Abstract.
The escalating burden of infections attributable to methicillin-resistant Staphylococcus aureus (MRSA) in East African countries is calling for interventional strategies to control the spread of this strain. The present study aimed at determining the prevalence, antimicrobial profiles, and staphylococcal cassette chromosome mec (SCCmec) typing of MRSA strains. This was a cross-sectional laboratory-based study involving 226 non-duplicated S. aureus isolates from different clinical samples of patients attending a referral hospital in Kigali. Kirby–Bauer disk diffusion method was used for drug susceptibility testing. Methicillin-resistant S. aureus were confirmed using polymerase chain reaction (PCR) assay for the mecA gene and SCCmec type PCR assay was used for genotyping. Of 138 S. aureus, 39 (31.2%) were found to be MRSA strains. The mean age of the patients was 21.9 years. The incidence of MRSA increases with age and was 27.1% in patient age group younger than 18 years, 33.3% in the age group between 19 and 65 years, and 66.7% in patient age group older than 65 years. There was a significant association between geographic regions and incidence of MRSA (P = 0.02) with the high MRSA isolates from Northern (61.5%) and Western (50%) provinces. Methicillin-resistant S. aureus strains were found to be mostly susceptible to linezolid (93.5%). Among the MRSA strains, SCCmec type I and SCCmec type IV were the most prevalent at 56.4% and 17.9%, respectively. A high prevalence of MRSA was found in Rwanda. Staphylococcal cassette chromosome mec type I (52.2%) was the most predominant. A continuous surveillance of MRSA strains, particularly in the hospital settings, should be an enduring exercise in Rwanda.
INTRODUCTION
Staphylococcus aureus remains a major public health problem worldwide as it causes local infections such as impetigo, folliculitis, cellulitis, wound sepsis, and invasive diseases such as bacteremia, necrotizing pneumonia, osteomyelitis, meningitis, endocarditis, toxic shock syndrome, and sepsis.1
The evolution of resistance in S. aureus has a profound historical background since penicillin was introduced in the market in the early 1940s as the treatment of choice for S. aureus.2 Twenty years later, S. aureus isolates account for 80% of penicillin resistance, thus development of methicillin (β-lactamase–resistant penicillins) as an alternative solution to this problem.2,3 In 1961, 2 years following introduction of methicillin, the first strain of methicillin-resistant S. aureus (MRSA) was reported in the United Kingdom and shortly after, the strain spread all over the world and became pandemic in various health facilities.4,5 Resistance to methicillin is due to the presence of chromosomal mecA gene, which encodes for low-affinity penicillin-binding protein 2a responsible for the resistance to methicillin and all other β-lactam antibiotics.4,6 The mecA gene is found on a mobile genetic element known as staphylococcal cassette chromosome mec (SCCmec).7 Depending on molecular size, five main types of SCCmec (type I–V) have been identified. Types I, II, and III are associated with hospital-acquired MRSA (HA-MRSA), whereas types IV and V are associated with community-acquired MRSA (CA-MRSA).4 In addition, SCCmec harbor genes responsible for β-lactam and non-β-lactam antimicrobial agents.4 Multiplex PCR which can detect the SCCmec types has been shown to be relatively simple and less costly to be used in developing countries compared with other molecular typing methods such as multilocus sequence typing and pulsed-field gel electrophoresis (PFGE).8–10
Methicillin-resistant S. aureus is a global health threat both in developed and developing countries, and its burden is projected to escalate and confer negative health impact across regions.1,4,11 A study conducted in eight African countries in 2003 showed an overall prevalence of 15%.12 Moreover, a significant genotypic diversity has been recently shown by a systematic review across African countries, necessitating the need to have country-specific surveillance for effective MRSA-associated infections prevention and control.1 In Eastern African Community Region (EAC), the proportion of MRSA among S. aureus isolates has been reported at a rate of 15%, 38%, and 84% in Tanzania, Uganda, and Kenya, respectively.10,13–15 Despite the prevailing information on the magnitude of MRSA and genotypic diversity in EAC and other African countries,1 limited information exists regarding MRSA in Rwanda. Therefore, the present study aimed at determining the prevalence, antimicrobial resistance patterns, and SCCmec genotypic of MRSA strains from clinical specimens among patients attending a University teaching hospital in Kigali, Rwanda, in order to have a baseline information for the management of MRSA-associated infections as well as infection prevention and control.
MATERIAL AND METHODS
Study design, sites, and sampling methods.
This was a cross-sectional laboratory-based study conducted from April to May 2014 using 226 archived S. aureus isolates. The study was conducted at Center Hospitalier Universtaire de Kigali (CHUK), Rwandan Biomedical Center National Reference Laboratory Division (RBC/BIOS-NRL), and Molecular Biology Laboratory Makerere University College of Health Sciences (MakCHS).
Center Hospitalier Universtaire de Kigali is a 441-bed referral hospital located in Kigali city; it receives approximately three-fourth of all referral cases in Rwanda, particularly from Kigali city, Northern, Eastern, and some districts of Western province. Collected S. aureus were from various clinical specimens obtained from inpatients and outpatients who attended this hospital from June 2013 to April 2014 and were kept at −80°C freezer.
The RBC/BIOS-NRL Division is one of the divisions of Rwanda Biomedical Center and is located at Boulevard de la Revolution in Kigali, Rwanda. The laboratory is responsible of developing policies regulating laboratories in Rwanda, training laboratory personnel, supervising laboratories, and providing external quality control of health facilities in Rwanda.
The Molecular Biology Laboratory is one of the teaching and research laboratories of the Department of Medical Microbiology in MakCHS, located at Mulago Hill in Kampala, Uganda. This laboratory helped in performing the mecA gene and SCCmec typing molecular assays.
Data collection and laboratory procedures.
Achieved non-duplicated S. aureus isolates were recorded into the study log book by using codes and were subcultured on blood agar supplemented with 5% sheep blood. Phenotypic reidentification of S. aureus was based on catalase, slide, and tube coagulase tests.16 Following identification, antimicrobial susceptibility testing (AST) was performed by Kirby–Bauer disk diffusion method on Muller–Hinton agar plate according to Clinical and Laboratory Standards Institute recommendations.17
Antimicrobial agents used to determine S. aureus drug susceptibility patterns included penicillin (10 units), cefoxitin (30 μg), erythromycin (15 μg), clindamycin (2 μg), trimethoprim–sulfamethaxazole (1.35/23.75 μg), tetracycline (30 μg), linezolid (30 μg), rifampin (5 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamycin (10 μg), and oxacillin (1 μg).18 All S. aureus strains with zone inhibition ≤ 21 mm of diameter to cefoxitin, were considered as MRSA and all strains with zone inhibition ≥ 22 mm were considered as MSSA.19 Oxacillin disk was also used in phenotypic detection of MRSA as previously described.16
Pure colonies of S. aureus isolates were inoculated into labeled cryovials of brain heart infusion supplemented with 20% glycerol and were triple-packaged during transportation to the Molecular Biology Laboratory in MakCHS into a cool box containing ice plastic bottles to maintain low temperature required for survival of isolates. A Material Transfer Agreement was signed between the RBC/BIOS-NRL Division and the Molecular Biology Laboratory, MakCHS, before shipment of isolates to Uganda for mecA gene and SCCmec typing assays.
Molecular assays.
DNA was extracted from S. aureus isolates using the crude DNA extraction method as previously described.18
Forward primer P4: 5′-TCCAATTACAACTTCACCAGG-3′ and reverse primer P7: 5′-CCACTTCATATCTTGTAACG-3′ were used to amplify segment of mecA gene by PCR method as described before.10 The mecA gene detection is universally considered as a golden standard in the diagnosis of MRSA as it detects the gene which encodes for methicillin resistance and other β-lactam antibiotics.
Staphylococcal cassette chromosome mec gene was amplified by using four forward and four reverse primers shown in the following table to detect the five major types of SCCmec (type I–V) using a multiplex PCR method as previously described.8 Two multiplex PCR were performed; the first multiplex amplified CCrC (CCrC F and R) and mecA-IS431 (5RmecA and 5R431), whereas the second multiplex amplified ccrA2-B (beta and alpha-3) and IS1272 (1272F1 and 1272R1).10 In case no band was observed, the respective isolate was regarded as “non-typeable.”
| Name of primer | Sequence of primer (5′ 3′) | Band size in bp | Target gene |
|---|---|---|---|
| β and α3 | ATTGCCTTGATAATAGCCYTCTTAAAGGCATCAATGCACAAACACT | 937 | ccrA2-B |
| ccrCF and ccrCR | CGTCTATTACAAGATGTTAAGGATAAT | 518 | ccrC |
| CCTTTATAGACTGGATTATTCAAAATAT | |||
| 1272F1 and 1272R1 | GCCACTCATAACATATGGAA | 415 | IS1272 |
| CATCCGAGTGAAACCCAAA | |||
| 5RmecA and 5R431 | TATACCAAACCCGACAACTAC | 359 | mecA-IS431 |
| CGGCTACAGTGATAACATCC |
The amplicons were analyzed by electrophoresis using a 2% TAE agarose gel in 1× TAE (Tris-acetate-EDTA) buffer run at constant voltage of 120 for 1 hour. The images were visualized and captured using the Bio-imager (UVP, LLC, Upland, CA).
Quality control.
Staphylococcus aureus ATCC 25923 and Staphylococcus epidermidis ATCC 12944 were used as positive control and negative control for identification tests. For AST, S. aureus ATCC 25923 was used to quality control antibiotic disks.17 Staphylococcus aureus ATCC 25923 and S. aureus ATCC 43300 were used as negative control and positive control for mecA gene detection, respectively.17
Data analysis.
Sociodemographic and clinical information of included isolates were retrieved from laboratory registers and hospital information management system and transferred to the Microsoft Excel for consistency check. The Statistical Package for the Social Sciences software word version 20.0 was used to analyze the data.
Study permission and ethical considerations.
Ethical permission was obtained from Rwanda National Ethics Committee and Rwanda Ministry of Education, Science, Technology and Research.
RESULTS
Description of study population and incidence of MRSA in subgroups.
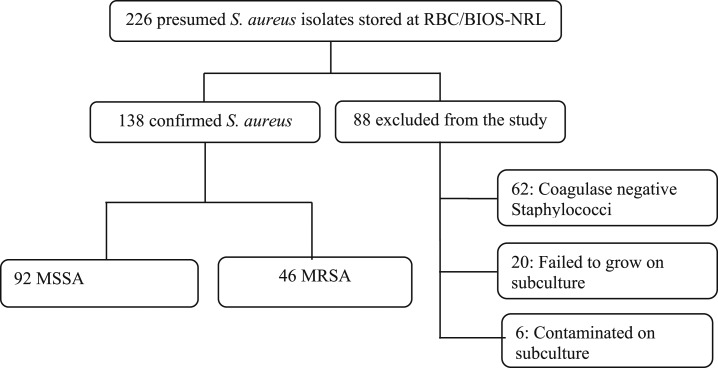
The study involved 138 confirmed S. aureus isolates from a total of presumed 226 non-duplicated S. aureus isolates; other isolates were excluded for various reasons (Figure 1). Demographic characteristics of the study participants were available in 125 individuals (90.6%) and are described in the Table 1. Of total isolates, 39 isolates (31.2%) were found positive for MRSA (Table 1). The study population comprised 70 (56%) males, with 31.4% MRSA prevalence, and 55 (46%) females, with 30.8% MRSA prevalence. The mean age of the patients was 21.9 years (standard deviation, 18.9 years), with a range of 8 months to 73 years. The incidence of MRSA increases with age and was 27.1% in patient age group less than 18 years, 33.3% in the age group between 19 and 65 years, and 66.7% in patient age group older than 65 years. However, no significant difference was seen between MRSA and age groups (P = 0.309). The majority of isolates (80.8%) were retrieved from pus swabs followed by blood sample (11, 2%). There was a significant association between province and incidence of MRSA (P = 0.02) with the high MRSA isolates from Northern (61.5%) and Western (50%) provinces.
Figure 1.
Enrollment of isolates. RBC/BIOS-NRL = Rwanda Biomedical Center National Reference Laboratory; MRSA = methicillin-resistant S. aureus; MSSA = methicillin-sensitive Staphylococcus aureus.
Table 1.
Distribution of MRSA in different groups
| Total (%) (N = 125) | N (%) MRSA (N = 39) | P value | |
|---|---|---|---|
| Sex | |||
| Male | 70 (56.0) | 22 (31.4) | 0.950 |
| Female | 55 (44.0) | 17 (30.9) | – |
| Age, years, mean = 21.9 ± (18.9) | |||
| ≤ 18 | 59 (47.2) | 16 (27.1) | 0.308 |
| 19–64 | 63 (50.4) | 21 (33.3) | – |
| ≥ 65 | 3 (22.4) | 2 (66.7) | – |
| Specimen | |||
| Pus | 101 (80.8) | 28 (27.7) | 0.208 |
| Blood culture | 14 (11.2) | 7 (50) | – |
| Nasal swab | 2 (1.6) | 0 (0) | – |
| Skin swab | 2 (1.6) | 1 (50) | – |
| *Others | 6 (4.8) | 3 (50) | – |
| Ward | |||
| Surgical | 47 (37.6) | 14 (29.8) | 0.723 |
| Pediatrics | 28 (22.4) | 7 (25) | – |
| Internal medicine | 15 (12.0) | 6 (40) | – |
| Gynecology Obstetrics | 9 (7.2) | 4 (44) | – |
| Outpatient department | 4 (3.2) | 2 (50) | – |
| Theater | 5 (4.0) | 2 (40) | – |
| Others† | 17 (13.6) | 4 (23.5) | – |
| Province | |||
| Kigali city | 53 (42.4) | 11 (20.8) | 0.020 |
| Eastern | 26 (20.8) | 9 (34.6) | – |
| Northern | 13 (10.4) | 8 (61.5) | – |
| Western | 14 (11.2) | 7 (50) | – |
| Southern | 19 (42.4) | 4 (21.1) | – |
MSSA = methicillin-sensitive Staphylococcus aureus.
Other specimens included alveolar punction, ascite punction, eye pus, skin pus, synovial punction, and urine.
Other wards included emergency ear, nose, and throat; neonatology; dermatology; and ophthalmology.
Susceptibility of S. aureus to different antibiotics.
The majority of the S. aureus isolates were resistant to penicillin (96.4%), trimethoprim–sulfamethaxazole (34.1%), and tetracycline (34.1%). Most of the S. aureus isolates were susceptible to chloramphenicol (83.3%), ciprofloxacin (85.5%), gentamicin (87%), and linezolid (94.9%) (Table 2).
Table 2.
Staphylococcus aureus antimicrobial susceptibility pattern (N = 138)
| Antimicrobials | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Penicillin | 5 (3.6) | NA | 133 (96.4) |
| Oxacillin | 89 (64.5) | 13 (9.4) | 36 (26.1) |
| Erythromycin | 98 (71.0) | 15 (10.9) | 25 (18.1) |
| Clindamycin | 107 (77.5) | 13 (9.4) | 18 (13.0) |
| Trimethoprim–sulfamethaxazole | 86 (62.3) | 5 (3.6) | 47 (34.1) |
| Tetracycline | 87 (63.0) | 4 (2.9) | 47 (34.1) |
| Chloramphenicol | 115 (83.3) | 6 (4.3) | 17 (12.3) |
| Ciprofloxacin | 118 (85.5) | 8 (5.8) | 12 (8.7) |
| Gentamicin | 120 (87.0) | NA | 18 (13.0) |
| Linezolid | 131 (94.9) | NA | 7 (5.1) |
MRSA = methicillin-resistant Staphylococcus aureus; NA = not applicable.
Prevalence of MRSA among S. aureus isolates.
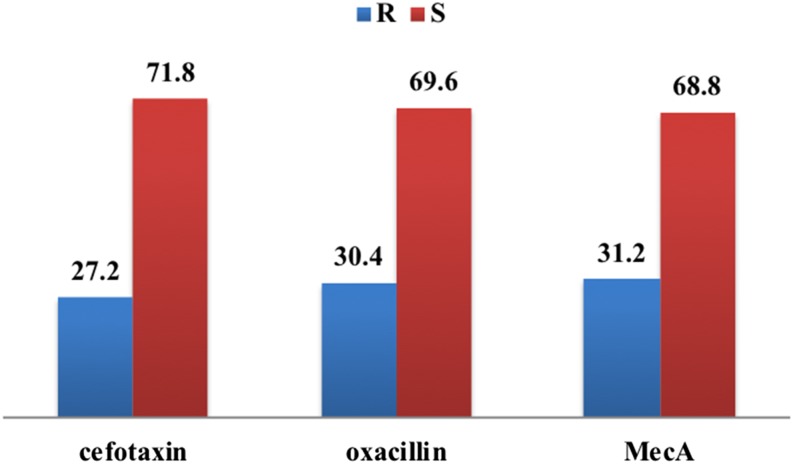
Based on the presence of mecA gene which is a gold standard, the proportion of MRSA among S. aureus isolates was 31.2% (39/138), whereas the proportion of MRSA by both phenotypic tests (cefoxitin and oxacillin disks) was 27.2% (34/138). Thus, the gold standard test detected three more MRSA isolates compared with phenotypic methods (Figures 2 and 3).
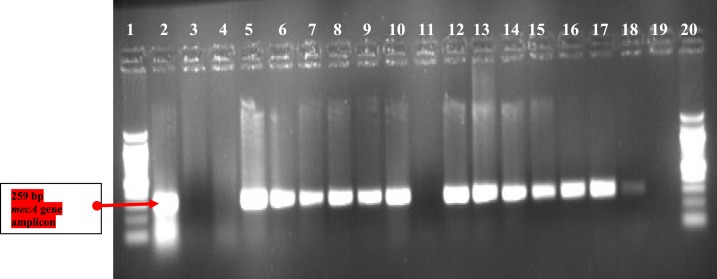
Figure 2.
mecA PCR agarose gel electrophoresis. Lanes 1 and 20 are 100-bp DNA ladder. Lanes 2 and 19 show the positive and negative controls. Lanes 3, 4, and 11 show mecA negative, whereas the rest show mecA-positive Staphylococcus aureus isolates. This figure appears in color at www.ajtmh.org.
Figure 3.
Prevalence of methicillin-resistant Staphylococcus aureus based on different methods. This figure appears in color at www.ajtmh.org.
Resistance of MRSA to other antibiotics.
Methicillin-resistant S. aureus strains showed a high resistance to trimethoprim–sulfamethaxazole 47.8%, erythromycin 41.3%, and tetracycline 39.1%, but low resistance to linezolid (6.5%) (Table 3).
Table 3.
Resistance of methicillin-resistant Staphylococcus aureus (N = 46) to antibiotics
| Antimicrobials | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Trimethoprim–sulfamethaxazole | 24 (52.2) | 0 (0.0) | 22 (47.8) |
| Erythromycin | 20 (43.5) | 7 (15.2) | 19 (41.3) |
| Tetracycline | 25 (54.3) | 3 (6.5) | 18 (39.1) |
| Gentamicin | 29 (63.0) | 0 (0.0) | 17 (37.0) |
| Clindamycin | 24 (52.2) | 9 (19.6) | 13 (28.3) |
| Chloramphenicol | 31 (67.4) | 2 (4.3) | 13 (28.3) |
| Ciprofloxacin | 31 (67.4) | 4 (8.7) | 11 (23.9) |
| Rifampin | 36 (78.3) | 2 (4.3) | 8 (17.4) |
| Linezolid | 43 (93.5) | 0 (0.0) | 3 (6.5) |
Genetic diversity of SCCmec in MRSA isolates.
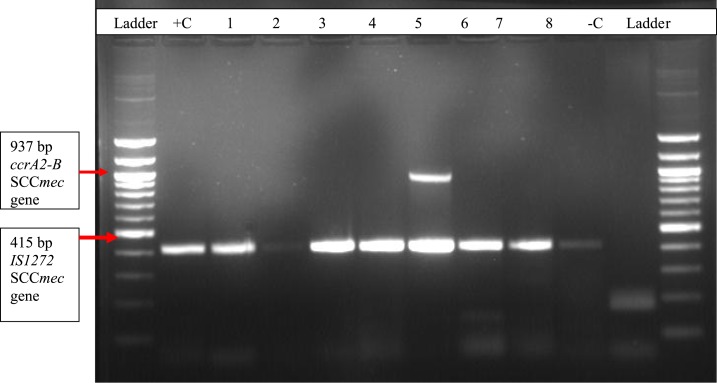
A multiplex PCR targeting four different MRSA genes was performed to differentiate the strains into HA-MRSA and CA-MRSA using SCCmec typing (Figure 4). Staphylococcal cassette chromosome mec type I was found to be the most prevalent with 52.2% (24/46, followed by SCCmec type IV 15.4% (7/46) and SCCmec type II was not found among MRSA strains in Rwanda. Non-typeable MRSA strains also have a high proportion of 26.6% (12/46).
Figure 4.
IS1272 and ccrA2-B genes on PCR agarose gel electrophoresis; 100-bp DNA ladder. Lanes +C and –C show the positive and negative controls. Lanes 1, 2, 3, 4, 6, 7, and 8 show IS1272 positive staphylococcal cassette chromosome mec (SCCmec type I), whereas lane 5 shows IS127 and ccrA2-B–positive MRSA (SCCmec type IV). This figure appears in color at www.ajtmh.org.
Distribution of SCCmec types among MRSA and in different groups is shown in Table 4. The majority of the MRSA strains belonged to SCCmec type I (22 stains, 56.4%). Staphylococcal cassette chromosome mec type IV a SCCmec type III were found in seven strains (17.9%) and in one strain (2.6%), respectively. Staphylococcal cassette chromosome mec type V was not confirmed in any of the MRSA strains, whereas nine strains (23.1%) could not carry any of the previously described SCCmec types. Social demographic data showed a disproportional distribution of SCCmec type in the different groups. The occurrence of SCCmec type I and type IV was similar in the age groups of less than 18 years (10 strains, 58.8% and three strains, 17.6%, respectively) and 19–64 years (11 strains, 52.4% and four strains, 19%, respectively). Females had high proportion of SCCmec type I 63.6% (14/22) compared with males 47.1 (8/17). Staphylococcal cassette chromosome mec type I was more predominantly carried out in MRSA strains from surgical department (81.3% [13/16]) and pus specimens (60.7 [7/28]), and Eastern Province (70% [7/10]).
Table 4.
Prevalence of SCCmec type in different groups (N = 39)
| SCCmec types | |||||
| Subgroup (n) | Non-typeable 9 (23.1) | Type I 22 (56.4) | Type III 1 (2.6) | Type IV 7 (17.9) | |
| Age group | ≤ 18 (17) | 3 (17.6) | 10 (58.8) | 1 (6) | 3 (17.6) |
| 19–64 (21) | 6 (28.6) | 11 (52.4) | 0 (0.0) | 4 (19) | |
| ≥ 65 (1) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | |
| Gender | Female (22) | 5 (27.7) | 14 (63.6) | 1 (4.6) | 2 (9.1) |
| Male (17) | 4 (23.5) | 8 (47.1) | 0 (0.0) | 5 (29.4) | |
| Specimen | Pus (28) | 5 (17.9) | 17 (60.7) | 0 (0.0) | 6 (21.4) |
| Blood culture (7) | 2 (28.6) | 3 (42.8) | 1 (14.3) | 1 (14.3) | |
| Others* (4) | 2 (50) | 2 (50) | 0 (0.0) | 0 (0.0) | |
| Ward | Surgical (16) | 1 (6.3) | 13 (81.3) | 0 (0.0) | 3 (18.6) |
| Pediatrics (7) | 2 (28.6) | 2 (28.6) | 1 (14.2) | 2 (28.6) | |
| Internal medicine (5) | 1 (20) | 3 (60) | 0 (0.0) | 1 (20) | |
| Gynecology obstetrics (4) | 2 (50) | 1 (25) | 0 (0.0) | 1 (25) | |
| Outpatient department (2) | 1 (50) | 1 (50) | 0 (0.0) | 0 (0.0) | |
| Others† (4) | 2 (50) | 2 (50) | 0 (0.0) | 0 (0.0) | |
| Province | Kigali city (10) | 4 (40) | 5 (50) | 0 (0.0) | 1 (10) |
| Eastern (10) | 1 (10) | 7 (70) | 1 (10) | 1 (10) | |
| Northern (9) | 2 (22.2) | 4 (44.4) | 0 (0.0) | 3 (33.4) | |
| Western (6) | 1 (16.7) | 4 (66.6) | 0 (0.0) | 1 (16.7) | |
| Southern (4) | 1 (25) | 2 (50) | 0 (0.0) | 1 (25) | |
SCCmec = staphylococcal cassette chromosome mec.
Other specimens included alveolar punction, ascite punction, eye pus, skin pus, synovial punction, and urine.
Other wards included emergency ear, nose, and throat; neonatology; dermatology; and ophthalmology.
DISCUSSION
Staphylococcus aureus causes local and invasive infections of public health importance both in the community and hospital settings.19–21 For the past six decades, this species has progressively developed resistance to β-lactams and other antibiotics posing treatment challenges, which in turn necessitates a need for continuous surveillance strategies to curb its spread both in the community and hospital settings.1,2,4 To our knowledge, this is the first study from Rwanda that provides the local epidemiological data and genetic diversity of MRSA. Although, not statistically significant, MRSA prevalence seems to increases with age, which is consistent with previous report showing an association between age and both the rate of MRSA.12 Kigali Province contributed to almost half of isolates (42.4%), most likely as it is the most populated province and the majority of patient admitted at CHUK are from the province. Northern Province had the highest prevalence of MRSA (61.5%). There are no clear reasons for this geographic variation of MRSA in this study; however, this might be explained by geographic variation in the prevalence of organisms in the hospitals across the country.
Although penicillin is among the cheapest antibiotics and therefore the most affordable for developing countries with limited resources, this study proved it to be ineffective in Rwanda health facilities as almost 100% of S. aureus isolates were resistant to the drug; these findings are similar to another study conducted in Uganda.10 The finding about high rates of penicillin resistance is not surprising and is also in line with several studies carried out in Africa that also reported a greater than 90% resistance to penicillin.1 Interestingly, S aureus isolates in the present study were susceptible to other non-β lactam antibiotics such as gentamicin, ciprofloxacin, and chloramphenicol which can be potential treatment options in Rwanda. The susceptibility to gentamicin (87.5%) is similar to another study in Uganda,22 but in contrary to other studies which showed low susceptibility to gentamicin and ciprofloxacin in approximately 15.9% and 25.6% respectively.13,23 The present study found a high prevalence of MRSA (33.3%) which may present the management challenges of infections associated with this strain because the readily available and cheap β-lactam drugs cannot be used, whereas the cost of the second-line drugs such as vancomycin is prohibitive. The MRSA prevalence in Rwanda is similar to the proportions ranging from 25% to 37.5% found in three studies conducted in Uganda10,22,24 but higher than 10% reported from Tunisia, Malta, and Algeria.15 Moreover, the prevalence of MRSA in this study is low compared with 46%, 59.8%, and 84% found in northern India, China, and Kenya, respectively.13,23,25 There is also variation in the rate of MRSA isolates resistant to vancomycin, refered as vancomycin-intermediate S. aureus (VISA), which is the last drug of choice. Although, this study could provide data VISA, the moderate rate of MRSA may suggest similar observation with reports of other studies in Tanzania, Kenya, and Uganda where no resistance to vancomycin was reported.10,13,15 The difference on the resistance rates may be due to different antimicrobial use policies, infection control practices, and different studied populations in these countries, but the finding about VRSA in Rwanda calls for further analysis with reliable antibiotic susceptibility method to ascertain its occurrence, source, and promptly control and prevent its spread.
Previous evidences suggest that SCCmec types I, II, and III tend to be more related to HA-MRSA, whereas SCCmec types IV and V are associated with CA-MRSA. This study found that SCCmec type I (56.4%) was the most predominant among MRSA strains, whereas SCCmec type II and V were not found among mecA gene-positive isolates in Rwanda. The majority of these isolates were collected from surgical wards, which accounted for 81.3% of the SCCmec type I followed by type IV (18.6%). Similar to other studies in Uganda, the predominance of SCCmec type I in surgical ward may indicate the possibility of transmission of these strains in the hospital setting10,26 and thus a need to emphasize on strengthening of infection control practices in Rwanda to reduce MRSA nosocomial infections. As opposed to SCCmec type I, the predominance of SCCmec type III (57.6%) and SCCmec type II (22.0%) was found in another study conducted in China.25 The low proportion of CA-MRSA may pin point proper usage of antibiotics in community settings in Rwanda.
However, 26% of MRSA isolates could not be assigned to any SCCmec types, and this may probably reflect the low discriminatory power of the method and thus calling for the need to use other cost-effective methods with high discriminatory power such as spa sequence typing.10
Limitations.
This study did not use the gold standard for molecular typing of strains which is PFGE; despite this, a baseline SCCmec typing to delineate nosocomial versus community-associated MRSA strains circulating in Rwanda has been conducted.
CONCLUSION
For the first time, high prevalence of MRSA (31.2%) in Rwanda has been documented and is associated with certain age groups and geographic regions, which has an important implication for developing prevention programs.
Staphylococcus aureus isolates from Rwanda are more susceptible to gentamicin, ciprofloxacin, chloramphenicol, and linezolid, whereas MRSA are mostly susceptible to linezolid. Staphylococcal cassette chromosome mec type I (52.2%) was found to be the most predominant MRSA genotype and mostly isolated from surgical pus specimens suggesting nosocomial MRSA transmission. A continuous surveillance of MRSA strains, particularly in the hospital settings, should be an enduring exercise in Rwanda so as to curb the transmission of MRSA strains in this setting.
Acknowledgments:
We thank Thadee Bwanakweri (RBC/BIOS-NRL Bacteriology Section); Edgar Kigozi and Fred Ashaba Gatabazi (Molecular Biology/Department of Medical Microbiology, Makerere University); and other staff members in Rwanda and Uganda for their excellent technical assistance and support.
REFERENCES
- 1.Abdulgader SS, Shittu AO, Nicol MP, Kaba M, 2015. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol 6: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG, 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci USA 99: 7687–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreillon P, Que Y, Glauser MP, 2005. Staphylococcus aureus (including staphylococcal toxic shock). Mandell GL, Bennett JE, Dolin R, eds. Principles and Practice of Infectious Diseases, 6th edition. London, United Kingdom: Churchill Livingstone, An Imprint of Elsevier. [Google Scholar]
- 4.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE, 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 13: 222–235. [DOI] [PubMed] [Google Scholar]
- 5.Deurenberg RH, Stobberingh EE, 2008. The evolution of Staphylococcus aureus. Infect Genet Evol 8: 747–763. [DOI] [PubMed] [Google Scholar]
- 6.Palavecino E, 2007. Clinical, epidemiological, and laboratory aspects of methicillin-resistant Staphylococcus aureus (MRSA) infections. Methods Mol Biol 391: 1–19. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K, 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat 6: 41–52. [DOI] [PubMed] [Google Scholar]
- 8.Boye K, Bartels MD, Andersen IS, Moller JA, Westh H, 2007. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I–V. Clin Microbiol Infect 13: 725–727. [DOI] [PubMed] [Google Scholar]
- 9.Struelens MJ, Hawkey PM, French GL, Witte W, Tacconelli E, 2009. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin Microbiol Infect 15: 112–119. [DOI] [PubMed] [Google Scholar]
- 10.Seni J, Bwanga F, Najjuka CF, Makobore P, Okee M, Mshana SE, Kidenya BR, Joloba ML, Kateete DP, 2013. Molecular characterization of Staphylococcus aureus from patients with surgical site infections at Mulago Hospital in Kampala, Uganda. PLoS One 8: e66153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Astagneau P, Rioux C, Golliot F, Brucker G, 2001. Morbidity and mortality associated with surgical site infections: results from the 1997–1999 INCISO surveillance. J Hosp Infect 48: 267–274. [DOI] [PubMed] [Google Scholar]
- 12.Kesah C, Ben Redjeb S, Odugbemi TO, Boye CS, Dosso M, Ndinya Achola JO, Koulla-Shiro S, Benbachir M, Rahal K, Borg M, 2003. Prevalence of methicillin-resistant Staphylococcus aureus in eight African hospitals and Malta. Clin Microbiol Infect 9: 153–156. [DOI] [PubMed] [Google Scholar]
- 13.Maina EK, Kiiyukia C, Wamae CN, Waiyaki PG, Kariuki S, 2013. Characterization of methicillin-resistant Staphylococcus aureus from skin and soft tissue infections in patients in Nairobi, Kenya. Int J Infect Dis 17: e115–e119. [DOI] [PubMed] [Google Scholar]
- 14.Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE, 2010. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr 10: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moremi N, Mshana SE, Kamugisha E, Kataraihya J, Tappe D, Vogel U, Lyamuya EF, Claus H, 2012. Predominance of methicillin resistant Staphylococcus aureus -ST88 and new ST1797 causing wound infection and abscesses. J Infect Dev Ctries 6: 620–625. [DOI] [PubMed] [Google Scholar]
- 16.Broekema NM, Van TT, Monson TA, Marshall SA, Warshauer DM, 2009. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in Staphylococcus aureus in a large-scale study. J Clin Microbiol 47: 217–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cockerill F, 2012. Performance Standards for Antimicro bial Susceptibility Testing: Twenty-Second Informational Supplement. Clinical and Laboratory Standards Institute (CLSI document M100-MS22). Wayne, PA: CLSI.
- 18.Kateete DP, Kimani CN, Katabazi FA, Okeng A, Okee MS, Nanteza A, Joloba ML, Najjuka FC, 2010. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann Clin Microbiol Antimicrob 9: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA, 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 355: 666–674. [DOI] [PubMed] [Google Scholar]
- 20.Lowy FD, 1998. Staphylococcus aureus infections. N Engl J Med 339: 520–532. [DOI] [PubMed] [Google Scholar]
- 21.McCaig LF, McDonald LC, Mandal S, Jernigan DB, 2006. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg Infect Dis 12: 1715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anguzu JR, Olila D, 2007. Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda. Afr Health Sci 7: 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arora S, Devi P, Arora U, Devi B, 2010. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in a tertiary care hospital in northern India. J Lab Physicians 2: 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ojulong J, Mwambu TP, Joloba M, Bwanga F, Kaddu-Mulindwa DH, 2009. Relative prevalence of methicilline resistant Staphylococcus aureus and its susceptibility pattern in Mulago Hospital, Kampala, Uganda. Tanzan J Health Res 11: 149–153. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, et al. 2013. Molecular and phenotypic evidence for the spread of three major methicillin-resistant Staphylococcus aureus clones associated with two characteristic antimicrobial resistance profiles in China. J Antimicrob Chemother 68: 2453–2457. [DOI] [PubMed] [Google Scholar]
- 26.Kateete DP, Namazzi S, Okee M, Okeng A, Baluku H, Musisi NL, Katabazi FA, Joloba ML, Ssentongo R, Najjuka FC, 2011. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res Notes 4: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]