SUMMARY
DNA binding by numerous transcription factors including the p53 tumor suppressor protein constitutes a vital early step in transcriptional activation. While the role of the central core DNA binding domain (DBD) of p53 in site-specific DNA binding has been established, the contribution of the sequence-independent C-terminal domain (CTD) is still not well understood. We investigated the DNA-binding properties of a series of p53 CTD variants using a combination of in vitro biochemical analyses and in vivo binding experiments. Our results provide several unanticipated and interconnected findings. First, the CTD enables DNA binding in a sequence-dependent manner that is drastically altered by either its modification or deletion. Second, dependence on the CTD correlates with the extent to which the p53 binding site deviates from the canonical consensus sequence. Finally, the CTD enables stable formation of p53-DNA complexes to divergent binding sites via DNA-induced conformational changes within the DBD itself.
INTRODUCTION
The p53 tumor suppressor protein is a DNA sequence-dependent transcription factor that controls the expression of myriad genes implicated in cell cycle, cell death, , DNA repair, metabolism and other outcomes (Vousden and Prives, 2009). The active tetrameric p53 protein comprises four domains: the N-terminal transactivation (NTD), the central DNA binding (DBD), the oligomerization (OD) and the C-terminal (CTD) domain. Of these, only two, the DBD and the OD, have been amenable to structural analysis (Joerger and Fersht, 2008). The natively unfolded regions that span the NTD, the CTD and the linker between the DBD and the OD, represent at least 40% of the total monomer (Joerger and Fersht, 2008). Both, the highly acidic NTD and the highly basic CTD may undergo disorder-to-order transitions upon binding to their protein or nucleic acid binding partners (Friedler et al., 2005; Lee et al., 2000; Mujtaba et al., 2004; Rustandi et al., 2000).
There is a wealth of knowledge about the structure and functions of the conserved central “core” DBD of the p53 protein wherein the vast majority of tumor derived missense mutations are located ((Olivier et al., 2010). This domain has been subjected to extensive biochemical analysis (Laptenko and Prives, 2006) and its structure has been solved (Cañadillas et al., 2006; Cho et al., 1994; Kitayner et al., 2006; Petty et al., 2006). Additionally, multiple studies have revealed and refined the consensus binding site (BS), comprising two copies of the sequence RRRCWWGYYY spaced by 0–13 base pairs, to which this region of the protein binds (Botcheva et al., 2011; el Deiry et al., 1992; Funk et al., 1992; Hoh et al., 2002; Nikulenkov et al., 2012; Wei et al., 2006). In contrast to myriad studies on the central DBD which are in general agreement, we still do not understand the roles and functions of the p53 CTD, whose several lysines when unmodified allow it to bind non-specifically to DNA and RNA ((Laptenko and Prives, 2006). Published reports have implicated CTD involvement in regulation of DNA binding (Anderson et al., 1997; Gu and Roeder, 1997; Luo et al., 2004; McKinney et al., 2004), p53 stability (Li et al., 2002; Nakamura et al., 2000; Rodriguez et al., 2000), p53 cellular localization (Gu et al., 2001; Lohrum et al., 2001; Nie et al., 2007; Stommel et al., 1999) and co-factor recruitment (An et al., 2004; Barlev et al., 2001; Chuikov et al., 2004; Lee et al., 2000; Mujtaba et al., 2004). Unfortunately, because of its unstructured nature, NMR and X-ray crystallography have been unable to dissect the role(s) of the CTD within the full length p53 tetramer. Similarly, while fascinating and provocative, in vivo studies such as cell culture and mouse models have not succeeded in dissecting the often interdependent roles of the CTD (Feng et al., 2005; Hamard et al., 2013; Krummel et al., 2005; Simeonova et al., 2013).
The function of the unmodified CTD in regulation of sequence specific binding by the central DNA binding domain has been the most extensively studied and perhaps the most controversial. Initially described as a negative regulator of sequence specific DNA binding (Anderson et al., 1997; Ayed et al., 2001; Hupp et al., 1992; Hupp and Lane, 1994; Jayaraman and Prives, 1995), the CTD was later credited with positive regulatory features such as facilitating p53 binding to long naked DNA molecules as well as to chromatin (Espinosa and Emerson, 2001) and promoting p53 linear diffusion on DNA (McKinney et al., 2004; Tafvizi et al., 2011). These features likely depend on low affinity electrostatic interactions between the multiple C-terminal lysines and the DNA phosphate backbone (Friedler et al., 2005). Two studies have presented evidence that implicates the CTD in site-specific binding of p53 although in depth mechanistic information is still lacking (Hamard et al., 2012; Kim et al., 2012).
Here using a combination of biochemical and computational approaches we have undertaken an exhaustive analysis of the CTD employing C-terminally modified versions of p53 and investigated their DNA binding properties in vitro and in cells. We discuss biological implications linked to our findings.
RESULTS
The C-terminal Domain is an Important Determinant of Site-Specific p53 DNA Binding
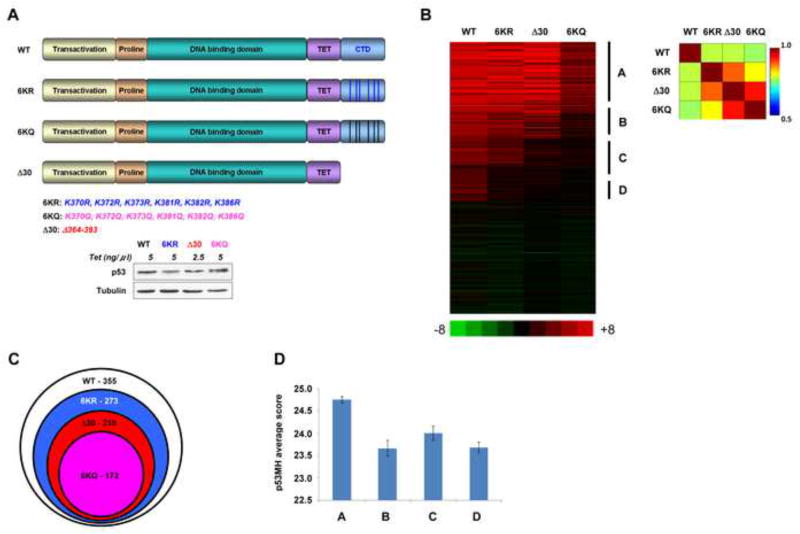
H1299 (p53 null) engineered to inducibly express wild-type (WT) p53, p53 lacking the C-terminal 30 amino acids (Δ30 p53) or p53 where the last 6 lysines were changed to arginine (6 KR) to maintain charge, or to glutamine (6KQ) to potentially mimic acetylation (Figure 1A; Poyurovsky et al., 2010) were used for a ChIP-on-chip analysis (Huggins et al, 2011). The C-terminal mutation positions and induced protein levels are shown in Figure 1A. Chromatin-bound DNA isolated from these cell lines was analyzed using a p53 focused array containing ~600 known p53 binding sites followed by bioinformatic analysis. The number of sites recognized by each p53 CTD variant was notably reduced when compared to WT p53 that bound 355 sites (Figure 1B and C). The p53 6KR mutant bound to 278 sites, while the acetylation-mimicking 6KQ and the Δ30 mutant forms of p53 bound to even less sites (172 and 210 sites respectively) and their association with these target sites was often not as strong as seen with WT p53, indicating apparent binding affinity defects. There were no targets bound by the CTD variants that were not bound by WT p53. Grouping the bound sites into categories A, B, C and D according to their binding preferences (with A bound by all p53 forms and D bound by WT only) (see Figure 1B) revealed both the superior binding of WT p53 across all the 355 targets as well as the higher dependency of p53 on its C-terminal lysines in binding to sites from B, C and D groups of targets. We then used a de-novo motif analysis tool, GLAM2 to identify and compare the binding motifs originated from the 4 groups of target sequences (Frith et al., 2008). Versions of the consensus p53 binding site were present in all groups, with only subtle over-all differences in the group-specific p53 binding motifs (Figure S1A). But by concentrating on the individual nucleotide positions within the p53 binding sites in each group and assessing their degree of deviation from the canonical 20 bp p53 sequence on a position-specific basis, we determined that sites from group A are significantly more similar to the consensus p53 binding site than groups C-D (Figure 1D and Figure S1B). In general, positions R and Y within the consensus binding site differed the most between the groups (Figure S1B). This analysis indicated that the CTD is required for the central core DBD to recognize the full repertoire of binding sites in vivo including ones that deviate the most from the consensus site.
Figure 1. The p53 CTD is Required for Binding to Targets that Deviate from the Consensus Binding Sequence.
(A). Schematic representation of p53 CTD variants expressed in H1299 cells. The positions of C-terminal lysines mutated to either arginine or glutamine are indicated below. Relative expression levels of p53 proteins used in the ChIP-on chip experiment are shown below. (B). Heat-map representation of ChIP-on-chip results. The vertical lines indicate the clustering of four groups of p53 target sites that were bound by the proteins indicated above each group. Indicated on the right of the heat map are group A, representing the sites bound by all p53 forms; B representing the sites bound by all p53 proteins other than the 6KQ; C representing the sites bound by WT and 6KR p53 variants; D representing the sites bound exclusively by WT p53. On the right is the correlation matrix corresponding to the heat map. (C). Venn diagram showing the total number of sites bound by each p53 variant and their relationship. (D). Average and standard error of p53 binding site scores (calculated by p53MH algorithm) for each distinct group. Group A score is significantly different from those of the other groups (P<0.006, 0.04 and 0.0002; two sides Student T test for groups B, C and D, respectively). See also Figure S1.
SELEX Reveals Substantial Differences in the DNA Binding Selection Properties of Wild-Type and CTD-Modified Versions of p53
To reveal the mechanism(s) behind the observed DNA binding deficiencies of the p53 CTD mutants we exploited several biochemical approaches using highly purified proteins (Figure S2A). In addition to the aforementioned versions of p53 (6KR, 6KQ and Δ30) we also produced WT p53 where the C-terminal lysines were extensively acetylated by p300 histone acetylase (referred to as WT Ac) as well as acetylated 6KR mutant p53 (6KR Ac) (Piluso et al., 2005). The modification status of these different p53 variants was anti-pan-acetyl lysine or anti acetyl K373 p53 antibodies (Figure S2D). When we subjected these proteins to native polyacrylamide gel electrophoresis either in the absence (Native PAGE) or presence (Blue Native PAGE) of Coomassie Brilliant Blue R-250 (Figure S2, panel B and C) differences in the mobility patterns of the CTD variants were observed. WT p53 migrated as multiple distinct forms and substitution of the CTD lysines with arginines (6KR and 6KR Ac) produced a similar pattern with even more of the slowly migrating fractions. By contrast, lysine-to-glutamine substitution (6KQ), extensive acetylation with p300 or deletion of the CTD resulted in near complete elimination of these slower migrating species. These both confirmed the highly acetylated status of the WT Ac p53 protein and suggested that WT Ac p53, 6KQ and Δ30 proteins share characteristics that are distinct from unacetylated WT p53. Analysis of these proteins along with a known dimeric p53 mutant (E343A) demonstrated that all the p53 variants used in our study are tetramers as well as higher-order oligomers (Figure S2, panel C).
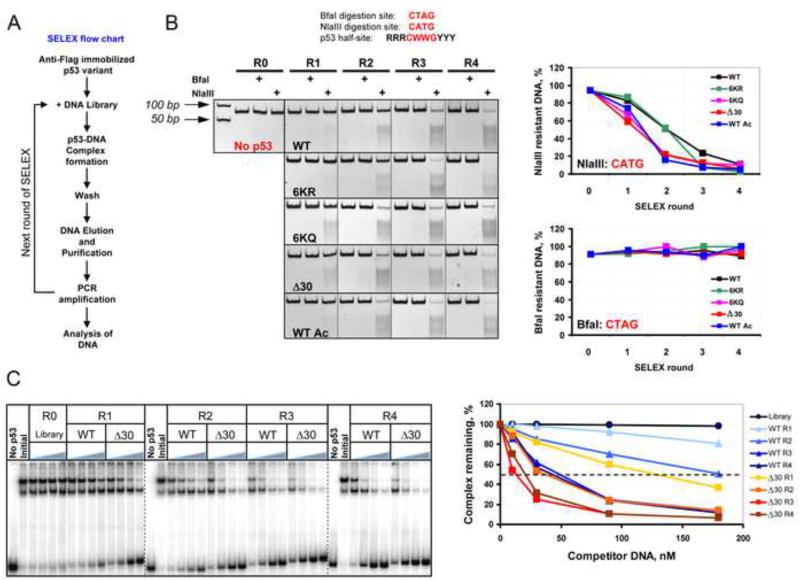
To confirm the sequence preferences of wild-type p53 vs. the C-terminally altered versions of p53, we employed the Systematic Evolution of Ligands by Exponential enrichment (SELEX) protocol (Gopinath et al., 2007). The p53 proteins were bound to a library of 66 bp DNA oligonucleotides in which centrally positioned random 20 base pairs were flanked on either side by constant sequences (Figure 2). Four rounds of SELEX were performed for WT, WT Ac, 6KR, 6KQ and Δ30 p53 variants and the fraction of DNA selected in each round of p53 variant-specific SELEX was PCR-amplified.
Figure 2. SELEX Reveals Substantial Differences in DNA Binding Site Selection Preferences of Wild Type and CTD-Modified Versions of p53.
(A). Flow chart depicting the SELEX protocol used to generate the pool of DNA targets selected by p53 CTD variants. (B). Upper: BfaI and NlaIII restriction endonuclease cleavage sites compared with the core element of the p53 consensus BS. Lower: DNA pools selected by p53 CTD modified or mutated proteins (See also Figure S2) in rounds 1–4 (R1, R2, R3, and R4) of SELEX were digested with NlaIII and BfaI and the reaction products were separated by 10% TBE PAGE. The gel boundaries are indicated with dotted lines. R0 - the initial degenerate pool of DNA targets. Graphs on right show the fraction of enzyme-resistant DNA specific to each p53 variant after each round of SELEX. The fraction of NlaIII- and BfaI-sensitive DNA in R0 was found to be ~1–2%. (C). Δ30 p53 was bound to 66 bp 32P-labeled mdm2 DNA in the presence of increasing concentration of unlabeled DNA that was pre-selected by either WT or Δ30 p53 in the different rounds of SELEX. The left panel shows a compilation of 3 representative PhosphorImager scans of 4 % native 0.5X TBE polyacrylamide gels. The gel boundaries are indicated with dashed lines. No p53 lanes: no Δ30 p53 in the reaction. Initial - no unlabeled DNA competitor in the reactions. The fraction of the remaining Δ30 p53-mdm2 DNA complexes in each condition expressed as a function of competitor DNA is shown on the right.
Within the canonical p53 half site (RRRCWWGYYY), the central core sequence CWWG (where W=A or T) is the most important element and the C and G nucleotides are critical, if not invariant. Having the two WW nucleotides between them is also a conserved feature with CATG being the preferred sequence (Beno et al., 2011; Nagaich et al., 1997). To determine whether the CTD contribution to p53 specific DNA binding depends on the exact sequence within the CWWG motif the selected pools of DNA were cleaved with NlaIII or BfaI site-specific restriction endonucleases, whose recognition sequences match the central portion of the p53 half-site, namely CATG and CTAG, respectively (Figure 2B). The results showed a very strong bias of CTD-altered p53 proteins for CATG-containing DNA sequences that was most evident within the first 2 rounds of selection. Neither WT nor the CTD p53 mutants favored the CTAG sequence, likely due its low degree of torsional flexibility and relatively high dependence on inter-DBD cooperativity (Beno et al., 2011). A similar experiment with NspI that cleaves RCTAGY that matches a longer portion of the p53 consensus half-site sequence revealed an even larger difference between WT p53 and the CTD-altered p53 variants, showing that the 6KQ and Δ30 C-terminal mutants were defective in binding stably to a more diverse population of p53 sites (Figure S3). The DNA binding profile of acetylated wild-type p53 in this assay more closely resembled Δ30 and 6KQ p53 proteins than either WT or 6KR p53 proteins. By the end of the 4th round more than 90% of the DNA was enriched with the sequence RCATGY regardless of the status of the CTD of the p53 used for selection. DNA selected in the absence of p53 showed virtually no overall increase in the enzyme- sensitive DNA by the final round of selection (Figure S3, sample M2R4).
We compared WT- and Δ30 p53 SELEX-selected DNAs as to their ability to compete for the binding of Δ30 p53 protein to DNA in an EMSA experiment (Figure 2C). The superior binding characteristics of Δ30-selected DNA over WT p53-selected DNA were especially evident when comparing the corresponding DNA derived from SELEX rounds 1 and 2 (see the corresponding graphical analysis in Figure 2, panel C). Together the ChIP-on-chip and SELEX-based experiments reveal a role of the C-terminal domain in allowing p53 to bind to those sites that are more divergent from the consensus p53 binding sequence.
The p53 C-terminal Domain is Required for Stable p53/DNA Complex Formation
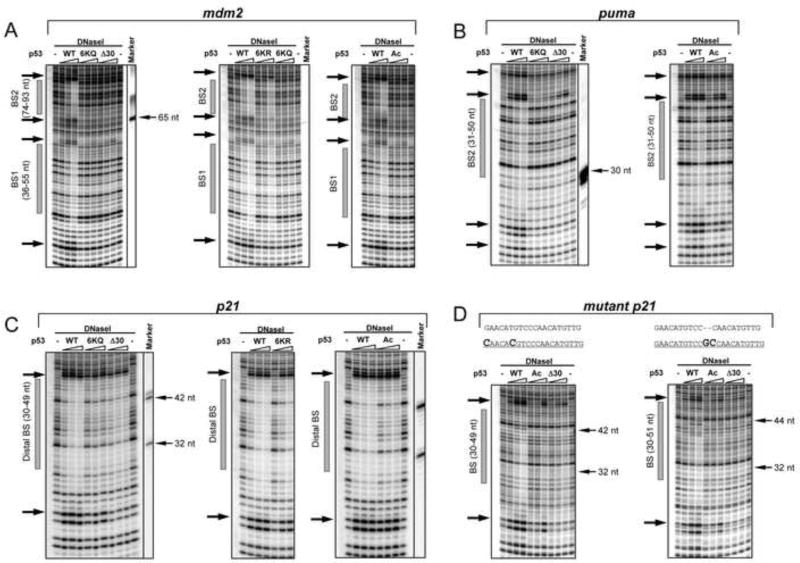
Protection from DNase cleavage can reveal the relative affinity of protein-DNA interactions, the relative stability of such complexes, and possible protein-induced conformational changes within the bound DNA. DNaseI footprinting experiments were performed using the purified CTD p53 variant proteins bound to DNA templates chosen to represent 3 types of bona fide p53 binding sites: (i) the high affinity “distal” binding site within the p21/ CDKN1 (hereafter referred as p21) promoter at −2.3 kb that has little divergence from the canonical p53 binding site, (ii) the medium affinity distal binding site within the puma/BBC3 (hereafter referred to as puma) that diverges somewhat from the p53 consensus site and (iii) the closely spaced low affinity bindings sites 1 and 2 within the mdm2 P2 promoter whose sequences deviate the most from the p53 consensus sequence (Figure S2, panel E).
Strikingly, in contrast to WT p53 protein, C-terminally truncated, p300 acetylated or 6KQ mutated p53 proteins were completely unable to protect the binding sites in mdm2 DNA from DNaseI cleavage (Figure 3A and Figure S4). The mutant 6KR p53 protein protected both mdm2 binding sites to a somewhat lesser extent than WT p53 indicating that unmodified lysines provide more than simple charge effects in specific p53-DNA interactions. Results with the puma p53 BS were similar and also demonstrated WT p53 superiority over the rest of the p53 CTD variants (Figure 3B). Only the high-affinity p21 distal p53 BS was protected from DNase I cleavage by all the p53 variants (Figure 3C). Even so, both 6KQ and Δ30 p53 proteins were significantly less effective than WT p53 in protecting the p21 site. Moreover, p53 binding-induced DNaseI hypersensitivity on both ends of the p21 distal site was diminished in the presence of 6KQ, 6KR and especially Δ30 p53 proteins.
Figure 3. The p53 CTD is Required for Stable p53-DNA Complex Formation in a Sequence-Dependent Manner.
DNAseI footprint experiments with DNA fragments containing the BS from mdm2 (BS1 and BS2 weak sites, panel A), puma (intermediate site, panel B), or the p21 (strong distal BS, panel C) or mutated p21 distal BSs (panel D). The sequences of the original and two mutated p21 distal BS with changes in bold are shown on the top of gels in panel D. Each panel represents a portion of a high-resolution PhosphorImager scan of the 11 % sequencing PAAG with the positions of the corresponding BSs shown as gray bars on the left side of each scan. The arrows on the left side of each gel denote p53-specific DNaseI hypersensitive sites. Full site scanning densitometry analysis specific to each p53 variant at its maximal concentration is shown in Figure S4. Sequence-specific DNA markers were prepared and run on the gels along with the experimental reaction mixture (indicated by thin arrows on the right of side of the gel).
Excluding possible bias due to flanking sequences within the p21 DNA-spanning fragment, mutation of the p21 binding sequence to become more divergent from the canonical binding sequence resulted in a marked decrease in protection from DNaseI in the presence of any CTD p53 variant (Figure 3D).
As a second approach, neither Δ30 nor WT Ac could protect even the high affinity p21 distal RE from ExoIII digestion, suggesting that the highly processive nature of ExoIII-dependent degradation of DNA may take advantage of even relatively small reductions in the stability of p53-DNA complexes (Figure S5) .
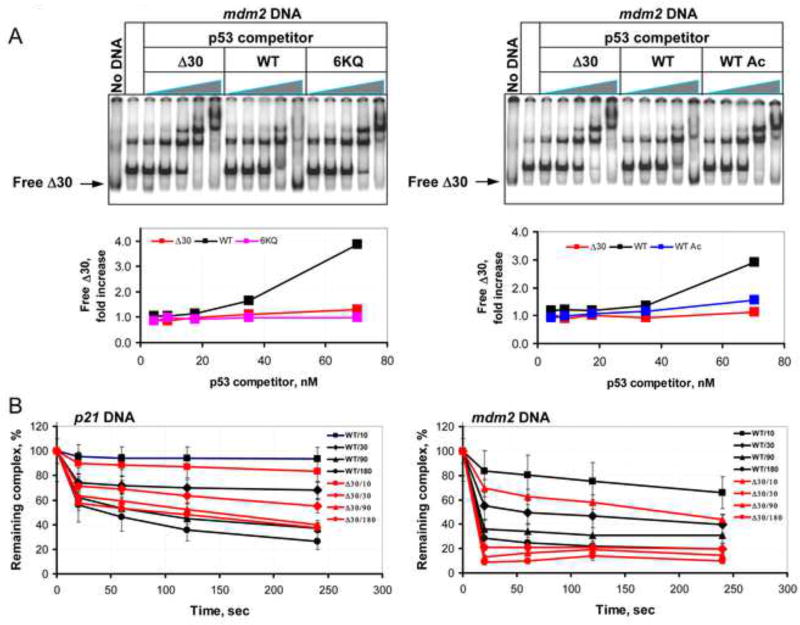
To compare the stability of different CTD variants in complexes with cognate DNA sequences, Δ30 p53 was engineered to contain a protein kinase A (PKA) site at the N-terminus. After phosphorylation with PKA the resulting 32P-lableled protein was bound to unlabeled DNA. The stability of that complex was assessed by adding increasing amounts of unlabeled p53 proteins (Δ30, WT, WT Ac or 6KQ) as competitors (Figure 4 A). WT p53 efficiently competed off the CTD -deleted p53 mutant protein from the DNA template at concentrations significantly lower than either Δ30 or 6KQ p53 variants (see the corresponding graphs in Figure 4A). Finally, EMSA DNA competition assays with 32P-end-labeled mdm2 and p21 DNA constructs in the presence of either unlabeled WT or Δ30 p53 protein showed that deletion of the CTD resulted in a significant decrease in the half-life of mdm2-p53 but not p21-p53 binary complexes (Figure 4B).
Figure 4. The p53 CTD Regulates Stability of p53-Cognate DNA Complexes.
(A). Representative gel scan of a competition binding assay that was performed with 32P-labeled Δ30 p53 protein in presence of unlabeled mdm2 BS-containing DNA and increasing amounts of unlabeled p53 WT or CTD-altered proteins as specified. The labeled Δ30 p53 protein was visualized using a PhosphorImager and relative amounts of displaced Δ30 p53 in each reaction mixture (indicated with arrow) were estimated using ImageQuant 5.2 software and presented as fold increase over the values obtained with no p53 competitor in the corresponding graphs below each scan. (B). Graphs show the quantification of the competition binding experiments performed with 32P-end-labeled p21 (left panel) or mdm2 (right panel) BS-containing DNA fragments in the presence of either unlabeled WT or Δ30 p53 protein as indicated. The experiment was performed as described in Experimental Procedures.
The CTD Regulates Base-Specific Interactions Between the Sequence Specific DNA Binding Domain and DNA
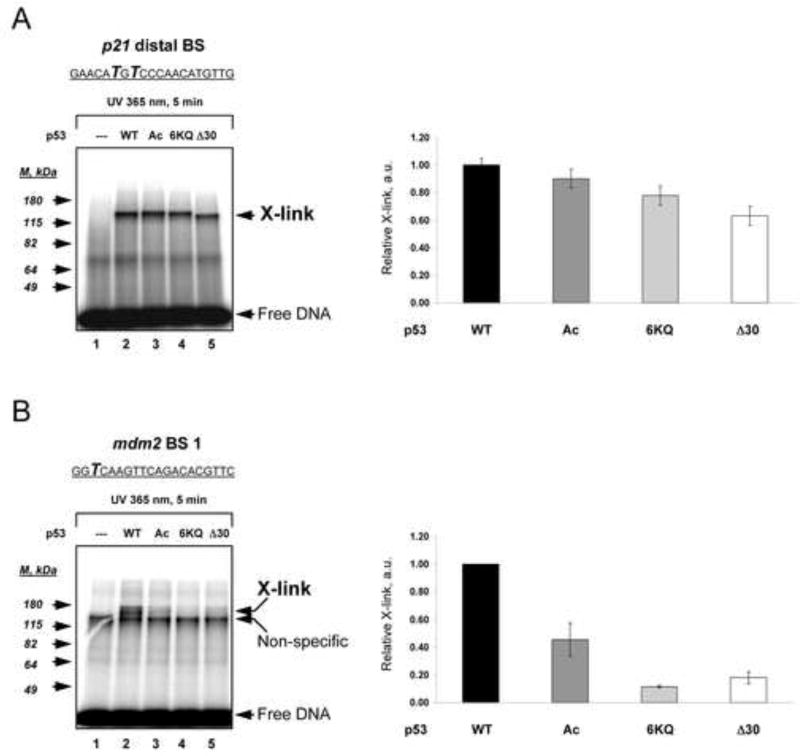
A function of the CTD might be to allow the central DBD to make stable base-specific complexes with weaker and divergent DNA binding sites. To test this, we employed UV-induced DNA photo cross-linking (Temiakov et al., 2003) with DNA templates that carried the photo cross-linkable UTP analogue (4-thio-dTMP) within either the p21 distal binding site or mdm2 binding site 1 (Figure 5A and B top of panels). . Efficient cross-linking requires close proximity between interacting molecules (equal to or less than 3 Å). Control experiments performed in the absence of p53 (Figure 5A and B lanes 1) or in the presence of temperature-inactivated WT p53 (data not shown) ensured the specificity of the cross-linked p53-DNA complexes. Relatively small differences were observed in the abilities of the WT and C-terminally modified p53 proteins to form cross-links with 4-thio-dTMP within the p21 binding site, with Δ30 p53 protein being the least efficiently cross-linked (~1.5 fold less than with WT p53; Figure 5A). By contrast, there was a marked reduction in cross-linked complexes especially with the 6KQ and Δ30 p53 variants when the low affinity mdm2 DNA template was used, where the relative cross-link yield for Δ30 and 6KQ p53 proteins were lower than WT p53 by factors of 6 and 10, respectively (Figure 5B). These results provided the impetus to further elucidate the mode by which the CTD regulates specific interactions of the DBD with DNA.
Figure 5. C-Terminally Deleted or Modified p53 Variants can be Cross-Linked to the p21 more efficiently than the mdm2 Binding Site.
WT, p300-acetylated p53 (Ac), 6KQ and Δ30 p53 proteins were bound to 32P end-labeled DNA fragments containing either p21 (A) or mdm2 (B) binding sites in which the UV cross-linkable nucleotide analogue 4-thio-dTTP was present at the indicated positions (shown in bold italics within the corresponding BS sequences). Following DNA-p53 complex formation, the reaction mixtures were subjected to UV-irradiation and then separated by 8% SDS PAGE. p53-DNA cross-linked products (X-link) were visualized using a PhosphorImager. Each panel shows a portion of the gel image (at left) and the corresponding graphical analyses (at right) of the relative intensities of each cross-link. The standard deviations were calculated from three parallel experiments.
The CTD Modulates Structural Changes Within the p53 Central DNA Binding Domain that are Important for Cooperative and Stable Binding of p53 to its Cognate DNA
The CTD could regulate specific p53-DNA complexes by two alternative but not mutually exclusive possibilities. First, the positively charged lysine residues within the CTD provide multiple non-specific p53-DNA interactions that stabilize the binary complex. In this model, the tetrameric CTD would function simply as an anchor set on each side of the corresponding binding site in order to provide extra stabilization. The second scenario posits that the CTD is necessary for conformational changes within the p53 tetramer upon binding to the p53 site, i.e. the induced fit mechanism (Spolar and Record, 1994).
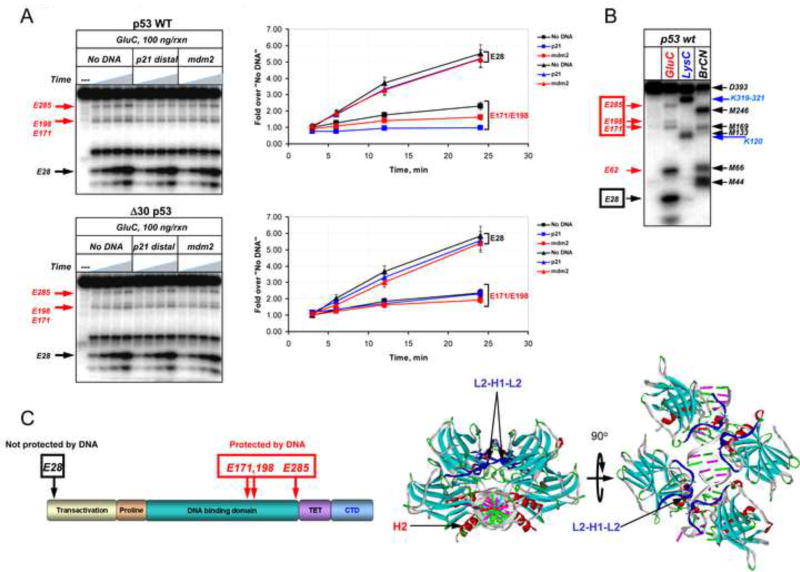
Evidence for the second possibility was provided when the C-terminal variants were subjected to residue -specific limited proteolysis in the presence or absence of DNA (Figure 6 and Figure S6). GluC (Staphylococcus aureus Protease V8) cleaves peptide bonds C-terminal to glutamic acid (glu) residues and 30 glu residues are well distributed throughout the p53 polypeptide, the majority of which are expected to be protected from cleavage due to structural constraints. We used recombinant p53 proteins N-terminally labeled with 32P by PKA to determine GluC cleavage patterns in the presence or absence of DNA spanning either the p21 or mdm2 binding sites. WT p53 produced two sets of cleavage products: low molecular weight (LMW) fragments whose accumulation in the presence of DNA was not detectably altered (e.g. E28) and high MW (HMW) fragments whose accumulation was significantly inhibited (e.g. E171/E198 and E285) by the presence of specific DNA sequences (Figure 6A, upper gel image and corresponding graph). These cleavage products were identified by a combination of chemical and enzymatic mapping experiments using p53 proteins radioactively labeled with 32P at either their N- or C-termini (mapping of the N-terminal domain-labeled p53 is shown in Figure 6 panel B and summarized in panel C). Accumulation of the LMW products (E28, E62) was very rapid, most likely due to relatively high exposure of the cleavage sites within the unstructured p53 N-terminal domain (NTD). On the other hand, site-specific degradation within the p53 DBD (E171/E198, E285) in the absence of DNA was significant but relatively, probably due to the compact structure of the DBD and relative inaccessibility of its glu residues. WT p53 demonstrated a markedly specific protective effect of DNA on the accumulation of HMW but not LMW cleavage products. Notably, the degree of protection from GluC cleavage correlated with the affinity (or degree of divergence from consensus sequence) of the corresponding DNA, such that p21 DNA was more efficient than mdm2 DNA in protecting the p53 DBD from cleavage with GluC. In striking contrast, we did not detect any significant DNA-dependent protection within the E171/E198 or E285 sites by Δ30 p53 (Figure 6A, lower image and corresponding graph) or by the CTD-mutated (6KQ) or acetylated WT p53 (Figure S6). Accumulation of the LMW degradation products was not affected by the presence of DNA regardless of the p53 CTD status.
Figure 6. The p53 CTD is Important for DNA-Induced Conformational Changes within the Central Specific DNA Binding Domain.
(A). WT or Δ30 p53 proteins N-terminally labeled with 32P were incubated in the presence or absence of the indicated DNA fragments and then subjected to limited proteolysis with GluC endopeptidase for 3, 6, 12 or 24 min. The labeled cleavage products were separated by 10–20% gradient SDS TG PAGE and visualized using a Phosphorimager (left panels). The intensity of the indicated specific cleavage products of p53 was quantified using ImageQuant 5.2 (graphs on the right). (B). GluC cleavage products were identified by chemical and enzymatic mapping experiment whose details are described in Experimental Procedures using GluC, or LysC, a highly specific endopeptidase that cleaves predominantly at Lys-X; BrCN, a chemical protease that cleaves at Met-X where X is any amino acid. (C) Left: Position of the GluC cleavage sites in (A) within the full length p53 are schematically summarized in the left panel and are shown on the p53 structure (PDB 2AC0) on the right. See also Figure S6.
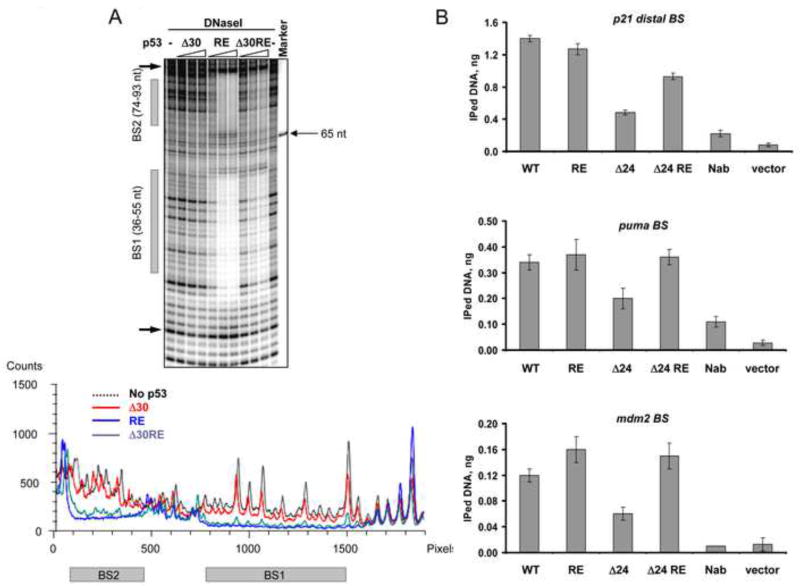
While E285 is localized within the H2 helix and could be directly protected by bound DNA (via R280-DNA interaction), the E171/E198 doublet band falls within the L2-H1-L2 region of the p53 DBD (Cho et al., 1994). Recent elegant structural and functional studies have linked this region to cooperative interactions of DBDs within the p53 tetramer and ensuing p53-DNA complex stability (Kitayner et al., 2006; Schlereth et al., 2010; Schlereth et al., 2013). Within that region a “super-stable” mutant p53 (E180R/R181E, RE) increases cooperative interactions between p53 DBDs and greatly stabilizes p53-DNA complexes (Schlereth et al., 2010). We generated a derivative of Δ30p53 (Δ30/180R/181E, referred to as Δ30RE here and in Figure 7 and Figure S7) and compared the relative abilities of Δ30 and Δ30RE p53 proteins to bind to the mdm2 (Figure 7A) or puma (Figure S7A) p53 binding sites by DNaseI footprinting as well as by DNA competition EMSA (Figure S7B) experiments. Δ30RE p53 displayed dramatically improved protection of both mdm2 and puma p53 binding sites from DNase I cleavage as compared to Δ30, at least as efficient as typically obtained with WT p53 (compare with the DNase I footprint data in Figure 3A). This improved binding of Δ30RE p53 to weak mdm2 or relatively weak puma sites was not accompanied by the appearance of DNaseI hypersensitive regions on both sides of the protected BS seen with WT and RE p53 proteins, suggesting that the CTD might be important for binding-accompanied structural changes within the DNA. We derived apparent Kd values for several p53 CTD mutants on different p53 binding sequences which are listed in Table S1. The results of the EMS A competition experiment also demonstrated dramatically higher stability of Δ30RE complexes when compared to Δ30p53 complexes assembled on mdm2 DNA (Figure S7, panel B).
Figure 7. The E180R/R181E (RE) Double Mutation within the Central DNA Binding Domain Restores Interactions of CTD-Deleted p53 with Low Affinity Binding Sites in vitro and in vivo.
(A). DNaseI footprinting analysis was performed on 32P-labeled mdm2 DNA (non-template strand labeled) with either Δ30 p53, full length RE or Δ30 RE p53 proteins. On top is shown a portion of the PhosphorImager scan of the sequencing gel around the indicated p53 binding sites and the corresponding densitometry analysis specific to each p53 variant indicated by different colors at their maximal concentration is shown below. (B). Empty vector or constructs expressing full length WT, full length RE, Δ24 and Δ24 RE p53 proteins under control of the endogenous p53 promoter as indicated were transfected into H1299 cells. 24 h post- transfection the cells were subjected to the ChIP protocol (see Experimental Procedures). p53-DNA complexes were immunoprecipitated with MAbs 1801 and DO.1. p53 binding to its sites within the p21, puma and mdm2 promoters was evaluated by real time quantitative PCR and expressed as amount of immunoprecipitated DNA. See also Figure S7.
To ask if the RE mutation may rescue binding by CTD-deleted p53 in vivo, we took advantage of constructs generated by Hamard et al (Hamard et al., 2012) where the relatively weaker Tp53 gene promoter drives the expression of p53 proteins. In that study the relative ability of WT p53 and p53 in which the last 24 amino acids were deleted (creating Δ24 p53) were compared. It is highly likely that Δ30 p53 and Δ24 p53 are phenotypically equivalent since both lack the full set of C-terminal lysines and previous studies examining either Δ30 p53 (McKinney et al., 2004; Kim et al., 2012) or Δ24 p53 (Hamard et al., 2012) provided virtually identical results i.e. that lack of either 24 or 30 C-terminal amino acids significantly impairs p53 binding and transactivation ability. Here we compared the relative abilities of either WT p53, WT p53 with the RE mutation (WT RE), Δ24 p53 or Δ24 RE p53 to bind to several target sites in vivo by chromatin immunoprecipitation (Figure 7B and Figure S7C Note that the extents of WT p53 binding as measured by quantification of the amount of immunoprecipitated DNA under these conditions approximated the results of our in vitro experiments (p21>puma>mdm). In the context of full length p53, with the exception of binding to the mdm2 promoter where a modest but reproducible increase in binding was observed, the RE mutation did not affect binding to any of the other sites assayed. By contrast, loss of the CTD drastically impaired binding to several sites as previously shown. Possible explanations for why CTD deletion impacted binding to both the strong and weak sites are provided in the Discussion. Importantly, however, in each case binding was rescued when Δ24 p53 also harbored the RE mutation. This strongly supports the conclusions of our experiments performed in vitro showing that the p53 CTD regulates specific DNA binding by the core domain via stabilizing cooperative interactions between individual DBDs within the tetramer.
DISCUSSION
By combining in vivo and in vitro experiments with biochemical and computational approaches we made unexpected observations on the role of the p53 CTD in regulation of sequence specific DNA binding. The ChIP-on-chip analysis provided both qualitative differences between subsets of binding sites preferentially recognized by each p53 CTD variant in cells and the relative degree of negative impact of the CTD truncation or 6KR and 6KQ mutants on DNA sequence-specific p53 binding. Our results suggest that binding of p53 to a significant number of sites within the genome might be greatly dependent on the availability of the unmodified CTD. Interestingly, the total number of sites bound by WT p53 upon treatment with Nutlin-3 is six times greater than upon treatment with the DNA damaging drug doxorubicin (Menendez et al., 2013) and, unlike Nutlin-3, doxorubicin (as well as many other p53-activating agents) is known to induce significant acetylation of p53 C-terminal lysines (Ito et al., 2001; Sakaguchi et al., 1998). Damage-induced modifications within the p53 CTD may be partially responsible for “filtering” of the binding process and the relatively lower number of p53 bound sites upon DNA damage inducing stess signals. Given the complexity of different p53 CTD modfications, deconstructing this will be a challenge to be met in future studies.
Cooperativity between individual DBDs within the p53 tetramer depends on E180 and R181 within the H1 helix and is required for the stability of p53-DNA complexes as shown by recent studies (Kitayner et al., 2006; Schlereth et al., 2010; Schlereth et al., 2013). Relevantly, it was reported that cooperativity-defective p53 mutants bind only to high-affinity consensus-like sequences, e.g. the p21 distal binding site (Schlereth et al., 2010; Schlereth et al., 2013). Binding of p53 to relatively rigid or highly divergent sequences in this regard could especially benefit from contributions of the CTD that we have identified herein. Having the core RE mutation along with the deletion of the CTD reversed the binding defect caused by lack of the CTD in vitro as well as in vivo.. But in cells loss of the CTD abrogated binding to both a strong (p21 distal ) or weak (mdm2 or puma) sites and in each case binding was recovered with the RE mutation. There are two possible and not mutually exclusive explanations for the lack of discrimination between p21 and other sites in vivo. First, while binding as measured by DNaseI footprinting in vitro of CTD-altered p53 was only detectable with the strong p21 site (in contrast to weak sites) it was still less than seen with WT p53, and even then binding to p21 by the altered CTD proteins as assessed by Exo III footprinting was undetectable. Therefore, intrinsic differences in the stability of binary complexes formed by such p53 variants may become more obvious when analyzed by techniques other than DNaseI footprinting. Second, in cells there is a nucleosome present at the p21 distal site while the mdm2 binding site region is relatively nucleosome-free (Laptenko et al, 2011). Since we observed that CTD-altered p53 variants are somewhat impaired in binding to the p21 site in the context of nucleosomal DNA in vitro (data not shown) this could even out the differences seen with purified proteins when we measured binding in vivo. Binding-associated structural changes within sequence-specific transcription factors have been documented and thought to be an important feature of specific interactions with DNA (Frankel and Kim, 1991; Spolar and Record, 1994). They represent the central concept of the induced fit theory. But only recently has this phenomenon been shown to accompany p53 binding to its cognate DNA, where the major conformational change within the DBD was thought to involve the L1 loop which, depending on the presence of specific DNA, could adapt either extended or recessed conformations (Petty et al., 2011). All published p53 structures to date, with the exception of Okorokov et al (Okorokov et al., 2006) who used cryo-EM, lack information on the position of the CTD and its possible inter-domain contacts in the context of a p53 oligomer. This is due to the physical absence of the unstructured domains within the p53 constructs used for crystallization. Therefore, fitting our biochemical data into currently existing structural models of the p53-DNA binary complex is virtually impossible. Nevertheless, to a first approximation, our data are in agreement with the model proposed by Fersht and colleagues (Tidow et al., 2007). For example our DNaseI footprint performed on the mutated p21 binding site (Figure 3D, left panel) clearly shows partial protection over the mutated part of the binding site as well as significant hypersensitivity signals flanking the binding site 20-mer and not a 10-mer. This is in accord with crystal structures that show all four DBDs in the p53 tetramer exist within the same plane, making contacts with the corresponding quarter-sites of the response element. X-ray crystal structures also indicate L2-H1-L2 mediated interactions between the DBDs when bound to the DNA, which has been confirmed by a number of studies. Regardless, the model proposed by the Fersht group cannot explain cross-talk between the CTD and the L2-H1-L2 region within the DBD.
The specificity of the interaction between any given sequence-specific transcription factor and DNA is determined by the combined rates of their association and complex dissociation. Non-specific molecular interactions (that constitute the majority of the intracellular encounters) are characterized by extremely short half-lives, and are therefore not productive. Specific complexes are relatively stable with half-lives that range from seconds to minutes as measured by photo-bleaching experiments (Hager et al., 2009; Larson, 2011). Substantial increases in complex half-life would allow more time for binding of the many transcriptional co-factors known to associate with p53 and the subsequent changes within chromatin that ultimately lead to transcription activation or repression. Our data imply that the unstructured positively charged C-terminal domain of p53 increases the stability of p53-DNA complexes and in doing so represents a newly discovered mechanism for specific and dynamic regulation of transcription by p53. Extensive site-specific modifications of the C-terminal lysine residues such as acetylation may lead to destabilization of p53-DNA complexes and/or binding discrimination. We propose that acetylation of the p53 CTD does not necessarily promote DNA binding but facilitates one or more post-binding steps that depend on the initial stable specific p53 interaction with its binding sites. Alternatively, depending on the nature of the p53 activating stress, lysine residue- and time-specific acetylation marks may function as a “filter” against certain highly divergent p53 target sites. We finally speculate that extensive accumulation of acetylation marks within the CTD may signify the final stage in p53-dependent activation of transcription leading to complex destabilization and p53 dissociation from the target promoter. While this manuscript was in preparation, Wu et al described TAF1-dependent phosphorylation of Thr55 within the N-terminus of p53 in the late DNA damage response phase and subsequent p53 dissociation from the p21 promoter (Wu et al., 2014). Interestingly, these authors reported that both site-specific phosphorylation and p53 promoter dissociation require acetylation of the CTD lysine residues.
EXPERIMENTAL PROCEDURES
ChIP-on-chip Experiments
Cell lines
H1299 cells expressing tetracycline regulated (“tet-off”) wild-type p53 and Δ30 p53 (Δ364–393) were previously described (Chen et al., 1996; McKinney et al., 2004). Mammalian expression constructs in the pTRE2 backbone (Life Technologies) expressing p53 6KR (K370R/K372R/K373R/K381R/K382R/K386R), p53 6KQ (K370Q/K372Q/K373Q/K381Q/K382Q/K386Q) were generated using the QuikChange protocol (Agilent Technologies) according to the manufacturer’s specifications. H1299-derived inducible cell lines expressing p53 6KR and p53 6KQ were created using a two-step tetracycline-regulated system and clonally selected with 400 µg/ml Hygromycin B (Invitrogen) as previously described (Chen et al., 1996).
ChIP-on chip
ChIP-on-chip experiments were performed essentially as described (Huggins et al., 2011). Details on data analysis and bioinformatics used in the study are given in Supplemental information.
p53 Proteins
All mutations within p53 were introduced with a QuikChange II site-directed mutagenesis kit (Agilent Technologies). All DNA constructs were verified by sequencing (GeneWiz, USA). Subsequent baculoviruses were generated with Bac-to-Bac system (Life Technologies). Unmodified and p300 acetylated N-terminally Flag-PKA-double-tagged wild-type and the CTD mutant p53 proteins and N-terminally Flag- C-terminally-PKA-double-tagged wild type p53 protein were affinity purified from recombinant baculovirus-infected Sf9 insect cells as described (Laptenko et al., 2011; Piluso et al., 2005).
Antibodies
In-house produced MAb DO.1 and MAb 1801 mouse monoclonal antibodies were used for detection of total p53 and corresponding ChIP experiments. To confirm site-specific acetylation of p53 by p300, we used Acetylated-Lysine Polyclonal Ab (Cell Signaling Technology, USA), Anti-Acetyl K373 p53 Ab and Anti-Acetyl K373/K382 p53 Ab (EMD Millipore, USA).
DNA Binding Assays
Electrophoretic Mobility Shift Assay (EMSA)
EMS A with 32P-labeled DNA was performed as previously described (Laptenko et al., 2011). In a modified version of the EMSA assay shown in Figure 4, the initial reaction mixture contained 32P-labeled Δ30 recombinant affinity purified p53 in the complex with the unlabeled mdm2 171 bp long DNA fragments. Increasing amounts of the indicated unlabeled p53 variants were used for competing off the 32P-labeled Δ30 p53.
ELEX (Systematic Evolution of Ligands by Exponential Enrichment)
Details on the initial library design, preparation, per-round procedure, PCR amplifications and analysis by sequence-specific endonucleases can be found in Supplemental information.
DNase I footprinting and Exonuclease III cleavage experiments
Sequences of the DNA templates used in nuclease footrpinting experiments can be found in Supplemental information.
DNaseI Footprinting
The DNaseI footprinting was done essentially as described (Laptenko et al., 2011). The primary data for calculation of the corresponding Kd values were obtained from DNaseI footprinting experiments as described (Trauger and Dervan, 2001). They were fitted by a nonlinear least-squares fitting procedure (Sigma Plot 11.0, Systat Software, Inc).
Exonuclease III Footprintin
The reaction set-up was as described in the DNaseI footprinting protocol. 5 or 10 units of Exonuclease III (NEB, USA) were added and the reaction was allowed to proceed for 90 sec at room temperature. All subsequent steps were as described for the DNaseI assays.
UV Cross-linking with 4-Thio-TTP-containing DNA
DNA templates with UV crosslinkable 4-Thio-TTP (TriLink Biotechnologies, USA) specifically positioned within either p21 distal or mdm2 BS1 p53 binding sites were prepared by primer extension according to (Temiakov et al., 2003) under conditions of very dim light. The DNA templates were column purified, 32P-radiolabeled and re-purified using Qiagen MiniElute PCR purification Kit (Qiagen, Germany). Purified wild-type or CTD mutant p53 proteins were mixed with labeled UV cross-linkable p21 or mdm2 DNA and unlabeled mutant DNA competitor, immediately placed into iced water and UV irradiated at 365 nm for 5 min. UV irradiated samples were mixed with SDS-containing PAGE sample buffer and separated by 8% TG SDS PAGE. The gels were fixed, dried, autoradiographed and quantified using a Molecular Dynamics Typhoon 9410 Variable Mode Imager.
GluC Proteolysis Experiments
N-terminally PKA-tagged p53 was radiolabeled with 32P and purified as described (Poyurovsky et al., 2010). Purified 32P-labeled p53 variants were incubated in 1X EMSA buffer supplemented with 0.4 mM spermidine in the presence or absence of the indicated DNA. GluC endopeptidase was added to the reaction for the indicated time periods. The reaction was stopped by adding SDS-containing PAGE sample buffer. The N-terminally labeled cleavage products were separated on a 10–20% gradient SDS TG PAAG and visualized by PhosphorImager.
In order to identify the GluC cleavage sites within p53, we subjected p53 proteins that were either N- or C-terminally radiolabeled to chemical cleavage at methionine residues by BrCN (Sigma-Aldrich, USA) under denaturing conditions according to a published protocol (Laptenko et al., 2003) or to proteolytic cleavage at Lys residues by Lys-C (Roche Applied Science, Switzerland) under native conditions.
Binding of p53 E180R/R181E Variants to Select p53 Targets in vivo: Chromatin Immunoprecipitation
The pDNAs expressing WT and Δ24 p53 under control of the endogenous p53 promoter were described (Hamard et al., 2012). The E180R/R181E (RE) double mutations were introduced into the corresponding cDNAs with a QuikChange II site-directed mutagenesis kit. All DNA constructs were verified by sequencing. Constructs expressing p53, p53 RE, Δ24 p53 and Δ24 p53 RE proteins were transiently transfected using Lipofectamine 2000 into H1299 cells. The cells were grown for 20 hours post-transfection and then treated with 10 µM Nutlin-3 for an additional 4 hours. Cross-linking, lysis, sonication, immunoprecipitation, DNA purification and qRT-PCR were performed as previously described (Laptenko et al., 2011). Equal amounts of each p53 variant (WT, RE, Δ24 or Δ24 RE) were determined and then used for each chromatin immunoprecipitation. Primer sequences are available upon request.
Supplementary Material
Acknowledgments
We thank members of the Prives laboratory for valuable comments and suggestions. We are particularly grateful to Dr. S. Borukhov (Rowan University, NJ) for valuable advice and stimulating discussions. This work was supported by NCI grant CA77742. The work in the lab of I. Simon was supported by the German-Israeli Foundation for Scientific Research and Development (grant 998/2008), the Israel Cancer Society, the Israel Cancer Research Foundation and the the Weinkselbaum Family Medical Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Anderson ME, Woelker B, Reed M, Wang P, Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol. Cell. Biol. 1997;17:6255–6264. doi: 10.1128/mcb.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Beno I, Rosenthal K, Levitine M, Shaulov L, Haran TE. Sequence-dependent cooperative binding of p53 to DNA targets and its relationship to the structural properties of the DNA targets. Nucleic Acids Res. 2011;39:1919–1932. doi: 10.1093/nar/gkq1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botcheva K, McCorkle SR, McCombie WR, Dunn JJ, Anderson CW. Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle. 2011;10:4237–4249. doi: 10.4161/cc.10.24.18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadillas JM, Tidow H, Freund SM, Rutherford TJ, Ang HC, Fersht AR. Solution structure of p53 core domain: structural basis for its instability. Proc. Natl. Acad. Sci. U S A. 2006;103:2109–2114. doi: 10.1073/pnas.0510941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Kim PS. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- Friedler A, Veprintsev DB, Freund SM, von Glos KI, Fersht AR. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure. 2005;13:629–636. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Frith MC, Saunders NF, Kobe B, Bailey T. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath SC. Methods developed for SELEX. Anal. Bioanal. Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- Gu J, Nie L, Wiederschain D, Yuan ZM. Identification of p53 Sequence Elements That Are Required for MDM2-Mediated Nuclear Export. Mol. Cell. Biol. 2001;21:8533–8546. doi: 10.1128/MCB.21.24.8533-8546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard PJ, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, Carvajal LA, Senturk E, Gillespie V, Aaronson SA, Merad M, Manfredi JJ. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard PJ, Lukin DJ, Manfredi JJ. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J. Biol. Chem. 2012;287:22397–22407. doi: 10.1074/jbc.M111.331298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc. Natl. Acad. Sci. U S A. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins P, Zhong S, Shiff I, Beckerman R, Laptenko O, Prives C, Schulz MH, Simon I, Bar-Joseph Z. DECOD: fast and accurate discriminative DNA motif finding. Bioinformatics. 2011;27:2361–2367. doi: 10.1093/bioinformatics/btr412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Lane DP. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 2001;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L, Prives C. Activation of p53 sequence-specific DNA bindingby short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim K, Choi J, Heo K, Baek HJ, Roeder RG, An W. p53 Requires an Intact C-Terminal Domain for DNA Binding and Transactivation. J. Mol. Biol. 2012;415:843–854. doi: 10.1016/j.jmb.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Beckerman R, Freulich E, Prives C. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc. Natl. Acad. Sci. U S A. 2011;108:10385–10390. doi: 10.1073/pnas.1105680108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Larson DR. What do expression dynamics tell us about the mechanism of transcription? Curr. Opin. Genet. Dev. 2011;21:591–599. doi: 10.1016/j.gde.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. C-Terminal Ubiquitination of p53 Contributes to Nuclear Export. Mol. Cell. Biol. 2001;21:8521–8532. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol. Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41:7286–7301. doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Appella E, Harrington RE. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J. Biol. Chem. 1997;272:14842–14849. doi: 10.1074/jbc.272.23.14842. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Roth JA, Mukhopadhyay T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Sasaki M, Maki CG. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J. Biol. Chem. 2007;282:14616–14625. doi: 10.1074/jbc.M610515200. [DOI] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, Selivanova G. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, Orlova EV. The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J. 2006;25:5191–5200. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty TJ, Emamzadah S, Costantino L, Petkova I, Stavridi ES, Saven JG, Vauthey E, Halazonetis TD. An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity. EMBO J. 2011;30:2167–2176. doi: 10.1038/emboj.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piluso LG, Wei G, Li AG, Liu X. Purification of acetyl-p53 using p300 co-infection and the baculovirus expression system. Protein Expr. Purif. 2005;40:370–378. doi: 10.1016/j.pep.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, Byeon IJ, Gabizon R, Mattia M, Zupnick A, Brown LM, Friedler A, Prives C. The C terminus of p53 binds the N-terminal domain of MDM2. Nat. Struct. Mol. Biol. 2010;17:982–989. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B (betabeta) Nat. Struct. Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vasilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth K, Beinoraviciute-Kellner R, Zeitlinger MK, Bretz AC, Sauer M, Charles JP, Vogiatzi F, Leich E, Samans B, Eilers M, Kisker C, Rosenwald A, Stiewe T. DNA binding cooperativity of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell. 2010;38:356–368. doi: 10.1016/j.molcel.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, Jarek M, Leich E, Rosenwald A, Stiewe T. Characterization of the p53 cistrome-DNA binding cooperativity dissects p53’s tumor suppressor functions. PLoS Genet. 2013;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonova I, Jaber S, Draskovic I, Bardot B, Fang M, Bouarich-Bourimi R, Lejour V, Charbonnier L, Soudais C, Bourdon JC, Huerre M, Londono-Vallejo A, Toledo F. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell Rep. 2013;27:2046–2058. doi: 10.1016/j.celrep.2013.05.028. [DOI] [PubMed] [Google Scholar]
- Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafvizi A, Huang F, Fersht AR, Mirny LA, van Oijen AM. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. U S A. 2011;108:563–568. doi: 10.1073/pnas.1016020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temiakov D, Anikin M, Ma K, Jiang M, McAllister WT. Probing the organization of transcription complexes using photoreactive 4-thio-substituted analogs of uracil and thymidine. Methods Enzymol. 2003;371:133–143. doi: 10.1016/S0076-6879(03)71009-X. [DOI] [PubMed] [Google Scholar]
- Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc. Natl. Acad. Sci. U S A. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauger JW, Dervan PB. Footprinting methods for analysis of pyrrole-imidazole polyamide/DNA complexes. Methods in Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wu Y, Lin JC, Piluso LG, Dhahbi JM, Bobadilla S, Spindler SR, Liu X. Phosphorylation of p53 by TAF1 Inactivates p53-Dependent Transcription in the DNA Damage Response. Mol. Cell. 2014;53:63–74. doi: 10.1016/j.molcel.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Johnston K, Clements A, Marmorstein R. Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution. J Biol Chem. 2001;276:12120–7. doi: 10.1074/jbc.M011644200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.