Abstract
In most mammals, including mice and humans, meiotic recombination is determined by the meiosis specific histone methytransferase PRDM9, which binds to specific DNA sequences and trimethylates histone 3 at lysine-4 and lysine-36 at the adjacent nucleosomes. These actions ensure successful DNA double strand break formation and repair that occur on the proteinaceous structure forming the chromosome axis. The process of hotspot association with the axis after their activation by PRDM9 is poorly understood. Previously, we and others have identified CXXC1, an ortholog of S. cerevisiae Spp1 in mammals, as a PRDM9 interactor. In yeast, Spp1 is a histone methyl reader that links H3K4me3 sites with the recombination machinery, promoting DSB formation. Here, we investigated whether CXXC1 has a similar function in mouse meiosis. We created two Cxxc1 conditional knockout mouse models to deplete CXXC1 generally in germ cells, and before the onset of meiosis. Surprisingly, male knockout mice were fertile, and the loss of CXXC1 in spermatocytes had no effect on PRDM9 hotspot trimethylation, double strand break formation or repair. Our results demonstrate that CXXC1 is not an essential link between PRDM9-activated recombination hotspot sites and DSB machinery and that the hotspot recognition pathway in mouse is independent of CXXC1.
Author summary
Meiotic recombination increases genetic diversity by ensuring novel combination of alleles passing correctly to the next generation. In most mammals, the meiotic recombination sites are determined by histone methyltransferase PRDM9. These sites are proposed to become associated with the chromosome axis with the participation of additional proteins and undergo double strand breaks, which are repaired by homologous recombination. In budding yeast, Spp1 (ortholog of CXXC1) binds to methylated H3K4 and connects these sites with the chromosome axis promoting DSB formation. However, our data suggest that even though CXXC1 interacts with PRDM9 in male germ cells, it does not play a crucial role in mouse meiotic recombination. These results indicate that, unlike in yeast, a recombination initiation pathway that includes CXXC1 could only serve as a non-essential pathway in mouse meiosis.
Introduction
Meiotic recombination ensures production of fertile gametes with a correct haploid chromosome number and genetic diversity [1]. In most of mammals, the meiotic recombination sites are restricted to 1–2 kb regions, termed recombination hotspots, locations of which are determined by the DNA binding histone methyltransferase PRDM9 [2–4]. Recombination initiates when PRDM9 binds to hotspots sequences with its zinc finger domain and trimethylates histone 3 at lysine 4 (H3K4me3) and lysine 36 (H3K36me3) resulting in formation of a nucleosome-depleted region [2–6], probably by the action of nucleosome motors [7]. DNA double strand breaks (DSB) are created at the nucleosome-depleted regions of activated hotspots [8–11], and eventually repaired as either crossovers or non-crossover conversions. Cytological staining for several proteins associated with DSB processing in early meiotic prophase show that, from the earliest time of their detection, DSB are associated with a proteinaceous structure known as chromosome axis [12–15]. We have previously shown that PRMD9 is associated, but not directly interacting, with chromosome axis elements such as phosphorylated REC8 (pREC8) and SYCP3 in spermatocytes [16]. Efficient H3K4 trimethylation at hotspots is crucial for normal DSB formation and repair that occurs on the chromosome axis [17], even though PRDM9 binding presumably occurs on the open chromatin loops [18]. However, we currently do not have detailed knowledge of the proteins and molecular mechanisms participating in hotspot association with the chromosome axis.
In Saccharomyces cerevisiae, which has no PRDM9, the PHD zinc finger protein Spp1, a member of the COMPASS complex (Complex associated with Set1, the protein catalyzing trimethylation of histone 3 at lysine-4), acts as a histone H3K4 methyl reader and promotes meiotic DSB formation at the existing H3K4me3 sites, such as promoters [19, 20]. Spp1 is predominantly located on the chromosome axes and connects H3K4me3 sites with the axis protein Mer2 to stimulate Spo11 dependent DSB formation [19, 20]. Recent study showed that Spp1 function in tethering DSB sites to chromosome axes and ensuring efficient DSB formation is independent of its function as a COMPASS complex member [21].
CxxC finger protein 1 (CXXC1, also known as CFP1 and CGBP) is an ortholog of S. cerevisiae Spp1 in mammals [21]. In somatic cells, CXXC1 binds to both unmethylated CpGs and SETD1, which is required for trimethylation of H3K4 at CpG islands [22]. CXXC1 is crucial for embryonic stem cell maintaining and development [23, 24]. Knocking out Cxxc1 in mice results in lethality at the early embryonic stages [25]. We have reported that CXXC1 interacts with PRDM9 in yeast two-hybrid assay and in vitro [16]. This interaction has recently been confirmed by another group, which also reported that CXXC1 interacts with the chromosome axis element IHO1 by yeast two-hybrid assay [26]. IHO1 is considered to be the ortholog of yeast Mer2 and is known to be essential for ensuring efficient DSB formation [27], therefore it is possible that CXXC1-IHO1 interaction serves the same function in mammalian meiosis as their orthologs Spp1-Mer2 in yeast. However, the function of CXXC1 in mammalian meiosis has not been characterized so far. It has been unclear whether CXXC1 binds to PRDM9 in germ cells and whether it participates in meiotic recombination initiation in any way, either as a partner of PRDM9 or as a methyl reader of H3K4me3/H3K36me3 marks that PRDM9 imposes at the nucleosomes surrounding the recombination hotspots.
Here we confirmed that CXXC1 is co-expressed with PRDM9 and indeed interacts with it in spermatocytes. To address whether and how CXXC1 functions in meiotic recombination, we created two Cxxc1 conditional knockout mouse models and deleted Cxxc1 in all germ cells and in late spermatogonia just before the onset of meiosis. In both models, loss of CXXC1 did not affect normal meiotic recombination process. Our study demonstrates that the presence of CXXC1 in mouse meiosis is not essential, and unlike its S. cerevisiae ortholog Spp1, CXXC1 does not appear to be a key factor for the DSB formation.
Results
CXXC1 interacts with PRDM9 in spermatocytes
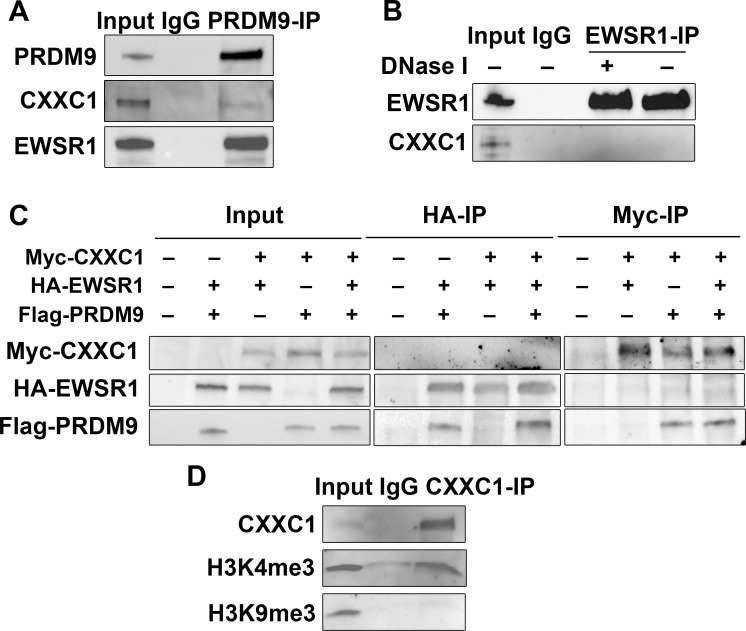
We tested whether CXXC1 interacts with PRDM9 in vivo by co-immunoprecipitation (co-IP) from spermatocytes isolated from 14 dpp B6 testis using antibody against PRDM9 [16]. We found that CXXC1 indeed interacts with PRDM9 in spermatocytes (Fig 1A). However, the interaction was not as strong as with PRDM9’s predominant interactor EWSR1 (Fig 1A) [16], which raised the possibility that the interaction between CXXC1 and PRDM9 could be mediated by stronger PRDM9 interactors. To test whether this is the case, we performed co-IP with EWSR1 and did not detect any interaction with CXXC1 in testicular extract (Fig 1B). To further test the interactions between the three proteins, we co-expressed Myc-tagged mouse CXXC1, HA-tagged EWSR1 and Flag-tagged PRDM9 proteins in human embryonal kidney 293 (HEK293) cells, and performed co-IP with antibodies against HA or Myc tags. Both EWSR1 and CXXC1 immunoprecipitated PRDM9 under these conditions, but there was no interaction between CXXC1 and EWSR1 in the presence or absence of PRDM9 (Fig 1C).
Fig 1. CXXC1 interacts with PRDM9 in spermatocytes.
(A) PRDM9 interacts with CXXC1 and EWSR1 in spermatocytes. Co-IP with PRDM9 from 14 dpp B6 testicular extract. Staining for each protein is indicated on left. In each blot, lane 1- input; lane 2 –co-IP with non-immune IgG; lane 3 –co-IP with antibody against PRDM9. (B) EWSR1 does not interact with CXXC1 in spermatocytes. Co-IP with EWSR1 from 14 dpp B6 testicular extract. Staining for EWSR1 and CXXC1. In each blot, lane 1- input; lane 2 –co-IP with non-immune IgG; lane 3 –co-IP with anti-EWSR1 after DNase I treatment; co-IP with anti-EWSR1 without DNase I treatment. (C) CXXC1 interacts with PRDM9 but not with EWSR1 in cell culture. Myc tagged CXXC1, HA tagged EWSR1 and Flag tagged PRDM9 were transfected into HEK293 cells. Input controls (left panel), co-IP with HA antibody (middle panel), and IP with Myc antibody (right panel). In each row, staining for the indicated proteins is shown. (D) CXXC1 binds to H3K4me3 but not to H3K9me3. Co-IP with CXXC1 from 14 dpp B6 testicular extract. Top row, staining for CXXC1; middle row, staining for H3K4me3; bottom row, staining for H3K9me3.
Several reports have shown that Spp1, the yeast ortholog of CXXC1, binds to H3K4me3 and tethers H3K4-trimethylated recombination hotspots to the chromosome axis [19–21]. To test whether CXXC1 binds to H3K4me3 in mouse spermatocytes, we performed CXXC1 co-IP from 14-dpp B6 testicular extract. Indeed, we detected CXXC1 interaction with H3K4me3 but not with the closed chromatin mark H3K9me3 (Fig 1D).
These results indicate that CXXC1 and EWSR1 form separate complexes with PRDM9. They also indicate that although CXXC1 interacts with PRDM9 in vivo, it is not a predominant interactor of PRDM9, and that their interaction could be mediated by other proteins such as histone 3 trimethylated at lysine 4.
CXXC1 is present in nuclei of all meiosis stages
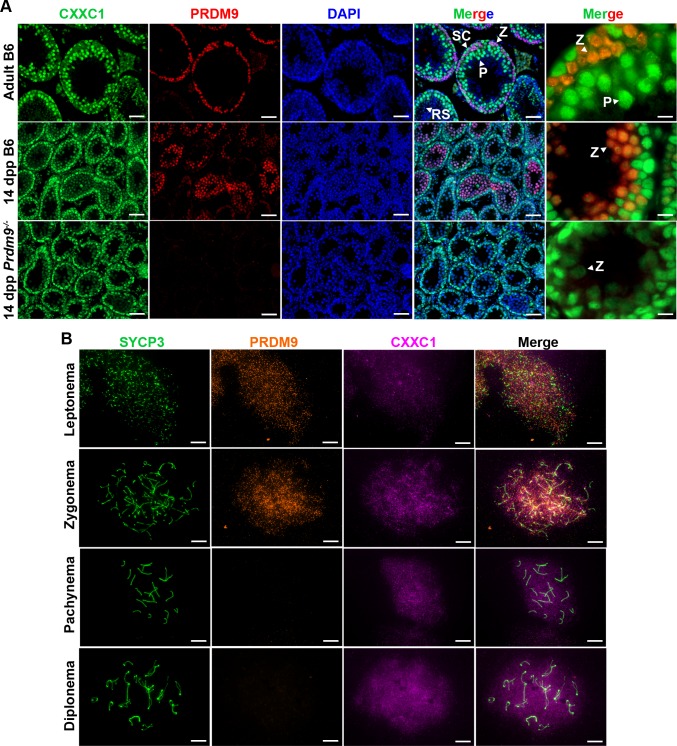
In seminiferous tubules of mouse testis, CXXC1 is expressed in both germ cells and Sertoli cells (Fig 2A, top panel). CXXC1 showed high expression in spermatogonia, low expression in leptonema and zygonema, and then again high expression in pachynema and diplonema, decreasing to undetectable levels in spermatids (Fig 2A, top panel).
Fig 2. CXXC1 is expressed in the spermatocytes in the presence or absence of PRDM9.
(A) Immunofluorescence staining for CXXC1 and PRDM9 in adult B6, 14-dpp B6 and 14-dpp Prdm9-/- seminiferous tubule cross sections. Green, CXXC1; red, PRDM9; blue, DAPI. SC, Sertoli cell; Z, zygonema; P, pachynema; RS, round spermatid. Scale bars, first 4 columns: 50 μm, last column: 10 μm. (B) Immunofluorescence staining for CXXC1 and PRDM9 on chromosome spreads from adult B6. Green, SYCP3; orange, PRDM9; magenta, CXXC1. Scale bars, 10 μm.
Previous reports showed that PRDM9 is present only in leptonema and zygonema during meiosis [28]. Double staining for PRDM9 and CXXC1 showed co-expression of these two proteins in nuclei from stage X seminiferous tubules (Fig 2A, top panel) and in 14-dpp (days post partum) testis (Fig 2A, middle panel), when the majority of spermatocytes are at leptotene and zygotene stages. Since CXXC1 interacts with PRDM9 in vivo (Fig 1A), we performed CXXC1 staining in Prdm9 knockout mouse testis (Prdm9-/-) to determine whether CXXC1 localization could be affected by the absence of PRDM9. In this mutant, CXXC1 showed the same localization pattern as in controls (Fig 2A, bottom panel). The pattern of PRDM9 and CXXC1 in leptonema and zygonema was further confirmed by chromosome spreads, where CXXC1 showed diffused signal over the entire nuclear region from leptonema through diplonema (Fig 2B).
These results show that CXXC1 and PRDM9 are both present in leptonema and zygonema nuclei, and that CXXC1 expression and localization are not affected by the presence or absence of PRDM9.
Male Cxxc1 knockout mice are fertile
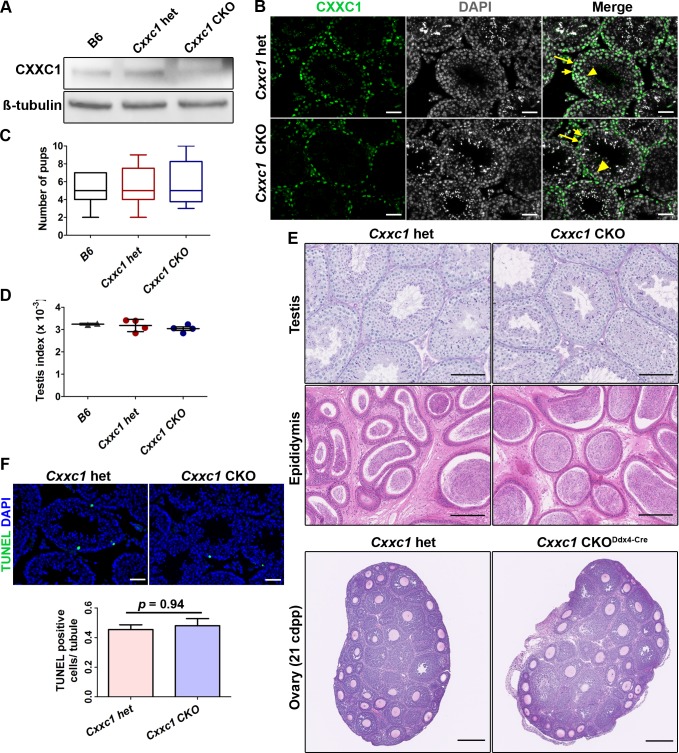
To test whether CXXC1 is involved in spermatogenesis, we generated a conditional knockout (CKO) model using CRISPR/Cas9 to insert loxP sites flanking exon 2 and 3 of Cxxc1 in C57BL/6J. The strategy of obtaining the CKO mutants is shown on S1 Fig. We bred late spermatogonia-specific knockout mice (Cxxc1loxP/Δ;Stra8-iCre, hereafter Cxxc1 CKO) by crossing the Cxxc1loxP/loxP mice with Stra8-iCre mice [29] and germ cell-specific knockout mice (Cxxc1loxP/Δ;Ddx4-Cre, hereafter Cxxc1 CKODdx4-Cre) with Ddx4-Cre mice [30]. Western blot confirmed that in knockout testes, the protein level of CXXC1 is reduced (Fig 3A). CXXC1 was absent in spermatocytes of the CKO, but present in spermatogonia and Sertoli cells in CKO testes (Fig 3B, short and long arrows) and only in Sertoli cells in Cxxc1 CKODdx4-Cre (S2A Fig, long arrows).
Fig 3. Knocking out CXXC1 does not affect male fertility or testis histology.
(A) CXXC1 is depleted in Stra8-cre CKO testes. Western blot of CXXC1 from adult B6, Cxxc1 het and CKO testicular extract. ß-tubulin was used as internal loading control. (B) CXXC1 is present in Sertoli cells but not in spermatocytes of Stra8-cre CKO mice. Immunostaining of CXXC1 on Cxxc1 het and CKO seminiferous tubule cross sections. Green, CXXC1; grey, DAPI. Long arrows, Sertoli cells; short arrows, spermatogonia; arrowhead, spermatocytes. Scale bar, 50 μm. (C) No change in fertility tests in CKO mice compared to B6 and Cxxc1 heterozygous controls. The number of viable pups for each genotype is shown. (D) Testis index (testis weight/body weight) is not changed in CKO mice compared to B6 and Cxxc1 heterozygous controls. (E) Normal histology of testis, epididymis and ovary is observed in both Cxxc1 control and CKO mice. Top panels, PAS staining of seminiferous tubule sections; scale bar, 100 μm. Middle panels, H&E staining of epididymis sections; scale bar, 200 μm. Bottom panels, H&E staining of 21 dpp ovary sections, Scale bar, 250 μm. Left panels, het control; right panels, Cxxc1 CKO with Stra8-Cre in male mice, and Ddx4-Cre in females. (F) No increased apoptosis is observed in testes of CKO mice. TUNEL staining in Cxxc1 het and CKO. Top panels, scale bar, 50 μm. Bottom panels, the apoptotic cell number is quantified as TUNEL positive cell number per seminiferous tubule. Data represent as mean ± SD, p = 0.94 by Student t-test.
We performed fertility test with two Cxxc1-deleted CKO mice of each model. To our surprise, in both models the male mice were fertile and produced similar number of viable progeny compared to the heterozygous (het) and B6 controls (Figs 3C and S2B). Testis index (testis weight/body weight) was the same in CKO as in het and wild type B6 controls (Fig 3D). Histology of testis and epididymis from CKO mice showed no detectable spermatogenesis defects (Figs 3E and S2C). No increased apoptosis in germ cells was detected using TUNEL assay (Figs 3F and S2D).
In contrast, Cxxc1 germ cell-specific knockout female mice with Ddx4-Cre (reduced protein level shown in S1E Fig) were sterile—no viable pups were produced from homozygous knockout Cxxc1 CKODdx4-Cre mating test, while the heterozygous control (Cxxc1loxP/+;Ddx4-Cre) mating produced normal number of pups (5.3 ± 1.7). However, the histology of CKO ovaries from 21 dpp and adult female mice both showed normal ovary morphology and follicle formation (Figs 3E and S2F). Thus, their sterility is most likely due to early embryonic developmental deficiency caused by the lack of maternal genome activation at zygotic stage as reported before [31] and not meiotic defects.
Expression and function of PRDM9 remains normal in Cxxc1 CKO
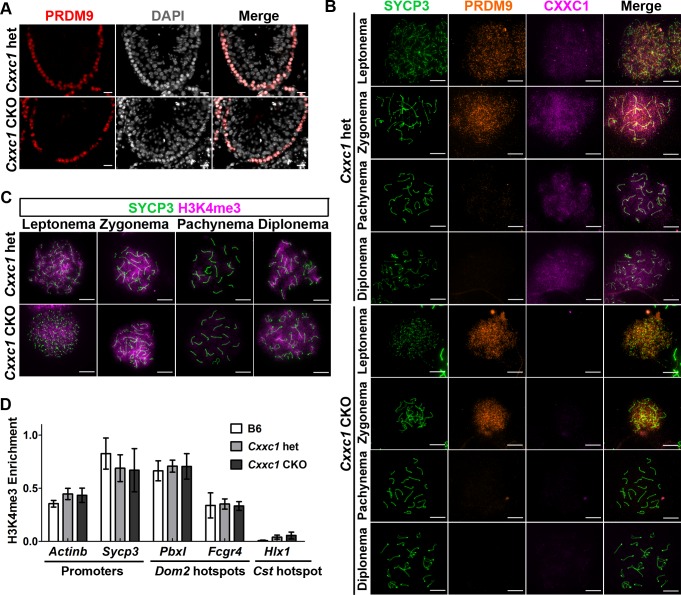
We further tested whether CXXC1 affects the localization, the expression pattern, or the function of PRDM9. Localization of PRDM9 in seminiferous tubules was preserved in CKO (Figs 4A and S3A, right panel). In addition, the expression pattern of PRDM9 in leptonema and zygonema was not affected in the absence of CXXC1 (Fig 4B).
Fig 4. PRDM9 expression and catalytic function are not impaired in Cxxc1 CKO.
(A) Immunostaining of PRDM9 shows unchanged pattern in adult CKO seminiferous tubules compared to the heterozygous control. Red, PRDM9; grey, DAPI. Scale bar, 20 μm. (B) PRDM9 and SYCP3 expression patterns are not changed in CXXC1 CKO chromosome spreads compared to the heterozygous control. Co-immunostaining of CXXC1 and PRDM9 on chromosome spreads from adult Cxxc1 het and CKO mice. Green, SYCP3; orange, PRDM9; magenta, CXXC1. First 4 rows, het control; last 4 rows, Cxxc1 CKO. Scale bar, 10 μm. (C) Meiosis progression occurs normally in Cxxc1 CKO testes. Immunostaining of H3K4me3 in adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; magenta, H3K4me3. Scale bar, 10 μm. (D) Testis-specific gene expression is not changed in Cxxc1 CKO testes. H3K4me3 ChIP-qPCR with chromatin isolated form Cxxc1 het and CKO mice. Promoter regions form Actinb and Sycp3, Dom2 hotspots PbxI and Fcgr4 were amplified. Cst hotspot HlxI was used as a negative control. Bars present mean ± SD of three biological replicates.
To test whether lack of CXXC1 affects PRDM9 methyltransferase function, we first compared H3K4me3 patterns in control and Cxxc1 CKO mice. Both control and CKO chromosome spreads showed abundant H3K4me3 signal in leptonema and zygonema, lower signal in pachynema and increased signal in diplonema (Fig 4C). In addition, the H3K4me3 staining on cross sections of CKO testis showed no decrease (S3B Fig). These data indicate that the hotspot trimethylation and transcriptional activation in spermatocytes are not affected by the loss of CXXC1.
Second, we tested whether loss of CXXC1 affects PRDM9 binding and its methytransferase activity at individual hotspots by H3K4me3 ChIP-qPCR. We found that H3K4me3 enrichment at hotspots PbxI and Fcgr4, which are regulated by Prdm9Dom2, the Prdm9 allele present in B6 mice, was not different in B6 control, Cxxc1 heterozygous and CKO. We measured as a control the H3K4me3 enrichment at promoter regions of the housekeeping gene Actinb and the meiosis specific gene Sycp3, which are not PRDM9-dependent. These were not changed as well (Fig 4D).
These results suggest that loss of CXXC1 does not affect PRDM9 expression, binding to hotspots, or its catalytic function. Therefore, CXXC1 is not required for PRDM9-dependent hotspot activation.
Double strand breaks occur normally at PRDM9 dependent sites in the absence of CXXC1
To test whether lack of CXXC1 affects DSB formation process, we next determined the number, position and activities of DSBs in the Cxxc1 CKO. During DSB formation, the single stranded DNA tail is initially coated by the replication protein A (RPA), and then RPA is gradually replaced by the RecA family members RAD51 and DMC1 [32–35]. When measured by the number of foci of DMC1 (Figs 5A and S4A), RAD51 (Figs 5B and S4B) and RPA (Figs 5C and S4C) in early and late zygonema, these numbers in Cxxc1 CKO were not statistically different from control samples (Fig 5A, 5B and 5C, lower panels). Also, we did not detect increased DMC1, RAD51 or RPA foci in CKO pachynema (Figs 5A, 5B and 5C and S4A, S4B and S4C). These data indicate that the number of DSB per meiosis and DSB repair process is not affected in the loss of CXXC1.
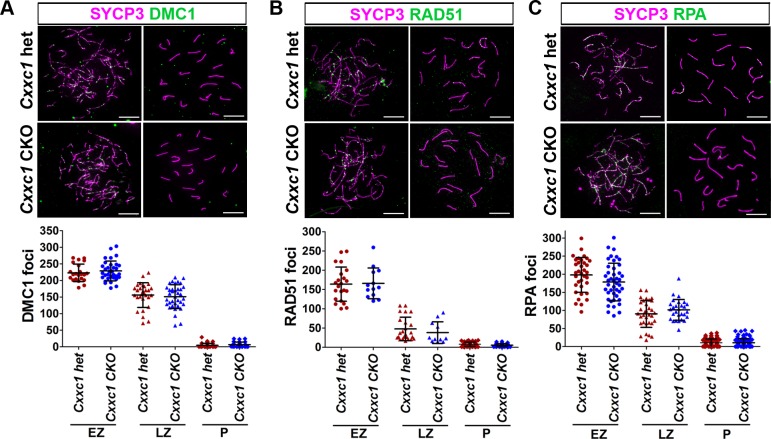
Fig 5. DSB number is not affected in Cxxc1 CKO.
The DSB number was determined by three markers reflecting different stages of their processing. (A) DMC1 staining on Cxxc1 control and CKO chromosome spread. Bottom panels, distribution plot of DMC1 foci in early zygotene (n = 22 in het, n = 34 in CKO), late zygotene (n = 29 in het, n = 36 in CKO) and pachytene (n = 36 in het, n = 26 in CKO) spermatocytes. (B) RAD51 staining on Cxxc1 control and CKO chromosome spread. Bottom panels, distribution plot of RAD51 foci in early zygotene (n = 22 in het, n = 13 in CKO), late zygotene (n = 24 in het, n = 11 in CKO) and pachytene (n = 52 in het, n = 36 in CKO) spermatocytes. (C) RPA staining on Cxxc1 control and CKO chromosome spread. Bottom panels, distribution plot of RPA foci in early zygotene (n = 35 in het, n = 43 in CKO), late zygotene (n = 33 in het, n = 28 in CKO) and pachytene (n = 89 in het, n = 92 in CKO) spermatocytes. For A-C, two individuals per genotype were measured. Bars represent mean ± SD. Scale bar, 10 μm.
To determine whether the locations of DSB sites in Cxxc1 CKO are affected, we performed ChIP-seq for DMC1 [8, 9] (S5A Fig). We detected 8,233 DMC1 peaks in control spermatocytes and 8,569 DMC1 peaks in CKO spermatocytes, in which 7,501 peaks are shared in heterozygous control and CKO samples (Fig 6A). We plotted the frequency distribution of DMC1 activity of the 732 unique peaks from control (Fig 6B, left) or 1068 CKO unique peaks (Fig 6B, right), and found that these virtually unique peaks were not unique, but had low DMC1 activity which prevented them from being detected by the peak calling algorithm. We found that 91.8% of shared DMC1 peaks (6,886 peaks), 78.8% of control unique peaks (577 peaks) and 84.3% of CKO unique peaks (900 peaks) contain a PRDM9 binding site at their centers (Fig 6C as an example). The aggregation plots also confirmed that DSBs contain PRDM9 binding motifs at their centers in both control and CKO spermatocytes (Fig 6D). Also, in the 615 shared, 155 het unique and 168 CKO unique DMC1 peaks which do not contain detectable PRDM9 binding motifs, only 18, 5 and 5 peaks, respectively, overlapped with transcription start sites. Therefore, unlike in Prdm9 knockout mice [9], promoter sites are not predominantly used for DSB formation in Cxxc1 CKO.
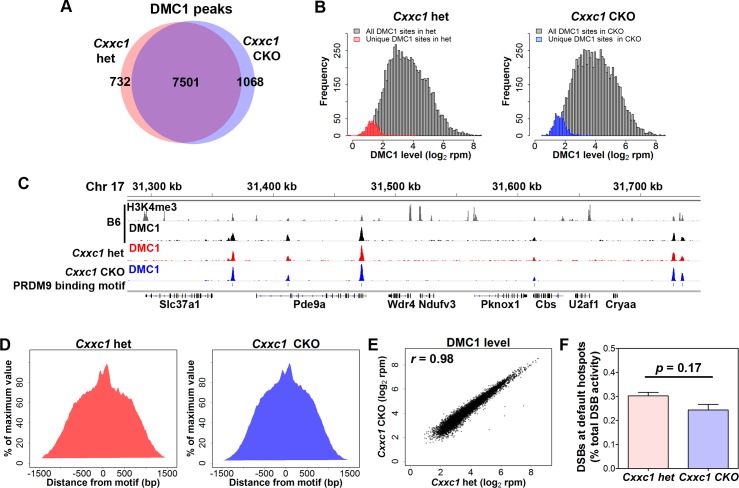
Fig 6. DSB occur at PRDM9 dependent sites in Cxxc1 CKO.
ChIP-seq data determining the number and positioning of DSB sites in CKO and heterozygous controls. (A) Venn diagram of DMC1 peak number in Cxxc1 het and CKO. 7,501 peaks are shared in Cxxc1 het and CKO. (B) Frequency distribution plot of DMC1 activity in Cxxc1 het (left panel) and CKO (right panel). All DMC1 peaks (n = 7,501) were shown in grey in both panels; unique peaks in het controls (n = 732) were shown in red; unique peaks in CKO samples (n = 1,068) were shown in blue. (C) Coverage profiles of published B6 H3K4me3 (grey), B6 DMC1 (black), Cxxc1 het DMC1 (red) and CKO DMC1 (blue) from a representative region on chromosome 17. PRDM9 binding motif sites are shown in line 5. (D) Aggregation plot of DMC1 signal in het control (left) and CKO (right). The signal was normalized to the maximum signal. (E) Plot of activity of DSBs from Cxxc1 CKO and control spermatocytes. Correlation coefficient r = 0.98. (F) Percentage of DMC1 activity in default sites contributes to total activity.
The activity of DMC1 signal in control and CKO is highly correlated (r = 0.98, Fig 6D), indicating the activity of DSB formation is not affected in the CKO spermatocytes. Furthermore, the activity of default DSB sites, which do not contain PRDM9 binding motif in the center, only contributes to 0.30% and 0.24% of total DMC1 activity in control and CKO samples, respectively (Figs 6E and S5B), similar as reported default DSB activity in male germ cells [36].
These data suggest that loss of CXXC1 does not affect DSB number or positions. Therefore, CXXC1 is not essential for PRDM9 dependent DSB initiation pathway.
Meiotic DSB repair is normal in Cxxc1 CKO
To further investigate whether the DSB repair process and chromosomal synapsis are impaired in CKO, we used staining for phosphorylated H2AX (γH2AX), which marks unrepaired DNA lesions and sex body in pachynema, to test for the processing of recombination repair. The pattern of γH2AX staining was not changed in CKO compared to the heterozygous control spermatocytes, showing γH2AX signal throughout the nucleus in leptonema when DSBs occur, which was then restricted to the sex body in pachynema when the autosomal breaks are repaired (Figs 7A and S4D). We also measured spermatocyte proportion based on the staining, and did not detect difference in cell proportion (Figs 7B and S4E). These results indicate that the sex body formation and DSB repair are not affected in the Cxxc1 CKO, and there is no major cell loss or arrest in CKO meiosis.
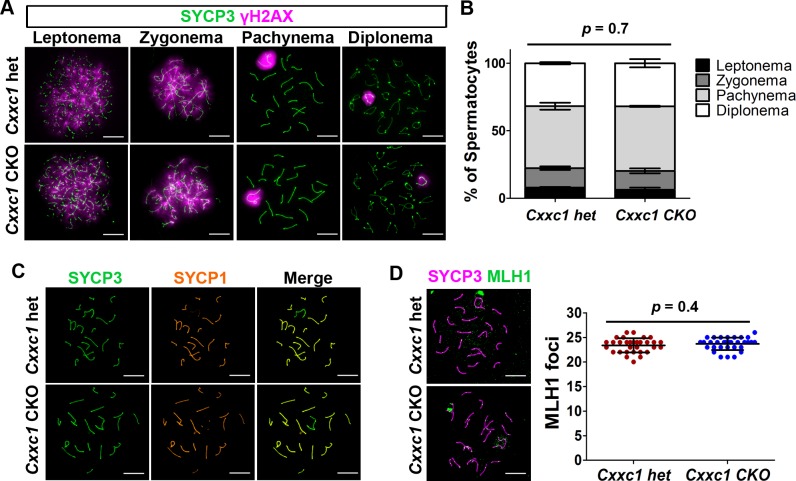
Fig 7. No major meiotic DSB repair or chromosome synapsis defects are observed in Cxxc1 CKO testis.
(A) Immunostaining of SYCP3 and γH2AX on adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; magenta, γH2AX. Scale bar, 10 μm. (B) Spermatocyte stage proportion in adult Cxxc1 het (n = 1,062 from two individuals) and CKO (n = 1,105 from two individuals) spermatocytes based on SYCP3/SYCP1/γH2AX staining. p = 0.7 by Chi-square test. (C) Immunostaining of SYCP3 and SYCP1 on adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; orange, SYCP1. Scale bar, 10 μm. (D) Crossover number measured by MLH1 staining on chromosome spreads of adult Cxxc1 het and CKO spermatocytes. Left, magenta, SYCP3; green, MLH1. Scale bar, 10 μm. Right, number of MLH1 foci per late pachynema in Cxxc1 het (n = 32 from two individuals) and CKO (n = 33 from two individuals). Bars represent mean ± SD. p = 0.4 by Student’s t-test.
Co-staining of SYCP1 and SYCP3 confirmed normal synapsis in all autosomes in CKO spermatocytes at pachynema (Figs 7C and S4F). 98.5% of Cxxc1 CKO pachynema with Stra8-Cre and 98.23% of CKO pachynema with Ddx4-Cre showed full synapsis, compared with 97.5% of fully synapsed pachynema in control (p = 0.15 and 0.91, respectively). Therefore, there is no increased chromosome asynapsis in the CKO spermatocytes compared to controls.
Finally, we examined whether loss of CXXC1 affects crossover resolution. Using MLH1 as a marker of crossover sites, we did not find any significant change of crossover number in the CKO spermatocytes compared to the het controls (Fig 7D).
Taken together, these results suggest that even though CXXC1 interacts with PRDM9 and H3K4me3 in spermatocytes, it is not required for PRDM9 binding at hotspots, their subsequent activation by PRDM9-dependent H3K4 trimethylation, DSB formation, repair, or crossover formation, and is therefore not essential for meiotic recombination events.
Discussion
In this study, we demonstrate that CXXC1 interacts with PRDM9 in spermatocytes. However, this interaction does not seem to be important for any cell function necessary for germ cell development or recombination processes. The germ cell specific Cxxc1 knockout male mice are fertile. In the knockout spermatocytes, the expression and function of PRDM9 are unchanged. The loss of CXXC1 does not affect DSB formation and repair, chromosome pairing and synapsis, and crossover numbers. Together, these results convincingly show that CXXC1 is not essential for normal meiotic recombination events and generally for spermatogenesis and oogenesis.
The yeast CXXC1 ortholog Spp1 is reported to be a key player in recombination by linking H3K4me3 sites with the chromosome axis and connecting them with the recombination protein Mer2 [19–21]. This suggested that CXXC1 might play similar function in mammalian meiosis. However, our results show that CXXC1 is not an essential player in mammalian recombination where PRDM9 controls the initial recognition and activation of recombination hotspots. In the absence of CXXC1, hotspot activation, axis integrity, DSB formation and crossover resolution occur normally, showing that DSB formation and recombination determination in most of mammals, which use the PRDM9 dependent pathway, differs from that in the budding yeast. In species with functional PRDM9, the function of the RMM complex consisting of orthologs of the yeast Rec114, Mei4 and Mer2 (REC114, MEI4 and IHO1 in mice) is still conserved [27, 37–39], and the association between hotspots and chromosome axis is crucial for efficient DSB formation [27, 40, 41]. However, the interaction between CXXC1 and IHO1 does not seem to play the same functional role as the one between Spp1 and Mer2 in yeast. One important difference is that in organisms that do not use PRDM9, DSB occur at H3K4me3 sites, whereas in those that use PRDM9, DSB occur at hotspots where surrounding nucleosomes are methylated at both H3K4 and H3K36 [5, 6]. This raises the likelihood that proteins with H3K36me3 methyl-reading activity, such as PWWP domain containing proteins [42], or with both H3K4me and H3K36me binding capability, such as Tudor domain containing proteins [43], might be involved in hotspot recognition in these species. Alternatively, activated hotspots may be recruited to the chromosome axis and DSB machinery without assistance of an H3K4me3/H3K36me3 reader. A recent study demonstrated that randomized DSBs induced by radiation in Spo11 mutant spermatocytes are associated with chromosome axis and can successfully recruit DSB repair proteins such as DMC1/RAD51 complex [44]. Other direct PRDM9 interactors, such as EWSR1, EHMT2, CDYL [16], PIH1D1 [26] and CTCF [45], could also be involved in hotspot association with the chromosome axis.
An alternative, PRDM9-independent pathway, can explain the fraction of DSB detected at promoters in wild type mice, and all DSB in PRDM9 mutant mice [9, 46]. Although we did not detect any substantial reduction of PRDM9-independent hotspot activity in the absence of CXXC1, this pathway could still, to some degree, involve CXXC1 as part of the SETD1 complex, known to bind H3K4me3 at promoters, in a way similar to Spp1-Mer2 role in yeast meiosis. It is not an essential pathway in most organisms using PRDM9 as hotspot determinant, but might play a major role in those lacking PRDM9, such as canids [47–49], where recombination hotspots are enriched in CpG-rich regions with a preference for unmethylated CpG islands [9, 17, 47, 50], similar feature as CXXC1 binding sites [22, 51]. One recent report of a woman having no active PRDM9 but completely fertile suggests that this pathway may become activated and ensure proper recombination even in organisms using PRDM9 as a recombination regulator [52].
Methods
Ethics statement
The animal care rules used by The Jackson Laboratory are compatible with the regulations and standards of the U.S. Department of Agriculture and the National Institutes of Health. The protocols used in this study were approved by the Animal Care and Use Committee of The Jackson Laboratory (Summary #04008). Euthanasia for this study was done by cervical dislocation.
Mouse models
All wild-type mice used in this study were in the C57BL/6J (B6) background. The conditional-ready mutants were produced by flanking exons 2 and 3 of Cxxc1 with loxP sites using CRISPR-Cas9. The Cxxc1 conditional knockout mice used in this study were produced by a two-step deletion scheme. Mice that harbor two conditional Cxxc1 alleles (Cxxc1loxp/loxp) were mated to Tg(Sox2-cre)1Amc/J mice (stock #004783) to generate one Cxxc1 allele deleted mice. The Cxxc1 hemizygous mice (Cxxc1Δ/+) were mated to Tg(Stra8-icre)1Reb/J (stock #08208) and Tg(Ddx4-cre)1Dcas/KnwJ (stock #018980) to obtain Cxxc1Δ/+;Stra8-iCre and Cxxc1Δ/+;Ddx4-Cre mice. Those Ewsr1Δ/+;Stra8-iCre and Cxxc1Δ/+;Ddx4-Cre mice were then mated to homozygous Cxxc1 loxp mice to generate heterozygous control mice (Cxxc1loxP/+;Stra8-iCreand Cxxc1loxP/+;Ddx4-Cre designated as Cxxc1 controls) or conditional knockout mice (Cxxc1loxP/Δ;Stra8-iCreand Cxxc1loxP/Δ;Ddx4-Cre designated as Cxxc1 CKO).
B6;129P2-Prdm9tm1Ymat/J mice (Prdm9-/-) have been previously described [53]. All animal experiments were approved by the Animal Care and Use Committee of The Jackson Laboratory (Summary #04008).
Co-immunoprecipitation assays
The co-immunoprecipitation assays for PRDM9 and EWSR1 with testicular extract were carried out using our reported protocol [16]. Total protein was extracted from testes of twenty 14-dpp B6 mice homogenized in 1 ml of Pierce IP buffer (Thermo Fisher Scientific, 87787). 10% of extract was set apart as input. Co- immunoprecipitation was performed by incubating extract with protein G Dynabeads conjugated with antibodies against PRDM9 [18, 54] or guinea pig IgG overnight at 4°C. The beads were washed three times with 1 ml of Pierce IP buffer, eluted with 200 μl of GST buffer (0.2 M glycine, 0.1% SDS, 1% Tween 20, pH 2.2) for 20 min at room temperature and neutralized with 40 μl of 1 M Tris-HCl, pH 8. After heated at 95°C for 5 min, 10 μg of IP and input samples were then subjected to electrophoresis and western blotting for detection of PRDM9 (1:1000, custom made), EWSR1 (1:1000, Abcam, ab54708) and CXXC1 (1:1000, Abcam, ab198977). The co-IP experiment is performed in two replicates.
The co-immunoprecipitation assays for PRDM9, CXXC1 and EWSR1 in cell culture were carried out using our reported protocol [55]. The vector expressing the PRDM9, CXXC1 and EWSR1 proteins were constructed by cloning mouse Prdm9, Cxxc1 and Ewsr1 cDNA into pCEP4-Flag, pCMV-Myc and pCMV-HA vectors, respectively. 2.5 μg of plasmids were transfected into HEK293 cells by X-tremeGENE HP DNA Transfection Reagent (Roche, 6366244001) in 6-well plates. At 2 days after transfection, cells were harvested and mixed with 600 μl Pierce IP buffer. 10% of extract was set apart as input. Co-immunoprecipitation was performed by incubating extract with protein G Dynabeads conjugated with antibodies against HA (Sigma, H9658) or Myc (Sigma, M5546) overnight at 4°C. After washing the beads and eluting with GST buffer, 10 μg of IP and input samples were then subjected to electrophoresis and western blotting for detection of HA, Myc and Flag (Sigma, F1804). All the blots were processed together with the same exposure.
Co- immunoprecipitation for CXXC1 was performed similarly to those for PRDM9 and EWSR1 with the following changes. The seminiferous tubules were digested with liberase and the germ cells were isolated. Then, the nuclei were isolated by incubation germ cells in hypotonic lysis buffer (10 mM Tris-HCL pH 8.0, 1 mM KCl, 1.5 mM MgCl2) for 30 min at 4°C and spinning down at 10,000 g for 10 min. The nuclear extract was obtained by incubation with the nuclear lysis buffer (50 mM HEPES, pH 7.8, 3 mM MgCl2, 300 mM NaCl, 1 mM DTT and 0.1 mM PMSF), 5 U/μl DNaseI and 2 U/ μl TurboNuclease for 30 min at 4°C. 10% of extract was saved as input. The co-IP was perform by incubating extract with protein G Dynabeads conjugated with antibodies against CXXC1 (Abcam, ab198977) or guinea pig IgG overnight at 4°C. After wash and elution, the IP and input samples were then subjected to electrophoresis and western blotting for detection of CXXC1 (1:1000, Abcam, ab198977), H3K4me3 (1:1000, Millipore, #07–473) and H3K9me3 (1:1000, Active Motif, 39766).
Measurement of testis index
Testicular weight and body weight of adult B6 (n = 3), Cxxc1 het (n = 3) and CKO (n = 4) mice were measured. Testis index was calculated as testis weight/body weight. Student’s t-test was used to determine the statistical significance.
Fertility test
Male fertility test was performed with 3 Cxxc1 het control and 5 CKO male mice. Each mouse was mated with at least two B6 females for at least two to five month period. Female fertility test was performed with 2 Cxxc1 control and 2 CKO female mice. Each one was mated with one B6 male for 3 month period. Litter size and viable pup number were recorded.
Histology
Testis, epididymis, ovaries from adult or 21 dpp Cxxc1 het control or CKO mice were dissected out. Testis and epididymis were fixed with Bouin’s solution, and ovaries were fixed in 2% PFA. and the tissues were embedded in paraffin wax, and sectioned at 5 μm. Sections of testis were stained with Periodic acid–Schiff–diastase (PAS), and section of epididymis and ovaries were stained with haematoxylin and eosin (H&E) using standard techniques.
Chromosome spread cytology
The drying-down technique [56] was used for preparation of chromosome spreads from spermatocytes of 14-dpp and 8-weeks B6, Cxxc1 control or CKO mice. Chromosome spread slides were immunolabeled with anti-PRDM9 (1:200), CXXC1 (1:1000), SYCP1 (1:300, Novus, NB300-229), SYCP3 (1:400, Novus, NB300-231), γH2AX (1:1000, Abcam, ab26350), DMC1 (1:200, Santa Cruz, sc-8973), RAD51 (1:200, Santa Cruz, H-92), RPA (1:300, Abcam, ab87272) or MLH1 (1:100, BD Pharmingen, 550838) antibodies.
Immunofluorescence stainings
For protein immunolocalization on tissue sections, testicular tissues from 8 week old B6, Prdm9-/-, Cxxc1 control and CKO mice were dissected out, fixed with 4% paraformaldehyde solution overnight, embedded in paraffin wax. 5-μm sections were prepared. For antigen retrieval, sections were heated in a microwave in Tris-EDTA buffer (10mM Tris, 1mM EDTA and 0.05% Tween 20, pH 9.0) for 10 min and cooled down to room temperature. Then, sections were treated with PBS containing 0.1% Triton X-100, blocked with 10% normal donkey serum, and stained with antibodies against PRDM9 (1:200), CXXC1 (1:1000) or H3K4me3 (1:1000, Millipore, #07–473).
H3K4me3 chromatin immunoprecipitation and real-time PCR
Chromatin immunoprecipitation (ChIP) was performed as previously described [57]. Briefly, spermatocytes were isolated from 14-dpp B6, Cxxc1 het and CKO spermatocytes, and crosslinked using 1% formaldehyde. Nuclei were isolated using hypotonic lysis buffer (10 mM Tris-HCL pH 8.0, 1 mM KCl, 1.5 mM MgCl2) and digested by MNase. The ChIP was done using antibody against H3K4me3. Real-time PCR was performed with purified ChIP DNA using Quantifast SYBR Green PCR Kit (Qiagen) Primer sequences used for real-time PCR are: Pbx1_F: AGAAACTGACATATGAAGGCTCA; Pbx1_R: GCTTTTGCTCCCTTAAACTGG; Fcgr4_F: CAAGGTGCATTCTTAGGAGAGA; Fcgr4_R: TTAATGCTTGCCTCACGTTC; Hlx1_F: GGTCGGTGTGAGTATTAGACG; Hlx1_R: GGCTACTATACCTTATGCTCTG; Actinb_promoter_F: GCCATAAAAGGCAACTTTCG; Actinb_promoter_R: TTTCAAAAGGAGGGGAGAGG; Sycp3_promoter_F: AAGGCGCCACAACCAAGG; Sycp3_promoter_F: TGCCTGGATGCCCAACTC.
DMC1 ChIP-seq and data analysis
DMC1 ChIP was performed from spermatocytes of 8 weeks old Cxxc1 het and CKO (Stra8-Cre) using an established method [58] in two replicates. The testes were cross-linked with 1% paraformaldehyde solution for 10 min, and then homogenized. After that, the nuclei were isolated, the chromatin was sheared to ~1000 bp by sonication and incubated with antibody against DMC1 overnight at 4°C, and then with protein G Dynabeads (Thermo Fisher Scientific, 10004D) for 4 hrs at 4°C. The beads were washed once with wash buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), wash buffer 2 (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mMTris-HCl, pH 8.0, 500 mM NaCl), then wash buffer 3 (0.25 M LiCl, 1% NP-40, 1mM EDTA, 10mMTris-HCl, pH 8.0, 1% Deoxycholic acid), and finally twice with TE buffer. The chromatin was eluted with dilution buffer (1% SDS, 0.1 M NaHCO3 pH 9.0) at 65°C for 30 min and then reverse-crosslinked by adding 200 mM NaCl and incubation overnight at 65°C. The libraries were then prepared according to the currently established method [58], and sequenced on an Illumina HiSeq 2500 platform, with 75 bp paired-end reads.
Fastq files for sequenced DMC1 libraries were trimmed using Trimmomatic (v0.32) and subsequently parsed for detection and selection of paired reads having homology at the 5’ and 3’ ends [9, 59] as established by protocols for single strand DNA enrichment to generate the files that contain only the detectable single strand reads, and then, these files were aligned to mm10 mouse genome using BWA (v.0.5.10-tpx). Bam files were parsed for detection and selection of reads containing true genomic sequence versus fill-in sequence at the homologous region. These reads were selected from the original paired-end fastq files, and then single-end fastq files were created that contained only the true genomic sequences of single strand DNA reads. All genomic data are available at NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE116336 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116336). 1,688,630 and 1,349,516 aligned DMC1 reads in Cxxc1 het controls, 1,590,925 and 1,961,645 aligned DMC1 reads in Cxxc1 CKO spermatocytes were obtained from the two replicate libraries. The correlation between the two biological replicates in each experiment was high (r = 0.96 in Cxxc1 het controls; r = 0.99 in Cxxc1 CKO. S5A Fig); thus, the data from each pair of replicates were merged. The DMC1 activity was normalized to reads per million (rpm). Peak calling was performed using MACS (v.2.0.9) with a FDR value 0.01. PRDM9 dependent or default sites were determined using bedtools (v2.22.0) intersects compared with unknown PRDM9 binding sites (GEO number: GSE61613) [18]. Analyses for the aggregation plots were carried out using the ACT [60], of which parameters were: nbins = 500, mbins = 0, radius = 1500.
Supporting information
Top line, wild type allele of Cxxc1; middle line, loxP allele of Cxxc1 by flanking exon 2 and 3 with loxP sites; bottom line, deletion allele of Cxxc1 after crossing the Cxxc1loxP/loxP mice with Cre mice. Orange boxes, coding regions; blue boxes, 3 or 5 prime untranslated regions; black lines, intron regions; red triangles, loxP sites; red arrow, a stop codon generated by frame shift in the deletion allele; grey boxes, untranslated regions after stop codon in the deletion allele. Exon numbers are indicated as E1 to E15.
(TIF)
(A) Immunostaining of CXXC1 on Cxxc1 het and CKODdx4-Cre seminiferous tubule cross sections. Green, CXXC1; grey, DAPI. Long arrows, Sertoli cells; short arrows, spermatogonia. Scale bar, 50 μm. (B) Fertility tests in Cxxc1 het and CKODdx4-Cre mice. The number of viable pups was shown. (C) PAS stating of seminiferous tubules in CKODdx4-Cre. Scale bar, 100 μm. (D) TUNEL staining (left) and quantification of apoptotic germ cells (right) in Cxxc1 CKODdx4-Cre. Scale bar, 50 μm. Data represented as mean ± SD, p = 0.94 by Student t-test.(E) Western blot of CXXC1 with adult B6, Cxxc1 het and CKODdx4-Cre whole ovary extract. ß-tubulin was used as internal loading control. (F) H&E staining of secondary follicles in 4-month old Cxxc1 het and CKODdx4-Cre. Scale bar, 50 μm.
(TIF)
(A) Immunostaining of PRDM9 in Cxxc1 het and CKODdx4-Cre. Red, PRDM9; grey, DAPI. Scale bar, 20 μm. (B) Immunofluorescence staining of H3K4me3 on adult Cxxc1 het and CKO with Stra8- and Ddx4-Cre seminiferous tubule cross sections. Magenta, H3K4me3; gray, DAPI. Scale bars: 50 μm.
(TIF)
The DSB number was determined by three markers reflecting different stages of their processing. (A) DMC1 staining on Cxxc1 control and CKO chromosome spread. Lower panel, distribution plot of DMC1 foci in early zygotene (n = 20for each genotype), late zygotene (n = 25) and pachytene (n = 34) spermatocytes. (B) RAD51 staining on Cxxc1 control and CKO chromosome spread. Lower panel, distribution plot of RAD51 foci in early zygotene (n = 8), late zygotene (n = 18) and pachytene (n = 53) spermatocytes. (C) RPA staining on Cxxc1 control and CKO chromosome spread. Lower panel, distribution plot of RPA foci in early zygotene (n = 15), late zygotene (n = 18) and pachytene (n = 48 i) spermatocytes. For A-C, cells from two individuals per genotype were measured. Bars represent mean ± SD. Scale bars, 10 μm. (D) Immunostaining of SYCP3 and γH2AX on adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; magenta, γH2AX. Scale bars, 10 μm. (E) Spermatocyte stage proportion in adult Cxxc1 het and CKO (n = 1,066 from two individuals) spermatocytes based on SYCP3/SYCP1/γH2AX staining. p = 0.7 by Chi-square test. (F) Immunostaining of SYCP3 and SYCP1 on adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; orange, SYCP1. Scale bars, 10 μm.
(TIF)
(A) Plots of activity of DSBs in two replicates of Cxxc1 CKO and control DMC1 ChIP-seq samples. Correlation coefficient r = 0.96 in het controls, r = 0.99 in CKO samples. (B) Plot of activity of DSBs from Cxxc1 CKO and control spermatocytes. Black dots, PRDM9-dependent sites; yellow dots, PRDM9-independent sites.
(TIF)
Acknowledgments
We thank all members of Petkov and Paigen labs, Michael Walker for providing the data analysis pipelines, Mary Ann Handel for sharing antibodies and useful suggestions, Ewelina Bolcun-Filas for helpful comments, and Anita Adams for technical help.
Data Availability
ChIP-seq data are available at Gene Expression Omnibus data repository under GEO number GSE116336 with secure token ilszsgyyfhglbel. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institutes of Health Grant R01 GM078452 to PMP and Cancer Core Grant CA34196 to the Jackson Laboratory. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kleckner N (1996) Meiosis: How could it work? Proceedings of the National Academy of Sciences of the United States of America 93: 8167–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parvanov ED, Petkov PM, Paigen K (2010) Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835 10.1126/science.1181495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, et al. (2010) PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 42: 859–863. 10.1038/ng.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. (2010) PRDM9 Is a Major Determinant of Meiotic Recombination Hotspots in Humans and Mice. Science 327: 836–840. 10.1126/science.1183439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, et al. (2016) The Meiotic Recombination Activator PRDM9 Trimethylates Both H3K36 and H3K4 at Recombination Hotspots In Vivo. PLoS Genet 12: e1006146 10.1371/journal.pgen.1006146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker CL, Walker M, Kajita S, Petkov PM, Paigen K (2014) PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res 24: 724–732. 10.1101/gr.170167.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, et al. (2011) Determinants of nucleosome organization in primary human cells. Nature 474: 516–520. 10.1038/nature10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, et al. (2011) Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. 10.1038/nature09869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV (2012) Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. 10.1038/nature11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada S, Kim S, Tischfield SE, Jasin M, Lange J, et al. (2017) Genomic and chromatin features shaping meiotic double-strand break formation and repair in mice. Cell Cycle 16: 1870–1884. 10.1080/15384101.2017.1361065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimitou EP, Yamada S, Keeney S (2017) A global view of meiotic double-strand break end resection. Science 355: 40–45. 10.1126/science.aak9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zickler D, Kleckner N (2015) Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter N (2015) Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, et al. (2011) Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. 10.1016/j.cell.2011.07.003 [DOI] [PubMed] [Google Scholar]
- 15.Kohler S, Wojcik M, Xu K, Dernburg AF (2017) Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proc Natl Acad Sci U S A 114: E4734–E4743. 10.1073/pnas.1702312114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parvanov ED, Tian H, Billings T, Saxl RL, Spruce C, et al. (2017) PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol Biol Cell 28: 488–499. 10.1091/mbc.E16-09-0686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diagouraga B, Clement JAJ, Duret L, Kadlec J, de Massy B, et al. (2018) PRDM9 Methyltransferase Activity Is Essential for Meiotic DNA Double-Strand Break Formation at Its Binding Sites. Mol Cell. [DOI] [PubMed] [Google Scholar]
- 18.Walker M, Billings T, Baker CL, Powers N, Tian H, et al. (2015) Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8: 31 10.1186/s13072-015-0024-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, et al. (2013) The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. 10.1126/science.1225739 [DOI] [PubMed] [Google Scholar]
- 20.Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V (2013) Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54. 10.1016/j.molcel.2012.11.008 [DOI] [PubMed] [Google Scholar]
- 21.Adam C, Guerois R, Citarella A, Verardi L, Adolphe F, et al. (2018) The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet 14: e1007223 10.1371/journal.pgen.1007223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, et al. (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086. 10.1038/nature08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clouaire T, Webb S, Bird A (2014) Cfp1 is required for gene expression-dependent H3K4 trimethylation and H3K9 acetylation in embryonic stem cells. Genome Biol 15: 451 10.1186/s13059-014-0451-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, et al. (2012) Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 26: 1714–1728. 10.1101/gad.194209.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlone DL, Skalnik DG (2001) CpG binding protein is crucial for early embryonic development. Mol Cell Biol 21: 7601–7606. 10.1128/MCB.21.22.7601-7606.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai Y, Baudat F, Taillepierre M, Stanzione M, Toth A, et al. (2017) The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanzione M, Baumann M, Papanikos F, Dereli I, Lange J, et al. (2016) Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nature Cell Biology 18: 1208-+ 10.1038/ncb3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun F, Fujiwara Y, Reinholdt LG, Hu J, Saxl RL, et al. (2015) Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE (2008) Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46: 738–742. 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallardo T, Shirley L, John GB, Castrillon DH (2007) Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45: 413–417. 10.1002/dvg.20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu C, Fan X, Sha QQ, Wang HH, Li BT, et al. (2017) CFP1 Regulates Histone H3K4 Trimethylation and Developmental Potential in Mouse Oocytes. Cell Rep 20: 1161–1172. 10.1016/j.celrep.2017.07.011 [DOI] [PubMed] [Google Scholar]
- 32.Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, et al. (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1: 697–705. [DOI] [PubMed] [Google Scholar]
- 33.Barlow AL, Benson FE, West SC, Hulten MA (1997) Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J 16: 5207–5215. 10.1093/emboj/16.17.5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plug AW, Peters AH, Xu Y, Keegan KS, Hoekstra MF, et al. (1997) ATM and RPA in meiotic chromosome synapsis and recombination. Nat Genet 17: 457–461. 10.1038/ng1297-457 [DOI] [PubMed] [Google Scholar]
- 35.Gray S, Cohen PE (2016) Control of Meiotic Crossovers: From Double-Strand Break Formation to Designation. Annu Rev Genet 50: 175–210. 10.1146/annurev-genet-120215-035111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brick K, Thibault-Sennett S, Smagulova F, Lam K, Pu Y, et al. (2018) Extensive sex differences at the initiation of genetic recombination. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libby BJ, Reinholdt LG, Schimenti JC (2003) Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci U S A 100: 15706–15711. 10.1073/pnas.2432067100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar R, Ghyselinck N, Ishiguro K, Watanabe Y, Kouznetsova A, et al. (2015) MEI4—a central player in the regulation of meiotic DNA double-strand break formation in the mouse. J Cell Sci 128: 1800–1811. 10.1242/jcs.165464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar R, Bourbon HM, de Massy B (2010) Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24: 1266–1280. 10.1101/gad.571710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, et al. (2011) Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13: 599–610. 10.1038/ncb2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, et al. (2012) Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev 26: 958–973. 10.1101/gad.187559.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin S, Min J (2014) Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci 39: 536–547. 10.1016/j.tibs.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 43.Myrick LK, Hashimoto H, Cheng XD, Warren ST (2015) Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Human Molecular Genetics 24: 1733–1740. 10.1093/hmg/ddu586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carofiglio F, Sleddens-Linkels E, Wassenaar E, Inagaki A, van Cappellen WA, et al. (2018) Repair of exogenous DNA double-strand breaks promotes chromosome synapsis in SPO11-mutant mouse meiocytes, and is altered in the absence of HORMAD1. DNA Repair (Amst) 63: 25–38. [DOI] [PubMed] [Google Scholar]
- 45.Grey C, Clement JA, Buard J, Leblanc B, Gut I, et al. (2017) In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites. Genome Res 27: 580–590. 10.1101/gr.217240.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV (2016) The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev 30: 266–280. 10.1101/gad.270009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auton A, Li YR, Kidd J, Oliveira K, Nadel J, et al. (2013) Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs. Plos Genetics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Axelsson E, Webster MT, Ratnakumar A, Consortium L, Ponting CP, et al. (2012) Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res 22: 51–63. 10.1101/gr.124123.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munoz-Fuentes V, Di Rienzo A, Vila C (2011) Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One 6: e25498 10.1371/journal.pone.0025498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell CL, Bherer C, Morrow BE, Boyko AR, Auton A (2016) A Pedigree-Based Map of Recombination in the Domestic Dog Genome. G3-Genes Genomes Genetics 6: 3517–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tate CM, Lee JH, Skalnik DG (2010) CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J 277: 210–223. 10.1111/j.1742-4658.2009.07475.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, et al. (2016) Health and population effects of rare gene knockouts in adult humans with related parents. Science 352: 474–477. 10.1126/science.aac8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayashi K, Yoshida K, Matsui Y (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. 10.1038/nature04112 [DOI] [PubMed] [Google Scholar]
- 54.Baker CL, Kajita S, Walker M, Saxl RL, Raghupathy N, et al. (2015) PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS genetics 11: e1004916 10.1371/journal.pgen.1004916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker CL, Petkova P, Walker M, Flachs P, Mihola O, et al. (2015) Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots. PLoS Genet 11: e1005512 10.1371/journal.pgen.1005512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters AH, Plug AW, van Vugt MJ, de Boer P (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5: 66–68. [DOI] [PubMed] [Google Scholar]
- 57.Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, et al. (2013) DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol 14: R35 10.1186/gb-2013-14-4-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV (2012) Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res 22: 957–965. 10.1101/gr.130583.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV (2012) Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome research 22: 957–965. 10.1101/gr.130583.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jee J, Rozowsky J, Yip KY, Lochovsky L, Bjornson R, et al. (2011) ACT: aggregation and correlation toolbox for analyses of genome tracks. Bioinformatics 27: 1152–1154. 10.1093/bioinformatics/btr092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top line, wild type allele of Cxxc1; middle line, loxP allele of Cxxc1 by flanking exon 2 and 3 with loxP sites; bottom line, deletion allele of Cxxc1 after crossing the Cxxc1loxP/loxP mice with Cre mice. Orange boxes, coding regions; blue boxes, 3 or 5 prime untranslated regions; black lines, intron regions; red triangles, loxP sites; red arrow, a stop codon generated by frame shift in the deletion allele; grey boxes, untranslated regions after stop codon in the deletion allele. Exon numbers are indicated as E1 to E15.
(TIF)
(A) Immunostaining of CXXC1 on Cxxc1 het and CKODdx4-Cre seminiferous tubule cross sections. Green, CXXC1; grey, DAPI. Long arrows, Sertoli cells; short arrows, spermatogonia. Scale bar, 50 μm. (B) Fertility tests in Cxxc1 het and CKODdx4-Cre mice. The number of viable pups was shown. (C) PAS stating of seminiferous tubules in CKODdx4-Cre. Scale bar, 100 μm. (D) TUNEL staining (left) and quantification of apoptotic germ cells (right) in Cxxc1 CKODdx4-Cre. Scale bar, 50 μm. Data represented as mean ± SD, p = 0.94 by Student t-test.(E) Western blot of CXXC1 with adult B6, Cxxc1 het and CKODdx4-Cre whole ovary extract. ß-tubulin was used as internal loading control. (F) H&E staining of secondary follicles in 4-month old Cxxc1 het and CKODdx4-Cre. Scale bar, 50 μm.
(TIF)
(A) Immunostaining of PRDM9 in Cxxc1 het and CKODdx4-Cre. Red, PRDM9; grey, DAPI. Scale bar, 20 μm. (B) Immunofluorescence staining of H3K4me3 on adult Cxxc1 het and CKO with Stra8- and Ddx4-Cre seminiferous tubule cross sections. Magenta, H3K4me3; gray, DAPI. Scale bars: 50 μm.
(TIF)
The DSB number was determined by three markers reflecting different stages of their processing. (A) DMC1 staining on Cxxc1 control and CKO chromosome spread. Lower panel, distribution plot of DMC1 foci in early zygotene (n = 20for each genotype), late zygotene (n = 25) and pachytene (n = 34) spermatocytes. (B) RAD51 staining on Cxxc1 control and CKO chromosome spread. Lower panel, distribution plot of RAD51 foci in early zygotene (n = 8), late zygotene (n = 18) and pachytene (n = 53) spermatocytes. (C) RPA staining on Cxxc1 control and CKO chromosome spread. Lower panel, distribution plot of RPA foci in early zygotene (n = 15), late zygotene (n = 18) and pachytene (n = 48 i) spermatocytes. For A-C, cells from two individuals per genotype were measured. Bars represent mean ± SD. Scale bars, 10 μm. (D) Immunostaining of SYCP3 and γH2AX on adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; magenta, γH2AX. Scale bars, 10 μm. (E) Spermatocyte stage proportion in adult Cxxc1 het and CKO (n = 1,066 from two individuals) spermatocytes based on SYCP3/SYCP1/γH2AX staining. p = 0.7 by Chi-square test. (F) Immunostaining of SYCP3 and SYCP1 on adult Cxxc1 het and CKO chromosome spreads. Green, SYCP3; orange, SYCP1. Scale bars, 10 μm.
(TIF)
(A) Plots of activity of DSBs in two replicates of Cxxc1 CKO and control DMC1 ChIP-seq samples. Correlation coefficient r = 0.96 in het controls, r = 0.99 in CKO samples. (B) Plot of activity of DSBs from Cxxc1 CKO and control spermatocytes. Black dots, PRDM9-dependent sites; yellow dots, PRDM9-independent sites.
(TIF)
Data Availability Statement
ChIP-seq data are available at Gene Expression Omnibus data repository under GEO number GSE116336 with secure token ilszsgyyfhglbel. All other relevant data are within the paper and its Supporting Information files.