Abstract.
Although Plasmodium vivax has been assumed to be absent from sub-Saharan Africa because of the protective mutation conferring the Duffy-negative phenotype, recent evidence has suggested that P. vivax cases are prevalent in these regions. We selected 292 dried blood spots from children who participated in the 2013–2014 Demographic and Health Survey of the Democratic Republic of the Congo (DRC), to assess for P. vivax infection. Four P. vivax infections were identified by polymerase chain reaction, each in a geographically different survey cluster. Using these as index cases, we tested the remaining 73 samples from the four clusters. With this approach, 10 confirmed cases, three probable cases, and one possible case of P. vivax were identified. Among the 14 P. vivax cases, nine were coinfected with Plasmodium falciparum. All 14 individuals were confirmed to be Duffy-negative by sequencing for the single point mutation in the GATA motif that represses the expression of the Duffy antigen. This finding is consistent with a growing body of literature that suggests that P. vivax can infect Duffy-negative individuals in Africa. Future molecular and sequencing work is needed to understand the relationship of these isolates with other P. vivax samples from Asia and South America and discover variants linked to P. vivax virulence and erythrocyte invasion.
INTRODUCTION
Of the five species causing malaria in humans, Plasmodium falciparum, Plasmodium malariae, and Plasmodium ovale are the only species of Plasmodium that have been considered to be endemic to sub-Saharan Africa. Individuals of western, central, and eastern African descent have long been considered refractory to infection by Plasmodium vivax. Resistance to P. vivax was demonstrated through a series of observations and experiments that found that the absence of the Duffy-antigen blood group, also known as the Duffy antigen/receptor for chemokines (DARC), prevented P. vivax merozoites from invading reticulocytes and erythrocytes.1–6 This Duffy-negative phenotype results from a single nucleotide polymorphism within the promoter region of the DARC gene (nucleotide position −33 T:C) that disrupts the GATA-1 transcription factor binding process and negates DARC gene expression on the erythroid surface.3
Despite a high prevalence of Duffy negativity in western, central, and eastern African populations, the risk of autochthonous P. vivax infections has become more appreciated across Africa and incorporated into P. vivax prevalence maps.7–9 Transmission of P. vivax in these high Duffy-negative regions has been attributed to P. vivax strains circulating among Duffy-positive residents and travelers, or possibly an ape reservoir.10–12 However, recent studies have demonstrated P. vivax infections in Duffy-negative individuals across Africa, including Angola and Equatorial Guinea,13 Benin,14 Botswana,15 Cameroon,16,17 Ethiopia,18,19 Western Kenya,20 Madagascar,6 Mauritania,21 and central Sudan22 (reviewed recently by Zimmerman et al.).23 In addition, standard blood smear microscopy has demonstrated ring-stage formation in infected reticulocytes from Duffy-negative patients, confirming that the P. vivax detected in some cases was invasive and not merely transient merozoites in the blood.6,19 Vivax malaria has also been identified in mosquitoes, suggesting active transmission.13 Given these findings, a better understanding of vivax biology in Africa is needed.
Using samples from a large population representative survey, the 2013–2014 Demographic Health Survey (DHS), we present evidence of P. vivax infection among Duffy-negative individuals in the Democratic Republic of the Congo (DRC). These individuals represent asymptomatic children found to be parasitemic by PCR-based methods within tested DHS clusters. We found that P. vivax infection was not uncommon in the cluster and that it most often presents as a coinfection with P. falciparum.
MATERIAL AND METHODS
Description of study participants and sample processing.
As part of the DRC DHS-II, clinical samples were collected from children surveyed across 536 distinct geographic clusters between November 2013 and February 2014 as part of DHS.24 Identified household participants were consented and then provided clinical samples that were evaluated for malaria infection. This evaluation included a rapid diagnostic test targeting P. falciparum histidine-rich protein II antigen (HRP2; SD BIOLINE Malaria Ag P.f., Standard Diagnostics, Inc.©, Gyeonggi-do, Republic of Korea) and light microscopy. Microscopy was conducted by two expert microscopists; however, species-level reporting was not included in the DHS. Each clinical sample was also used to prepare a dried blood spot, which were shipped to the University of North Carolina-Chapel Hill for DNA extraction and laboratory analysis. DNA was extracted using Chelex® and tested for P. falciparum with a real-time PCR assay targeting the P. falciparum lactate dehydrogenase gene (pfldh) as previously described.25,26
Approval for this study was granted by the institutional review boards of the University of North Carolina-Chapel Hill and the University of Kinshasa. Informed consent was obtained for all participants from a parent or guardian.
Plasmodium vivax initial screening.
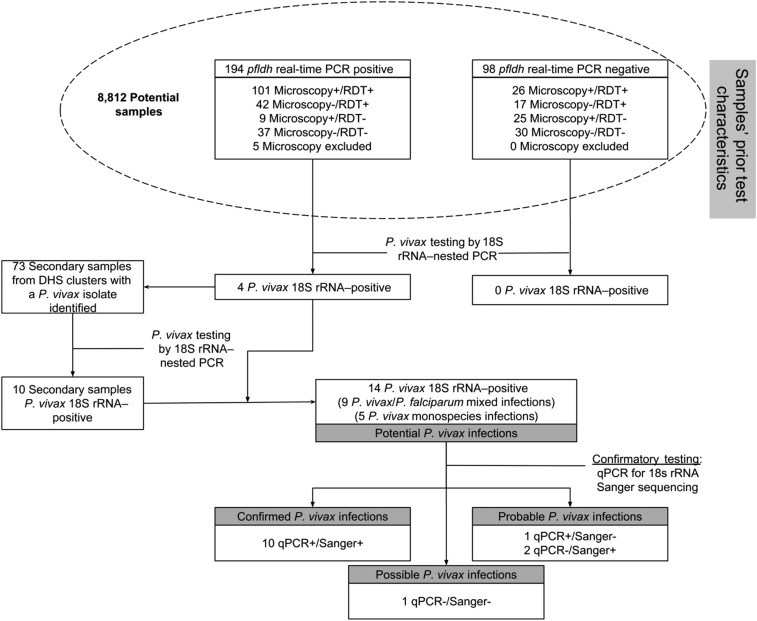
Using the DRC DHS-II as the source population, we wanted to test a subset of samples for P. vivax infection across the country.24 In addition, based on previous work comparing the sensitivity and specificity of the various detection methods for P. falciparum27 and screens for P. ovale and P. malariae in the same sample bank,28 we hypothesized that the prevalence of P. vivax would vary among the different strata of P. falciparum diagnostic results (i.e., pfldh real-time PCR, HRP2-rapid diagnostic test [RDT], and microscopy). As a result, to maximize potential identification of both monospecies P. vivax infections and P. vivax/P. falciparum coinfections, we selected 292 primary samples across 208 clusters from the DRC DHS-II with a range of P. falciparum detection results (Figure 1; for a map with all DRC DHS-II clusters, see Supplemental Figure 1).
Figure 1.
Prior sample testing characteristics for included samples and diagnostic algorithm for Plasmodium vivax infections. To screen for potential P. vivax infections that were potentially coinfections with both Plasmodium falciparum and P. vivax, we selected 292 samples with various prior results by three different detection methods. Samples first underwent 18s rRNA–nested PCR to identify potential cases. Cases were confirmed by both qPCR of the 18s rRNA gene and Sanger sequencing of the 18s rRNA fragment from the nested PCR.
These 292 primary samples then underwent testing for P. vivax using a diagnostic nested PCR that targeted the small subunit (18S) ribosomal ribonucleic acid (rRNA) gene. In the first round, a Plasmodium genus-specific 18S rRNA region was amplified, which then underwent a species-specific second round of PCR (protocol modified from Singh et al.;29 Supplemental Table 1). Amplification was confirmed by visualization of the nested product on gel electrophoresis.
In addition, for clusters that contained P. vivax potential infections, all remaining samples within that cluster were screened as secondary samples using the nested PCR approach described previously. The testing characteristics and algorithm for the samples included in this study are outlined in Figure 1.
Plasmodium vivax confirmatory screening.
To define the likelihood that a potential infection represented a true vivax infection, we used a multistep testing approach with two confirmatory assays after the initial screening by 18S rRNA–nested PCR. The confirmatory tests included a real-time PCR assay targeting the P. vivax 18S rRNA gene (protocol modified from Veron et al.;30 Supplemental Table 1) and Sanger sequencing of the 18S rRNA–nested PCR products. Potential infections were considered “Confirmed” if they were positive by both confirmatory assays, “Probable” if positive by one, and “Possible” if negative by both confirmatory assays.
The quantitative PCR was conducted on a Bio-Rad CFX384™ real-time PCR detection system (Bio-Rad, Hercules, CA) in 25-μL reactions (Supplemental Table 1). Samples were run alongside appropriate controls, to ensure only P. vivax was amplified (Table 1). Samples were considered positive if they had a cycle threshold of less than or equal to 38 cycles.
Table 1.
Summary of the Plasmodium vivax potential and confirmed cases identified in the Democratic Republic of the Congo during the 2013 DHS
| Sample name | Cluster | Pfldh | Microscopy | RDT | P. vivax–nested 18s rRNA PCR | P. vivax by RT-PCR | P. vivax by Sanger sequencing | Duffy −33 T:C mutation |
|---|---|---|---|---|---|---|---|---|
| B5A7N | 69 | N | N | N | P | P | P | P |
| J6V8T | 69 | P | P | N | P | P | P | P |
| M2B5U* | 69 | P | N | P | P | P | P | P |
| T6B8J | 69 | P | P | P | P | P | P | P |
| Z6W9M | 69 | P | N | P | P | P | P | P |
| X3M5S* | 144 | P | N | N | P | N | P | P |
| Q8J6O | 144 | N | N | N | P | P | P | P |
| D9U3K | 144 | N | N | N | P | P | P | P |
| G0R3B | 238 | P | P | P | P | N | N | P |
| U1P9U* | 238 | P | P | P | P | P | N | P |
| O6Y4X | 238 | N | N | N | P | P | P | P |
| P8A6E* | 298 | P | N | P | P | N | P | P |
| I4G3V | 298 | P | N | P | P | P | P | P |
| N4M7L | 298 | N | N | P | P | P | P | P |
DHS = Demographic Health Survey; pfldh = P. falciparum lactate dehydrogenase; RDT = rapid diagnostic test; * = index cases; P = positive; N = negative. The columns above provide information on the DHS cluster the case was identified in, whether the case was coinfected with Plasmodium falciparum as identified with real-time PCR for the pfldh gene, and whether the patient was considered positive for malaria by microscopy and RDT. In addition, each isolate has results for nested 18s rRNA P. vivax PCR, P. vivax real-time PCR, and P. vivax Sanger sequencing. Finally, the host genotype was sequenced for the −33 T:C mutation that predicts the Duffy-negative phenotype.3
Nested PCR products of the 18S rRNA assay from the potential P. vivax infections were submitted for Sanger sequencing (Eton Bioscience, Inc., Durham, NC) with approximately 3 μL of 20 μM of the rViv 1 primer (Supplemental Table 1) and analyzed using Geneious® 10.1.3 (Biomatters Limited, Auckland, New Zealand). Sequences underwent pairwise alignment with ClustalW (IUB cost matrix, gap open cost: 15, gap extend cost: 6.66) and the ends of all sequences were trimmed for low-quality bases using the Geneious default settings. Two sequences did not align with the other samples in regions where base pairs could be accurately called, and on visual inspection, the sequences were determined to have too much noise and low-quality bases to accurately call a genotype. As such, these two samples were excluded from subsequent analysis (and considered negative by Sanger confirmation). After trimming the ends for low-quality bases, the remaining 12/14 samples had sequence lengths ranging from 28 to 83 bp (Supplemental Table 2). These sequences were aligned to the P. vivax and P. falciparum 18S rRNA genes (GenBank accession nos. U03079.1 and M19173.1, respectively) using the Highest Sensitivity/Slow Geneious mapper with up to five iterations to confirm identity.
Duffy genotyping.
To determine host susceptibility to P. vivax infection, the promoter region of the DARC gene was amplified with a hemi-nested PCR approach (Supplemental Table 1). Nested PCR products were then submitted to Sanger sequencing (Eton Bioscience, Inc.) with approximately 3 μL of 360 nM of the Duffy GATA1 reverse primer (Supplemental Table 1). The resulting Sanger sequences were trimmed and then aligned with the Highest Sensitivity/Slow Geneious mapper to the Homo sapien DARC gene (GenBank accession no. X85785.1) using Geneious 10.1.3 (Biomatters Limited).
Descriptive statistics and spatial analyses.
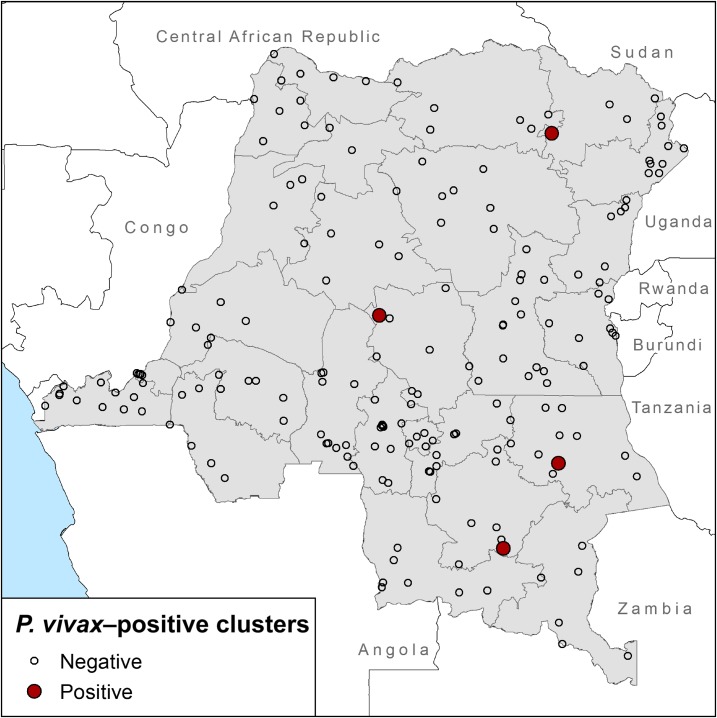
Finally, we calculated descriptive statistics for P. vivax potential infections (hereafter called cases), P. falciparum cases (previously identified in Meshnick et al.24), and non-cases. All covariates were taken from the DRC DHS-II dataset and used the DHS coding/categorization structure.31,32 Categorical covariates were summarized as counts and within-case-status percentages, whereas continuous covariates were summarized with mean and standard deviation calculations. Analyses were conducted using R-statistical software (version 3.4.1) with the base and tidyverse packages.33,34 We then used ArcGIS (version 10.4.1; ESRI, Redlands, CA) and the global positioning system coordinates for each cluster from the DHS to create a map of the DRC to examine the spatial distribution of the P. vivax–positive clusters (Figure 2). In addition, nonhuman ape territories downloaded from the International Union for Conservation of Nature were plotted to determine the overlap between nonhuman ape ranges and P. vivax cases.35
Figure 2.
Map of the Democratic Republic of the Congo (DRC) with clusters that screened negative (unfilled) and positive (red) for Plasmodium vivax. In total, 208 geographical clusters were screened for P. vivax; however, 19 clusters (N = 32 samples) were missing coordinate data from the Demographic Health Survey database and are not displayed on the above map. Shapefiles for the DRC were downloaded from the Database of Global Administrative Areas (http://www.gadm.org/) and plotted with ArcGIS (version 10.4.1). This figure appears in color at www.ajtmh.org.
RESULTS
Of the initial 292 samples tested for P. vivax, we identified four potential infections by 18S rRNA–nested PCR, each from a different geographical cluster (Table 1, Figure 1). Geographical clusters were dispersed across the central western region of the country with largely varying latitudes (Figure 2). These four potential infections were used as an index to screen the rest of the samples from each respective cluster. This secondary screen resulted in the identification of 10/73 additional potential infections.
To confirm P. vivax infections, we used two confirmatory approaches: qPCR for 18S rRNA and Sanger sequencing of the 18S rRNA fragment from the nested PCR. Of the 12 Sanger sequences aligned to the P. vivax 18S rRNA reference gene (GenBank accession no. U03079), 10/12 sequences demonstrated 100% identity, whereas the other two sequences had 1–2 single nucleotide polymorphisms (SNPs) at the end of their alignments. These SNPs were likely because of conservative end trimming. By contrast, all 12 of the Sanger sequences aligned to the P. falciparum 18S rRNA reference gene (GenBank accession no. M19173.1) as a “negative control” showed approximately 65–40% identity because of numerous SNPs throughout the sequence. As a result, the 12 Sanger sequences preferentially aligned to the P. vivax 18S rRNA reference gene, which is strong evidence that these are true P. vivax infections. Overall, based on our testing algorithm (Figure 1), we found 10 confirmed infections, three probable infections, and one possible infection of P. vivax (Table 1). Among the P. vivax cases, coinfection by P. falciparum based on pfldh-PCR was common (9/14, 64%). However, microscopy, RDT, and pfldh real-time PCR results demonstrated several discordant results from both each other and pfldh real-time PCR (Table 1).
We then evaluated host, behavioral, and environmental characteristics associated with P. vivax infection and P. falciparum infection, respectively (Table 2). Among the 14 hosts of the P. vivax cases, all were homozygous for the −33 T:C mutation, suggesting they all expressed a Duffy-negative phenotype.3 All of the P. vivax cases occurred in rural households, and only 5/14 cases were recorded as using long-lasting insecticidal bed nets (Table 2). In terms of socioeconomic status, 9/14 P. vivax cases were classified in the poorest wealth index bracket. Of the 11 P. vivax cases with hemoglobin data, three were classified with moderate anemia and two were classified with mild anemia, whereas the remaining six were not anemic. Overall, 9/14 P. vivax cases were coinfected with P. falciparum. Finally, among the four clusters that screened positive for P. vivax, two clusters were within known nonhuman ape ranges (Supplemental Figure 1). In addition, a third cluster that screened positive for P. vivax in the southeast region of the DRC was located near known nonhuman ape ranges.
Table 2.
Characteristics of Plasmodium vivax cases, Plasmodium falciparum cases, and non-cases from the 365 individuals sampled
| P. vivax cases | P. falciparum cases | Non-cases | |
|---|---|---|---|
| Observations | 14 | 221 | 139 |
| Urban setting (%) | 0 (0) | 40 (18.10) | 19 (13.67) |
| Cluster altitude (SD) | 648.86 (88.27) | 664.24 (287.62) | 726.15 (297.68) |
| Socioeconomic status (%) | |||
| Poorest | 9 (64.29) | 73 (33.03) | 50 (35.97) |
| Poorer | 0 (0) | 47 (21.27) | 42 (30.22) |
| Middle | 4 (28.57) | 58 (26.24) | 28 (20.14) |
| Rich | 1 (7.14) | 35 (15.84) | 14 (10.07) |
| Richer | 0 (0) | 8 (3.62) | 5 (3.60) |
| Female (%) | 7 (50.0) | 111 (50.23) | 70 (50.36) |
| Age in months (SD) | 40.27 (16.13) | 36.28 (14.08) | 34.09 (14.80) |
| Missing | 3 | 45 | 9 |
| Anemia status (%) | |||
| Severe | 0 (0) | 11 (4.98) | 5 (3.60) |
| Moderate | 3 (21.43) | 81 (36.65) | 47 (33.81) |
| Mild | 2 (14.29) | 36 (16.29) | 35 (25.18) |
| Not anemic | 6 (42.86) | 48 (21.72) | 43 (30.94) |
| Missing | 3 (21.43) | 45 (20.36) | 9 (6.47) |
| Long-lasting insecticidal net use (%) | 5 (35.71) | 117 (52.94) | 72 (51.80) |
SD = standard deviation. Given that 9/14 of the P. vivax cases were coinfected with P. falciparum, these individuals are considered in both the P. vivax and the P. falciparum columns. Individuals were only considered as non-case if they were not infected by P. vivax or P. falciparum. For categorical covariates, the count and percentage (%) were reported, whereas the mean and SD were reported for continuous covariates.
Among the 221 P. falciparum cases, 19 occurred in rural households, 117 used long-lasting insecticidal nets, 111 were female, and only 73 were in the poorest wealth index bracket. When data were available, 11 individuals were classified with severe anemia, 81 were classified with moderate anemia, and 36 were classified with mild anemia, whereas the remaining 48 were not anemic.
DISCUSSION
We identified 14 potential infections of P. vivax in Duffy-negative individuals residing in the DRC. To our knowledge, this is the first report of P. vivax infection in Duffy-negative subjects from the DRC. Plasmodium vivax infections were first identified with a nested PCR and confirmed with qPCR and Sanger sequencing.29,30 We identified 10 confirmed infections, three probable infections, and one possible infection among the 14 isolates. Given that speciation information and parasite characterization by light microscopy were not available, it is possible that the P. vivax detected by PCR methods in this study were transient pre-erythrocytic parasites.36 Future work will need to rely on microscopy data to confirm parasite invasion, as has been done previously in Madagascar.6
The relatively few infections and sampling strategy used in this study prohibited robust analysis of epidemiologic factors associated with P. vivax infection in the DRC. However, a minority of participants with P. vivax infection in this study were using long-lasting insecticidal nets. Most were in the poorest income strata and, when data were available, none had severe anemia. A similar behavioral and demographic profile was found for participants infected with P. falciparum, although the distribution of wealth appeared to be wider among the P. falciparum cases as compared with the P. vivax cases.
Of the clusters that screened positive for P. vivax, three occurred within or near known nonhuman ape ranges. This suggests that there may be potential zoonotic transmission, as has been previously suggested in the Central African Republic.12 However, DNA sequencing is needed to determine if our samples cluster with the proposed nonhuman ape P. vivax lineages or fall within the monophyletic human clade and associate more closely with contemporary, circulating strains of P. vivax.10–12 As such, this high degree of spatial overlap is interesting but cannot be considered causative.
This study was largely limited by its sampling strategy, which was designed to optimize P. vivax detection among potential monoclonal P. vivax infections and coinfection with P. falciparum, and not with external validity and generalizability in mind. As a result, our study sample may suffer from selection bias and the results are not necessarily generalizable to the entire DRC population. Given this, we only presented descriptive statistics and did not perform association analyses. The main strength of this study lies with its robust approach for confirming the P. vivax cases with multiple modalities.
Future work is needed to screen the DRC for additional cases of P. vivax. A large survey will provide a systematic estimation of the prevalence of P. vivax in the DRC and allow for a robust risk factor analysis. In addition, incorporation of next-generation sequencing efforts to determine the geographical and etiological origins of these infections is needed. Given the breadth of publicly available contemporary P. vivax whole-genome sequences from Asia and South America, future work will be able to readily identify the population genetic structure and origin of these sub-Saharan Africa P. vivax cases.37–39 Next-generation sequencing can also be leveraged to identify potential selective sweeps or variants associated with red blood cell invasion that may shed light on the mechanism of Duffy-negative erythrocyte invasion.19,40 This work will be important to characterize the P. vivax population in sub-Saharan Africa and to assess the risk of infection among Duffy-negative individuals. If P. vivax has returned to Africa, malaria elimination campaign efforts will need to shift to include this restored threat.
Supplementary Material
Supplemental tables and figure
Acknowledgments:
We would like to thank our excellent field team in the Democratic Republic of the Congo and the numerous participants in the Demographic Health Surveys.
Note: Supplemental tables and figure appear at www.ajtmh.org.
REFERENCES
- 1.Livingstone FB, 1984. The Duffy blood groups, vivax malaria, and malaria selection in human populations: a review. Hum Biol 56: 413–425. [PubMed] [Google Scholar]
- 2.Miller LH, Mason SJ, Clyde DF, 1976. The resistance factor to Plasmodium vivax in blacks: the Duffy-blood-group genotype, FyFy. N Engl J Med 295: 302–304. [DOI] [PubMed] [Google Scholar]
- 3.Tournamille C, Colin Y, Cartron JP, Van Kim CL, 1995. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy–negative individuals. Nat Genet 10: 224–228. [DOI] [PubMed] [Google Scholar]
- 4.Hadley TJ, 1986. Invasion of erythrocytes by malaria parasites: a cellular and molecular overview. Annu Rev Microbiol 40: 451–477. [DOI] [PubMed] [Google Scholar]
- 5.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK, 1975. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189: 561–563. [DOI] [PubMed] [Google Scholar]
- 6.Ménard D, et al. 2010. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA 107: 5967–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howes RE, Battle KE, Mendis KN, Smith DL, Cibulskis RE, Baird JK, Hay SI, 2016. Global epidemiology of Plasmodium vivax. Am J Trop Med Hyg 95: 15–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battle KE, Gething PW, Elyazar IRF, Moyes CL, Sinka ME, Howes RE, Guerra CA, Price RN, Baird KJ, Hay SI, 2012. The global public health significance of Plasmodium vivax. Adv Parasitol 80: 1–111. [DOI] [PubMed] [Google Scholar]
- 9.Guerra CA, et al. 2010. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis 4: e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, et al. 2014. African origin of the malaria parasite Plasmodium vivax. Nat Commun 5: 3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, Sharp PM, Hahn BH, 2016. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol 47: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prugnolle F, et al. 2013. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci USA 110: 8123–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, do Rosário VE, Benito A, Berzosa P, Arez AP, 2011. Duffy negative antigen is no longer a barrier to Plasmodium vivax—molecular evidences from the African west coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis 5: e1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirier P, et al. 2016. The hide and seek of Plasmodium vivax in west Africa: report from a large-scale study in Beninese asymptomatic subjects. Malar J 15: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motshoge T, et al. 2016. Molecular evidence of high rates of asymptomatic P. vivax infection and very low P. falciparum malaria in Botswana. BMC Infect Dis 16: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngassa Mbenda HG, Das A, 2014. Molecular evidence of Plasmodium vivax mono and mixed malaria parasite infections in Duffy-negative native Cameroonians. PLoS One 9: e103262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo G, et al. 2017. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, west Cameroon. Malar J 16: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woldearegai TG, Kremsner PG, Kun JFJ, Mordmüller B, 2013. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans R Soc Trop Med Hyg 107: 328–331. [DOI] [PubMed] [Google Scholar]
- 19.Gunalan K, Lo E, Hostetler JB, Yewhalaw D, Mu J, Neafsey DE, Yan G, Miller LH, 2016. Role of Plasmodium vivax Duffy-binding protein 1 in invasion of Duffy-null Africans. Proc Natl Acad Sci USA 113: 6271–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan JR, et al. 2006. Evidence for transmission of Plasmodium vivax among a Duffy antigen negative population in western Kenya. Am J Trop Med Hyg 75: 575–581. [PubMed] [Google Scholar]
- 21.Wurtz N, et al. 2011. Vivax malaria in Mauritania includes infection of a Duffy-negative individual. Malar J 10: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdelraheem MH, Albsheer MMA, Mohamed HS, Amin M, Mahdi Abdel Hamid M, 2016. Transmission of Plasmodium vivax in Duffy-negative individuals in central Sudan. Trans R Soc Trop Med Hyg 110: 258–260. [DOI] [PubMed] [Google Scholar]
- 23.Zimmerman PA, 2017. Plasmodium vivax infection in Duffy-negative people in Africa. Am J Trop Med Hyg 97: 636–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meshnick S, Janko M, Doctor S, Anderson O, Thwai K, Levitz L, Emch M, Mwandagalirwa K, Tshefu A, Ntuku H, 2015. Democratic Republic of the Congo Demographic Health Survey 2013–2014 Supplemental Malaria Report Available at: http://dhsprogram.com/pubs/pdf/FR300/FR300.Mal.pdf. Accessed August 25, 2017.
- 25.Doctor SM, et al. 2016. Malaria surveillance in the Democratic Republic of the Congo: comparison of microscopy, PCR, and rapid diagnostic test. Diagn Microbiol Infect Dis 85: 16–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE, 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 52: 565–568. [DOI] [PubMed] [Google Scholar]
- 27.Doctor SM, et al. 2016. Malaria surveillance in the Democratic Republic of the Congo: comparison of microscopy, PCR, and rapid diagnostic test. Diagn Microbiol Infect Dis 85: 16–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doctor SM, et al. 2016. Low prevalence of Plasmodium malariae and Plasmodium ovale mono-infections among children in the Democratic Republic of the Congo: a population-based, cross-sectional study. Malar J 15: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh B, Bobogare A, Cox-Singh J, Snounou G, 1999. A genus-and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg 60: 687–692. [DOI] [PubMed] [Google Scholar]
- 30.Veron V, Simon S, Carme B, 2009. Multiplex real-time PCR detection of P. falciparum, P. vivax and P. malariae in human blood samples. Exp Parasitol 121: 346–351. [DOI] [PubMed] [Google Scholar]
- 31.Methodology DHS The Demographic Health Survey Program Available at: http://dhsprogram.com/What-We-Do/Survey-Types/DHS-Methodology.cfm. Accessed September 1, 2017.
- 32.Ministère du Plan et Suivi de la Mise en œuvre de la Révolution de la Modernité (MPSMRM), Ministère de la Santé Publique (MSP) et ICF International , 2014. Enquête Démographique et de Santé en République Démocratique du Congo 2013–2014. Rockville, MD: MPSMRM, MSP and ICF International.
- 33.Wickham H, 2017. Tidyverse: easily install and load ‘tidyverse’ packages. R Package Version 1(1). Available at: https://tidyverse.tidyverse.org/. Accessed September 10, 2018.
- 34.R Core Team , 2018. R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org/. Accessed Septmeber 6, 2018.
- 35.The IUCN Red List of Threatened Species, IUCN , 2016. Available at: http://www.iucnredlist.org. Accessed April 21, 2018. Version 2016-1.
- 36.Abkallo HM, et al. 2014. DNA from pre-erythrocytic stage malaria parasites is detectable by PCR in the faeces and blood of hosts. Int J Parasitol 44: 467–473. [DOI] [PubMed] [Google Scholar]
- 37.Hupalo DN, et al. 2016. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet 48: 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson RD, et al. 2016. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 48: 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parobek CM, et al. 2016. Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc Natl Acad Sci USA 113: E8096–E8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hester J, Chan ER, Menard D, Mercereau-Puijalon O, Barnwell J, Zimmerman PA, Serre D, 2013. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl Trop Dis 7: e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables and figure