Abstract
Objective:
The efficacy of therapeutic vaccines against HIV-1 infection has been modest. New inerts to redirect responses to vulnerable sites are urgently needed to improve these results.
Design:
We performed the first-in-human clinical trial with naked mRNA (iHIVARNA) combining a dendritic cell activation strategy (TriMix:CD40L+CD70+caTLR4 RNA) with a novel HIV immunogen sequences (HTI immunogen).
Methods:
A dose escalation, phase I clinical trial was performed in 21 chronic HIV-1-infected patients under ART who received three intranodal doses of mRNA (weeks 0, 2 and 4) as follow: TriMix-100 g, TriMix-300 g, TriMix-300 g with HTI-300 g, TriMix-300 g with HTI-600 g, TriMix-300 g with HTI-900 g. Primary end-point was safety and secondary-exploratory end-points were immunogenicity, changes in viral reservoir and transcriptome.
Results:
Overall, the vaccine was secure and well tolerated. There were 31 grade 1/2 and 1 grade 3 adverse events, mostly unrelated to the vaccination. Patients who received the highest dose showed a moderate increase in T-cell responses spanning HTI sequence at week 8. In addition, the proportion of responders receiving any dose of HTI increased from 31% at w0 to 80% postvaccination. The intervention had no impact on caHIV-DNA levels, however, caHIV-RNA expression and usVL were transiently increased at weeks 5 and 6 in the highest dose of iHIVARNA, and these changes were positively correlated with HIV-1-specific-induced immune responses.
Conclusion:
This phase I dose-escalating trial showed that iHIVARNA administration was safe and well tolerated, induced moderate HIV-specific T-cell responses and transiently increased different viral replication readouts. These data support further exploration of iHIVARNA in a phase II study.
ClinicalTrials.gov Identifier:
Keywords: HIV, HIVACAT immunogen, naked mRNA, therapeutic vaccine, TriMix
Introduction
Antiretroviral therapy (ART) has proven to be highly effective to prevent HIV-associated clinical progression and death [1] and to influence the AIDS pandemic as part of HIV prevention strategies [2,3]. Despite these successes, current ART has a number of public health, economical and clinical limitations. First, although 80% of HIV-infected individuals in United States and Europe know that they are HIV-infected, only 30 and 50%, respectively, are virally suppressed (hence with low probability of sexual transmission) [4,5]. Second, ART is unable to cure or eradicate the infection [6]. The high cost of lifelong treatment remains an important issue in the implementation of the the Joint United Nations Programme on HIV/AIDS (UNAIDS) strategy of universal treatment. Third, suboptimal treatment adherence can lead to the development of viral resistance [7]. Finally, the potential medium–long term adverse effects of ART have important clinical limitations.
A well tolerated, affordable and scalable cure could address both the individual and public health limitations that are associated with lifelong ART. The scientific community has acknowledged this position and there is a growing interest in developing curative strategies to tackle HIV persistence [8,9], among, which therapeutic vaccines represent one of the most promising approaches [10,11]. However, although, most immunogens have been able to induce HIV-specific immune responses in clinical trials, they have shown very limited efficacy to control viral replication [12]. The cytolytic T-lymphocyte (CTL) escape mutations and a poor antigen presentation by dendritic cells are some of the major hurdles that need to be addressed by rationally designed therapeutic vaccine candidates to improve their effectiveness. New inserts to redirect responses to vulnerable sites of HIV and vectors targeting dendritic-cell pathways could be necessary to achieve remission of HIV-1 infection [13].
Recently, direct administration of mRNA targeting dendritic cells has been proposed as an alternative to the classic immunogens [14–19]. Van Lint et al.[20] have designed mRNAs encoding a mixture of activation molecules functional in antigen-presenting cells (APC), including CD40L, a constitutively active variant of Toll-like receptor (TLR) 4 and CD70 (jointly referred as TriMix). The intention behind this strategy is to induce dendritic-cell maturation with CD40L and caTLR4 and to support activated T-cell survival and proliferation with CD70. dendritic cells modified in vitro or in vivo with TriMix mRNA have been shown to be significantly more potent and immunogenic than unmodified dendritic cell [20,21]. Complementing these advances in vector design, Mothe et al.[13] proposed a rational design for the selection of HIV antigens based on the viral targets of protective HIV-1 specific T-cell responses observed in three large cohorts of HIV-infected individuals [13]. This approach resulted in the design of the HIVACAT T-cell Immunogen (HTI) sequence constituting 16 joined fragments of 10–70 amino acids each, encoding critical HIV-1 target epitopes in Gag, Pol, Vif and Nef. These particular HIV-1 regions are more conserved and elicited responses of higher functional avidity and broader cross-reactivity than other segments in the HIV proteome [22].
Mice immunized with mRNA-encoding HTI in combination with TriMix [21] and mice and rhesus macaques immunized with DNA/MVA expressing HTI [22] showed broad and balanced T-cell responses to several segments within Gag, Pol, and Vif. These data demonstrate that it is possible to redirect responses to vulnerable sites of HIV-1 while avoiding the induction of responses to potential decoy targets that may divert effective T-cell responses towards variable and less protective viral determinants.
We have performed the first-in-human phase I dose-escalating clinical trial with naked mRNA containing dendritic cell activation signals (TriMix) and encoding a novel HIV immunogen sequence (HTI) to redirect T-cell immunity in HIV-infected individuals to the most vulnerable viral targets. The primary endpoint of this study included feasibility and safety of the immune intervention, while the secondary exploratory endpoints were immunogenicity, changes in the viral reservoir and in transcriptomic variations.
Patients and methods
Patients and samples
From June 2015 until October 2016 in Barcelona, we conducted a single-center, open-label, dose-escalating phase I clinical trial in 21 chronic HIV-1 infected patients under stable ART with plasma viral load (pVL) below 50 copies/ml and stable CD4+ T-cell counts above 450 cells/μl. Patients received three inguinal intranodal doses of mRNA by ultrasound-guided injections performed by a radiologist at weeks 0, 2 and 4 according to the following dose escalation scheme (see Fig. 1a and b):
Fig. 1.
(a) Dose escalation flow chart. (b) Assignation of cohorts.

DSMB, Data and Safety Monitoring Board; DLT, dose-limiting toxicity.
Group 1 (control): three patients received 100 μg of mRNA (i.e. 100 μg TriMix mRNA).
Group 2 (control): three patients received 300 μg of mRNA (i.e. 300 μg TriMix mRNA).
Group 3 (study): three patients received 600 μg of mRNA (i.e. 300 μg HTI mRNA and 300 μg TriMix mRNA).
In these three groups, if two or more of the three patients would have developed a dose-limiting toxicity (DLT), Data and Safety Monitoring Board (DSMB) would have been consulted and study could potentially have been terminated. If one or no patients would have had a DLT, three patients would be enrolled at the next dose level.
Group 4 (study): three patients received 900 μg of mRNA (i.e. 600 μg HTI mRNA and 300 μg TriMix mRNA). If two or more of the three first patients would have shown DLT, then three additional patients would have been enrolled at the previous dose level (600 μg of mRNA per vaccination). If one or no patients would have had a DLT, three additional patients would have been enrolled at the 900 μg of mRNA dose level. If two or more of the six patients receiving 900 μg of mRNA would have had a DLT, then three additional patients would have been enrolled at the previous level dose (600 μg of mRNA per vaccination). If one or no patients of the six patients would have had a DLT, six patients would be enrolled at the next dose level.
Group 5 (study): six patients received 1200 μg of mRNA (i.e. 900 μg HTI mRNA and 300 μg TriMix mRNA).
Within each group, each patient was observed for a minimum of 1 week before the next patient was treated. All patients within a group were observed for a minimum of 2 weeks after the third vaccination prior to administration of the vaccine to a patient at the next dose level (Fig. 1b). Patients could not change groups or dose assignments upon the initiation of the study protocol. The study treatment period was 4 weeks for each patient. All the patients were followed-up for 24 weeks (see supplementary M&M).
Immunogenicity evaluations
ELISPOT
Immunogenicity was assessed on cryopreserved peripheral blood mononuclear cells (PBMC) at baseline (i.e. day of first immunization) and weeks 4, 6, 8 and 24 of follow-up by the quantification of T-cell responses by a IFN-γ ELISPOT assay according to standardized operating procedures (SOP) in a single research laboratory. Briefly, cryopreserved PBMC were thawed and rested for overnight at 37 °C. Next, 100 000 PBMCs were stimulated with peptide pools (1 μg of each single peptide) in 100 μl of complete media (Roswell Park Memorial Institute with 10% fetal calf serum) in duplicate conditions. To evaluate the HIV-specific T-cell responses against the whole HIV proteome, we used sets of overlapping HIV peptide pools [10 different pools containing from 5 to 22 15-mer peptides, overlapping by 11-mer, which matched the HTI immunogen (’IN’) and 8 pools ranging from 62 to 105 15-mer peptides covering NIH consB HIV sequences not located within HTI (’OUT’ pools)]. Media alone in triplicate was used as negative control. Phytohemagglutinin PHA-P (1 μg/ml) and stimulation with CEF (cytomegalovirus, Epstein–Barr virus, and Flu) pool were used as positive controls. Results are expressed as the mean number of spot-forming cells (SFC)/106 cells from duplicate wells. The following criteria were used to define the technical validity and positive responses: PBMC viability had to be greater than 80% to be analyzed; the assay background (PBMCs with media alone) had to be less than 50 SFC/106 PBMC; positive responses against PHA-P had to be above 500 SFC/106 PBMC; and ELISPOT responses were considered positive in case of greater than 50 SFC/106 PBMC and number of SFC/106 PBMC at least two-fold over media control (Fig. 2).
Fig. 2.
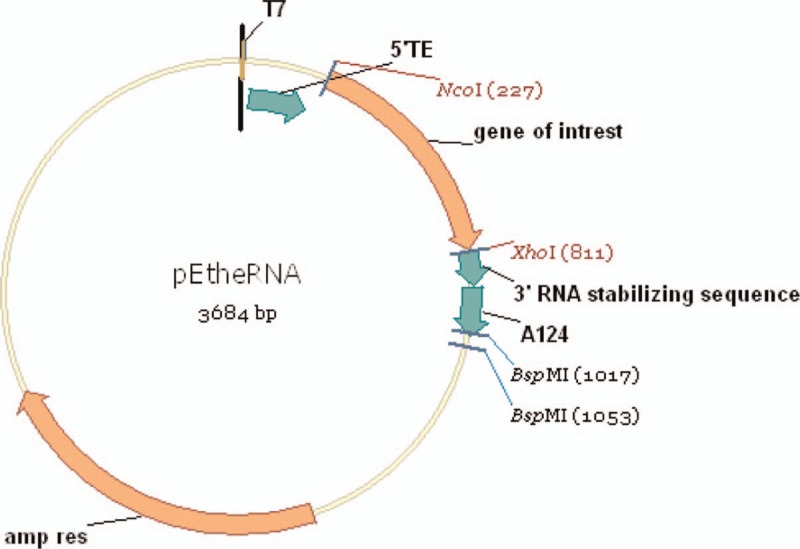
pETheRNA mRNA vector.
Flow cytometry assays
We evaluated the activation (or senescence) of T (CD4+ and CD8−), B (CD19+), natural killer cells (CD56+), and monocytes (CD14+) according to the expression of specific surface markers by flow cytometry (see Supplementary Table S1).
Viral reservoir and ultrasensitive plasma viral load assessment
To explore potential changes induced by the activation of HIV-1-specific latently infected CD4+ T cells, proviral HIV-DNA levels and cell-associated HIV-RNA expression were quantified at baseline and at weeks 4, 6, 8, and 24 of follow-up in peripheral CD4+ T cells. Additional determinations of caHIV-RNA at week 2 +1 day, week 3, week 4 +1 day, and week 5 were performed in groups 4 and 5.
Both the parameters were measured by digital droplet PCR (dd PCR) with two different sets of primers (5′ LTR and Gag loci) to avoid mismatching as previously described [23,24]. caHIV-RNA was calculated as HIV RNA copies relative to the housekeeping gene TATA- binding protein (TBP). Proviral HIV-DNA is expressed as copies of HIV-DNA per million of CD4+ T cells measured by the housekeeping gene RPP30.
In addition, the effect of vaccination on plasma viral load (pVL) at baseline and week 2 + 1 day, week 3, week 4, week 4 + 1 day, week 5, week 6, week 8, and week 24 was assessed in groups 4 and 5. In short, 5 ml of EDTA plasma was ultracentrifuged at 170 000 × g for 1 h at 4 °C. Tubes were equilibrated with Tris-buffered saline (50 mM Tris-Cl, pH 7.6; 150 mmol/l NaCl) to a final volume of 12 ml. After centrifugation, 11.20 ml of supernatant was carefully aspirated and discarded. The pellet was thoroughly resuspended in the remaining 800 μl and tested for viral load with the Cobas HIV-1 test on the Roche Cobas 4800 system. To account for the concentration of the virus, the obtained result was multiplied by a factor 0.16 (0.8/5).
Transcriptome profiling
Transcriptome profiles were obtained from whole blood samples collected in Tempus blood RNA tubes (Thermo Fisher Scientific, Waltham, Massachusetts, USA), at baseline and week 6. RNA was isolated according to the manufacturer's instructions and hybridized onto Affymetrix Human GenomeU133 Plus 2.0 microarray chips as previously described (http://dx.doi.org/10.1016/j.vaccine.2015.04.047). Samples were quantile normalized and summarized using median polish (i.e. the RMA method). During quality control, we had to remove a single sample as it had insufficient quality to be normalized. Batch effect removal, differential gene expression and gene set analysis were performed using limma. Principal component analysis (PCA) showed batch effects related to RNA processing and hybridization, but these could be effectively removed using limma. Gene set analysis was performed using limma and the REACTOME curated database gene set definitions [25,26].
Statistical analysis
The sample size is the minimum required to the study objectives as stated on Guideline on Requirements for First-in-man clinical trials for potential high-risk medicinal products (EMEA/CHMP/SWP/28367/2007). This was an exploratory study, and the safety analysis were descriptive. The safety endpoints were described and summarized by number and percentage of adverse events and grading. We also stratified the adverse events into related (possibly, probably and definitely related to vaccination) and unrelated to vaccination (unlikely to be related, unrelated). The total magnitude of HIV-1 specific IFN-γ T-cell responses was described as the sum of SFC/million input PBMC. Differences in breadth, magnitude of HIV-1 specific responses, caHIV-DNA, caHIV-RNA or usVL between two longitudinal determinations in the same individuals were assessed by Wilcoxon Signed-Rank Test.
Results
mRNA vaccination with TriMix or iHIVARNA in chronically antiretroviral-treated HIV-1-infected patients is well tolerated
All the 21 patients received the three doses of TriMix or the different doses of iHIVARNA (HTI with TriMix) given by inguinal intranodal route. All 21 participants completed the study as per protocol and were included in the safety analysis. Clinical characteristics of the patients are shown in Table 1. Overall, the vaccine was safe and well tolerated. No serious adverse events nor deaths were observed. A total of 32 adverse events were reported during follow-up (4, 7, 5, 11 and 5 in each group, respectively). Nineteen adverse events were classified as grade 1, twelve as grade 2 and one as grade 3. Half of them (n = 16/32) were not related to vaccination, 14 had a possible relationship and 2 had a definite relationship (Table 2). No laboratory abnormalities were observed during the follow-up.
Table 1.
Clinical characteristics of participants.
| n = 21 | Group 1 (n = 3) | Group 2 (n = 3) | Group 3 (n = 3) | Group 4 (n = 6) | Group 5 (n = 6) |
| Median age (IQR) | 48 (48–51) | 51 (48–51) | 45 (37–46) | 55 (53–57) | 47 (43–55) |
| Male | 2 | 2 | 2 | 4 | 6 |
| MSM | 2 | 2 | 2 | 4 | 6 |
| Heterosexual (HTSX) | 1 | 1 | 1 | 0 | 0 |
| Intravenous drug user (IDU) | 0 | 0 | 0 | 2a | 0 |
| Hepatitis C virus (HCV) infection | 0 | 0 | 0 | 2b | 0 |
| Median CD4+ cell count at baseline (IQR) | 762 (686–770) | 726 (716–824) | 821 (741–957) | 829 (680–1124) | 904 (868–1071) |
| Median CD4+ cell count at week 6 (IQR) | 672 (663–683) | 938 (784–993) | 643 (589–1040) | 819 (640–928) | 956 (805–1053) |
IQR: Interquartile range.
aFormer IDU.
bBoth HCV infections cured: 1 spontaneously (2009), 1 treated with sofobusvir/daclatasvir (2015).
Table 2.
Total adverse events classified by severity and relationship with the vaccination.
| Variable | Value | Val | I | II | III | IV | V | All |
| Grade 1 | N | 3 | 4 | 1 | 9 | 2 | 19 | |
| % | 75 | 57 | 20 | 82 | 40 | 59 | ||
| Severity | Grade 2 | N | 1 | 3 | 3 | 2 | 3 | 12 |
| % | 25 | 43 | 60 | 18 | 60 | 38 | ||
| Grade 3 | N | 0 | 0 | 1 | 0 | 0 | 1 | |
| % | 0 | 0 | 20 | 0 | 0 | 3 | ||
| All | N | 4 | 7 | 5 | 11 | 5 | 32 | |
| Definite relationship | N | 2 | 0 | 0 | 0 | 0 | 2 | |
| % | 50 | 0 | 0 | 0 | 0 | 6 | ||
| Probable relationship | N | 0 | 0 | 0 | 0 | 0 | 0 | |
| % | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Casual relationship | Possible relationship | N | 1 | 4 | 3 | 6 | 0 | 14 |
| % | 75 | 57 | 60 | 55 | 0 | 44 | ||
| No related | N | 1 | 3 | 2 | 5 | 5 | 16 | |
| % | 75 | 43 | 40 | 45 | 100 | 50 | ||
| Unknown | N | 0 | 0 | 0 | 0 | 0 | 0 | |
| % | 0 | 0 | 0 | 0 | 0 | 0 | ||
| All | N | 4 | 7 | 5 | 11 | 5 | 32 |
Increased frequencies of HIV-1-specific T cells after iHIVARNA vaccination
At week 8, patients who had received the highest iHIVARNA dose (group 5) showed a moderate increase in T-cell responses spanning HTI sequence (IN) at week 8 whereas no changes were observed in responses against the rest of the HIV-1 proteome (OUT) compare to baseline (Fig. 3). In addition, the proportion of responders receiving any dose of iHIVARNA (n = 15) increased from 31% (n = 5) at week 0 to 80% (n = 12) postvaccination. This increase was not observed in patients receiving TriMix alone (n = 6) from 50% (n = 3) to 67% (n = 4). The HIV-specific T-cell responses were mainly directed against the following peptide pools: p2 (Gag p17), p4 (Gag p24), p5 (Gag p15), p7 (RT) and p8 (INT), in the IN-peptide pools, whereas in the HIV OUT-peptide pools, the responses were mainly towards: p1 (Gag), p2 (Pol), p4 (Pol), p6 (Env), p7 (Vif, Nef) and p8 (Tat, Vpu, Vpr, Rev; data not shown).
Fig. 3.
Changes in the magnitude of total HIV-1-specific immune responses against IN and OUT peptide pools as measured by ELISPOT (at week 0, 4, 6, 8 and 24).
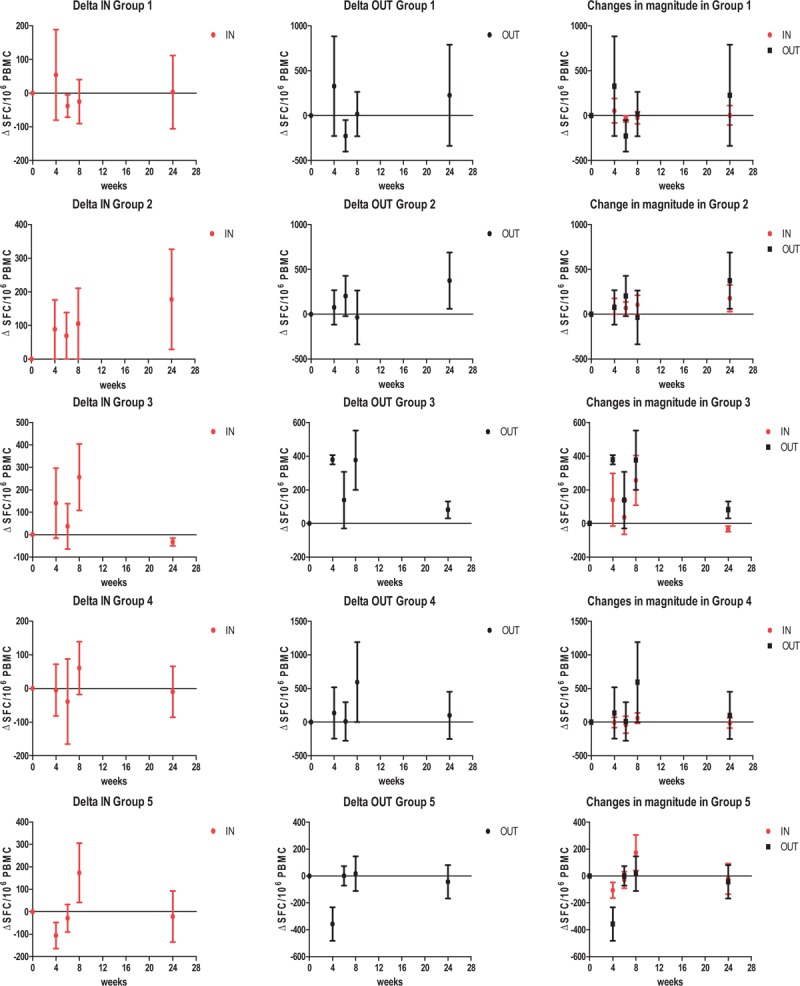
Results were considered positive if the number of SFC/106 cells in stimulated wells was two-fold higher than that in unstimulated control wells, and if there were at least 50 SFC/106 cells after background subtraction. Mean differences from baseline (Δ+/− SEM SFC/106 PBMC) are represented on the graphs.
Concerning the analysis of cell subsets, we only observed statistically significant decreases between baseline and week 8 (fourth weeks after the last immunization) in percentages of CD8+ CD38+HLA-DR+ T cells and CD8+ PD-1+ T cells in group 5 (data not shown).
Increased viral expression but stable proviral reservoir after iHIVARNA vaccination
Neither TriMix alone or different doses of iHIVARNA had an impact on proviral HIV-DNA in any of the studied arms (Fig. 4 a). However, there was a transient increase in caHIV-RNA expression at higher doses of iHIVARNA (arms 4 and 5) during weeks 4–6 [whereas this was not observed with TriMix alone (groups 1 and 2) or with low doses of iHIVARNA (group 3)], and subsequently normalized at weeks 8 and 24 (Fig. 4 b). Moreover, the ratio of ca-HIV RNA at week 6 as compared with week-4 levels was significantly higher in patients receiving any dose of iHIVARNA (groups 3–5 merged) vs. patients receiving TriMix alone (P = 0.0126). Finally, usVL also significantly increased at weeks 6 and 8 (P < 0.05) in groups 4 and 5 and returned to baseline values at week 24 (Fig. 4 c). In fact, further analysis of these two groups, showed a positive and significant correlation between the increase of the elicited T-cell immune responses against HTI sequence (IN) and the usVL at week 6 (P < 0.05). No significant correlation was observed between T-cell responses against the rest of the HIV-1 proteome (OUT) and the usVL (See Supplementary Fig. S1).
Fig. 4.
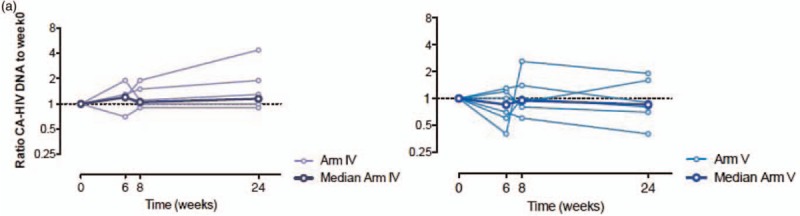
(a) Impact of the highest doses of iHIVARNA (Groups 4 and 5) in HIV-1 cell-associated total DNA. (b) Impact of TriMix (Groups 1 and 2) or the different doses of iHIVARNA (Groups 3–5) in HIV-1 cell-associated RNA. (c) Impact of the highest doses of iHIVARNA (Groups 4 and 5) in HIV-1 ultrasensitive plasma RNA
Fig. 4 (Continued).

(a) Impact of the highest doses of iHIVARNA (Groups 4 and 5) in HIV-1 cell-associated total DNA. (b) Impact of TriMix (Groups 1 and 2) or the different doses of iHIVARNA (Groups 3–5) in HIV-1 cell-associated RNA. (c) Impact of the highest doses of iHIVARNA (Groups 4 and 5) in HIV-1 ultrasensitive plasma RNA
iHIVARNA vaccination does not shift gene expression
We did not observe robust differentially expressed genes in any of the group-wise comparisons, although gene set analysis indicates some effect on pathways such as RNA metabolism and host response to viruses, but with very low significance levels (Fig. 5). There were no differentially expressed genes in any of the group-wise comparisons or in the immunologic responders vs. nonresponders, based on ELISPOT assays. Gene set analysis indicated some effect of the vaccine on pathways such as RNA metabolism and host response to viruses. These were related to the presence of a patient effect and an effect of the HIV reservoir size. However, these pathways had very low levels of significance (FDR >0.1), indicating that there was only a modest effect on most pathways.
Fig. 4 (Continued).
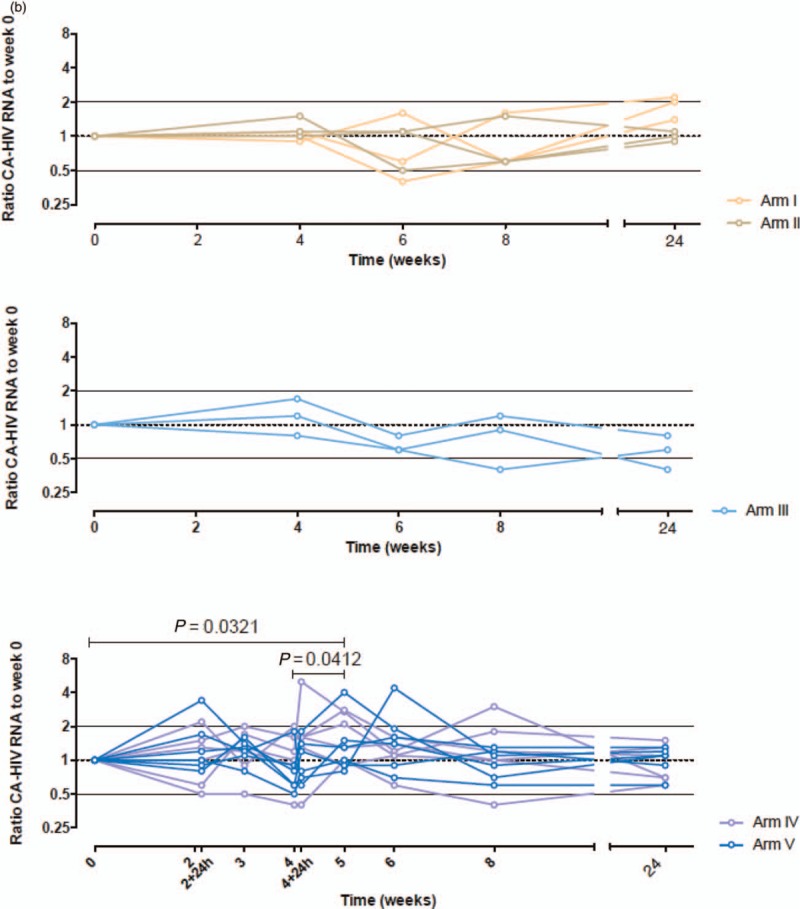
(a) Impact of the highest doses of iHIVARNA (Groups 4 and 5) in HIV-1 cell-associated total DNA. (b) Impact of TriMix (Groups 1 and 2) or the different doses of iHIVARNA (Groups 3–5) in HIV-1 cell-associated RNA. (c) Impact of the highest doses of iHIVARNA (Groups 4 and 5) in HIV-1 ultrasensitive plasma RNA
Fig. 5.
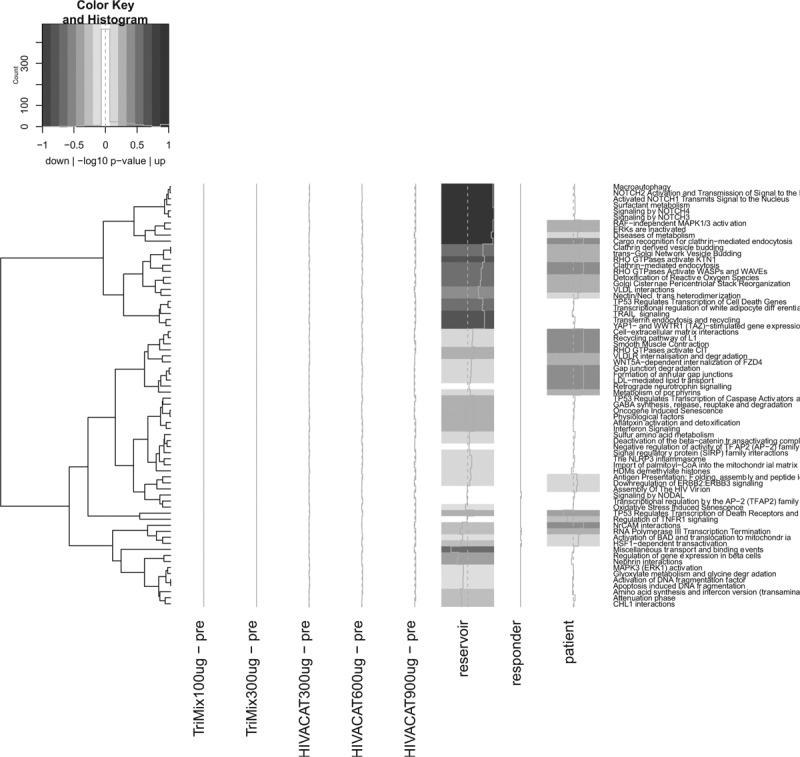
Reactome gene set analysis.
Discussion
Recently, direct intranodal administration of naked mRNA has been proposed as an alternative to the immunogens used so far in HIV vaccination trials. As compared with plasmid DNA and viral vectors, mRNA has a better safety profile, can be easily obtained by commercially available kits and stored at room temperature. Additionally, mRNA-mediated gene transfer occurs in nondividing cells and offers the advantage of not being restricted to a subject-specific human leukocyte antigen (HLA) allele [27]. Promising preliminary results have been reported in other infectious diseases and cancer with mRNA vaccines. Indeed, an mRNA influenza vaccine candidate has demonstrated similar efficacy to licensed vaccines in animal models [14], a prophylactic mRNA-based candidate vaccine against rabies was well tolerated and induced boostable functional antibodies [17] and direct administration of mRNA has entered clinical testing in cancer [15,16,18,19]. To our knowledge, this is the first in human clinical trial using direct intranodal administration of naked mRNA as a therapeutic vaccine against HIV-1 infection. We have shown that intranodal injection of the iHIVARNA vaccine was feasible, safe and well tolerated. No severe adverse events were observed even with the highest dose of the vaccine, namely group 5 constituting a total of 1200 μg of mRNA (900 μg HTI mRNA and 300 μg TriMix mRNA). Therefore, this dose has been selected for a currently ongoing phase II clinical trial.
The vast majority (98%) of latent viruses in chronic HIV-1-infected patients carry CTL escape mutations that render infected cells insensitive to CTLs directed at standard (canonical) epitopes [28]. It is likely that many of the therapeutic vaccines currently under evaluation expand preexisting clones, which are exhausted and target escape variants. There is specific interest in approaches that stimulate responses against novel, nondominant epitopes [13,29,30]. Mothe et al.[13] proposed a rational design for the selection of the HIV antigens based on the viral targets of protective HIV-1 specific T-cell responses from three large cohorts of HIV-infected individuals [13]. In addition, two other groups have hypothesized that T-cell vaccines targeting the most conserved regions of the HIV-1 proteome will induce more efficient immune response than whole protein-based T-cell vaccines [31,32]. Letourneau et al.[33] designed the HIVconsv immunogen by assembling the 14 most conserved regions of the HIV-1 proteome into one chimeric protein. When delivered in vaccines vectored by MVA and chimpanzee adenovirus, these vaccines were able to shift preexisting immune responses towards conserved, vaccine-encoded regions of HIV in early-treated HIV-infected individuals [29–33]. Our iHIVARNA candidate supports this strategy. The data presented indicate that the vaccine was able to induce moderate HIV-specific immune responses (increase in magnitude and breadth as well as increase in percentage of responders) against overlapping HIV peptide pools, which matched the HTI sequence (’IN’) whereas no augmented responses were observed against peptide pools covering HIV proteins not included in the HTI sequence (’OUT’). However, the Phase I results are not conclusive because of the limited number of patients included in each group.
The ELISPOT assay has been established for the direct ex-vivo quantification of peptide-reactive T lymphocytes from peripheral blood mononuclear cells (PBMC). However, it is true that the predictive power of this assay has been challenged because of the lack of efficacy of some HIV vaccine trials despite the induction of robust Elispot responses [34]. This finding and the emergence of new techniques that have the potential advantage of simultaneously quantifying numerous parameters, raises questions regarding the future role of IFN-γ Elispot in clinical trials of candidate vaccines. Nevertheless, the IFN-γ Elispot assay has been, unlike other techniques, evaluated and validated in several proficiency panels and is advantageous in cost-effectively detecting and mapping T-cell responses [35]. All these benefits are particularly important in a Phase I clinical trial where safety and tolerability were the major end-points.
There is evidence that HIV-1 vaccines are by themselves insufficient to fully harness the stimulatory potential of dendritic cells. It has been suggested that targeting in-vivo dendritic cells by co-stimulatory molecules improves the effectiveness of the vaccines [20,36]. This type of strategy has already been tested in humans with a vaccine co-expressing immune activator molecules. A clinical trial testing a recombinant fowl pox virus vector co-expressing HIV1Gag/Pol and human interferon-γ has been reported [37,38]. In addition, Van Lint et al.[20] designed mRNAs encoding a mixture of APC activation molecules, referred as TriMix. Dendritic cells modified in vitro or in vivo with TriMix mRNA have been shown to be significantly more immunogenic than unmodified dendritic cells [20,21].
The higher doses of iHIVARNA mRNA might have increased HIV expression as a transient increment in caHIV-RNA expression and usVL were observed. It is likely to be triggered by activation of the immune system through recognition of TLRs. However, the existence of a direct and significant association between the elicited HIV-1 immune response against epitopes included in the vaccine (and not to the rest of the proteome) and the usVL (1 or 2 weeks after the last dose) suggest that it could be secondary to an specific stimulus rather than to an ambiguous and unspecific reaction because of the mere addition of mRNA. Given the limited number of patients, this association needs to be further explored in the ongoing phase IIa clinical trial to be confirmed.
Using whole blood-derived transcriptome analyses, we only observed modest effects on inflammatory pathways. These effects were related to intrinsic differences in the activity of inflammatory pathways between individual patients, rather than to the effect of any of the vaccine formulations. Therefore, the data suggest that any immune activation induced by this vaccine is modest and not detectable by comprehensive transcriptome profiling of whole blood samples.
In conclusion, this phase I exploratory dose-escalating trial showed that iHIVARNA vaccination was feasible, harmless and well tolerated, was able to induce moderate HIV-specific immune responses and transiently increased caHIV-RNA expression and ultrasensitive plasma viremia. These data support further exploration of iHIVARNA in the currently ongoing phase II clinical trial.
Acknowledgements
This study was partially supported by grants: FP7-HEALTH-2013-INNOVATION-1 Proposal No: 602570–2, SAF2015–66193-R, FIS PI15/00641, FIS PI15/00480, Fondo Europeo para el Desarrollo Regional (FEDER), RIS∗∗.
∗∗RIS: The SPANISH AIDS Research Network RD16/0025/0002- ISCIII – FEDER.
This study was presented in part at the 2018 Conference on Retroviruses and Opportunistic Infections, Boston, USA.
iHIVARNA Consortium: Consorci Institut d’Investigacions Biomédiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain: Lorna Leal, Blanca Paño, Carlos Nicolau, Amparo Tricas, Marta Sala, Encarnación Moreno, Cristina Rovira, Carmen Hurtado, Constanza Lucero, Irene Fernández, Flor Etcheverry, Alberto Crespo, Manel Bargalló, Miriam García, Alexy Inciarte, Ismael Pérez, Laura Mensa, Laura Mendoza, Anna Vanesa Oliveira, Mª Jose Maleno, Agathe León, Maria Joyera, Judit Pich, Jose M Gatell, Joan A Arnaiz, Montserrat Plana, Felipe García.
Instituut voor Tropische Geneeskunde (ITM), Antwerp, Belgium: Guido Vanham, Eric Florence, Jozefien Buyze, Pieter Pannus.
Vrije Universiteit Brussel (VUB), Brussels, Belgium: Kris Thielemans, Joeri Aerts, Sabine Allard, Patrick Lacor, Nik Claesen, Elger Vercayie, Patrick Tjok.
eTheRNA BVBA (eTheRNA), Brussels, Belgium: Carlo Heirman, Sonja Van Meirvenne, Hilde Van Raemdonck, An Van Nuffel, Jacques Berlo, Inge Pettersson, Gust Schols.
Erasmus Universitair Medisch Centrum Rotterdam (EMC), Rotterdam, Netherlands: Rob Gruters, Marion Koopmans, Wesley de Jong, Henk-Jan van den Ham, Patrick Boers, Rachel Scheuer, Eric Van Gorp, Cynthia Lungu, Arno Andeweg, Ab Osterhaus.
irsiCaixa AIDS Research Institute, Badalona, Spain: Christian Brander, Bonaventura Clotet, Marta Marszalek, Sara Moron-López, Beatriz Mothe, Alex Olvera, Miriam Rosas, Maria Salgado, Javier Martinez-Picado, Mireia Manent, Judith Dalmau.
Synapse Research Management Partners S.L. (SYNAPSE), Barcelona, Spain: Carlos Díaz, Montse Camprubí.
Asphalion, S.L. (ASPHALION), Barcelona, Spain: Lídia Cánovas, Núria Coderch, Marta Rayo 9Lunar y Joel Montané
Authors contributions: L.L., J.P., J.A.A., J.M.G. and F.G. conducted the clinical trial. A.C.G. and M.P. conducted the immunogenicity analyses. B.M. and C.B. developed the HTI in the iHIVARNA vaccine. S.M.L., M.S. and J.M.P. were in charge of the reservoir study. C.H., S.V.M. and K.T. developed the Trimix in the iHIVARNA vaccine and quality control for the vaccine. G.V. and P.P. studied the changes in usVL. H.H., R.G. and A.A. were in charge of the transcriptome analyses. All authors contributed in writing and revising the manuscript.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
References
- 1.Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med 1998; 338:853–860. [DOI] [PubMed] [Google Scholar]
- 2.Lima VD, Lourenço L, Yip B, Hogg RS, Phillips P, Montaner JSG. AIDS incidence and AIDS-related mortality in British Columbia, Canada, between 1981 and 2013: a retrospective study. Lancet HIV 2015; 2:e92–e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrazzo JM, del Rio C, Holtgrave DR, Cohen MS, Kalichman SC, Mayer KH, et al. International Antiviral Society-USA Panel. HIV prevention in clinical care settings: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA 2014; 312:390–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner EM, McLees MP, Steiner JF, Del Rio C, Burman WJ. The spectrum of engagement in HIV care and its relevance to test-and-treat strategies for prevention of HIV infection. Clin Infect Dis 2011; 52:793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell C, Ambrosioni J, Miro JM, Esteve A, Casabona J, Navarro G, et al. The continuum of HIV care in Catalonia. AIDS Care 2015; 27:1449–1454. [DOI] [PubMed] [Google Scholar]
- 6.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–728. [DOI] [PubMed] [Google Scholar]
- 7.Beyrer C, Pozniak A. HIV drug resistance — an emerging threat to epidemic control. New Engl J Med 2017; 377:1605–1607. [DOI] [PubMed] [Google Scholar]
- 8.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012; 12:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, et al. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med 2016; 22:839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leal L, Lucero C, Gatell JM, Gallart T, Plana M, Garcia F. New challenges in therapeutic vaccines against HIV infection. Expert Rev Vaccines 2017; 16:587–600. [DOI] [PubMed] [Google Scholar]
- 11.Garcia F, Climent N, Guardo AC, Gil C, Leon A, Autran B, et al. A dendritic cell-based vaccine elicits T cell responses associated with control of HIV-1 replication. Sci Transl Med 2013; 5:166ra2. [DOI] [PubMed] [Google Scholar]
- 12.Papagno L, Alter G, Assoumou L, Murphy RL, Garcia F, Clotet B, et al. ORVACS Study Group. Comprehensive analysis of virus-specific T-cells provides clues for the failure of therapeutic immunization with ALVAC-HIV vaccine. Aids 2011; 25:27–36. [DOI] [PubMed] [Google Scholar]
- 13.Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, Zamarreno J, et al. Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med 2011; 9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol 2012; 30:1210–1216. [DOI] [PubMed] [Google Scholar]
- 15.Weide B, Carralot JP, Reese A, Scheel B, Eigentler TK, Hoerr I, et al. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 2008; 31:180–188. [DOI] [PubMed] [Google Scholar]
- 16.Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, et al. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother 2009; 32:498–507. [DOI] [PubMed] [Google Scholar]
- 17.Alberer M, Gnad-Vogt U, Hong HS, Mehr KT, Backert L, Finak G, et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, nonrandomised, prospective, first-in-human phase 1 clinical trial. Lancet 2017; 390: [DOI] [PubMed] [Google Scholar]
- 18.Rittig SM, Haentschel M, Weimer KJ, Heine A, Muller MR, Brugger W, et al. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther 2011; 19:990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017; 547:222–226. [DOI] [PubMed] [Google Scholar]
- 20.Van Lint S, Goyvaerts C, Maenhout S, Goethals L, Disy A, Benteyn D, et al. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res 2012; 72:1661–1671. [DOI] [PubMed] [Google Scholar]
- 21.Guardo AC, Joe PT, Miralles L, Bargallo ME, Mothe B, Krasniqi A, et al. iHIVARNA. Preclinical evaluation of an mRNA HIV vaccine combining rationally selected antigenic sequences and adjuvant signals (HTI-TriMix). AIDS 2017; 31:321–332. [DOI] [PubMed] [Google Scholar]
- 22.Mothe B, Hu X, Llano A, Rosati M, Olvera A, Kulkarni V, et al. A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J Transl Med 2015; 13:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Bonet M, Puertas MC, Fortuny C, Ouchi D, Mellado MJ, Rojo P, et al. Establishment and replenishment of the viral reservoir in perinatally HIV-1-infected children initiating very early antiretroviral therapy. Clin Infect Dis 2015; 61:1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moron-Lopez S, Puertas MC, Galvez C, Navarro J, Carrasco A, Esteve M, et al. Sensitive quantification of the HIV-1 reservoir in gut-associated lymphoid tissue. PLoS One 2017; 12:e0175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 2017; 18:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H, et al. Annotating cancer variants and anticancer therapeutics in reactome. Cancers 2012; 4:1180–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Youn H, Chung JK. Modified mRNA as an alternative to plasmid DNA (pDNA) for transcript replacement and vaccination therapy. Expert Opin Biol Ther 2015; 15:1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 2015; 517:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mothe B, Manzardo C, Coll P, Cobarsi P, Sanchez A, Escrig R, et al. Safety and immunogenicity of ChAd.HIVconsv and MVA.HIVconsv therapeutic vaccines in a cohort of early treated HIV—1 infected individuals. 8th IAS Conference on HIV Pathogenesis, Treatment & Prevention Vancouver, Canada. 2015 [Google Scholar]
- 30.Im EJ, Hong JP, Roshorm Y, Bridgeman A, Letourneau S, Liljestrom P, et al. Protective efficacy of serially up-ranked subdominant CD8+ T cell epitopes against virus challenges. PLoS Pathog 2011; 7:e1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borthwick N, Ahmed T, Ondondo B, Hayes P, Rose A, Ebrahimsa U, et al. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther 2014; 22:464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulkarni V, Valentin A, Rosati M, Alicea C, Singh AK, Jalah R, et al. Altered response hierarchy and increased T-cell breadth upon HIV-1 conserved element DNA vaccination in macaques. Plos One 2014; 9:e86254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letourneau S, Im EJ, Mashishi T, Brereton C, Bridgeman A, Yang H, et al. Design and preclinical evaluation of a universal HIV-1 vaccine. PLoS One 2007; 2:e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streeck H, Frahm N, Walker BD. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat Protoc 2009; 4:461–469. [DOI] [PubMed] [Google Scholar]
- 35.Schmittel A, Keilholz U, Bauer S, Kuhne U, Stevanovic S, Thiel E, et al. Application of the IFN-gamma ELISPOT assay to quantify T cell responses against proteins. J Immunol Methods 2001; 247:17–24. [DOI] [PubMed] [Google Scholar]
- 36.Flynn BJ, Kastenmuller K, Wille-Reece U, Tomaras GD, Alam M, Lindsay RW, et al. Immunization with HIV Gag targeted to dendritic cells followed by recombinant New York vaccinia virus induces robust T-cell immunity in nonhuman primates. Proc Natl Acad Sci U S A 2011; 108:7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emery S, Kelleher AD, Workman C, Puls RL, Bloch M, Baker D, et al. Influence of IFNgamma co-expression on the safety and antiviral efficacy of recombinant fowlpox virus HIV therapeutic vaccines following interruption of antiretroviral therapy. Hum Vaccin 2007; 3:260–267. [DOI] [PubMed] [Google Scholar]
- 38.Emery S, Workman C, Puls RL, Bloch M, Baker D, Bodsworth N, et al. Randomized, placebo-controlled, phase I/IIa evaluation of the safety and immunogenicity of fowlpox virus expressing HIV gag-pol and interferon-gamma in HIV-1 infected subjects. Hum Vaccin 2005; 1:232–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


