotides or unlabeled cyclic nucleotide for the competition assays) in GraFit (Figure 2C, Schmidpeter et al., 2018 ). Analysis was performed for every single titration and competition experiment separately, the resulting KD values were averaged and the standard error of the mean (SEM) was calculated. For each condition, at least three independent experiments were performed for different protein purifications ( Schmidpeter et al., 2018 ).
-
Analysis of the titration experiments
The titration of fluorescent cyclic-nucleotides with SthK is analyzed according to a two-state model (Equation 1) that directly defines the dissociation constant KD (Equation 2).
Eq. 1
Eq. 2
With [SthK] being the concentration of free SthK monomers, [cNMP] the concentration of free cyclic-nucleotide, [SthK·cNMP] the concentration of the resulting complex and KD the dissociation constant.
Given the conservation of mass the free concentrations can be expressed as function of the total concentration and the concentration of the complex (e.g., [SthK] = [SthK]0 - [SthK·cNMP]). Equation 2 can be re-written to Equation 3 leading to a quadratic equation that describes the complex formation (Equation 4).
Eq. 3
Eq. 4
Solving the quadratic equation gives Equation 5.
Eq. 5
As the concentration of the fluorescent cNMP is constant throughout the experiment Equation, 5 can be divided by [cNMP]0. The equation now describes the fraction of bound cNMP as function of [SthK]0. Furthermore, we assume that the fluorescence increase of cNMP upon interaction with SthK describes the complex formation leading to the final equation to analyze the binding data (Equation 6, Figure 2C).
Eq. 6
With Fmin and Fmax being the minimum and the maximum fluorescence, respectively (i.e., the fluorescence of [cNMP]0 alone and [cNMP]0 saturated with SthK).
The measured fluorescence values can be normalized using Fmin and Fmax as determined from this fitting procedure which then describe the fraction of bound cNMP according to the assumptions made before.
-
Analysis of the competition experiments
Analysis of the data obtained from the competition experiments is more complex as two ligands compete for one binding site according to Equation 7 and has been described before (Wang, 1995; Wilkinson, 2004; Cukkemane et al., 2007 ).
Eq. 7
where [SthK], [fcAMP] and [cNMP] denote for the concentrations of free SthK, 8-NBD-cAMP and cNMP (either cAMP or cGMP), respectively. [SthK·fcAMP] and [SthK·cNMP] describe the two possible complexes between SthK and 8-NBD-cAMP or SthK and cNMP.
Accordingly, two dissociation constants can be determined, Ka for the complex between SthK and 8-NBD-cAMP (Equation 8) and Kb for the complex between SthK and cNMP (Equation 9).
Eq. 8
Eq. 9
Similar to the titration experiment and considering mass conservation Equations 7-9 can be combined to a cubic equation (Equation 10).
Eq. 10
with a, b and c defined in Equation 11-13.
Eq. 11
Eq. 12
Eq. 13
where [fcAMP]0, [cNMP]0 and [SthK]0 denote for the total concentration of each compound.
The cubic function has only one meaningful solution for the specified problem and a detailed derivation of the analysis can be found in Wang (1995) with a more descriptive explanation in Wilkinson (2004). Again, the observed signal change is related to the complex formation between SthK and 8-NBD-cAMP and the data from the competition experiment can be analyzed according to Equation 14.
Eq. 14
withdefined in Equation 15.
Eq. 15
Note: This analysis enables the calculation of Ka and Kb simultaneously. However, the dissociation constant for f-cAMP as determined in the direct binding assay can also be set as constant during the fitting process to reduce the number of free parameters.
Notes
In general, both assays, direct binding of a fluorescent ligand and competition between two ligands, are very robust. In our experience, the biggest variability comes from the content of bound cAMP after gel filtration as determined by the final A260/A280 ratio of the protein sample.
Recipes
-
Amphipol stock solution
100 mg/ml A8-35 in ddH2O (hydrated overnight at 4 °C)
-
200 mM cAMP stock solution
Dissolve 0.66 g of cAMP in 9.5 ml ddH2O
Adjust pH to 7.4, fill up to 10 ml and check the exact concentration of the stock solution by measuring the absorbance at 260 nm (the extinction coefficient of cAMP is ε260 14,600 M-1 cm-1)
Store cAMP stock solution in 200 μl aliquots at -20 °C
-
Assay buffer
20 mM HEPES, 100 mM KCl, pH 7.4, sterile filtered (0.22 μm)
Acknowledgments
Work for this protocol was funded in part by the National Institutes of Health (R01GM124451 and R01GM088352) to CN and the Deutsche Forschungsgemeinschaft (SCHM3198/1-1) and the American Heart Association (18POST33960309) to PS. Similar studies have been performed before by Cukkemane et al. (2007) and the analysis was summarized by Wilkinson (2004).
Competing interests
The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Baumann A., Frings S., Godde M., Seifert R. and Kaupp U. B.(1994). Primary structure and functional expression of a Drosophila cyclic nucleotide-gated channel present in eyes and antennae . EMBO J 13(21): 5040-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biel M., Altenhofen W., Hullin R., Ludwig J., Freichel M., Flockerzi V., Dascal N., Kaupp U. B. and Hofmann F.(1993). Primary structure and functional expression of a cyclic nucleotide-gated channel from rabbit aorta. FEBS Lett 329(1-2): 134-138. [DOI] [PubMed] [Google Scholar]
- 3. Biel M., Zong X., Distler M., Bosse E., Klugbauer N., Murakami M., Flockerzi V., and Hofmann F.(1994). Another member of the cyclic nucleotide-gated channel family, expressed in testis, kidney, and heart. Proc Natl Acad Sci U S A 91(9): 3505-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Craven K. B. and Zagotta W. N.(2006). CNG and HCN channels: two peas, one pod. Annu Rev Physiol 68: 375-401. [DOI] [PubMed] [Google Scholar]
- 5. Cukkemane A., Gruter B., Novak K., Gensch T., Bonigk W., Gerharz T., Kaupp U. B. and Seifert R.(2007). Subunits act independently in a cyclic nucleotide-activated K+ channel . EMBO Rep 8(8): 749-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldschen-Ohm M. P., Klenchin V. A., White D. S., Cowgill J. B., Cui Q., Goldsmith R. H. and Chanda B.(2016). Structure and dynamics underlying elementary ligand binding events in human pacemaking channels. Elife 5: e20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higgins M. K., Weitz D., Warne T., Schertler G. F. and Kaupp U. B.(2002). Molecular architecture of a retinal cGMP-gated channel: the arrangement of the cytoplasmic domains. EMBO J 21(9): 2087-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. James Z. M., Borst A. J., Haitin Y., Frenz B., DiMaio F., Zagotta W. N. and Veesler D.(2017). CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel. Proc Natl Acad Sci U S A 114(17): 4430-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kapoor M.(2015). How to cross-link proteins.
- 10. Kaupp U. B. and Seifert R.(2002). Cyclic nucleotide-gated ion channels. Physiol Rev 82(3): 769-824. [DOI] [PubMed] [Google Scholar]
- 11. Lee C. H. and MacKinnon R.(2017). Structures of the human HCN1 hyperpolarization-activated channel. Cell 168(1-2): 111-120 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M., Zhou X., Wang S., Michailidis I., Gong Y., Su D., Li H., Li X., and Yang J.(2017). Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542(7639): 60-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maity S., Mazzolini M., Arcangeletti M., Valbuena A., Fabris P., Lazzarino M. and Torre V.(2015). Conformational rearrangements in the transmembrane domain of CNGA1 channels revealed by single-molecule force spectroscopy. Nat Commun 6: 7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mazzolini M., Arcangeletti M., Marchesi A., Napolitano L. M. R., Grosa D., Maity S., Anselmi C. and Torre V.(2018). The gating mechanism in cyclic nucleotide-gated ion channels. Sci Rep 8(1): 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rheinberger J., Gao X., Schmidpeter P. A. and Nimigean C. M.(2018). Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. Elife 7: e39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidpeter P. A. M., Gao X., Uphadyay V., Rheinberger J. and Nimigean C. M.(2018). Ligand binding and activation properties of the purified bacterial cyclic nucleotide-gated channel SthK. J Gen Physiol 150(6): 821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z. X.(1995). An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett 360(2): 111-114. [DOI] [PubMed] [Google Scholar]
- 18. Weyand I., Godde M., Frings S., Weiner J., Muller F., Altenhofen W., Hatt H. and Kaupp U. B.(1994). Cloning and functional expression of a cyclic-nucleotide-gated channel from mammalian sperm. Nature 368(6474): 859-863. [DOI] [PubMed] [Google Scholar]
- 19. Wilkinson K. D.(2004). Quantitative analysis of protein-protein interactions. Methods Mol Biol 261: 15-32. [DOI] [PubMed] [Google Scholar]
- 20. Yu W. P., Grunwald M. E. and Yau K. W.(1996). Molecular cloning, functional expression and chromosomal localization of a human homolog of the cyclic nucleotide-gated ion channel of retinal cone photoreceptors. FEBS Lett 393(2-3): 211-215. [DOI] [PubMed] [Google Scholar]
- 21. Zagotta W. N. and Siegelbaum S. A.(1996). Structure and function of cyclic nucleotide-gated channels. Annu Rev Neurosci 19: 235-263. [DOI] [PubMed] [Google Scholar]