Abstract
Objective
The phenotypic and pathological features of small cell cervical carcinoma (SMCC) and small small cell lung cancer (SCLC) are very similar; thus, the chemotherapy regimens used for the rare SMCC have been routinely based on regimens used for common SCLC. We set out to explore the protein expression profile similarities between these 2 cancers to prove that linking their therapeutic regimens is justified, with a secondary aim of finding tumor-specific proteins to use as additional biomarkers for more accurate diagnosis of SMCC, and potentially to use as therapeutic targets.
Methods
Protein expression analysis was performed for 3 cases of SMCC and 1 example each of SCLC, mucinous adenocarcinoma of the cervix (MACC), lung mucinous adenocarcinoma (MACL), and squamous cell carcinoma of the cervix (SCC). We used cancer tissue–originated spheroids (CTOS) and isobaric tags for relative and absolute quantitation (iTRAQ)–based comprehensive and quantitative protein expression profile analysis. Expression in corresponding clinical samples was verified by immunohistochemistry.
Results
Rather than organ of origin–specific patterns, the SMCC and SCLC samples revealed remarkably similar protein expression profiles—in agreement with their matching tumor pathology phenotypes. Sixteen proteins were expressed at least 2-fold higher in both small cell carcinomas (SMCC and SCLC) than in MACC or SCC. Immunohistochemical analysis confirmed higher expression of creatine kinase B-type in SMCC, compared with MACC and SCC.
Conclusions
We demonstrate a significant overlapping similarity of protein expression profiles of lung and cervical small cell carcinomas despite the significant differences in their organs of origin.
Key Words: Small cell carcinoma, Small cell lung cancer, Cervical cancer, iTRAQ, Cluster analysis
Although small cell carcinomas can arise in almost any organ, roughly 95% of those with a clinical significance arise in the lung; thus, the rare exceptions, which include small cell carcinoma of the uterine cervix (SMCC), are referred to as extrapulmonary small cell carcinomas.1 Like the rare small cell lung carcinoma (SCLC), SMCC’s counterpart in the lung, small cell tumors occur in only 0.9% of all cases of cervical carcinomas.
Small cell carcinomas of the uterine cervix are observed mostly in younger women of reproductive age. Small cell carcinoma of the uterine cervix has a much worse prognosis compared with other histological types of cervical carcinoma, such as squamous cell carcinoma of the cervix (SCC), adenocarcinoma, or adenosquamous cell carcinoma; the 5-year disease-specific survival of SMCC in stage I–IIA, IIB–IVA, and IVB disease was reported to be 36.8%, 9.8%, and 0%, respectively.2
Because of its rarity, a standard treatment strategy has not yet been established for SMCC. To date, the chemotherapy regimens used in SMCC have been largely based on regimens used for SCLC, because the pathological features of SCLC and SMCC are so similar. The chemotherapeutic agents most often used for SCLC are various combinations of cisplatin, carboplatin, etoposide, and irinotecan.3 However, because the prognosis for advanced stages of both SCLC and SMCC remains similarly poor, there is a tremendous demand for the development of newer and better precision therapeutic agents.4
The origins and mechanisms for generation of SCLC and SMCC are still not fully elucidated. Although lung SCLC is related to smoking, cervical SMCC is not. Conversely, although SMCC is clearly related to human papillomavirus infection, SCLC is not. Roughly 90% of SCLCs contain mutations of p53 and/or RB1; however, the molecular etiology of SMCC has not been as clear.5
To the best of our knowledge, there has been no previous report describing a comparison of SMCC to SCLC in terms of gene or protein expression profiles. One of our key objectives in this study was to conduct a proteomic comparison of SMCC to SCLC to find similarities that extend beyond simple histology, to discover important patterns of shared tumor-specific protein expression. Such results would give needed additional validation for our current practice of conducting therapeutic regimens for SMCC that are similar to those used for SCLC. In addition, we hoped to find tumor-specific proteins expressed in SMCC, which could be biomarkers for SMCC diagnosis and potential future targets for improved chemo-, immuno-, and nucleic acid-based therapies.
We conducted our protein expression comparisons using the isobaric tags for relative and absolute quantitation (iTRAQ)–based technique for comprehensive and quantitative protein expression profiling. A supervised hierarchical cluster analysis was performed to evaluate for similarities between the identified protein profiles. We extended this protein expression profiling to SMCC, SCC, mucinous adenocarcinoma of the cervix (MACC), SCLC, and lung mucinous adenocarcinoma (MACL). For an initial validation of these findings, we performed immunohistochemistry for 2 of the 16 proteins we found to be uniquely enriched in SMCC.
For our analysis, we used cancer tissue–originated spheroids (CTOS), an approach we recently developed as a novel method for primary culture of human cancer cells,6 as samples for iTRAQ profiling. With this approach, cancer cells with high purity can be feasibly isolated with high efficiency from patient samples. The CTOS and CTOS-derived xenografts preserve many of the essential characteristics of the original tumor epithelium cells, but without conflicting contamination from stromal or blood cells, making them ideally suited to conduct proteomic iTRAQ-based comprehensive and quantitative protein expression profiling.
MATERIAL AND METHODS
CTOS Preparation, Culture, and Storage
Our study was approved by the institutional ethics committees of the Osaka International Cancer Institute and the Osaka University Hospital. Fresh surgically removed cancer tissues were used for primary cultures of the CTOS. The SMCC and lung cancer CTOS used in this study were established previously.7,8 All CTOS were cultured in StemPro hESC media (Invitrogen, Carlsbad, CA). For storage, CTOS were first mildly digested with 0.25% trypsin-EDTA (Life Technologies, Carlsbad, CA), suspended with CELLBANKER 1 (Nippon Zenyaku Kogyo, Fukushima, Japan) containing 10 μM Y-27632 (Wako Pure Chemical Industries, Osaka, Japan), then frozen at −80°C. CTOS, along with Growth Factor Reduced Matrigel Matrix (BD Biosciences, San Jose, CA), were injected subcutaneously into nonobese diabetic/severe combined immunodeficiency mice. Cancer tissue–originated spheroids were prepared from xenograft tumors, as described before. Briefly, xenograft tumors were cut into pieces, digested with liberase DH (Roche Applied Science, Mannheim, Germany), then filtered through 100- or 40-μm cell strainers (BD Falcon, Franklin Lakes, NJ).
Cell Lines and Culture
The human MACC cell line TCO-2 and the human SMCC cell line TC-YIK were purchased from the RIKEN Cell Bank (RIKEN BRC, Tsukuba, Japan). TCO-2 was maintained in MEM with 10% fetal bovine serum; TC-YIK was maintained in RPMI 1640 with 10% fetal bovine serum. All cells were cultured with 100 U/mL penicillin and 100 μg/mL streptomycin. “Cell line samples” were prepared by combining equal amounts of protein extracted from the TCO-2 and TC-YIK tumor cell lines. The cell line samples were used as reference baselines for analysis of protein expression data.
iTRAQ Labeling
For protein extraction, cell line samples and CTOS samples were incubated in 9.8 M urea containing 1% phosphatase inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) and 1% protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) for 30 minutes at room temperature, before centrifugation at 15,000 rpm for 10 minutes at 10°C. Extracted proteins from CTOS samples and cell line samples were reduced, alkylated, digested with trypsin, and labeled with iTRAQ reagents (AB SCIEX, Framingham, CA), according to the manufacturer’s instructions. Cell line samples were labeled with iTRAQ reagent 113. The SCLC-CTOS LC26 was labeled with iTRAQ reagent 114. The MACL-CTOS LC193 was labeled with iTRAQ reagent 115. The SMCC-CTOS cerv23, cerv5, and cerv9 were labeled with iTRAQ reagents 116, 117, and 118, respectively. The SCC-CTOS cerv44 was labeled with iTRAQ reagent 119, and the MACC-CTOS cerv46 was labeled with iTRAQ reagent 121. Labeled samples were pooled, then desalted and fractionated using strong cation exchange chromatography, as previously described.9,10
Mass Spectrometric Analysis
Nano liquid chromatography–mass spectrometry/mass spectrometry analysis was performed using an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA), as previously described.9,10
iTRAQ Data Analysis
Protein identification and quantification for iTRAQ analysis were carried out using Proteome Discoverer software (v.1.3, Thermo Fisher Scientific) and the SwissProt human protein database. Taxonomy was set to Homo sapiens. Search parameter conditions were as previously described.8,9 Protein expression data from the CTOS samples were obtained using the reagent 113–labeled cell line sample as a reference. Next, we performed median normalization in all samples and identified 1152 proteins variably expressed across the samples, based on SD values of 0.2. Unsupervised hierarchical cluster analysis was performed using Pearson correlation and average linkage.
Immunohistochemistry
Sections for immunohistochemistry were prepared from formalin-fixed paraffin-embedded tissues from surgical specimens. The diagnosis of each specimen was confirmed by central pathology review. The specimens were sliced at 4-μm thickness, deparaffinized, and then rehydrated in graded alcohols. Immunohistochemical stainings for creatine kinase B-type (CKB) and serine/threonine-protein kinase VRK1 (VRK1) were performed using the avidin-biotin-peroxidase complex (ABC) method, with a primary reaction with rabbit polyclonal anti-CKB antibody (Atlas Antibodies, Stockholm, Sweden) or mouse polyclonal anti-VRK1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), respectively. As secondary antibody, the Vectastain ABC Rabbit IgG KIT or the Vectastain ABC Mouse IgG Kit (Vector Laboratories, Burlingame, CA) was used according to the manufacturer’s protocol. Immunostained sections were photographed with an Olympus FSX100 (Olympus, Tokyo, Japan). Immunostainings were scored for the extent of stained cells. Cases with 71% to 100% of the tumor cells staining positively were considered strongly positive (score 3), cases with 41% to 70% cells were considered medium positive (score 2), those with 10 to 40% were considered weakly positive (score 1), and cases with 0% to 9% were considered negative (score 0). Three independent gynecologic oncologists (K.Y., Y.U., and S.M.), who were blinded to the histological data, analyzed the stained sections using an Olympus BH2 microscope. In cases of disagreement, the staining results were reevaluated by central pathology review, followed by careful discussion—until a consensus was reached.
Statistical Analysis
Statistical analysis was completed using GraphPad Prism (version 7.01, GraphPad Software, Inc., San Diego, CA). The Kruskal-Wallis test, followed by Dunn multiple comparisons test, was used for analyzing immunohistochemistry. P values less than 0.05 were considered statistically significant.
RESULTS
Protein Expression Analysis and Unsupervised Hierarchical Cluster Analysis
Seven CTOS lines were used: 3 SMCC and 1 each of SCC, MACC, SCLC, and MACL (Table 1). In 7 CTOS cell lines, a total of 3109 tumor-expressed proteins were identified by the iTRAQ method. Of these, 1152 proteins were variably expressed across all samples, with SD values greater than 0.2. These were included in an unsupervised hierarchical cluster analysis (Fig. 1, Table 2).
TABLE 1.
Clinical characteristics of tumors used to create CTOS
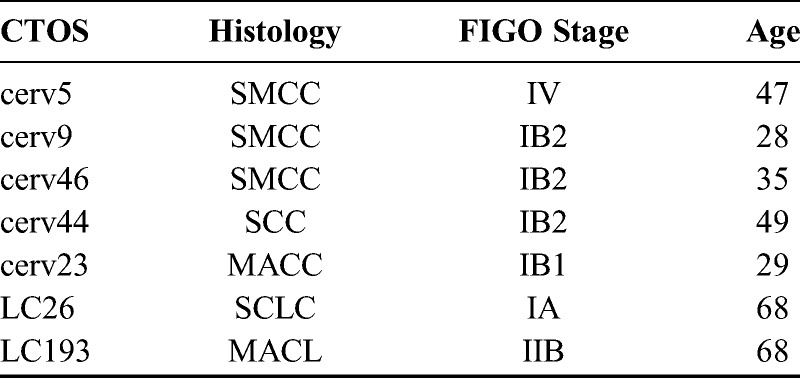
FIGURE 1.
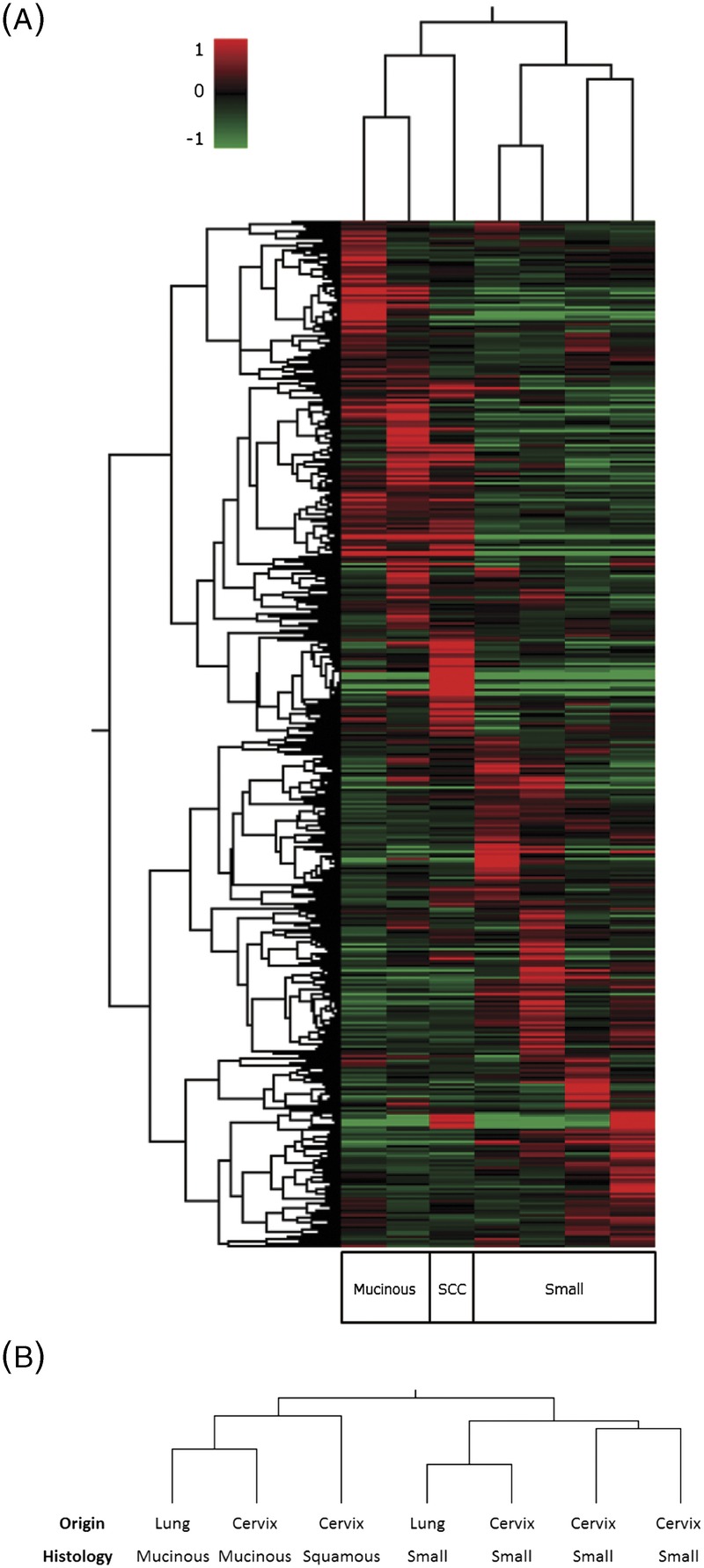
A, Differential protein expression profiles of 4 small cell carcinomas, 2 mucinous adenocarcinomas, and 1 squamous cell carcinoma. The expression level of each protein is colored; red represents expression above the mean, and green indicates expression below the mean. B, Dendrogram produced by unsupervised hierarchical cluster analysis of CTOS. Two major clusters were identified in the dendrogram. All SMCC and SCLC were classified into the right-side cluster. MACC, MACL, and SCC were classified into the left-side cluster.
TABLE 2.
Proteins overexpressed in SMCC

All 4 of the small cell carcinoma CTOS cell lines (3 SMCC, 1 SCLC) showed similar protein expression profiles and were classified into the same right-side cluster (Fig. 1A). The tumors of other histological types were classified into the left-side cluster (Fig. 1A). On the other hand, when focusing on organ of origin, the 2 non-small cell uterine cervical cancers were not in the same cluster with the 3 SMCC (Fig. 1B). Similarly, lung cancers were not all in the same cluster either (Fig. 1B).
The protein expression profiles obtained in the present study demonstrated a close similarity between SMCC and SCLC. This means that carcinomas sharing the histological type of small cell carcinoma are much more likely to also share similar protein expression profiles than carcinomas that share merely an organ-specific origin but little other phenotype.
Among the 3109 cervical tumor–expressed proteins, 44 were expressed more than 2-fold higher in small cell carcinomas (SMCC) compared with the mucinous adenocarcinoma (MACC), and 36 proteins were expressed more than 2-fold higher in the small cell carcinomas compared with the squamous cell carcinoma (SCC). There were 16 proteins in common between the groups of 44 and 36 higher expressed SMCC proteins (Table 2). As expected, gamma-enolase (NSE), which has long been known to be overexpressed in small cell carcinomas of many types, was included in the 16 proteins in common. The other 15 proteins are listed in Table 2.
Immunohistochemistry
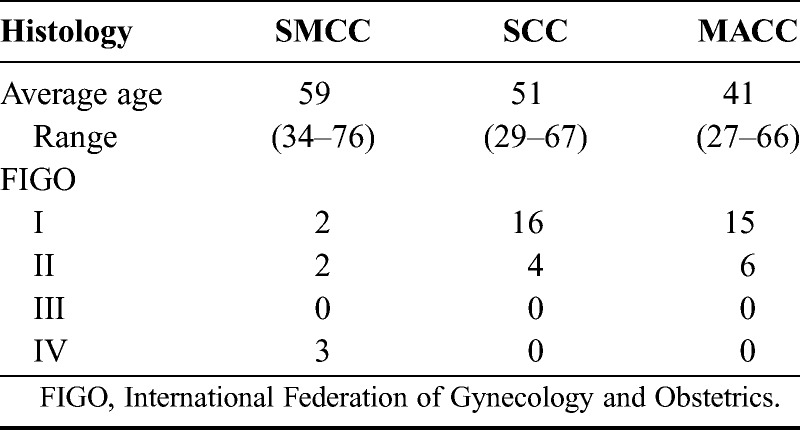
We conducted immunohistochemistry for CKB and VRK1 expression in clinical samples of uterine cervical cancer Fig. 2. Characteristics of the clinical samples are listed in Table 3.
FIGURE 2.
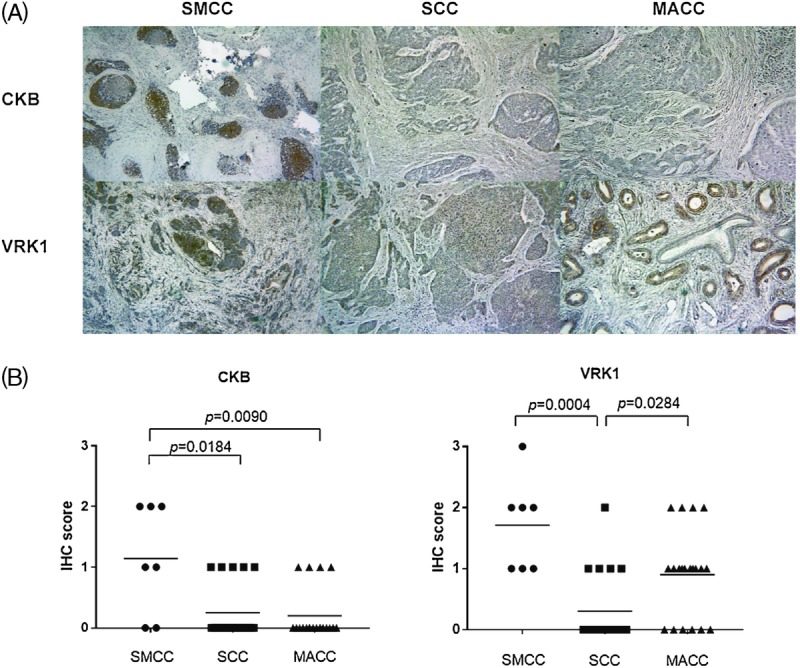
A, Representative immunohistochemical staining for CKB or VRK1 in SMCC, SCC, and MACC. Immunostained sections were counterstained with hematoxylin and photographed (magnification, ×100). The CKB was stained more intensely in SMCC than in SCC or MACC. The VRK1 was stained more intensely in SMCC and MACC than in SCC. B, IGFBP2, VRK1, and CKB immunoreactivity in SMCC, SCC, and MACC. Immunohistochemical scoring was performed according to the extent of cells staining positively.
TABLE 3.
Characteristics of clinical samples
The CKB was found to be expressed significantly higher in SMCC than in SCC or MACC (P = 0.0184, P = 0.0090, respectively). Serine/threonine-protein kinase VRK1 were also expressed significantly higher in SMCC than in SCC (P = 0.0004); however, the expressions of VRK1 in MACC were not significantly different when compared with SMCC (p = 0.1470). The expression of VRK1 in MACC was significantly higher than in SCC (p = 0.0284).
DISCUSSION
In the present study we have used iTRAQ-based quantitative proteomic analysis and unsupervised hierarchical cluster analysis to demonstrate a strong similarity between the protein expression profiles of small cell carcinomas of the lung and cervix. To our knowledge, this is the first such in-depth proteomics report demonstrating the strong similarities of protein expression profiles of small cell carcinomas originating from multiple organs.
We had previously discovered protein expression profile similarities between high-grade ovarian and endometrial serous carcinomas.10 Likewise, Zorn et al reported similarities in gene expression patterns between renal, endometrial, and ovarian clear cell carcinomas,11 showing that overt histological phenotypic similarities are often linked to underlying gene and protein expression similarities.
The cell origins of SMCC and SCLC in humans have not yet been fully elucidated. However, in mouse models, SCLC arises from either neuroendocrine or alveolar type 2 cells.12 Because of similar protein expression profiles of SMCC and SCLC previously found in much more limited studies, it has been hypothesized that SMCC in humans might also arise from neuroendocrine cells of the cervix13; we have now shown that this shared cell type of origin is to be quite likely.
For the first time, the novel CTOS method was used for this type of comparative proteomic analysis. Each CTOS consists solely of cancer cells; it is absent stromal and blood cells that could adversely influence proteomic analysis. In a previous study, of high-grade serous carcinomas, we used laser microdissection of the clinical samples to select only cancer cells.10 However, the CTOS method does not require laser microdissection to remove unnecessary cells, thus making it more convenient for conducting proteomic analyses.
Using iTRAQ-based quantitative proteomic analysis, we found a number of proteins that are more highly expressed in SMCC than in 2 more common types of cervical carcinomas: SCC and MACC. Two proteins, CKB and VRK1, were chosen from the list of 16 as most likely to be informative for further study, after referring to results from our previous RNA array studies. Those studies were conducted using 7 SMCC-CTOS cell lines and the cell line TC-YIK. We chose these 2 proteins for validation because they were differentially highly expressed in all 8 cell lines (data not shown) and were previously reported to relate to tumorigenesis. Another consideration was which proteins on the list had commercially available antibodies, because we intended to validate the unique overexpression of the proteins in SMCC by immunohistochemistry.
By iTRAQ analysis, the expression of CKB was significantly higher in cervical SMCC than in SCC or MACC; this was validated by immunohistochemistry. The CKB is an enzyme that reversibly catalyzes the transfer of phosphate between adenosine triphosphate (ATP) and various other phosphogens and plays a key role in cellular energy metabolism. The CKB has been shown previously to be overexpressed in various types of cancers, including breast, colorectal, and ovarian.14–16 Recently, it has been reported that CKB promotes metastatic survival by modulating intra- and extracellular energetics of colorectal cancer, suggesting that CKB could be a potent therapeutic target.16 However, our current finding is the first to report significant overexpression of CKB in rare SMCC uterine cervical cancers, especially compared with SCC and MACC. Immunohistochemistry for CKB may thus be a highly useful diagnostic marker for SMCC, as it is often difficult to differentially diagnose the rare SMCC from the much more common SCC and adenocarcinoma.
Next, iTRAQ-based analysis showed that the expression of VRK1 was 2-fold higher in SMCC than in SCC or MACC. In addition, by immunohistochemistry, the expression of VRK1 was significantly higher in SMCC than in SCC; however, the iTRAQ-based analysis and the immunohistochemical findings for VRK1were discordant in MACC. By immunohistochemistry, the expression of VRK1 in MACC (like SMCC) was significantly higher than in SCC. This discordance is likely to result from the low sample numbers used; only 1 sample each of SCC and MACC was used for the iTRAQ-based analysis, which was a limitation of our study. With that noted, the prognosis of MACC is still worse than SCC,17 so, if validated by further studies, VRK1 could become a useful prognostic marker for these 2 rare tumor types.
Our report is the first known report concerning VRK1’s overexpression in human uterine cervical cancers, in this case, in both SMCC and MACC. In mammals, VRK1 is a member of the Ser/Thr kinase family and acts through phosphorylation of several important cancer-related substrates, including histone H3 and p53. There are reports that expression of VRK1 is correlated with cancer progression in breast, head-and-neck squamous cell, lung, and hepatocellular cancers.19–21 Expression of the VRK1 gene has been shown in a cell line of a high-grade serous carcinoma of the ovary,22 but there have been no similar reports of VRK1 protein expression in cervical or endometrial cancers.
In summary, we have performed protein expression profiling of SMCC, SCC, SCLC, MACC and MACL using iTRAQ-based proteomic analysis of CTOS isolates. Using an unsupervised hierarchical cluster analysis, we have demonstrated a strong similarity between cervical SMCC and lung SCLC. This similarity between 2 types of small cell carcinoma disregards their disparate organs of origin. The level of proteomic overlap we have found strongly supports the practice of conducting similar cancer treatments, that is, chemotherapy regimens, for both SMCC and SCLC. In addition, with this high-throughput proteomic approach, we have identified multiple proteins that could represent potential future therapeutic targets for SMCC.
ACKNOWLEDGMENTS
We would like to express our deepest appreciation to Ms Kanako Sakiyama, Hazuki Abe, and Asami Yagi for their secretarial and technical help. We thank also Mr Kosei Yamaura and Ms Fumika Mori for their experimental support. We would also like to thank Dr G. S. Buzard for his constructive critiquing and editing of our manuscript.
Footnotes
This work was supported by MEXT KAKENHI (grant no. JP26861326) from the Japanese Ministry of Education, Culture, Sports, Science and Technology.
The authors declare no conflicts of interest.
REFERENCES
- 1.Zheng X, Liu D, Fallon JT, et al. Distinct genetic alterations in small cell carcinoma from different anatomic sites. Exp Hematol Oncol. 2015;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen JG, Kapp DS, Shin JY, et al. Small cell carcinoma of the cervix: treatment and survival outcomes of 188 patients. Am J Obstet Gynecol. 2010;203:347. e1–347. e6. [DOI] [PubMed] [Google Scholar]
- 3.Pillai RN, Owonikoko TK. Small cell lung cancer: therapies and targets. Semin Oncol. 2014;41:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadducci A, Carinelli S, Aletti G. Neuroendrocrine tumors of the uterine cervix: a therapeutic challenge for gynecologic oncologists. Gynecol Oncol. 2017;144:637–646. [DOI] [PubMed] [Google Scholar]
- 5.Rickman DS, Beltran H, Demichelis F, et al. Biology and evolution of poorly differentiated neuroendocrine tumors. Nat Med. 2017;23:1–10. [DOI] [PubMed] [Google Scholar]
- 6.Kondo J, Endo H, Okuyama H, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci U S A. 2011;108:6235–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakajima A, Endo H, Okuyama H, et al. Radiation sensitivity assay with a panel of patient-derived spheroids of small cell carcinoma of the cervix. Int J Cancer. 2015;136:2949–60. [DOI] [PubMed] [Google Scholar]
- 8.Endo H, Okami J, Okuyama H, et al. Spheroid culture of primary lung cancer cells with neuregulin 1/HER3 pathway activation. J Thorac Oncol. 2013;8:131–139. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama T, Enomoto T, Serada S, et al. Plasma membrane proteomics identifies bone marrow stromal antigen 2 as a potential therapeutic target in endometrial cancer. Int J Cancer. 2013;132:472–484. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Yoshino K, Serada S, et al. Similar protein expression profiles of ovarian and endometrial high-grade serous carcinomas. Br J Cancer. 2016;114:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zorn KK, Bonome T, Gangi L, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11:6422–6430. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. [DOI] [PubMed] [Google Scholar]
- 13.Guadagno E, De Rosa G, Del Basso De Caro M. Neuroendocrine tumours in rare sites: differences in nomenclature and diagnostics-a rare and ubiquitous histotype. J Clin Pathol. 2016;69:563–574. [DOI] [PubMed] [Google Scholar]
- 14.Zarghami N, Giai M, Yu H, et al. Creatine kinase BB isoenzyme levels in tumour cytosols and survival of breast cancer patients. Br J Cancer. 1996;73:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramani M, Day BW, Schoen RE, et al. Altered expression and localization of creatine kinase B, heterogeneous nuclear ribonucleoprotein F, and high mobility group box 1 protein in the nuclear matrix associated with colon cancer. Cancer Res. 2006;66:763–769. [DOI] [PubMed] [Google Scholar]
- 16.Li XH, Chen XJ, Ou WB, et al. Knockdown of creatine kinase B inhibits ovarian cancer progression by decreasing glycolysis. Int J Biochem Cell Biol. 2013;45:979–986. [DOI] [PubMed] [Google Scholar]
- 17.Loo JM, Scherl A, Nguyen A, et al. Extracellular metabolic energetics can promote cancer progression. Cell. 2015;160:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara H, Yokota H, Monk B, et al. Gynecologic Cancer InterGroup (GCIG) consensus review for cervical adenocarcinoma. Int J Gynecol Cancer. 2014;24:S96–S101. [DOI] [PubMed] [Google Scholar]
- 19.Santos CR, Rodriguez-Pinilla M, Vega FM, et al. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol Cancer Res. 2006;4:177–185. [DOI] [PubMed] [Google Scholar]
- 20.Salzano M, Vazquez-Cedeira M, Sanz-Garcia M, et al. Vaccinia-related kinase 1 (VRK1) confers resistance to DNA-damaging agents in human breast cancer by affecting DNA damage response. Oncotarget. 2014;5:1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee N, Kwon JH, Kim YB, et al. Vaccinia-related kinase 1 promotes hepatocellular carcinoma by controlling the levels of cell cycle regulators associated with G1/S transition. Oncotarget. 2015;6:30130–30148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baratta MG, Schinzel AC, Zwang Y, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc Natl Acad Sci U S A. 2015;112:232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]


