Synopsis (100 words or fewer)
The use of next-generation sequencing and multi-omic analysis reveals new insights on the identity of microbes in the lower airways blurring the lines between commensals and pathogens. Microbes are not found in isolation, rather they form complex meta-communities where microbe-host and microbe-microbe interactions play important roles on the host susceptibility to pathogens. Additionally, the lower airway microbiota exert significant effects on host immune tone. Thus, this review highlights the roles that microbes in the respiratory tract play in the development of pneumonia.
Keywords: lung, microbiome, antibiotics, immune responses, inflammation, bacterial taxa
Until recently, the purpose of studying microbes in pneumonia was the identification of an organism that could assume the role of “pathogen” in disease. Common findings, using culture techniques designed to isolate these possible pathogens, often identify these microbes as “confounders”. An example is the frequent identification of oral flora in lower airway samples from clinical cultures obtained in patients with pneumonia.1 This finding is frequently disregarded as contamination. However, with recent advances in sequencing techniques, new insights on the role of these oral flora are being discovered. Indeed, the lower airways of healthy individuals are not sterile but rather frequently visited by varying degrees of these microbes, predominantly from sources in the upper airways (Figure 1) Exposure of the lower airways to microbes commonly occurs among healthy individuals, such as microaspiration of oral secretions containing high concentrations of microbes or inhalation of airborne microbes (low biomass but constant exposure). In many airway diseases, epidemiological evidence suggests that some of these events occur more often in illness than in health. Examples include the association between gastroesophageal reflux (GERD) and microaspiration with chronic obstructive pulmonary disease (COPD), bronchiectasis, asthma, and cystic fibrosis.2–4 With the use of culture independent approaches to study the lower airway microbiota (the collection of microbes present in the lower airways) we have gained new insights about the complex microbial community that exist in the pulmonary environment. In this review, we highlight the existing evidence that supports a potentially critical role for the lower airway microbiota in patients with pneumonia as well as in chronic respiratory diseases with an increased prevalence of pneumonia.
Figure 1: Schema of the change in pathophysiological view of pneumonia in the era of culture independent approaches used to study microbial communities.
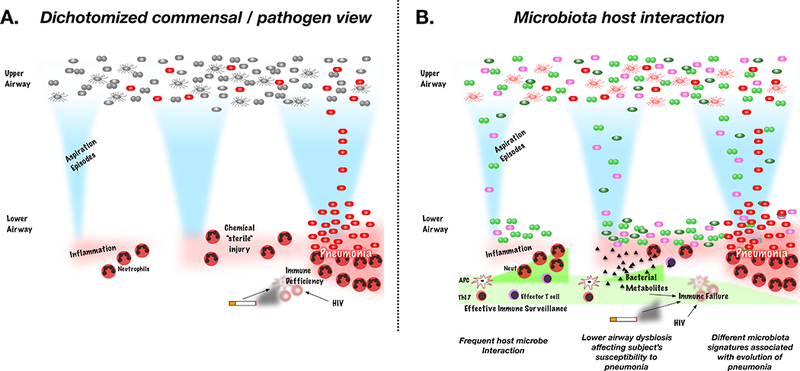
A. Previous conceptualization of pneumonia stratified microbes into commensals (grey bacteria) and pathogens (red bacteria). Identification of a commensal in the lower airways was deemed a contaminant. Aspiration episodes were recognized as cause of lower airway injury thought to be “sterile” chemical noxious stimuli. Aspiration of “pathogens” was the key event leading to the development of pneumonia. B. A more complex view of microbes in the upper airways where multiple different types of bacteria coexist. Aspiration events brings microbes to the lower airways that may be cleared by the host immune response or may persist leading to lower airway dysbiosis (bacterial seeding). Effector T cells, Th17 cells and antigen presenting cells (APC) will be determinant of the effectiveness of the immune surveillance. These host immune cell are likely affected by frequent interactions with microbes. Some of the bacterial products have significant effects on the inflammatory tone such as short chain fatty acids. The dynamics of the aspiration events, lower airway microbiota and lower airway immune tone will be determinant of the conditions that may favor the development of pneumonia.
Why should culture independent techniques be considered in the setting of pneumonia?
The paradigmatic view of microorganisms in pneumonia focuses on microbes pre-assigned to a pathogenic role. Typical pathogens associated with pneumonia include Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae, and Hemophilus influenzae, while atypical microbes include Chlamydophila psittaci, Mycoplasma pneumoniae, and Legionella pneumophilia (Figure 1A). However, culture based methods identify positive cultures in approximately half of patients with community acquired pneumonia (CAP).5,6 Rates of identification of microorganisms in hospital associated pneumonia (HAP) and Ventilator Associated Pneumonia (VAP) can also vary greatly.7–11 A major limitation of pathogen identification is related to difficulties growing microorganisms using culture-dependent techniques, such as Legionella and Mycobacterium that require specialized media and conditions.12,13 In addition, misidentification of pathogens may have significant effects on treatment, such as the selection of inappropriate antibiotics14–17 leading to increased morbidity and mortality.12,18,19 Moreover, subjects with culture-negative pneumonia may represent a different group of patients than those with positive culture pneumonia. In a study of patients with culture-negative pneumonia, subjects had lower mortality and less severity of illness than their culture positive counterparts.8 These findings suggest that culture-negative pneumonia may be a “milder” form of local and systemic injury. Another example of commonly considered culture-negative lung injury is aspiration pneumonitis. In subjects that suffer aspiration, the dogma has been that the nature of the lung injury present in this condition is related to “sterile” chemical injury (Figure 1A), despite the large number of bacteria present in the upper airways and the upper gastrointestinal tract. Thus, therapeutic recommendations do not include the use of antibiotics except for: a) presence of poor dentition, b) alcohol use, and c) evidence of abscess on chest imaging. The vast majority of microbes responsible for pneumonia come from the upper airway. Thus, periodic exposure of the lower airways to upper airway microorganisms represents an important seeding mechanism that may influence microbial selection in the lower airways. As an example of this selection pressure, S. pneumoniae, a minor component of both the upper and lower airway microbiomes, causes more than half of all cases of CAP (Figure 1A).
Among the cases of pneumonia with a pathogen identified by culture, institution of accurate antimicrobial therapy results in favorable clinical outcomes.18 Data from these cultures have shown frequent isolation of oral microorganisms in samples from the lower airways.1,20–23 However, the techniques used to sample the lower airways require passage through the upper airways and sterile surgical lung biopsies are often not feasible. Thus, contamination of samples with oral flora is commonly blamed for these results.24–26 Other factors that limit the use of culture-based techniques include: a) the time required to grow organisms; b) low bacterial burden in the lungs; c) difficulties growing fastidious bacteria; and d) the inability to describe complex microbial communities.27-29 New culture-independent techniques may hold the advantage of earlier identification without the need to grow microbes.30,31 Some of these techniques are not new and targeted culture-independent techniques have been used to identify specific microbes suspected of having a pathogenic role in the lung. Examples of this include: a) the screening for Streptococcus antigens in oral swabs, b) the search for Legionella antibodies in serum, and c) the identification of DNA from mycobacteria using PCR.32–34 These techniques are based on approaches biased toward a suspected agent and aim to provide an expedited diagnosis of a possible pathogenic microbe.
Newer sequencing technology takes advantage of the ability to identify multiple microbial products in a high-throughput approach and to process large amounts of data. This allows for the identification of microbes using an ‘unbiased’ approach. For example, a technique widely used in research is the amplification and sequencing of the 16S rRNA gene.35 The 16S rRNA gene, a constituent gene of the bacteria domain, contains genomic signatures (defined by hypervariable regions), and allows for specific taxonomic identification and description of complex mixtures of microbes. In addition, it provides semi-quantitative data about each microbe present in the sample expressed as relative abundance. The presence of microbial DNA of an organism with a potential pathogenic role, especially if present in high relative abundance, can be seen as supporting a causative role in the correct clinical context.30 Culture-independent methods, including next-generation sequencing coupled to microbial reference databases, represent a powerful new technology that may have significant clinical impact on the identification of microbes in pneumonia.
The use of high throughput approaches provides a view of the intricate landscape of microbes present in the lower airways without an a priori bias towards specific pathogen identification. This is changing our understanding of the complex mixture of microbes in the lower airways and poses new scientific dilemmas to consider for the pathogenesis of pneumonia: a) How does a microbe become a pathogen? b) What are the main sources of microbes into the lower airways? c) How does host and microbe interaction affect the immunological tone of the lower airways? d) How does upper and lower airway dysbiosis increase susceptibility to pneumonia? and e) How do distinct microbiota signatures in pneumonia affect the natural history of this disease?
How does a microbe become a pathogen in the new era of the lung microbiome?
Classifying microbes as commensals or pathogens has been the foundation of a dichotomized view of infection (Figure 1A). In recent years, an evolution from this view occurred suggesting that pathogenicity and commensalism may fall across a spectrum based on host-microbial interactions. Several tenets describe the pathogenic role of microbes: 1) pathogenesis is the result of both host and microbe; 2) the pathological or clinical outcome is determined by the damage to the host; and 3) this damage can result from the host immune response and/or the effects of virulence factors from the microbe.36,37 Therefore, microorganisms frequently considered commensals can have a major role in the pathogenesis of pneumonia. For example, Staphylococcus epidermidis is a common inhabitant of the upper airways, but can cause disease under certain conditions.37,38 Complex interactions between microorganisms and the host are determined by multiple factors, likely non-canonical, that will define the pathogenic role, a clear evolution from the classical Koch’s postulates.39
Our current understanding of the lung microbiome has contributed to the complexity of the host-microbe interactions, introducing another factor in the debate: what is the role of the complex microbial community that exists in the lower airway? Many studies of the lung microbiota now show that we can frequently find multiple species of oral commensals in the lower airways and the implications for the host may further obscure the distinction between pathogens and commensals.40 The presence of these bacteria frequently regarded as commensals from the oral cavity in the lower airways impacts how other co-occurring microbes respond to the environment and host.41 In the schematic in Figure 1B, the complex community of microbes in the lower airway may increase or hinder a microorganism’s ability to cause infection. Microorganisms that regularly inhabit certain mucosae (e.g. oral) may contribute to the pathogenic process by inducing inflammation when found in other sites (e.g. lung mucosa).
The presence of oral commensals in the lung microbiome allows us to ask: what is the role of these microorganisms in the lower airways? Do these microbes affect other microbes, especially those classically identified as the responsible pathogen? Figure 1B depicts a more complex cross pollination of microbes between the upper and lower airway leading to dysbiosis of the lower airway microbiota and affecting not only microbial-host interaction but also microbe-microbe interaction, both of which may contribute to the pathogenic process. It is reasonable to postulate, that in the healthy lower airway microbiota, some microbes outcompete others with greater pathogenic potential. This could be due to different factors such as sequestering vital nutrients and byproducts necessary for growth and promoting the host immune defense to enhance recognition and killing of pathogens. Alternatively, some of these host immune mechanisms may be impaired due to lower airway dysbiosis, increasing an individual’s susceptibility to pneumonia.
What are the sources of microbes to the lower airways?
Culture data in subjects with acute lung infections and chronic airway inflammatory conditions, such as COPD and cystic fibrosis, have shown that the upper airway is the most common contributor of microbes to the lower airways.42–45 Culture independent data also suggest that ‘microaspiration’ is frequently observed in normal subjects, leading to episodic seeding of oral microbes into the lower airways, 2,3,46 and with a higher prevalence in several lung diseases including COPD, asthma, obstructive sleep apnea, cystic fibrosis, and lung infections.3,4,47–50 Microaspiration occurs more frequently while sleeping due to reduced coordination of breathing with swallowing and GERD.2–4,47 Several chronic pulmonary diseases are characterized by impairment of airway clearance, such as in COPD and cystic fibrosis, which likely favors the seeding of micro-aspirated organisms.51,52 Environmental exposures, frequent antibiotic and/or anti-inflammatory use, or diet may also contribute to the selection pressure on the lower airway microbiome.53 The current understanding of the dynamics that determine the lung microbiota are best explained by an adapted island model and complex adaptive lung ecosystem, processes present in both health and disease.40 We now know that when the airway microbiota is characterized topographically, the greatest similarity with the upper airway microbiota occurs in areas with the greatest potential for deposition of microaspiration (e.g. carina and main stem bronchi and alveolar spaces), evidence that supports that the main route of enrichment for the lower airways remains microaspiration of oropharyngeal secretions.40 The rate of elimination of the aspirated microorganisms will depend on the environment present in the lower airways (e.g. protein and nutrients available, pH, oxygen tension, biofilms, etc.) and active immune clearance.
The role of host-microbe interaction in the mucosal immunological tone
Humans evolved to co-exist with microorganisms. Since the discovery of single celled organisms by Antoni van Leeuwenhoek, multiple mucosae within the human body were found to be colonized with microorganisms. However, the lungs were believed to be sterile despite being in direct communication with other mucosae with very high bacterial burden.54 In the past ten years, with the utilization of culture independent techniques, we have identified a complex community of microorganisms on multiple mucosal surfaces that coexist in the body.55 Indeed, the sum of these microbes that inhabit our bodies can be considered a subject-specific superorganism that carries genetic information more diverse than our own human DNA.55–58
In mucosal surfaces other than the lungs, examples of the co-evolution of microbes and host include the intricate functions performed by microbes that are needed for immune modulation59,60 and immune maturation and host homeostasis.61–65 We now know that there are microbes in the lower airways of humans40 and experimental models,66 challenging the anachronistic dogma that the lower airways are sterile. 16S rRNA gene sequencing techniques revealed complex microbial communities in the lower airways associated with distinct host immune tone. The distinct immunological homeostasis of the lung mucosa may be from either viable and metabolically active bacteria or from exposure to bacterial by-products.67
Multiple studies demonstrated that the bronchoalveolar lavage (BAL) and lung tissue of healthy subjects and smokers frequently contain an enrichment with bacteria commonly considered oral ommensals.40,68–70 The enrichment of the lower airway microbiota with oral commensals, such as Prevotella, Streptococcus, Fusobacterium, Rothia, and Veillonella is associated with sub-clinical inflammation.40,70 The inflammatory signal is characterized by an increase in neutrophils and lymphocytes. Further endotyping of the lower airway inflammatory tone supports that exposure to these microbes is associated with a Th17 phenotype, characterized by increase in CD4+ IL-17+ lymphocytes, increased STAT3 expression, Fractalkine, and IL-1α.69,70 Again, it is unclear if the inflammatory signal is due to viable and metabolically active bacteria, dead bacteria, or by-products of bacterial metabolism.67 It is also likely that microorganisms shape our immune system as much as our immune system shapes our microbiome. Studies done in large European cohorts show that the exposure to diverse microbes during childhood, such as growing up on a farm, is protective against asthma and allergies.71–73 House dust mite exposure from households with large canines attenuate Th2 cytokine production, decrease activated T-cells, and leads to an enrichment with Lactobacillus johnsonii in the nasal microbiome.74 This observation is coincident with gut microbiota data where early exposure to bacteria is needed for immune maturation in early life.65,75 This is commonly referred to as the “hygiene hypothesis”, under which restricted microbial exposure in early life may lead to inadequate “priming” of the immune system during maturation resulting in Th1/Th2 cell subset imbalances,76 Treg cell deficiency,77 and innate immune abnormalities.78
Changes in diet, improved sanitary conditions, and use of antibiotics may limit the exposure to environmental microbes and be responsible for the increase in autoimmune diseases observed in recent decades.71,79–84 In childhood asthma, two observations about the microbial exposure in early life highlight the importance of the microbiome in the development of the immune system. First, childhood exposure to a diverse microbial environment, either by farm habitation or pet exposure is protective and reduces the risk of asthma.71,85 Second, the acquisition of airway microbiota enrichment with pathogenic microorganisms (e.g. S. pneumoniae, M. catarrhalis, H. influenzae) in infancy increases susceptibility to asthma.86 More recently, nasal carriage with Streptococcus was found to be a strong asthma predictor.33 Importantly, while the pro-inflammatory role pathogenic bacteria such as S. pneumoniae, M. catarrhalis, and H. influenzae is well defined, less is known about “healthy” microbial exposure responsible for an anti-inflammatory role suggested by the “hygiene hypothesis”. In mouse models, nasal inhalation of an innocuous strain of Escherichia coli leads to re-programming dendritic cells and macrophages in the lungs and results in protection against allergic responses.87 This model suggests that direct exposure of the airways to certain bacteria is sufficient to elicit a protective effect. In addition, gastrointestinal microbiota trigger immunological cross-talk between the gut and lung.74 For example, children colonized in the stomach with H. pylori are 40% to 60% less likely to develop asthma than children who are not carriers.88,89 Lessons from animal models show that disruption of the gastrointestinal microbiota may lead to abnormal immune responses that affect the airway mucosa.90–94 Ultimately, both the gut and lung mucosa may function as a single aerodigestive immune system and share the physiological role of immune surveillance that shape the host immune tone locally and systemically.
How does upper and lower airway dysbiosis increase susceptibility to pneumonia?
The upper airways are a microbial reservoir and the main source of microbes to the lower airways. It is not unexpected that the composition of the upper airway microbiota has direct effects on an individual’s risks for pneumonia. Recent data using culture independent approaches suggest that reduction in nasal microbiome diversity and domination by Rothia, Lactobacillus, and Streptococcus increased the risk of pneumonia.95 Among neonates, nasal colonization with Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus occurs frequently.96 Importantly, enrichment of the nasal microbiota with Moraxella, Streptococcus, and Haemophilus was associated with an increase of acute respiratory infections.33
The composition of the lower airway microbiota may also affect a subjects’ susceptibility to pneumonia. The lung microbiome of advanced HIV subjects show dysbiosis with increasing Prevotella and Veillonella that persists for years despite treatment.97 These treatment naïve participants also have decreased diversity and greater inter-sample diversity than those subjects uninfected by HIV. Thus, this dysbiotic signature may be associated with increased pneumonia susceptibility seen in HIV patients.98 The lower airway microbiota may also affect a subject’s susceptibility to pneumonia through immunological regulation mediated by bacterially derived metabolites. For example, short chain fatty acids (SCFAs), an end-product of bacterial anaerobic metabolism, is associated with an increase of an individual’s susceptibility to tuberculosis in an HIV cohort.99 One possible mechanism is that SCFAs, such as butyrate, have direct inhibitory effects on T cell function by suppressing INF-γ and IL-17 production.99
In cystic fibrosis and COPD, decrease in α diversity (a measure of within sample diversity or how many different types of taxa are in a sample) of the lower airway microbiota is associated the severity of disease.45 Considering that advanced-stage cystic fibrosis and COPD are associated with increased risk of developing pneumonia and that the prognosis of pneumonia is worse in these conditions than in healthier population,100,101 it is possible that changes to the lung microbiota may impact the natural course of pneumonia in these diseases.102,103 It is also possible that the associated changes to the lung microbiome may assist to evaluate the prognosis in these patients. Those patients who have a lower α diversity may have worse outcomes and an accelerated declination in their disease.
The changes to the upper or lower airway microbiome may modulate the immune response increasing host susceptibility to the development of pneumonia (see Figure 1B). For example, the presence of anaerobic taxa in the nasal microbiome was correlated with increased nasal IgA against the influenzae virus in the nasal microbiome after inoculation with live-attenuated influenza vaccine. Prevotella melaninogenica positively correlated with increased influenzae-specific IgA antibodies.104 In a cohort of asthmatic subjects with clinically stable but sub-optimally controlled asthma, bronchial hyper-responsiveness was associated with increased bacterial burden and microbial diversity in airway brush samples. Perturbations of the commensal microbial community may influence the clinical phenotype in asthma and highlights the potential “pathogenic” role of commensal bacteria possibly resulting in increased risk for lower airway infections.
In advanced COPD, increased bacterial colonization and recurrent infections are associated with increased risk of exacerbations and accelerated loss of lung function.105 In moderate to advanced COPD, there is reduced bacterial diversity as compared with healthy or mild COPD.106 Exacerbations often occur after infection with a new bacterial strain or change in bacterial load107 and dysbiosis of the microbiome has been associated with increased inflammation.108 In severe COPD, exacerbations requiring mechanical ventilation, there is a diverse bacterial community suggesting a poly-microbia cause.109 This highlights the potential for ecological interaction of different bacterial strains during exacerbations. The core of this bacterial community may be comprised of previously unrecognized lung pathogens such as oropharyngeal bacterial species that are part of the lung microbiome during health, but in periods of dysbiosis with microbe-microbe interaction may result in increased frequency of COPD exacerbations and risk of pneumonia (Figure 1B).
In cystic fibrosis, reduction in bacterial diversity is associated with disease progression and colonization with pathogens.45 Low microbiota diversity also precedes the development of cystic fibrosis exacerbations.42 It is likely that dynamic changes of airway microbiota occur over time, where a change from a “healthy” well-balanced poly-microbial microbiome to an “unhealthy” restricted, less-diverse airway microbiota renders the airway susceptible to a dominant pathogen (e.g. Pseudomonas or Burkholderia) and consequent lung injury.
The risk for developing HAP increases with the recent use of antibiotics.110 Differences in exposures, environments, fomites, colonization, host factors, host-microbe interactions, and hospital antibiotic nomograms influence patients’ susceptibility to pneumonia.110 It is plausible that some of the increased risk is due to the selection pressure by antibiotics, leading to upper/lower airway dysbiosis once a subject is admitted to a hospital and increases the chance for a “pathogen” to bloom. These selection pressures may affect healthy microbes in the upper and lower airways, interrupt immune surveillance, and encourage development of a lower airway microenvironment supportive for pathogens (Figure 1B). Host immune characteristics are obviously determinant of the selection pressure to the microbiota. In subjects with immunodeficiency due to HIV and no lung disease, the lung microbiome is enriched with Tropheryma whipplei as compared with controls.111 The increased relative abundance of this taxon in the lung, as compared with paired upper airway samples, suggests that the lung may constitute a true niche of T. whipplei. In addition, antiretroviral medication leads to changes in the lung microbiome with enrichment with Prevotella and Veillonella.97 The persistence of the dysbiosis despite the use of anti-retroviral medications may be responsible for the increased susceptibility to inflammatory lung diseases as well as to pneumonia among HIV subjects fully reconstituted with normal CD4 counts.
While we focused on changes observed in either the upper and lower airway microbiota that might be linked to increased risk for pneumonia, there are data suggesting that the microbiota of distant mucosal sites may impact pneumonia. For example, in allogenic hematopoietic cell transplantation patients, changes in gut microbiota are associated with pulmonary complications.112 Pulmonary complications, defined in that study as abnormal parenchymal findings on chest imaging with respiratory symptoms, were found in the majority of participants. The use of antibiotics, low baseline gut microbiome diversity, and Gammaproteobacteria enrichment in the gut microbiome predicted pulmonary complications.112 It is possible that changes in gut microbiota may affect systemic immune tone. Moreover, the epithelial barrier in these subjects is frequently disrupted due to intensive immunosuppressive treatment allowing for bacterial translocation to the lung. Future investigations must consider evaluating interactions between different mucosae by carefully sampling the involved mucosae and the systemic compartments.
Even less in known about the roles of viruses or fungi on the susceptibility to pneumonia. Non-bacterial microbes are mostly neglected from current lung microbiome studies due to technical difficulties but should receive further attention. Viruses play a major role in chronic inflammatory diseases of the lung such as asthma, COPD, and cystic fibrosis. However, few studies have evaluated the airway virome.113,114 Infection with rhinovirus in COPD patients has been shown to be associated with increased bacterial load and change in microbiota composition.115 Rhinovirus infection leads to a change in the relative abundances of many pathogenic and non-pathogenic bacteria with an increase in Hemophilus and Neisseriaceae species at day 15. These data support that inter-kingdom interactions (in this case viruses with bacteria) may affect subjects’ susceptibility to acquire microbes with potential pathogenic relevance and could explain the propensity to develop pneumonia among patients with chronic inflammatory airway diseases such as COPD or cystic fibrosis.
How distinct microbiota signatures in pneumonia affect the natural history of the disease.
For the last 50 years, research has focused on pathogen-host interactions that occur when patients develop pneumonia. The current understanding of the complex microbial communities existing in the lower airways invite us to broaden this view to uncover the role of microbiota-host interactions during pneumonia. Studies in HIV-infected patients in Uganda and the United States117 demonstrate that the oral and lung microbiome in HIV-infected patients treated with antimicrobials changes during acute pneumonia.116 The lower airway microbiota exhibited significantly higher relative abundance of multiple members of the Proteobacteria phyla, including several pathogens such as Klebsiella pneumoniae and Pseudomonas species, and these distinct microbiota signatures may contribute to the natural history of the disease. In another study performed with the Ugandan cohort of HIV subjects admitted to a local hospital for pneumonia, distinct lung microbiota signatures were associated with disease progression.118 Using a clustering approach on the 16S rRNA gene sequencing data, the lower airway samples from HIV subjects with pneumonia organized into distinct groups. One group was dominated by Pseudomonaceae (group MCS1). The second group was subdivided into two sub-clusters enriched with Streptococcaeae (MCS2A) or Prevotellaceae (MCS2B).118 Enrichment with Prevotellaceae trended toward an increase in mortality at 1 week after bronchoscopy (MSC1 0.0% mortality vs. MSC2B 7.4%) and 70 days after bronchoscopy (MSC1 13% mortality compared to MSC2A 16%, and MSC2B 22%).118 The clusters were also associated with distinct immune profiles based on metabolomics.118 These data suggest that lung microbiota signatures among subjects with pneumonia may play a role in the pathogenesis and may help us understand differences in outcomes when patients develop pneumonia. It is possible that a “healthier” microbiome enriched with Pseudomonaceae may suppress virulence of potential pathogens and promote the restoration of a ‘healthy’ lung microbiome. Conversely, a lower airway microbiota enriched with Streptococacceae or Prevotellaceae may favor a more pro-inflammatory endotype that may promote the persistence and blooming of pathogens by driving nutrients to the alveolar space or promote virulence factors (Figure 1B).119 In transplant, the lung microbiota of subjects diagnosed with pneumonia was found to have decreased diversity and was dominated with Pseudomonas, Staphylococcus, and Streptococcus.120 Difficulties obtaining samples prior to the development of pneumonia are a significant limitation for studying the lung microbiome during CAP. Although confounded by multiple issues, we can gain insight by studying intubated patients prior to the development of ventilator associated pneumonia (VAP). In a small study where samples were obtained longitudinally from the upper and lower respiratory tract, there was a significant decrease over time in α diversity their upper airways (although not in lower respiratory samples) associated with the development of pneumonia.16 The reduction in diversity prior to the development of pneumonia may be an important step reflecting dysbiosis along the airway microbiome.
Lung microbiome: what can we expect from future investigations?
A better understanding of the lung microbiota in pneumonia is needed to uncover important microbiota-host and microbe-microbe interactions that will likely yield improvements in prevention, diagnosis, and treatment of pneumonia. The microbial dynamics across mucosal membranes (i.e. upper/lower airways and gut) in different disease states likely affects an individual’s susceptibility to pneumonia. Defining the pathways that dictate microbe-microbe interactions, microbe-host interactions, and selection pressure differences using unbiased, culture independent methods allows us to characterize the complex microbial community dynamics of the lower airways. The gut microbiota may also shape the immune system and ‘spillover’ affecting the lower airway deserves careful consideration. In addition, other mucosal locations and/or specific timing (e.g. early childhood) may shape the immune tone and will be critical to our understanding of an individual’s susceptibility to pneumonia. By studying the microbial reservoirs to the lower airways (e.g. oropharynx and nasopharynx) we may be able to identify potentially more accessible therapeutic targets that will indirectly affect the lower airway microbiota. Existing examples of this already exist such as decontamination of nasal carriage with MRSA with mupirocin,121 oral hygiene to prevent HAP,122 and oral decontamination with chlorhexidine for intubated patients.123 These have been based on culture based understanding of microbes and the approach has been targeting specific pathogens or a “sledgehammer” antimicrobial approach. Better understanding of the complexity of the existing microbial communities will likely lead to a more targeted approach tailored to multiple microbes and keystone species personalized for each individual patient.
Currently, there is no unbiased, high-throughput culture-independent technique widely available to guide individualized patient care. This is an area of active research and relevant to the care of patients at the bedside. As sequencers become smaller and even attachable to a USB port on a laptop, major limitations and challenges for these approaches remains the bioinformatic power and time needed to perform analysis. As these techniques are entering the phase of possible clinical bedside application, it will be important to start testing these approaches in large cohort studies, where reproducibility and feasibility can be best assessed.30
Among the potential therapeutic options for either prevention or treatment of pneumonia, a better understanding of the lung microbiota may shift our current “pathogen-killing” focus to include the use of probiotics (e.g. living bacteria intended to benefit health), prebiotics (e.g. diet ingredients that confer specific changes in the microbiome and lead to beneficial effects in the host), or selective antibiotics (e.g. eradication of specific strains of bacteria not necessarily identified as pathogen but may augment the pathogenic process).124 Other therapies attempting to modify the composition of the airway microbiota may include the use of anti-bacterial conjugate vaccines or focused bacteriophages eliminating individual strains of a single species125,126 and replacing the entire community with a new intact airway microbiota (following the example of fecal transplantation in Clostridium difficile colitis). Similar to the rationale for using probiotics in diet, it might be feasible to nurture and promote a “healthier” airway microbiota by inhaling a specific mixture of microbial species or microbial metabolites tailored to an individual’s microbiota to restore or promote airways health.
As research of the microbiota in pneumonia grows, we identify the following major challenges: a) lack of animal models developed to study microbe-host and microbe-microbe interactions that accounts for the complexities of microbial communities existing in humans; b) difficulties examining virome and mycobiome due to limitation with current gene marker approaches and reference libraries; c) limited access to lower airway samples; d) difficulties studying the events that occur at early time points of pneumonia or pre-clinical disease; e) heterogeneity of pneumonia as a pathogenic condition and clinical diagnosis; and f) multiple confounders present at the time of diagnosis such as comorbidities, environmental factors, and effect of different treatments.
Pre-clinical models have been key to our mechanistic understanding of pneumonia. However, these have been tailored to study the acquisition of a single organism without considering resident or microbial communities (beyond the use of either germ free or pathogen free models). Future investigations will need to design how to study complex microbial interactions in these models of disease. Experiments utilizing longitudinal, prospective cohorts may give us insight into the changes in the upper and lower airway that predict the development of pneumonia. Community acquired pneumonia, hospital acquired pneumonia, and ventilator acquired pneumonia may share some common pathophysiological events but are fundamentally different clinical entities that will require different study designs and approaches.
In summary, high-throughput sequencing enables more comprehensive characterization of airway microbial community composition and has the potential to detect more difficult-to-culture microbes that have significant relevance in the pathogenesis of pneumonia. The line between what we understand as a commensal and a pathogen has become more blurred with the discovery of the lung microbiome. The use of culture independent techniques to study the lung microbiome challenges our belief that the healthy lung is sterile and provides new insights into the importance of the microbiome for mucosal immune maturation and response that is relevant to the development and natural history of pneumonia. Rather than prescribing antibiotics, the evaluation of the airway microbiota and its immune interactions may allow for better-targeted and individualized approaches with antimicrobials as well as other non-antibiotic therapies intended to regulate microbial community composition, microbial metabolism, or enhance the efficacy of the immune response. The paradigm will move away from a sole pathogen causing disease, to that of a disrupted community of microorganisms that may enhance the pathogenic potential of each other (Figure 1B). The research efforts to understand the role of the lung microbiota in pneumonia require both preclinical models and rigorous and well-designed prospective cohort studies that the field currently lacks to understand the interactions between the host and the community of microorganisms in the airway to contribute to our understanding of the pathogenesis of pneumonia.
3–5 key points:
A significant research gap exists in the study of the lung microbiome and pneumonia.
Complex microbial communities exist in the upper and lower airway.
Microbe-host and microbe-microbe interactions blur the line between pathogen and commensal.
The use of next-generation sequencing with reference microorganism databases allows for an unbiased approach to identifying large communities of microbes and potential pathogens.
The microbial community of the lung may play an important role in pneumonia impacting susceptibility and the natural history of disease.
Acknowledgments
Sources of support: This work supported by K23 AI102970 (LNS) Flight Attendant Medical Research Institute (BGW) Stony Wold-Herbert Fund (BGW) T32 CA193111 (BGW) UL1TR001445 (BGW)
Footnotes
Financial Disclosure: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Yamasaki K, Kawanami T, Yatera K, et al. Significance of anaerobes and oral bacteria in community-acquired pneumonia. PLoS One. 2013;8(5):e63103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rascon-Aguilar IE, Pamer M, Wludyka P, et al. Role of gastroesophageal reflux symptoms in exacerbations of COPD. Chest. 2006;130(4):1096–1101. [DOI] [PubMed] [Google Scholar]
- 3.Cvejic L, Harding R, Churchward T, et al. Laryngeal penetration and aspiration in individuals with stable COPD. Respirology. 2011;16(2):269–275. [DOI] [PubMed] [Google Scholar]
- 4.Morse CA, Quan SF, Mays MZ, Green C, Stephen G, Fass R. Is there a relationship between obstructive sleep apnea and gastroesophageal reflux disease? Clin Gastroenterol Hepatol. 2004;2(9):761–768. [DOI] [PubMed] [Google Scholar]
- 5.Jain S, Self WH, Wunderink RG, et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. The New England journal of medicine. 2015;373(5):415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadsby NJ, Russell CD, McHugh MP, et al. Comprehensive Molecular Testing for Respiratory Pathogens in Community-Acquired Pneumonia. Clin Infect Dis. 2016;62(7):817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlueter M, James C, Dominguez A, Tsu L, Seymann G. Practice patterns for antibiotic de-escalation in culture-negative healthcare-associated pneumonia. Infection. 2010;38(5):357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labelle AJ, Arnold H, Reichley RM, Micek ST, Kollef MH. A comparison of culture-positive and culture-negative health-care-associated pneumonia. Chest. 2010;137(5):1130–1137. [DOI] [PubMed] [Google Scholar]
- 9.Webb BJ, Dangerfield BS, Pasha JS, Agrwal N, Vikram HR. Guideline-concordant antibiotic therapy and clinical outcomes in healthcare-associated pneumonia. Respir Med. 2012;106(11):1606–1612. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Nieto JM, Torres A, Garcia-Cordoba F, et al. Impact of invasive and noninvasive quantitative culture sampling on outcome of ventilator-associated pneumonia: a pilot study. American journal of respiratory and critical care medicine. 1998;157(2):371–376. [DOI] [PubMed] [Google Scholar]
- 11.Hayon J, Figliolini C, Combes A, et al. Role of serial routine microbiologic culture results in the initial management of ventilator-associated pneumonia. American journal of respiratory and critical care medicine. 2002;165(1):41–46. [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Haery C, Paladugu B, et al. The duration of hypotension before the initiation of antibiotic treatment is a critical determinant of survival in a murine model of Escherichia coli septic shock: association with serum lactate and inflammatory cytokine levels. J Infect Dis. 2006;193(2):251–258. [DOI] [PubMed] [Google Scholar]
- 13.Kumar R, Maynard CL, Eipers P, et al. Colonization potential to reconstitute a microbe community in patients detected early after fecal microbe transplant for recurrent C. difficile. BMC Microbiol. 2016;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dortet L, Legrand P, Soussy CJ, Cattoir V. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J Clin Microbiol. 2006;44(12):4471–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seki M, Gotoh K, Nakamura S, et al. Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. J Med Microbiol. 2013;62(Pt 5):801–803. [DOI] [PubMed] [Google Scholar]
- 16.Kelly BJ, Imai I, Bittinger K, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvat RT, El Atrouni W, Hammoud K, Hawkinson D, Cowden S. Ribosomal RNA sequence analysis of Brucella infection misidentified as Ochrobactrum anthropi infection. J Clin Microbiol. 2011;49(3):1165–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589–1596. [DOI] [PubMed] [Google Scholar]
- 19.McMenamin JD, Zaccone TM, Coenye T, Vandamme P, LiPuma JJ. Misidentification of Burkholderia cepacia in US cystic fibrosis treatment centers: an analysis of 1,051 recent sputum isolates. Chest. 2000;117(6):1661–1665. [DOI] [PubMed] [Google Scholar]
- 20.Monso E, Ruiz J, Rosell A, et al. Bacterial infection in chronic obstructive pulmonary disease. A study of stable and exacerbated outpatients using the protected specimen brush. American journal of respiratory and critical care medicine. 1995;152(4 Pt 1):1316–1320. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein EJ, Citron DM, Goldman PJ, Goldman RJ. National hospital survey of anaerobic culture and susceptibility methods: III. Anaerobe. 2008;14(2):68–72. [DOI] [PubMed] [Google Scholar]
- 22.Lagier JC, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28(1):208–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28(1):237–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chastre J, Fagon JY, Bornet-Lecso M, et al. Evaluation of bronchoscopic techniques for the diagnosis of nosocomial pneumonia. Am J Respir Crit Care Med. 1995;152(1):231–240. [DOI] [PubMed] [Google Scholar]
- 25.Fagon JY, Chastre J, Trouillet JL, et al. Characterization of distal bronchial microflora during acute exacerbation of chronic bronchitis. Use of the protected specimen brush technique in 54 mechanically ventilated patients. Am Rev Respir Dis. 1990;142(5):1004–1008. [DOI] [PubMed] [Google Scholar]
- 26.Sethi S Bacteria in exacerbations of chronic obstructive pulmonary disease: phenomenon or epiphenomenon? Proc Am Thorac Soc. 2004;1(2):109–114. [DOI] [PubMed] [Google Scholar]
- 27.Cabello H, Torres A, Celis R, et al. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: a bronchoscopic study. Eur Respir J. 1997;10(5):1137–1144. [DOI] [PubMed] [Google Scholar]
- 28.Monso E, Rosell A, Bonet G, et al. Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur Respir J. 1999;13(2):338–342. [DOI] [PubMed] [Google Scholar]
- 29.Riise GC, Andersson B, Ahlstedt S, et al. Bronchial brush biopsies for studies of epithelial inflammation in stable asthma and nonobstructive chronic bronchitis. Eur Respir J. 1996;9(8):1665–1671. [DOI] [PubMed] [Google Scholar]
- 30.Pendleton KM, Erb-Downward JR, Bao Y, et al. Rapid Pathogen Identification in Bacterial Pneumonia Using Real-Time Metagenomics. American journal of respiratory and critical care medicine. 2017;196(12):1610–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Q, Cui S, Chen C, et al. Metagenomic Analysis of Sputum Microbiome as a Tool toward Culture-Independent Pathogen Detection of Patients with Ventilator-associated Pneumonia. American journal of respiratory and critical care medicine. 2016;194(5):636–639. [DOI] [PubMed] [Google Scholar]
- 32.Xu L, Zhu Y, Ren L, et al. Characterization of the nasopharyngeal viral microbiome from children with community-acquired pneumonia but negative for Luminex xTAG respiratory viral panel assay detection. J Med Virol. 2017;89(12):2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell host & microbe. 2015;17(5):704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. The New England journal of medicine. 2001;344(1):11–16. [DOI] [PubMed] [Google Scholar]
- 35.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadevall A, Pirofski LA. The damage-response framework of microbial pathogenesis. Nat Rev Microbiol. 2003;1(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casadevall A, Pirofski LA. What is a pathogen? Ann Med. 2002;34(1):2–4. [DOI] [PubMed] [Google Scholar]
- 38.Otto M Staphylococcus epidermidis--the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7(8):555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrd AL, Segre JA. Infectious disease. Adapting Koch’s postulates. Science. 2016;351(6270):224–226. [DOI] [PubMed] [Google Scholar]
- 40.Dickson RP, Erb-Downward JR, Freeman CM, et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio. 2017;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pustelny C, Komor U, Pawar V, et al. Contribution of Veillonella parvula to Pseudomonas aeruginosa-mediated pathogenicity in a murine tumor model system. Infection and immunity. 2015;83(1):417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmody LA, Zhao J, Schloss PD, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc. 2013;10(3):179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang YJ, LiPuma JJ. The Microbiome in Cystic Fibrosis. Clin Chest Med. 2016;37(1):59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23(2):299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Schloss PD, Kalikin LM, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(15):5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest. 1997;111(5):1266–1272. [DOI] [PubMed] [Google Scholar]
- 47.Teramoto S, Ohga E, Matsui H, Ishii T, Matsuse T, Ouchi Y. Obstructive sleep apnea syndrome may be a significant cause of gastroesophageal reflux disease in older people. J Am Geriatr Soc. 1999;47(10):1273–1274. [DOI] [PubMed] [Google Scholar]
- 48.Field SK, Underwood M, Brant R, Cowie RL. Prevalence of gastroesophageal reflux symptoms in asthma. Chest. 1996;109(2):316–322. [DOI] [PubMed] [Google Scholar]
- 49.Scott RB, O’Loughlin EV, Gall DG. Gastroesophageal reflux in patients with cystic fibrosis. J Pediatr. 1985;106(2):223–227. [DOI] [PubMed] [Google Scholar]
- 50.Koh WJ, Lee JH, Kwon YS, et al. Prevalence of gastroesophageal reflux disease in patients with nontuberculous mycobacterial lung disease. Chest. 2007;131(6):1825–1830. [DOI] [PubMed] [Google Scholar]
- 51.Randell SH, Boucher RC. Effective mucus clearance is essential for respiratory health. Am J Respir Cell Mol Biol. 2006;35(1):20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor AE, Finney-Hayward TK, Quint JK, et al. Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J. 2010;35(5):1039–1047. [DOI] [PubMed] [Google Scholar]
- 53.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One. 2012;7(10):e47305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickson RP, Huffnagle GB. The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLoS Pathog. 2015;11(7):e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aagaard K, Petrosino J, Keitel W, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27(3):1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Group NHW, Peterson J, Garges S, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaiss MM, Rapin A, Lebon L, et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity. 2015;43(5):998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonseca DM, Hand TW, Han SJ, et al. Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell. 2015;163(2):354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawa S, Lochner M, Satoh-Takayama N, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12(4):320–326. [DOI] [PubMed] [Google Scholar]
- 62.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29(6):958–970. [DOI] [PubMed] [Google Scholar]
- 63.Farkas AM, Panea C, Goto Y, et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J Immunol Methods. 2015;421:104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ivanov II, Frutos Rde L, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4(4):337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pezzulo AA, Kelly PH, Nassar BS, et al. Abundant DNase I-Sensitive Bacterial DNA in Healthy Porcine Lungs and Its Implications for the Lung Microbiome. Appl Environ Microbiol. 2013;79(19):5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pradhan D, Segal LN, Kulkarni R, et al. Bronchial Reactivity In Early Emphysema May Be Associated With Local Neutrophilic Inflammation. Am J Respir Crit Care Med. 2013:A1110. [Google Scholar]
- 68.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. American journal of respiratory and critical care medicine. 2011;184(8):957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segal LN, Clemente JC, Tsay J-CJ, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nature Microbiology. 2016:16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ege MJ, Mayer M, Normand AC, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–709. [DOI] [PubMed] [Google Scholar]
- 72.Ege MJ, Bieli C, Frei R, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. The Journal of allergy and clinical immunology. 2006;117(4):817–823. [DOI] [PubMed] [Google Scholar]
- 73.Roduit C, Wohlgensinger J, Frei R, et al. Prenatal animal contact and gene expression of innate immunity receptors at birth are associated with atopic dermatitis. The Journal of allergy and clinical immunology. 2011;127(1):179–185, 185 e171. [DOI] [PubMed] [Google Scholar]
- 74.Fujimura KE, Demoor T, Rauch M, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111(2):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dimmitt RA, Staley EM, Chuang G, Tanner SM, Soltau TD, Lorenz RG. Role of postnatal acquisition of the intestinal microbiome in the early development of immune function. J Pediatr Gastroenterol Nutr. 2010;51(3):262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535(7610):65–74. [DOI] [PubMed] [Google Scholar]
- 79.van Nimwegen FA, Penders J, Stobberingh EE, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128(5):948–955 e941–943. [DOI] [PubMed] [Google Scholar]
- 80.Bager P, Melbye M, Rostgaard K, Benn CS, Westergaard T. Mode of delivery and risk of allergic rhinitis and asthma. J Allergy Clin Immunol. 2003;111(1):51–56. [DOI] [PubMed] [Google Scholar]
- 81.Marra F, Marra CA, Richardson K, et al. Antibiotic use in children is associated with increased risk of asthma. Pediatrics. 2009;123(3):1003–1010. [DOI] [PubMed] [Google Scholar]
- 82.Murk W, Risnes KR, Bracken MB. Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics. 2011;127(6):1125–1138. [DOI] [PubMed] [Google Scholar]
- 83.Depner M, Ege MJ, Genuneit J, et al. Atopic sensitization in the first year of life. J Allergy Clin Immunol. 2013;131(3):781–788. [DOI] [PubMed] [Google Scholar]
- 84.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355(21):2226–2235. [DOI] [PubMed] [Google Scholar]
- 85.Fujimura KE, Johnson CC, Ownby DR, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2):410–412, 412 e411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–1495. [DOI] [PubMed] [Google Scholar]
- 87.Nembrini C, Sichelstiel A, Kisielow J, Kurrer M, Kopf M, Marsland BJ. Bacterial-induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax. 2011;66(9):755–763. [DOI] [PubMed] [Google Scholar]
- 88.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167(8):821–827. [DOI] [PubMed] [Google Scholar]
- 89.Reibman J, Marmor M, Filner J, et al. Asthma is inversely associated with Helicobacter pylori status in an urban population. PLoS One. 2008;3(12):e4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73(1):30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72(9):4996–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. American journal of respiratory and critical care medicine. 2007;175(6):561–569. [DOI] [PubMed] [Google Scholar]
- 93.Ezendam J, van Loveren H. Lactobacillus casei Shirota administered during lactation increases the duration of autoimmunity in rats and enhances lung inflammation in mice. The British journal of nutrition. 2008;99(1):83–90. [DOI] [PubMed] [Google Scholar]
- 94.Kitagaki K, Businga TR, Kline JN. Oral administration of CpG-ODNs suppresses antigen-induced asthma in mice. Clinical and experimental immunology. 2006;143(2):249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 2016;10(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. American journal of respiratory and critical care medicine. 2013;188(10):1246–1252. [DOI] [PubMed] [Google Scholar]
- 97.Twigg Iii HL, Knox KS, Zhou J, et al. Effect of Advanced HIV Infection on the Respiratory Microbiome. American journal of respiratory and critical care medicine. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Selwyn PA, Feingold AR, Hartel D, et al. Increased risk of bacterial pneumonia in HIV-infected intravenous drug users without AIDS. AIDS. 1988;2(4):267–272. [DOI] [PubMed] [Google Scholar]
- 99.Segal LN, Clemente JC, Li Y, et al. Anaerobic Bacterial Fermentation Products Increase Tuberculosis Risk in Antiretroviral-Drug-Treated HIV Patients. Cell host & microbe. 2017;21(4):530–537 e534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ledson MJ, Gallagher MJ, Jackson M, Hart CA, Walshaw MJ. Outcome of Burkholderia cepacia colonisation in an adult cystic fibrosis centre. Thorax. 2002;57(2):142–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Restrepo MI, Mortensen EM, Pugh JA, Anzueto A. COPD is associated with increased mortality in patients with community-acquired pneumonia. Eur Respir J. 2006;28(2):346–351. [DOI] [PubMed] [Google Scholar]
- 102.Mullerova H, Chigbo C, Hagan GW, et al. The natural history of community-acquired pneumonia in COPD patients: a population database analysis. Respir Med. 2012;106(8):1124–1133. [DOI] [PubMed] [Google Scholar]
- 103.Flume PA, Mogayzel PJ Jr., Robinson KA, et al. Cystic fibrosis pulmonary guidelines: treatment of pulmonary exacerbations. American journal of respiratory and critical care medicine. 2009;180(9):802–808. [DOI] [PubMed] [Google Scholar]
- 104.Salk HM, Simon WL, Lambert ND, et al. Taxa of the Nasal Microbiome Are Associated with Influenza-Specific IgA Response to Live Attenuated Influenza Vaccine. PLoS One. 2016;11(9):e0162803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. The New England journal of medicine. 2008;359(22):2355–2365. [DOI] [PubMed] [Google Scholar]
- 106.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6(2):e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sethi S, Evans N, Grant BJ, Murphy TF. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med. 2002;347(7):465–471. [DOI] [PubMed] [Google Scholar]
- 108.Sze MA, Dimitriu PA, Suzuki M, et al. Host Response to the Lung Microbiome in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2015;192(4):438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang YJ, Kim E, Cox MJ, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. Omics. 2010;14(1):9–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kalil AC, Metersky ML, Klompas M, et al. Executive Summary: Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lozupone C, Cota-Gomez A, Palmer BE, et al. Widespread Colonization of the Lung by Tropheryma whipplei in HIV Infection. Am J Respir Crit Care Med. 2013;187(10):1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harris B, Morjaria SM, Littmann ER, et al. Gut Microbiota Predict Pulmonary Infiltrates After Allogeneic Hematopoietic Cell Transplantation. American journal of respiratory and critical care medicine. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Willner D, Haynes MR, Furlan M, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am J Respir Cell Mol Biol. 2012;46(2):127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Willner D, Furlan M, Schmieder R, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4547–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Molyneaux PL, Mallia P, Cox MJ, et al. Outgrowth of the Bacterial Airway Microbiome following Rhinovirus Exacerbation of Chronic Obstructive Pulmonary Disease. American Journal of Respiratory and Critical Care Medicine. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iwai S, Fei M, Huang D, et al. Oral and airway microbiota in HIV-infected pneumonia patients. Journal of clinical microbiology. 2012;50(9):2995–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iwai S, Huang D, Fong S, et al. The lung microbiome of Ugandan HIV-infected pneumonia patients is compositionally and functionally distinct from that of San Franciscan patients. PLoS One. 2014;9(4):e95726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shenoy MK, Iwai S, Lin DL, et al. Immune Response and Mortality Risk Relate to Distinct Lung Microbiomes in Patients with HIV and Pneumonia. American journal of respiratory and critical care medicine. 2017;195(1):104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. The Lancet Respiratory medicine. 2014;2(3):238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shankar J, Nguyen MH, Crespo MM, et al. L ooking Beyond Respiratory Cultures: Microbiome-Cytokine Signatures of Bacterial Pneumonia and Tracheobronchitis in Lung Transplant Recipient s. Am J Transplant. 2016;16(6):1766–1778. [DOI] [PubMed] [Google Scholar]
- 121.Perl TM, Cullen JJ, Wenzel RP, et al. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. The New England journal of medicine. 2002;346(24):1871–1877. [DOI] [PubMed] [Google Scholar]
- 122.Baker D, Quinn B. Hospital Acquired Pneumonia Prevention Initiative-2: Incidence of nonventilator hospital-acquired pneumonia in the United States. Am J Infect Control. 2018;46(1):2–7. [DOI] [PubMed] [Google Scholar]
- 123.Genuit T, Bochicchio G, Napolitano LM, McCarter RJ, Roghman MC. Prophylactic chlorhexidine oral rinse decreases ventilator-associated pneumonia in surgical ICU patients. Surg Infect (Larchmt). 2001;2(1):5–18. [DOI] [PubMed] [Google Scholar]
- 124.Bowater RJ, Stirling SA, Lilford RJ. Is antibiotic prophylaxis in surgery a generally effective intervention? Testing a generic hypothesis over a set of meta-analyses. Ann Surg. 2009;249(4):551–556. [DOI] [PubMed] [Google Scholar]
- 125.Schooley RT, Biswas B, Gill JJ, et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob Agents Chemother. 2017;61(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mitsi E, Roche AM, Reine J, et al. Agglutination by anti-capsular polysaccharide antibody is associated with protection against experimental human pneumococcal carriage. Mucosal Immunol. 2017;10(2):385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]


