Abstract.
A clinical, serological, and molecular investigation was performed to determine the presence of dengue virus (DENV) and other flaviviruses among residents of the city of Reynosa, Tamaulipas, on the Mexico–U.S. border in 2014–2016. The sample population consisted of 2,355 patients with suspected dengue, in addition to 346 asymptomatic individuals recruited during a household-based epidemiological investigation designed to identify flavivirus seroconversions. Sera were collected from patients with suspected dengue in the acute phase of illness and from asymptomatic individuals at enrollment and every 5–7 months for 19 months. Sera from suspected dengue patients were tested for DENV antigen by enzyme-linked immunosorbent assay (ELISA), and select antigen-positive sera were further tested using a serotype-specific, quantitative reverse transcription–polymerase chain reaction. Sera from the household cohort were tested for flavivirus-reactive antibodies by immunoglobulin (Ig) M and IgG ELISAs using DENV antigen. A total of 418 (17.7%) patients with suspected dengue had laboratory-confirmed DENV infections, including 82 patients who were positive for DENV RNA. The most frequently detected serotype was DENV-1 (61 patients), followed by DENV-2 (16 patients) and DENV-3 (five patients). A total of 217 (62.7%) asymptomatic individuals had flavivirus-reactive antibodies at enrollment, and nine flavivirus-naïve individuals seroconverted. Sera from a subset of dengue patients and household participants, including all those who seroconverted, were further tested by plaque reduction neutralization test, resulting in the detection of antibodies to DENV-1, DENV-2, and West Nile virus. In summary, we provide evidence for the co-circulation of multiple flaviviruses in Reynosa, Tamaulipas, on the Mexico–U.S. border.
INTRODUCTION
Dengue virus (DENV) is an Aedes-transmitted virus that belongs to the genus Flavivirus (family Flaviviridae). Dengue virus occurs as four serotypes (DENV-1 to DENV-4) and is hyperendemic in the tropics and subtropics.1–3 Dengue virus is the most important arthropod-borne virus in terms of its global impact on human health, with more than half of the world’s population living in areas where they are at risk of infection. An estimated 390 million new DENV infections occur worldwide each year, of which 96 million infections produce clinical manifestations.4 In Mexico, an estimated 139,000 clinical episodes occur each year.5
DENVs are the etiological agents of dengue. The World Health Organization (WHO) previously classified dengue using three disease categories: dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS).6 Dengue fever is an undifferentiated febrile illness, often accompanied by headache, retro-orbital pain, myalgia, arthralgia, rash, and nausea. Dengue hemorrhagic fever is characterized by hemorrhagic manifestations, thrombocytopenia, and hemoconcentration or other evidence of vascular leakage, and can progress to shock and death. Dengue hemorrhagic fever is divided into grades (designated as DHF grades I–IV) based on disease severity. One limitation of this classification system is that it does not always allow for the reliable identification of severe cases of dengue.7,8 To address this concern, WHO revised the guidelines for dengue diagnosis in 2009, with cases now classified as dengue without warning signs (DwoWS), dengue with warning signs (DWWS), and severe dengue (SD).9 Dengue without warning signs presents as an acute febrile illness with at least two of the following: nausea/vomiting, rash, aches and pains, leukopenia, and a positive tourniquet test. To fulfill the criteria of DWWS, at least one warning sign must be observed, with warning signs defined as abdominal pain, persistent vomiting, fluid accumulation, mucosal bleeding, lethargy, liver enlargement, and increasing hematocrit with decreasing platelets. Severe dengue is associated with severe plasma leakage, severe bleeding, or severe organ impairment. The Secretariat of Health (Secretaría de Salud; SS) in Mexico incorporated the most recent guidelines into their nationwide dengue surveillance program in 2016.
Other mosquito-transmitted flaviviruses that occur in Mexico include West Nile virus (WNV), St. Louis encephalitis virus (SLEV), and Zika virus (ZIKV), all of which produce clinical presentations that overlap substantially with those of dengue. West Nile virus infections can present as West Nile fever (WNF), a mild febrile illness often accompanied by a variety of nonspecific symptoms such as headache, myalgia, nausea, fatigue, weakness, and vomiting.10,11 West Nile virus infection can also lead to West Nile neuroinvasive disease, which manifests as encephalitis, meningitis, or acute flaccid paralysis. Clinical SLEV infections are usually characterized by a fever and various other nonspecific symptoms, with severe cases progressing to aseptic meningitis and encephalitis.12,13 Symptoms most commonly associated with ZIKV infection are fever, macular or papular rash, arthritis, and arthralgia, but serious manifestations, such as microcephaly and Guillain–Barré syndrome, can occur.14,15
In this study, we performed a clinical, serological, and molecular investigation to increase our understanding of the public health impact of dengue and other flaviviruses in Reynosa, a city in Tamaulipas on the Mexico–U.S. border. There are limited data in the PubMed database on the public health impact of flaviviruses in Reynosa.16,17 In the most comprehensive study described to date, 2,706 suspected cases of dengue were reported in Reynosa in 1995, with another 2,052 suspected cases reported elsewhere in Tamaulipas.17 Dengue virus-2, DENV-3, and DENV-4 were isolated from patients in Reynosa, and DENV-1 was isolated from patients in other locations, demonstrating concurrent circulation of all four serotypes in Tamaulipas in a single season. Other reports have described dengue outbreaks elsewhere on the Tamaulipas–Texas border.18–20 The sample population of the present study consisted of 1) patients who presented in January 2014 to December 2016 with suspected dengue at hospitals and clinics of the SS and 2) individuals recruited in 2014 and 2015 during a household-based seroepidemiological survey designed to estimate the seroprevalence of flaviviruses and identify seroconversions in the general population.
MATERIALS AND METHODS
Study area.
Study participants were residents of Reynosa, a city in the state of Tamaulipas, northern Mexico, on the Mexico–U.S. border (Figure 1). Reynosa is located in the binational Reynosa–McAllen Metropolitan Area, along with McAllen, a city in Hidalgo County, Texas. According to the 2013 census, Reynosa has a population of 672,183. The official estimated populations of McAllen and Hidalgo County in 2014 are 138,596 and 779,194, respectively. The climate in Reynosa is semiarid and the mean annual temperature and precipitation are 23.2°C and 532 mm3, respectively.
Figure 1.
Geographic location of the Reynosa–McAllen metropolitan area. The dashed rectangle indicates the neighborhood of Nuevo Amanecer.
Sample population and performance sites for the clinical investigation.
The sample population for the clinical investigation consisted of patients who presented in January 2014 to December 2016 with suspected dengue at hospitals and clinics of the SS in Reynosa. Seven hospitals that participated in the study are as follows: Hospital General de Reynosa, Hospital Materno Infantil de Reynosa, Hospital Baudelio Villanueva del Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado (Institute for Social Security and Services for State Workers), Hospital Regional de Pemex, Unidad de Medicina Familiar 33 del IMSS (Instituto Mexicano del Seguro Social; Mexican Social Security Institute), Unidad de Medicina Familiar 40 del IMSS, and Hospital General de Zona 15 del IMSS. Twenty first-level health units from Jurisdiction IV in Reynosa also participated.
To be eligible for inclusion, patients who presented in 2014 or 2015 had to meet the clinical criteria for DF, DHF, or DSS, following the guidelines established by WHO in 1999 for dengue diagnosis.6 Dengue hemorrhagic fever patients were not graded by severity. Patients who presented in 2016 were classified using the dengue guidelines established by WHO in 2009 and, therefore, had to meet the clinical criteria for DwoWS, DWWS, or SD.9 The most recent dengue disease classifications were not used in Mexico before 2016. Patients who did not live in Reynosa were not included in the sample population.
Five milliliters of whole blood was taken from each patient in the acute phase of illness (defined as 0–5 days post-illness onset).21 Blood was extracted by venipuncture without the use of anticoagulant, and sera were collected and stored at 2–8°C. Patients not willing to have blood drawn were excluded from the sample population. Demographic information was documented by health-care providers and select information (age, gender, and sometimes travel histories) was forwarded to us. On most occasions, travel histories were often defined as whether the patient had or had not ever left Tamaulipas. Clinical information was also collected, although we were not provided with information on specific symptoms (only disease classifications and whether the patient had been hospitalized). Informed consent to participate in the study was not required because it is obligatory to report dengue in Mexico. Sera were blind-coded and all identifying information was removed before receipt to protect patient identities. All research protocols were approved by the institutional review boards at the SS in Reynosa, Instituto Politécnico Nacional in Reynosa, Iowa State University, Universidad Autónoma de Yucatán, Universidad Autónoma de Ciudad Juárez, and Instituto Politécnico Nacional in the Ciudad de México.
Sample population for the seroconversion study.
The sample population consisted of individuals recruited during a household survey designed to detect evidence of flavivirus exposure and seroconversions. The study cohort was selected using a convenient sampling method. Individuals with flavivirus-like symptoms were not purposely excluded from the study, but all subjects were asymptomatic at the time of enrollment. The study was performed in the neighborhood of Nuevo Amanecer in Reynosa (Figure 1). Each house was visited on four occasions: March 2014, October 2014, April or May 2015, and October 2015. Sera were collected from study participants on each visit, unless they were not home or declined to further participate. Every participant was assigned a unique and randomly generated four-letter identification code. Sera collections were performed by registered nurses.
Written consent to participate was obtained from each subject after a full explanation of the study. If the person was < 18 years of age, written consent was obtained from a parent or legal guardian. Individuals aged 8–18 years were asked to assent. Adults who declined to sign the consent form and children who did not assent were not included in the study. Children < 2 years of age were ineligible for inclusion. One person from each household was asked to complete a questionnaire to obtain demographic information and other pertinent information (i.e., availability of air-conditioning). Data were handled confidentially and anonymously. All research protocols were approved by the institutional review boards at the participating universities.
Enzyme-linked immunosorbent assays.
Sera from all patients with suspected dengue were tested for DENV nonstructural protein 1 (NS1) by enzyme-linked immunosorbent assay (ELISA) using the Panbio Dengue Early ELISA (Inverness Medical Innovations Ltd., Australia [formerly Panbio Ltd.]) or Platelia™ Dengue NS1 Antigen ELISA (BioRad, Hercules, CA) assays. Both assays have been approved for use by the SS for dengue diagnosis.22 Sera were tested at a dilution of 1:2. When the Panbio Dengue Early ELISA was used, index values were calculated following the manufacturer’s instructions, with index values < 0.9, between 0.9 and 1.1, and > 1.1 considered to be negative, equivocal, and positive, respectively. When the Platelia Dengue NS1 Antigen ELISA was used, sample ratios were calculated following established protocols, with ratios < 0.5, between 0.5 and 1.0, and > 1.0 considered to be negative, equivocal, and positive, respectively. If DENV NS1 was detected, the patient was considered to have a laboratory-confirmed DENV infection. Nonstructural protein 1 ELISAs were performed in the Laboratorio Estatal de Salud Pública de Tamaulipas (State Laboratory of Public Health of Tamaulipas) and diagnostic findings were forwarded to health-care providers.
Sera from all participants sampled in the household survey were tested for flavivirus-specific immunoglobulin (Ig) M and IgG using the DENV Detect™ IgM Capture ELISA (Inbios International Inc., Seattle, WA) and DENV Detect IgG ELISA (Inbios International Inc.), respectively. For both assays, immune status ratios (ISRs) were calculated following the manufacturer’s instructions, with negative, equivocal, and positive results defined as those that generated ISR values of ≤ 1.65, > 1.65 and < 2.84, and ≥ 2.84, respectively. Immunoglobulin M and IgG ELISAs were performed at the Centro de Biotecnología Genómica del Instituto Politécnico Nacional in Reynosa, Tamaulipas.
Quantitative reverse transcription–polymerase chain reaction.
A subset of DENV NS1-positive sera was randomly selected and an aliquot of each was sent to the National Institute for Diagnosis and Epidemiological Reference (INDRE) in Mexico City. Total RNA was extracted using the QIAamp viral RNA extraction kit (Qiagen, Valencia, CA) and tested using a multiplex, quantitative reverse transcription–polymerase chain reaction (qRT-PCR) designed to detect and differentiate between all four serotypes of DENV. Assays were performed using standardized protocols developed by the U.S. Centers for Disease Control and Prevention and routinely used by the SS for dengue diagnosis.22,23 Samples that yielded quantitation cycle (Cq) values ≤ 35 and ≥ 39 were considered to be positive and negative, respectively, for DENV RNA. Samples that produced Cq values > 35 but < 39 were repeated in duplicate and, if the same result was obtained, the sample was considered to be positive.
Plaque reduction neutralization tests.
Sera were assayed by plaque reduction neutralization test (PRNT), following standard protocols.24 Plaque reduction neutralization tests were performed using DENV-1 (strain Hawaii), DENV-2 (strain NGC), DENV-3 (strain H-87), DENV-4 (strain 241), SLEV (strain TBH-28), WNV (strain NY99-35261-11), and ZIKV (strain PRVABC59). Viruses were obtained from the World Health Organization Center for Arbovirus Reference and Research maintained at the Centers for Disease Control and Prevention, Division of Vector-Borne Infectious Diseases (Fort Collins, CO). Plaque reduction neutralization tests were performed in six-well plates using African green monkey kidney (Vero) cells. Sera were tested in the presence of 8% labile serum factor.25 Sera were tested at a starting dilution of 1:40 and titers were expressed as the reciprocal of serum dilutions yielding ≥ 90% reduction in the number of plaques (PRNT90). For etiologic diagnosis, the PRNT90 antibody titer to the respective virus was required to be at least 4-fold greater than that of the other flaviviruses tested. If the PRNT90 titers for two or more viruses were ≥ 1,280, the individual was considered to have a secondary flavivirus infection. Plaque reduction neutralization tests were performed at Iowa State University.
RESULTS
Clinical investigation.
A total of 3,012 residents of Reynosa presented with suspected dengue in 2014–2016. Serum was collected from 2,355 (78.2%) patients; the remainder declined to have blood drawn and were excluded from the sample population (Table 1). Dengue virus NS1 was detected by ELISA in sera from 418 (17.7%) patients in the sample population. Most patients with confirmed dengue presented in 2014 (218 cases), followed by 2015 and 2016 (134 and 66 cases, respectively). There were 309 confirmed cases of DF (183 cases in 2014 and 126 cases in 2015) and 43 confirmed cases of DHF (35 cases in 2014 and eight cases in 2015). There were no suspected or confirmed cases of DSS. In 2016, which is when the most recent dengue classifications were adopted by the SS, there were 63 confirmed cases of DwoWS and three confirmed cases of DWWS. There were no confirmed cases of SD. Thirteen patients (two, nine, and two patients from 2014, 2015, and 2016, respectively) yielded equivocal ELISA results. Thirty-five patients were hospitalized, including 18 patients with confirmed dengue. Hospitalized patients with confirmed dengue presented in 2014 (17 patients) and 2015 (one patient), and all survived. The dengue patients hospitalized in 2014 were diagnosed with DHF (16 patients) and DF (one patient), and DENV-1 RNA was detected in one DHF patient. The dengue patient hospitalized in 2015 was diagnosed with DHF and was positive for DENV-2 RNA.
Table 1.
Numbers of suspected and confirmed cases of dengue in Reynosa, Tamaulipas, 2014–2016
| Year | Disease classification | |||||
|---|---|---|---|---|---|---|
| DF | DHF | DwoWS | DWWS | SD | Total | |
| *Number (%) of confirmed/suspected cases | ||||||
| 2014 | 183/703 (26.0) | 35/58 (60.3) | NA | NA | NA | 218/761 (28.6) |
| 2015 | 126/1,064 (11.8) | 8/26 (30.8) | NA | NA | NA | 134/1,090 (12.3) |
| 2016 | NA | NA | 63/491 (12.8) | 3/6 (50.0) | 0/7 (0.0) | 66/504 (13.1) |
| Total | 309/1,767 (17.5) | 43/84 (51.2) | 63/491 (12.8) | 3/6 (50.0) | 0/7 (0.0) | 418/2,355 (17.7) |
DF = dengue fever; DHF = dengue hemorrhagic fever; DwoWS = dengue without warning signs; DWWS = dengue with warning signs; NA = not applicable; SD = severe dengue.
Thirteen patients with suspected dengue (two, nine, and two patients from 2014, 2015, and 2016, respectively) yielded equivocal test results. Another 657 patients with suspected dengue (96, 37, and 524 patients from 2014, 2015, and 2016, respectively) were excluded from the sample population because acute sera were not collected and/or diagnostic testing was not performed.
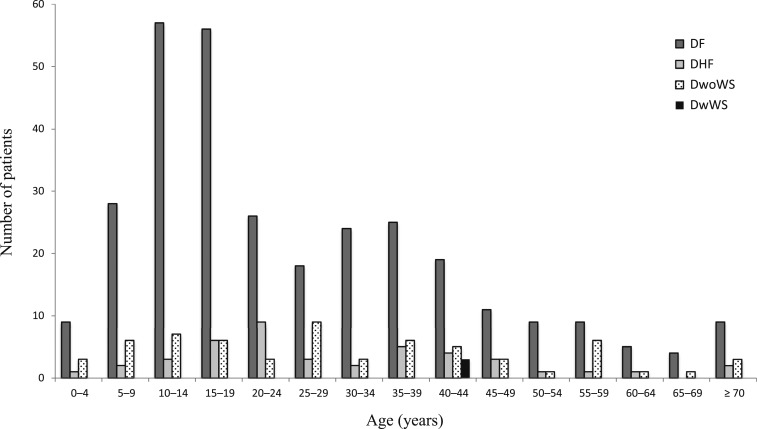
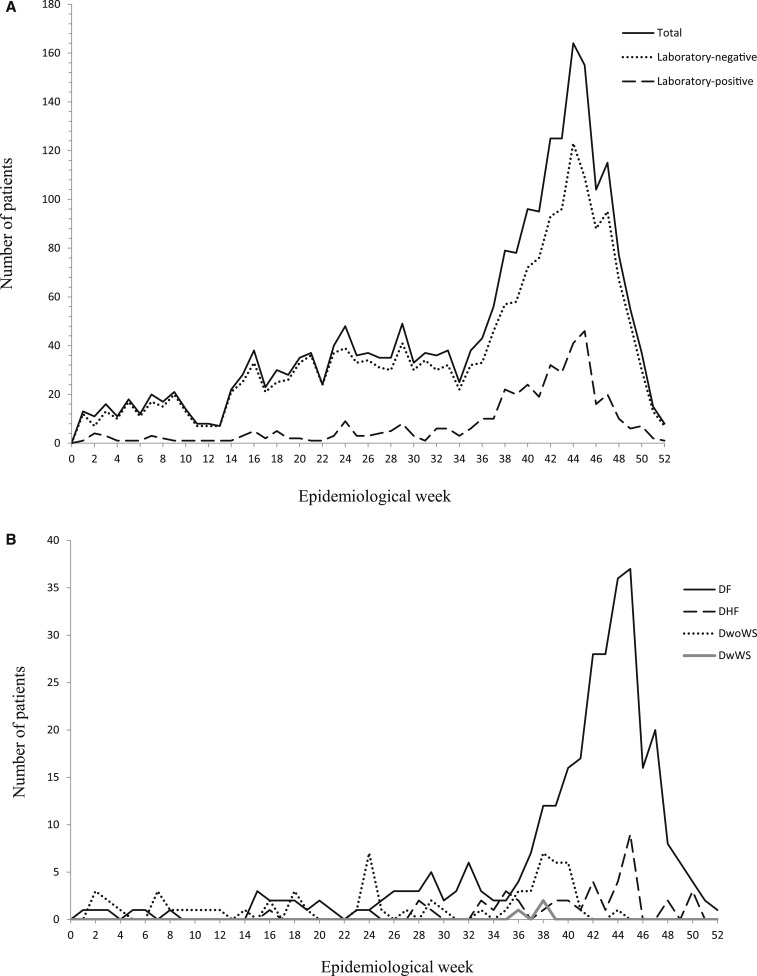
The study population consisted of 1,005 (42.7%) males and 1,350 (57.3%) females. There were 174 males (41.6%) and 244 females (58.4%) with confirmed dengue. The ages of patients with suspected and confirmed dengue ranged from < 1 to 98 and 1 to 87 years, respectively. Confirmed dengue occurred in patients of all age categories, with greatest numbers reported among patients aged 10–14 and 15–19 years (Table 2). Confirmed DF and DwoWS also occurred in patients of all age categories, and DHF occurred in patients of most age categories (Figure 2). The three cases of DWWS occurred in 40- to 44-year-old patients. Most of the patients with confirmed dengue presented in epidemiological weeks (EWs) 38–48, and cases peaked in EW 45 (Figure 3A). Similar findings were observed for patients without laboratory-confirmed dengue. Cases of confirmed DF, DHF, DwoWS, and DWWS also peaked late in the year (Figure 3B).
Table 2.
Age characteristics of confirmed and suspected dengue cases in Reynosa, Tamaulipas, 2014–2016
| Ages (years) | Number (%) of confirmed/suspected cases | |||
|---|---|---|---|---|
| 2014 | 2015 | 2016 | All years | |
| 5 | 9/67 (13.4) | 1/47 (2.1) | 3/31 (9.7) | 13/145 (9.0) |
| 5–9 | 21/132 (15.9) | 9/104 (8.7) | 6/55 (10.9) | 36/291 (12.4) |
| 10–14 | 44/153 (28.8) | 16/158 (10.1) | 7/70 (10.0) | 67/381 (17.6) |
| 15–19 | 38/110 (34.5) | 24/141 (17.0) | 6/59 (10.2) | 68/310 (21.9) |
| 20–24 | 26/48 (54.2) | 9/83 (10.8) | 3/30 (10.0) | 38/161 (23.6) |
| 25–29 | 13/44 (29.5) | 8/64 (12.5) | 9/41 (22.0) | 30/149 (20.1) |
| 30–34 | 13/41 (31.7) | 13/89 (14.6) | 3/39 (7.7) | 29/169 (17.2) |
| 35–39 | 20/51 (39.2) | 10/100 (10.0) | 6/39 (15.4) | 36/190 (18.9) |
| 40–44 | 13/39 (33.3) | 10/85 (11.8) | 8/39 (20.5) | 31/163 (19.0) |
| 45–49 | 8/23 (34.8) | 6/53 (11.3) | 3/30 (10.0) | 17/106 (16.0) |
| 50–54 | 3/19 (15.8) | 7/59 (11.9) | 1/24 (4.2) | 11/102 (10.8) |
| 55–59 | 3/13 (23.1) | 7/36 (19.4) | 6/18 (33.3) | 16/67 (23.9) |
| 60–64 | 3/7 (42.9) | 3/22 (13.6) | 1/8 (12.5) | 7/37 (18.9) |
| 65–69 | 0/5 (0.0) | 4/17 (23.5) | 1/8 (12.5) | 5/30 (16.7) |
| ≥ 70 | 4/9 (44.4) | 7/31 (22.6) | 3/12 (25.0) | 14/52 (26.9) |
| Not provided | 0/0 (0.0) | 0/1 (0.0) | 0/1 (0.0) | 0/2 (0.0) |
| Total | 218/761 (28.6) | 134/1,090 (12.3) | 66/504 (13.1) | 418/2,355 (17.7) |
Figure 2.
Age characteristics of patients from the hospital-based clinical investigation. Numbers of patients with confirmed dengue fever, dengue hemorrhagic fever, dengue without warning signs, and dengue with warning signs according to age.
Figure 3.
Weekly incidence of dengue in Reynosa, Tamaulipas, in 2014–2016. (A) Numbers of patients with suspected dengue and with or without laboratory-confirmed dengue according to epidemiological week. (B) Numbers of patients with confirmed dengue fever, dengue hemorrhagic fever, dengue without warning signs, and dengue with warning signs according to epidemiological week.
Sera from 85 patients with confirmed dengue were tested for DENV RNA by serotype-specific qRT-PCR. Viral RNA was detected in 82 patients (Table 3). The serotype most commonly detected was DENV-1 (N = 61), followed by DENV-2 (N = 16) and DENV-3 (N = 5). Dengue virus-4 RNA was not identified in any patient. The most common serotype in 2014 was DENV-1 (55 of 59 patients). The most common serotype in 2015 and 2016 was DENV-2 (11 of 20 and four of six patients, respectively). Dengue virus-1 RNA was not detected in any patients in 2016. Travel histories were provided for 15 DENV-2 RNA-positive patients, with 11 of 12 patients from 2015 and two of three patients from 2016 having never left Tamaulipas. Travel histories were also provided for seven DENV-3 RNA-positive patients, with all three patients from 2014, one of two patients from 2015, and both patients from 2016 having never left their state of residence.
Table 3.
Detection of dengue virus RNA in acute sera from dengue patients in Reynosa, Tamaulipas, 2014–2016
| Year | Disease classification | Total number of patients | Number of patients positive/tested by qRT-PCR | Serotype | |||
|---|---|---|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | ||||
| Number of qRT-PCR–positive patients | |||||||
| 2014 | DF | 183 | 53/53 | 51 | –* | 2 | – |
| DHF | 35 | 6/6 | 4 | 1 | 1 | – | |
| 2015 | DF | 126 | 14/17 | 6 | 8 | – | – |
| DHF | 8 | 3/3 | – | 3 | – | – | |
| 2016 | DwoWS | 63 | 6/6 | – | 4 | 2 | – |
| DWWS | 3 | – | – | – | – | – | |
| SD | – | – | – | – | – | – | |
| Total | 418 | 82/85 | 61 | 16 | 5 | – | |
DENV = dengue virus; DF = dengue fever; DHF = dengue hemorrhagic fever; DwoWS = dengue without warning signs; DWWS = dengue with warning signs; qRT-PCR = quantitative reverse transcription–polymerase chain reaction; SD = severe dengue.
0.
Sera from a subset of patients with confirmed dengue (N = 67) were tested by PRNT for antibodies to selected flaviviruses (data not shown). Fourteen patients were seropositive for DENV-1 and three patients were seropositive for DENV-2. Another 13 patients had secondary flavivirus infections, 33 patients had antibodies to an undetermined flavivirus, and three patients had no detectable neutralizing antibodies. Antibodies to DENV-4, SLEV, WNV, or ZIKV were not identified.
Seroconversion study.
The sample population consisted of 346 study participants from 114 houses. Another six houses were also visited but the residents declined to participate. Each participating household was visited on four occasions: March 2014, October 2014, April–May 2015, and October 2015. The mean number of study participants in each house was 3.0, with a range of 1–10, and none had symptoms consistent with a flavivirus infection at enrollment. Ages of the study participants ranged from 3 to 95 years. There were 143 males and 203 females. Sera were collected from 118 participants on all four occasions and from 61, 71, and 96 participants on three, two, or one occasion, respectively. Air-conditioning was available in 24 (21.1%) households.
All sera were tested by ELISA for flavivirus IgG, and sera collected at enrollment were also tested for flavivirus IgM. A total of 217 (62.7%) study participants were positive for flavivirus IgG at enrollment, including four participants who were also positive for flavivirus IgM. Seroprevalence was lowest among study participants < 20 years of age (Table 4). The male to female ratios for participants with and without flavivirus IgG at enrollment were similar (0.71 and 0.68, respectively).
Table 4.
Age characteristics of the household cohort
| Ages (years) | Number (%) positive for flavivirus IgG at baseline/total number tested |
|---|---|
| < 5 | 1/5 (20.0) |
| 5–9 | 6/25 (24.0) |
| 10–14 | 16/37 (43.2) |
| 15–19 | 16/43 (37.2) |
| 20–24 | 17/35 (48.6) |
| 25–29 | 14/17 (82.4) |
| 30–34 | 14/24 (58.3) |
| 35–39 | 16/21 (76.2) |
| 40–44 | 30/36 (83.3) |
| 45–49 | 18/24 (75.0) |
| 50–54 | 16/18 (88.9) |
| 55–59 | 19/22 (86.4) |
| 60–64 | 10/11 (90.1) |
| 65–69 | 7/8 (87.5) |
| ≥ 70 | 17/20 (85.0) |
| Total | 217/346 (62.7) |
Nine flavivirus-naive participants seroconverted during the study period (Table 5). Flavivirus IgG was first detected in sera collected in October 2014 (three subjects), April or May 2015 (two subjects), and October 2015 (four subjects). The mean age of participants who seroconverted was 23.7 years, with a range of 8–50 years. There were four males and five females. Seven individuals who seroconverted had never left Tamaulipas, whereas the other two individuals (denoted as MDRR and MLMM) had recently traveled. MDRR had visited family in Veracruz, eastern Mexico, and MLMM had traveled to Nuevo Leon, northern Mexico, during the study period.
Table 5.
Travel histories, clinical findings, and serologic data for study participants who seroconverted
| Human subject | Travel* | Illness† | Flavivirus IgG | PRNT90 titer | PRNT interpretation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date of serum collection | DENV-1 | DENV-2 | DENV-3 | DENV-4 | SLEV | WNV | ZIKV | |||||||
| March 2014§ | October 2014 | April–May 2015 | October 2015‡ | |||||||||||
| SACL | No | Yes | −‖ | NT¶ | +# | + | 80 | –** | – | – | – | – | – | DENV-1 |
| MBCO | No | No | − | + | + | + | 80 | 80 | – | – | – | – | – | Flavivirus |
| AMGO | No | No | − | + | NT | + | – | – | – | – | 40 | 640 | – | WNV |
| MDRR | Yes | No | − | + | + | + | – | – | – | – | – | 160 | – | WNV |
| MLMM | Yes | No | − | − | + | + | 640 | – | 160 | – | – | – | – | DENV-1 |
| MLLN | No | No | − | − | − | + | – | – | – | – | – | 640 | – | WNV |
| YFVO | No | No | − | − | − | + | – | – | – | – | – | – | – | Negative |
| IFCF | No | Yes | − | − | − | + | – | – | – | – | – | 320 | – | WNV |
| FCLG | No | No | − | − | − | + | – | 40 | – | – | – | – | – | Flavivirus |
DENV-1 = dengue virus 1; DENV-2 = dengue virus 2; DENV-3 = dengue virus 3; DENV-4 = dengue virus 4; IgG = immunoglobulin G; PRNT = plaque reduction neutralization test; SLEV = St. Louis encephalitis virus; WNV = West Nile virus; ZIKV = Zika virus.
Defined as study participants who have or have never left Tamaulipas.
SACL and IFCF developed symptoms consistent with DF and WNF, respectively.
Most PRNTs were performed using sera collected in October 2015.
Sera collected at baseline (March 2014) were also tested for flavivirus IgM but all were negative.
Negative.
Not tested (study participant unavailable for serum collection).
Positive.
< 40.
Sera from the study participants who seroconverted were further tested by PRNT (Table 5). Of the seven study participants who had never traveled, three individuals were seropositive for WNV, one individual was seropositive for DENV-1, two individuals were seropositive for an undetermined flavivirus, and one individual had no detectable neutralizing antibodies. The individual seropositive for DENV-1, a 31-year-old female denoted as SACL, developed symptoms consistent with DF. One individual seropositive for WNV, a 15-year-old female denoted as IFCF, developed symptoms consistent with WNF. MDRR and MLMM, the two individuals who had recently traveled, were seropositive for WNV and DENV-1, respectively.
Sera from the four study participants with flavivirus IgM and IgG at baseline were tested by PRNT (Table 6). Two participants were seropositive for DENV-1, whereas one each had antibodies to DENV-2 and an undetermined flavivirus. The two participants seropositive for DENV-1 and the participant with antibodies to an undetermined flavivirus had never traveled outside of Tamaulipas. Sera from five study participants with flavivirus IgG, but not IgM, at baseline were randomly selected and also tested by PRNT. Two participants had antibodies to DENV-2, whereas one each had antibodies to DENV-1, WNV, and an undetermined flavivirus. The participants seropositive for WNV and an undetermined flavivirus, in addition to one of the participants seropositive for DENV-2, had never traveled outside of Tamaulipas.
Table 6.
Serologic findings for select participants from the household-based investigation, including all with flavivirus IgM at baseline
| Human subject | Travel* | Illness | Baseline ELISA data† | PRNT90 titer‡ | PRNT interpretation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgM | IgG | DENV-1 | DENV-2 | DENV-3 | DENV-4 | SLEV | WNV | ZIKV | ||||
| BROO | No | No | +§ | + | ≥ 1,280 | 160 | 160 | 40 | –‖ | 40 | – | DENV-1 |
| GAPG | No | No | + | + | 640 | – | 80 | – | – | 320 | – | Flavivirus |
| MMPO | No | No | + | + | 640 | 160 | 160 | 40 | – | – | – | DENV-1 |
| ADTO | Yes | No | + | + | – | 80 | – | – | – | – | – | DENV-2 |
| GMJO | Yes | No | −¶ | + | 640 | – | 80 | 40 | – | 40 | – | DENV-1 |
| CDPG | No | No | − | + | – | 160 | 40 | – | – | – | 80 | Flavivirus |
| MILO | No | No | − | + | 160 | ≥ 1,280 | 40 | – | – | – | – | DENV-2 |
| CAZF | Yes | No | − | + | – | ≥ 1,280 | 80 | – | 40 | 320 | – | DENV-2 |
| MTRO | No | No | − | + | 160 | 160 | 160 | 40 | 80 | ≥ 1,280 | – | WNV |
DENV-1 = dengue virus 1; DENV-2 = dengue virus 2; DENV-3 = dengue virus 3; DENV-4 = dengue virus 4; ELISA = enzyme-linked immunosorbent assay; IgG = immunoglobulin G; IgM = immunoglobulin M; PRNT = plaque reduction neutralization test; SLEV = St. Louis encephalitis virus; WNV = West Nile virus; ZIKV = Zika virus.
Defined as study participants who have or have never left Tamaulipas.
All baseline sera were collected in March 2014, except for GAPG with their baseline serum collected in October 2014.
All PRNTs were performed using sera collected in October 2015.
Positive.
< 40.
Negative.
DISCUSSION
We provide evidence of autochthonous transmission of multiple flaviviruses in northern Mexico. In the household-based investigation, antibodies to DENV-1, DENV-2, and WNV were detected in study participants who had never traveled outside of Tamaulipas, consistent with local virus transmission. The detection of flavivirus IgM in study participants who had never left Tamaulipas suggests that some infections were recent. The detection of antibodies to DENV-1 and WNV in participants who seroconverted and had not recently traveled provides further evidence of recent autochthonous virus transmission. Dengue virus-1, DENV-2, and DENV-3 RNA were identified in dengue patients who presented at hospitals and clinics of the SS. Although information on travel histories was not provided for most cases, several patients positive for DENV-2 and DENV-3 RNA had never traveled. Several local cases of DENV-2 and DENV-3 occurred in 2015, the same year that the DENV-1 and WNV seroconversions were identified. In addition, we recently detected several cases of non–travel-acquired chikungunya virus (CHIKV; genus Alphavirus, family Togaviridae) in Tamaulipas in 2015.26 Taken together, we provide evidence for the co-circulation of five arboviruses of medical concern in a single season in Tamaulipas, northern Mexico.
A total of 418 (17.7%) patients with suspected dengue had laboratory-confirmed DENV infections, although this is likely an underestimate because DENV NS1-negative patients were not tested for flavivirus IgM which presumably would have resulted in the identification of additional dengue cases. Eighty-two patients had DENV RNA. The majority (74.4%) was positive for DENV-1; the remainder had been infected with DENV-2 (19.5%) and DENV-3 (6.1%). Dengue virus-1 was the dominant serotype in Nuevo Leon, northern Mexico, in 2010.27 Dengue virus-1 was also the dominant serotype in southern Texas in 2013.20 However, our data should be interpreted with caution if used to predict the identity of the dominant serotype in Tamaulipas. First, travel histories were not obtained for most patients and, therefore, some infections may have been acquired elsewhere. Second, much higher numbers of patients were tested for DENV RNA in 2014 (N = 59) compared with 2015 (N = 20) and 2016 (N = 6). Although most DENV RNA-positive patients had been infected with DENV-1, the serotype most frequently detected in both 2015 and 2016 was DENV-2, and there was no evidence of DENV-1 in 2016. These findings could indicate that serotype displacement had occurred. The phenomenon of serotype displacement is well documented. A review of the nationwide surveillance data from 2000 to 2011 revealed that DENV-2 was the dominant serotype in Mexico in 2000–2005, followed by DENV-1 in 2006–2010, with both serotypes detected in similar proportions in 2011.28
Simultaneous circulation of all four DENV serotypes has been reported in Tamaulipas. Dengue virus-2, DENV-3, and DENV-4 were isolated from patients in Reynosa in 1995, with DENV-1 recovered from patients elsewhere in Tamaulipas that same year.17 Concurrent circulation of all four serotypes has also been reported in South America and Asia.29–32 We provide evidence for the co-circulation of three serotypes. Dengue virus-4 was not detected, although nationwide surveillance data collected in 2000–2011 revealed that DENV-4 is the least common serotype in Mexico.28 Many other studies also report the concurrent circulation of three serotypes.33–37
Dengue cases peaked in September to November. Other studies performed in northern Mexico report similar findings.17,27,38,39 Most confirmed cases of dengue identified in southern Texas in 2013 also occurred during these months.20 The dengue season coincides with peak mosquito abundance which is dependent on climatic conditions favorable for mosquito breeding. Fifty-eight percent of patients with confirmed dengue were female. Other studies performed on the Mexico–U.S. border also report dengue in slightly more females than males, although it is important to note that often slightly more females than males also present with suspected dengue.19,20,38,39 In this regard, 57% of patients in our entire sample population were female. Dengue cases peaked during childhood; almost one-third of cases were 10–19 years old, and lowest numbers occurred in those older than 50 years of age. A similar age-group distribution pattern has been reported elsewhere in Mexico.27,40,41 Most of the patients with suspected dengue did not have laboratory-confirmed DENV infections, as observed in other dengue studies performed in Mexico.27,40,42 Taken together, the epidemiological characteristics of dengue in Reynosa in 2014–2016 are similar to those reported in many other dengue investigations.
Reynosa is located within the binational Reynosa–McAllen Metropolitan Area, along with McAllen, a city in Hidalgo County, Texas. The number of dengue cases identified in our study greatly exceeds the number of dengue cases reported in Hidalgo County over the same time interval according to data released by the U.S. Centers for Disease Control and Prevention. Two dengue cases (both imported) occurred in Hidalgo County in 2014, with no dengue cases reported in 2015 and 2016.43 Five dengue cases (all imported) occurred in Texas counties located in the lower Rio Grande Valley in 2014–2016.43 This is in sharp contrast to the 418 dengue cases identified in our hospital-based investigation.
Other studies performed at the Mexico–U.S. border have also reported a much higher incidence of dengue in Mexico compared with the United States.17–19,39,44,45 One example is a household-based seroepidemiologic study performed in 2005 in the contiguous border cities of Matamoros, Tamaulipas, and Brownsville, Texas.18 The study population consisted of 132 participants from 111 households in Matamoros and 141 participants from 118 households in Brownsville. Dengue virus-reactive antibodies were detected by ELISA in 101 (76.5%) participants from Matamoros and 49 (39.2%) participants from Brownsville. Forty-two (31.7%) participants from Matamoros and six (3.7%) participants from Brownsville had recent infections. Environmental and socioeconomic factors that affect mosquito–human contact, such as air-conditioning, appear to be the primary reasons for these differences.18,44 Air-conditioning was available in 29% of participating households in Matamoros and 85% of participating households in Brownsville.18 In another household-based study, air-conditioning was available in 24% and 82% of participating households in Nuevo Laredo, Tamaulipas, and Laredo, Texas, respectively.44 In all, 21% of households in our study were air-conditioned. Other factors associated with an increased risk of infection are the presence of mosquito breeding sites, nonuse of mosquito repellents, smaller lot sizes, and low family income.18,44–46
Three WNV seroconversions were identified in the household-based investigation, and one study participant developed clinical manifestations consistent with WNF. However, the case cannot be conclusively linked to WNV because a precise date of symptom onset was not obtained. There have been no documented cases of WNV in Tamaulipas, although two cases have occurred in the neighboring state of Nuevo Leon.47,48 The first evidence of WNV in Tamaulipas was provided in a serological investigation that identified antibodies to WNV in resident birds in Tamaulipas in 2013.49 Evidence of WNV infection has also been detected in mosquitoes and horses elsewhere in northern Mexico.47,50–54 Unlike dengue, the incidence of WNV disease is much lower in Mexico compared with that in the United States, potentially because preexisting immunity to DENV protects against WNV disease.10,42
Antibodies to SLEV or ZIKV were not detected in any participants in the household- or hospital-based investigations, although these findings are not unexpected. St. Louis encephalitis virus is not a common cause of infection in humans in Mexico.12 Zika virus was not present in Mexico for most of the study period, with the first case detected in November 2015 according to WHO.55 The first confirmed case of ZIKV in Tamaulipas occurred in June 2016 in the municipality of Madero (S. D. Carmona, unpublished data). The first confirmed case of ZIKV in Reynosa occurred in August 2016, with a total of 125 confirmed cases reported in Reynosa as of EW 14 of 2018.
In summary, we provide evidence for the concurrent circulation of four flaviviruses, DENV-1, DENV-2, DENV-3, and WNV, in Tamaulipas, northern Mexico. In addition, we recently reported autochthonous transmission of CHIKV, an Alphavirus, in Tamaulipas.26 All of the aforementioned viruses were associated with human disease in the study area, demonstrating the important need to continue performing vigilant arbovirus surveillance and diagnosis on the Mexico–U.S. border.
Acknowledgments:
We thank all the nurses who participated in the study, particularly Dinorah Muñoz-Gutiérrez, Fanny B. Hernández-Castan, Karla E. Mendez-Jiménez, and Leticia Guzmán-Hernández from the Secretaría de Salud, Jurisdicción IV in Reynosa, Tamaulipas. We also thank the residents of Nuevo Amanecer who participated in the study and the patients for providing sera.
REFERENCES
- 1.Brathwaite Dick O, San Martin JL, Montoya RH, del Diego J, Zambrano B, Dayan GH, 2012. The history of dengue outbreaks in the Americas. Am J Trop Med Hyg 87: 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB, 2016. Dengue infection. Nat Rev Dis Primers 2: 16055. [DOI] [PubMed] [Google Scholar]
- 3.Murray NE, Quam MB, Wilder-Smith A, 2013. Epidemiology of dengue: past, present and future prospects. Clin Epidemiol 5: 299–309.23990732 [Google Scholar]
- 4.Bhatt S, et al. 2013. The global distribution and burden of dengue. Nature 496: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Undurraga EA, et al. 2015. Economic and disease burden of dengue in Mexico. PLoS Negl Trop Dis 9: e0003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization , 1997. Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control, 2nd edition. Geneva, Switzerland: WHO. [Google Scholar]
- 7.Balmaseda A, Hammond SN, Perez MA, Cuadra R, Solano S, Rocha J, Idiaquez W, Harris E, 2005. Short report: assessment of the World Health Organization scheme for classification of dengue severity in Nicaragua. Am J Trop Med Hyg 73: 1059–1062. [PubMed] [Google Scholar]
- 8.Phuong CX, et al. 2004. Clinical diagnosis and assessment of severity of confirmed dengue infections in Vietnamese children: is the world health organization classification system helpful? Am J Trop Med Hyg 70: 172–179. [PubMed] [Google Scholar]
- 9.World Health Organization , 2009. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control, New Edition. Geneva, Switzerland: WHO. [PubMed] [Google Scholar]
- 10.Blitvich BJ, 2008. Transmission dynamics and changing epidemiology of West Nile virus. Anim Health Res Rev 9: 71–86. [DOI] [PubMed] [Google Scholar]
- 11.Petersen LR, Brault AC, Nasci RS, 2013. West Nile virus: review of the literature. JAMA 310: 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reisen WK, 2003. Epidemiology of St. Louis encephalitis virus. Adv Virus Res 61: 139–183. [DOI] [PubMed] [Google Scholar]
- 13.Oyer RJ, David Beckham J, Tyler KL, 2014. West Nile and St. Louis encephalitis viruses. Handb Clin Neurol 123: 433–447. [DOI] [PubMed] [Google Scholar]
- 14.Petersen LR, Jamieson DJ, Powers AM, Honein MA, 2016. Zika virus. N Engl J Med 374: 1552–1563. [DOI] [PubMed] [Google Scholar]
- 15.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N, 2016. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 130: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibanez-Bernal S, Briseno B, Mutebi JP, Argot E, Rodriguez G, Martinez-Campos C, Paz R, de la Fuente-San Roman P, Tapia-Conyer R, Flisser A, 1997. First record in America of Aedes albopictus naturally infected with dengue virus during the 1995 outbreak at Reynosa, Mexico. Med Vet Entomol 11: 305–309. [DOI] [PubMed] [Google Scholar]
- 17.Rawlings J, et al. 1996. Dengue fever at the US-Mexico border, 1995–1996 (reprinted from MMWR, vol 45, pg 841, 1996). JAMA 276: 1464–1465.8903248 [Google Scholar]
- 18.Ramos MM, et al. 2008. Epidemic dengue and dengue hemorrhagic fever at the Texas-Mexico border: results of a household-based seroepidemiologic survey, December 2005. Am J Trop Med Hyg 78: 364–369. [PubMed] [Google Scholar]
- 19.Brunkard JM, Robles Lopez JL, Ramirez J, Cifuentes E, Rothenberg SJ, Hunsperger EA, Moore CG, Brussolo RM, Villarreal NA, Haddad BM, 2007. Dengue fever seroprevalence and risk factors, Texas-Mexico border, 2004. Emerg Infect Dis 13: 1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas DL, et al. 2016. Reemergence of dengue in southern Texas, 2013. Emerg Infect Dis 22: 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Secretaría de Salud México, Boletín Epidemiológico SNdVESÚdI , 2015. Available at: https://www.gob.mx/cms/uploads/attachment/file/10564/sem27.pdf. Accessed January 1, 2018.
- 22.Secretaria de Salud , 2015. Lineamientos para la Vigilancia Epidemiológica de Dengue por Laboratorio (Guidelines for Epidemiological Surveillance of Dengue in the Laboratory) Available at: https://www.gob.mx/cms/uploads/attachment/file/23789/Lineamientos_para_la_vigilancia_epidemiologica_de_dengue.pdf. Accessed April 9, 2018.
- 23.Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJ, 2006. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J Clin Microbiol 44: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaty B, Calisher C, Shope R, 1995. Arboviruses. Lennette E, Lennette D, Lennette E, eds. Diagnostic Procedures for Viral, Rickettsial, and Chlamydial Infections. Washington, DC: American Public Health Association, 189–212. [Google Scholar]
- 25.Chappell WA, Sasso DR, Toole RF, Monath TP, 1971. Labile serum factor and its effect on arbovirus neutralization. Appl Microbiol 21: 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laredo-Tiscareño SV, et al. 2018. Arbovirus surveillance near the Mexico-U.S. border: isolation and sequence analysis of chikungunya virus from patients with dengue-like symptoms in Reynosa, Tamaulipas. Am J Trop Med Hyg 99: 191–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leduc-Galindo D, et al. 2015. Characterization of the dengue outbreak in Nuevo Leon state, Mexico, 2010. Infection 43: 201–206. [DOI] [PubMed] [Google Scholar]
- 28.Dantes HG, Farfan-Ale JA, Sarti E, 2014. Epidemiological trends of dengue disease in Mexico (2000–2011): a systematic literature search and analysis. PLoS Negl Trop Dis 8: e3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrivastava S, Tiraki D, Diwan A, Lalwani SK, Modak M, Mishra AC, Arankalle VA, 2018. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS One 13: e0192672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrade EH, et al. 2016. Spatial-temporal co-circulation of dengue virus 1, 2, 3, and 4 associated with coinfection cases in a hyperendemic area of Brazil: a 4-week survey. Am J Trop Med Hyg 94: 1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endy TP, Nisalak A, Chunsuttiwat S, Libraty DH, Green S, Rothman AL, Vaughn DW, Ennis FA, 2002. Spatial and temporal circulation of dengue virus serotypes: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am J Epidemiol 156: 52–59. [DOI] [PubMed] [Google Scholar]
- 32.Castonguay-Vanier J, et al. 2018. Molecular epidemiology of dengue viruses in three provinces of Lao PDR, 2006–2010. PLoS Negl Trop Dis 12: e0006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caron M, Grard G, Paupy C, Mombo IM, Bikie Bi Nso B, Kassa Kassa FR, Nkoghe D, Leroy EM, 2013. First evidence of simultaneous circulation of three different dengue virus serotypes in Africa. PLoS One 8: e78030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridde V, Agier I, Bonnet E, Carabali M, Dabire KR, Fournet F, Ly A, Meda IB, Parra B, 2016. Presence of three dengue serotypes in Ouagadougou (Burkina Faso): research and public health implications. Infect Dis Poverty 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Simone TS, Nogueira RM, Araujo ES, Guimaraes FR, Santos FB, Schatzmayr HG, Souza RV, Teixeira Filho G, Miagostovich MP, 2004. Dengue virus surveillance: the co-circulation of DENV-1, DENV-2 and DENV-3 in the state of Rio de Janeiro, Brazil. Trans R Soc Trop Med Hyg 98: 553–562. [DOI] [PubMed] [Google Scholar]
- 36.Humayoun MA, Waseem T, Jawa AA, Hashmi MS, Akram J, 2010. Multiple dengue serotypes and high frequency of dengue hemorrhagic fever at two tertiary care hospitals in Lahore during the 2008 dengue virus outbreak in Punjab, Pakistan. Int J Infect Dis 14: E54–E59. [DOI] [PubMed] [Google Scholar]
- 37.Lorono-Pino MA, Cropp CB, Farfan JA, Vorndam AV, Rodriguez-Angulo EM, Rosado-Paredes EP, Flores-Flores LF, Beaty BJ, Gubler DJ, 1999. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am J Trop Med Hyg 61: 725–730. [DOI] [PubMed] [Google Scholar]
- 38.Jones JM, et al. 2016. Binational dengue outbreak along the United States-Mexico border—Yuma County, Arizona, and Sonora, Mexico, 2014. MMWR Morb Mortal Wkly Rep 65: 495–499. [DOI] [PubMed] [Google Scholar]
- 39.Centers for Disease Control and Prevention , 2007. Dengue hemorrhagic fever–U.S.-Mexico border, 2005. MMWR Morb Mortal Wkly Rep 56: 785–789. [PubMed] [Google Scholar]
- 40.Lorono-Pino MA, et al. 2004. Introduction of the American/Asian genotype of dengue 2 virus into the Yucatan state of Mexico. Am J Trop Med Hyg 71: 485–492. [PubMed] [Google Scholar]
- 41.Gunther J, Ramirez-Palacio LR, Perez-Ishiwara DG, Salas-Benito JS, 2009. Distribution of dengue cases in the state of Oaxaca, Mexico, during the period 2004–2006. J Clin Virol 45: 218–222. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez Mde L, et al. 2010. Serologic surveillance for West Nile virus and other flaviviruses in febrile patients, encephalitic patients, and asymptomatic blood donors in northern Mexico. Vector Borne Zoonotic Dis 10: 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Centers for Disease Control and Prevention ArboNet, Dengue Virus (Locally Acquired and Imported) Available at: https://wwwn.cdc.gov/arbonet/maps/ADB_Diseases_Map/index.html. Accessed April 24, 2018.
- 44.Reiter P, et al. 2003. Texas lifestyle limits transmission of dengue virus. Emerg Infect Dis 9: 86–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark GG, 2008. Dengue and dengue hemorrhagic fever in northern Mexico and south Texas: do they really respect the border? Am J Trop Med Hyg 78: 361–362. [PubMed] [Google Scholar]
- 46.Brunkard JM, Cifuentes E, Rothenberg SJ, 2008. Assessing the roles of temperature, precipitation, and ENSO in dengue re-emergence on the Texas-Mexico border region. Salud Publica Mex 50: 227–234. [DOI] [PubMed] [Google Scholar]
- 47.Elizondo-Quiroga D, et al. 2005. West Nile virus isolation in human and mosquitoes, Mexico. Emerg Infect Dis 11: 1449–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rios-Ibarra C, Blitvich BJ, Farfan-Ale J, Ramos-Jimenez J, Muro-Escobedo S, Martinez-Rodriguez HR, OrtizLopez R, Torres-Lopez E, Rivas-Estilla AM, 2010. Fatal human case of West Nile disease, Mexico, 2009. Emerg Infect Dis 16: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Salas I, et al. 2003. Serologic evidence of West Nile virus infection in birds, Tamaulipas state, Mexico. Vector Borne Zoonotic Dis 3: 209–213. [DOI] [PubMed] [Google Scholar]
- 50.Blitvich BJ, et al. 2004. Phylogenetic analysis of West Nile virus, Nuevo Leon state, Mexico. Emerg Infect Dis 10: 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ibarra-Juarez L, Eisen L, Bolling BG, Beaty BJ, Blitvich BJ, Sanchez-Casas RM, Ayala-Sulca YO, Fernandez-Salas I, 2012. Detection of West Nile virus-specific antibodies and nucleic acid in horses and mosquitoes, respectively, in Nuevo Leon state, northern Mexico, 2006–2007. Med Vet Entomol 26: 351–354. [DOI] [PubMed] [Google Scholar]
- 52.Blitvich BJ, Fernandez-Salas I, Contreras-Cordero JF, Marlenee NL, Gonzalez-Rojas JI, Komar N, Gubler DJ, Calisher CH, Beaty BJ, 2003. Serologic evidence of West Nile virus infection in horses, Coahuila state, Mexico. Emerg Infect Dis 9: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deardorff E, et al. 2006. Introductions of West Nile virus strains to Mexico. Emerg Infect Dis 12: 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Estrada-Franco JG, et al. 2003. West Nile virus in Mexico: evidence of widespread circulation since July 2002. Emerg Infect Dis 9: 1604–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.World Health Organization Zika Virus Infection—Mexico Geneva, Switzerland: WHO. Available at: http://www.who.int/csr/don/03-december-2015-zika-mexico/en/. Accessed May 5, 2018.