We identify a cell type–specific modification of HLA-I using lamprey VLR antibodies as a new class of research reagents.
Abstract
Memory B cells and plasma cells are antigen-experienced cells tasked with the maintenance of humoral protection. Despite these prominent functions, definitive cell surface markers have not been identified for these cells. We report here the isolation and characterization of the monoclonal variable lymphocyte receptor B (VLRB) N8 antibody from the evolutionarily distant sea lamprey that specifically recognizes memory B cells and plasma cells in humans. Unexpectedly, we determined that VLRB N8 recognizes the human leukocyte antigen–I (HLA-I) antigen in a tyrosine sulfation–dependent manner. Furthermore, we observed increased binding of VLRB N8 to memory B cells in individuals with autoimmune disorders multiple sclerosis and systemic lupus erythematosus. Our study indicates that lamprey VLR antibodies uniquely recognize a memory B cell– and plasma cell–specific posttranslational modification of HLA-I, the expression of which is up-regulated during B cell activation.
INTRODUCTION
Memory B cells (Bmem) and plasma cells (PCs) serve a key function in providing long-lasting humoral protection to pathogenic challenge, both in the context of natural infections and following vaccinations (1, 2). Despite these central roles, many questions remain unanswered about these cells, including the functions of subpopulations of human Bmem and PC, regulatory mechanisms governing their activation, and the basis of their longevity. One impediment to answering these questions is the paucity of suitable markers for specific Bmem identification. Blood Bmem in humans are identified by the expression of CD27 (3, 4), whereas tissue-based Bmem are recognized as CD19+/IgD−/CD38− (5, 6). Although widely used as a marker for Bmem, CD27 is also expressed on germinal center (GC) B cells, PC, and many non–B lineage cells (7, 8), and several studies identified Bmem populations lacking CD27 expression (9–11). Consequently, Bmem are currently best defined as antigen-experienced B cells, which are not engaged in an ongoing immune reaction, rather than described by the expression of well-defined cell surface markers. To address this issue, we developed a structure-based approach for the identification of cell surface antigens on Bmem using the variable lymphocyte receptor (VLR) antibodies of the jawless vertebrate sea lamprey (Petromyzon marinus).
Despite observations of adaptive immune responses by jawless vertebrates—that is, lampreys and hagfish—over 50 years ago (12, 13), no homologs of conserved prototypical adaptive immune genes, such as human leukocyte antigen (HLA) or recombining B cell and T cell antigen receptors, could be identified in evolutionarily distant jawless vertebrates. Only recently, the molecular basis of adaptive immune responses in jawless vertebrates was defined with the discovery of clonally diverse VLR anticipatory receptors (14). This receptor system provides a potential repertoire exceeding 1014 clonotypes (14–16). Unlike antibodies of jawed vertebrates, which use the immunoglobulin (Ig) fold as the basic structural unit, the VLRB antibodies consist of β sheet–forming leucine-rich repeat (LRR) units. Structural analyses of monoclonal VLRB antibodies in complex with protein and carbohydrate antigens indicate that residues of VLRB antibodies located on the inner concave surface and a variable loop structure emanating from the C-terminal capping LRR unit interact with the antigen (17–19). We reasoned that the radically different protein architecture of VLR antibodies and the large evolutionary distance of jawless vertebrates from jawed vertebrates would allow the generation of monoclonal VLR antibodies to antigens, which conventional mammalian antibodies do not readily recognize because of structural or tolerogenic constraints.
In previous studies, we established monoclonal VLR antibodies as a new class of research reagents for discovery of biomarkers on human lymphocytes and PCs (20, 21). Here, we report the isolation of the monoclonal antibody VLRB N8, reactive with an epitope that is expressed specifically on HLA-I on Bmem and PC, is up-regulated on Bmem in autoimmune disorders and is dependent on tyrosine sulfation of HLA-I, indicating that HLA-I–dependent immune responses are subject to an as yet unexplored level of immune regulation.
RESULTS
VLRB N8 specifically recognizes human Bmem and PCs
In an effort to generate novel reagents targeting late stages in B lineage differentiation, we screened 628 monoclonal VLRB antibodies from a library generated from lymphocytes of sea lamprey larvae immunized with a cocktail of the plasmacytoma cell lines NCI-H929, U266, and RPMI 8226. Monoclonal VLRB antibodies that displayed reactivity to these cell lines were further evaluated for recognition of primary lymphocytes. One of these VLRB antibodies, VLRB N8, recognized human CD27+/IgD− Bmem, and CD27+/IgD+ marginal zone equivalent (MZe) cells (Fig. 1A, top), blood B lineage cells whose somatically mutated antigen receptor sequences are indicative of post-GC status (4, 22, 23). The VLRB N8 antibody did not react with T cells, non–B/T lineage cells, or monocytes (Fig. 1A, top). When tonsil samples were used to evaluate VLRB N8 reactivity with tissue-based lymphocytes, we found that VLRB N8 again strongly recognized Bmem and PCs (Fig. 1A, bottom). VLRB N8 only very weakly detected a small number of naïve B cells and detected no GC B cells or non–B lineage cells. Sequence analysis revealed that VLRB N8 contains four variable LRR units and is thus larger than average VLRB molecules (Fig. 1B) (24).
Fig. 1. VLRB N8 recognizes human Bmem and PC in blood and tonsils.
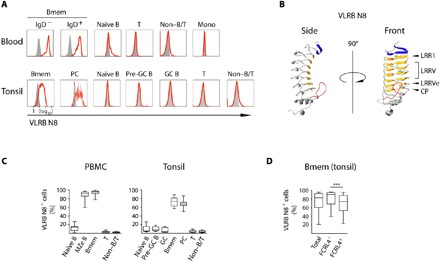
(A) Peripheral blood mononuclear cells (PBMCs) were separated into Bmem (CD19+/IgD−/CD27+) and marginal zone-equivalent (MZe) cells (CD19+/IgD+/CD27+), naïve B cells (CD19+/IgD+/CD27−), non–B/T cells (CD19−/CD3−), and T cells (CD19−/CD3+). A representative of 12 examined PBMC samples is shown. Human tonsillar lymphocytes were separated into the following subpopulations: Bmem (CD19+/IgD−/CD38−), PC (CD19+/IgD−/CD38++), naïve B cells (CD19+/IgD+/CD38−), pre-GC B cells (CD19+/IgD+/CD38+), GC B cells (CD19+/IgD−/CD38+), non–B/T cells (CD19−/CD3−), and T cells (CD19−/CD3+). A representative example of 14 analyzed tonsil samples is shown. VLRB N8 reactivity by flow cytometric analysis is indicated by solid red lines, and VLR4 reactivity (specific for the BclA antigen of the exosporium of Bacillus anthracis) as a negative control is shown as solid gray histogram. (B) Ribbon model of a monomeric antigen-binding unit of VLRB N8. Parallel β sheets lining the inner concave surface encoded by the N-terminal capping LRR are shown in blue, and sequences encoded by the LRR1, variable LRRV1-4, LRRVe and connecting peptide (CP) units are depicted in orange. A variable loop protruding from the capping C-terminal LRR is shown in red. The model was generated using the IntFOLD modeling platform (49). (C) Frequencies of VLRB N8–reactive cells for each analyzed cell population in healthy blood and tonsil samples. (D) Frequencies of VLRB N8–reactive cells between FCRL4+ and FCRL4− Bmem from 14 additional tonsil specimens. Statistical significance was assessed using a Wilcoxon signed-rank test, ***P < 0.001; n = 14.
Analysis of VLRB N8 reactive cell frequencies showed that nearly all circulating Bmem were reactive with the lamprey antibody (Fig. 1C). In contrast, VLRB N8 reacted strongly with 70 to 80% of tonsillar Bmem and PC (Fig. 1C). In tonsil, the immunoregulatory Fc receptor-like 4 (FCRL4) molecule characterizes a morphologically and functionally distinct subpopulation of CD20hi/CD21lo Bmem (25). Using FCRL4 as a marker to discriminate defined Bmem subpopulations, we determined on 14 additional tonsil specimens that FCRL4+ Bmem were found more frequently among VLRB N8lo/− Bmem than FCRL4− Bmem (Fig. 1D). This predominant Bmem/PC specificity observed for VLRB N8 has not been shown for any previously reported conventional antibody.
VLRB N8 reacts with the HLA-I antigen
To use VLRB N8 as an affinity reagent for antigen purification and identification, we initially screened panels of human cell lines for recognition by VLRB N8 (fig. S1). Analysis of VLRB N8 immunoprecipitates of cell surface–biotinylated VLRB N8–reactive KMS-11 plasmacytoma cells revealed a prominent band of approximately 40 kDa (fig. S2). We used tandem mass spectrometry to determine binding partners in VLRB N8 immunoprecipitates and tentatively identified peptides corresponding to sequences of several HLA-I alleles and the HLA-associated β2-microglobulin as the most prevalent protein signals (Table 1). In the immunoprecipitates, we also detected other molecules for which cis interactions with HLA-I have been reported, such as the transferrin receptor (CD71) (26) and HLA-II (27).
Table 1. Mass spectrometric analysis of VLRB N8 immunoprecipitates.
Displayed are identified proteins that remain following elimination of sequences associated with intracellular molecules and with VLR4 negative control coimmunoprecipitates. Mw, molecular weight.
| Rank | Identified proteins | Accession no. | Mw (kDa) |
| 1 | HLA class I histocompatibility antigen, A-24 α chain | 1A24_HUMAN | 41 |
| 2 | HLA class I histocompatibility antigen, B-55 α chain | 1B55_HUMAN | 40 |
| 3 | Galectin-1 | LEG1_HUMAN | 15 |
| 4 | Transferrin receptor protein 1 | TFR1_HUMAN | 85 |
| 5 | Protein S100-A9 | S10A9_HUMAN | 13 |
| 6 | 4F2 cell surface antigen heavy chain | B4E2Z3_HUMAN | 56 |
| 7 | Ubiquitin | B4DV12_HUMAN | 17 |
| 8 | HLA class II histocompatibility antigen, DRB1-16 β chain | 2B1G_HUMAN | 30 |
| 9 | HLA class I histocompatibility antigen, Cw-14 α chain | 1C14_HUMAN | 41 |
| 10 | β2-microglobulin | B2MG_HUMAN | 14 |
| 11 | HLA class I histocompatibility antigen, B-51 α chain | 1B51_HUMAN | 41 |
| 12 | HLA class II histocompatibility antigen, DR α chain | DRA_HUMAN | 29 |
To confirm the isolation of HLA-I from VLRB N8 immunoprecipitates, we used KMS-11 cells expressing exogenous HLA-I (A*24:02) green fluorescent protein (GFP) fusion proteins for VLRB N8 immunoprecipitation experiments, followed by detection of endogenous or exogenous HLA-I by Western blotting using anti-HLA or anti-GFP antibodies, respectively. These experiments confirmed the initial observation of HLA-I isolation using VLRB N8 as an affinity purification reagent (fig. S3). To further confirm the interaction of VLRB N8 with HLA-I, we transfected VLRB N8–reactive KMS-11 cells with small interfering RNA (siRNA) targeting β2-microglobulin, ablation of which interferes with HLA-I folding, stability, and cell surface expression (28). This inhibition resulted in substantial reduction of HLA-I detection using conventional pan–HLA-I antibodies and even stronger reduction of VLRB N8 binding compared with transfections with scrambled control siRNA (Fig. 2A). In contrast, binding of VLRB EHT46, recognizing an unknown but ubiquitously expressed antigen, was unaffected, as was the detection of syndecan-1 using a conventional anti-CD138 antibody. Interference of VLRB N8 binding by siRNA-mediated down-regulation of HLA-I was also observed for primary Bmem (Fig. 2B). In an independent experimental approach, we observed that VLRB N8 binding to KMS-11 cells was completely blocked by pretreatment with the w6/32 and G46-2.6 pan–HLA-I antibodies, but not the HC-10 anti–HLA-I antibody, reported to detect β2-microglobulin–free HLA-I α-chains (Fig. 2C) (29). On the other hand, anti–β2-microglobulin antibodies BBM.1 and BM-63 did not interfere with VLRB N8 binding to KMS-11 cells. As expected, pretreatment with anti-CD138 antibody had no effect on the binding of VLRB N8 to KMS-11 cells. Blocking of VLRB N8 recognition of HLA-I using anti–HLA-I antibody w6/32 was also observed for primary Bmem and PC (Fig. 2D). These experiments indicate that VLRB N8 recognizes a unique HLA-I epitope on Bmem/PC that is absent on other hemopoietic cells.
Fig. 2. VLRB N8 recognizes a Bmem/PC-specific epitope of HLA-I.

(A) KMS-11 cells and (B) primary human Bmem were transfected with siRNA targeting β2-microglobulin (Δβ2) or scrambled control siRNA (−) before analysis for VLRB N8 reactivity (top row). Modulation of HLA-I cell surface expression was assessed using conventional anti–HLA-I antibodies (bottom row). Off-target effects of transfected siRNA were assessed by staining with VLRB EHT46 and conventional anti-CD138, respectively. (C) Pan–HLA-I antibodies block the recognition of HLA-I by VLRB N8. KMS-11 cells were preincubated with anti–HLA-I antibodies G46-2.6 or w6/32, anti–β2-microglobulin antibodies BBM.1 or BM-63, free HLA-I heavy chain–detecting HC-10 antibodies, or anti-CD138 antibodies before the addition of VLRB N8. The binding of VLRB N8 (left) or the blocking antibodies (right) was assessed by flow cytometric analysis. MFIs normalized to negative control VLR4 or isotype-matched control antibodies ± SD (n = 5) are shown. Ab, antibody. (D) Pan–HLA-I antibody w6/32 blocks the recognition of HLA-I by VLRB N8. Tonsillar lymphocytes were preincubated with anti–HLA-I antibodies w6/32 or HC-10, followed by evaluation of VLRB N8 binding. MFIs normalized to negative control VLR4 or isotype-matched control antibodies ± SD (n = 12) are shown. Statistically significant differences of P < 0.05 were determined with Student’s t test (A and C) and Wilcoxon signed-rank test (B and D) and were indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
VLRB N8 reactivity does not correlate with HLA-I cell surface expression levels
The specific interaction of VLRB N8 with Bmem/PC contrasts with the ubiquitous expression pattern of HLA-I. Binding of VLRB N8 to panels of cell lines revealed that HLA-I recognition by VLRB N8 does not correlate with HLA-I cell surface expression levels (fig. S1). We then extended our investigation into the reactivity of VLRB N8 with primary circulating and tissue-based cells relative to HLA-I expression. Median fluorescence intensities (MFIs) of VLRB N8 observed for Bmem or PC were consistently increased over values observed with other cell populations (Fig. 3, top row). We found strongly increased VLRB N8 binding to Bmem for a subset of individuals diagnosed with the systemic lupus erythematosus (SLE) and multiple sclerosis (MS) autoimmune disorders. Increased VLRB N8 binding was also seen for class-switched CD27− atypical Bmem that have been observed in the circulation of patients with SLE and MS (9, 30). HLA-I expression was increased on some of the analyzed cell populations, although only to a modest degree (Fig. 3, bottom row), an observation resulting in characteristic increased ratios of VLRB N8 signals normalized to HLA-I (V/H), indicating independence of VLRB N8 recognition of HLA-I from HLA-I cell surface expression levels. This was most evident in comparisons of tonsillar Bmem and PC with GC cells, B lineage cell populations with comparable HLA-I expression levels that are either strongly VLRB N8 reactive (Bmem and PC) or nonreactive (GC) (Fig. 3 and fig. S4), respectively, and in comparative analyses of cell lines for HLA-I expression and VLRB N8 reactivity (fig. S1).
Fig. 3. VLRB N8 reactivity does not correlate with HLA-I cell surface expression levels.
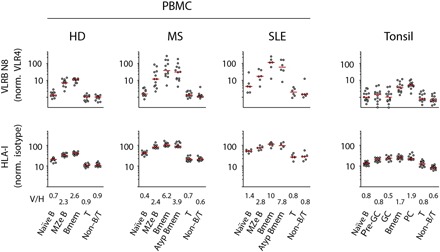
Blood from healthy donors (HDs) and individuals diagnosed with multiple sclerosis or SLE, or tonsillar lymphocytes were analyzed for VLRB N8 binding (top row) and HLA-I binding (bottom row). Cell populations were gated as described in Fig. 1. Atypical Bmem were defined as CD19+/CD3−/IgD−/CD27−. Median values for each population are indicated by red horizontal bars. V/H indicates the numerical values of the median of VLRB N8 signals normalized to the corresponding value of HLA-I.
VLRB N8 recognition of HLA-I is induced following B cell activation
Since Bmem and PC are antigen-experienced B cells, we investigated whether antigen encounter could promote VLRB N8 recognition of B lineage cells. We stimulated cells of the VLRB N8 nonreactive BJAB Burkitt’s lymphoma cell line with phorbol 12-myristate 13-acetate (PMA) and ionomycin as a model to simulate the response to antigen receptor signaling. Under these conditions, BJAB cells became VLRB N8 reactive without changes in HLA-I expression levels (Fig. 4A). VLRB N8 reactivity was acquired with relatively slow kinetics and maintained for the duration of the experiment (120 hours; Fig. 4B, top), although VLRB N8 binding decreased toward the end of the time course. HLA-I expression levels remained unchanged for each of the examined time points (Fig. 4B, middle), thereby resulting in strong increases of VLRB N8/HLA-I ratios (Fig. 4B, bottom). Similar to PMA/ionomycin stimulation, ligation of the antigen receptor with anti-Ig antibodies induced VLRB N8 reactivity of BJAB cells (Fig. 4C, top). Interferon (IFN) gene signatures are frequently observed in individuals with autoimmune disorders such as MS and SLE (31–34). In light of our observation of the strongly increased VLRB N8 binding to Bmem from MS and SLE samples, we were interested in the potential of IFN to induce the VLRB N8 reactivity. Stimulation of BJAB cells with type I or with type II IFN resulted in the induction of VLRB N8 binding to these cells (Fig. 4C, middle panels). Moreover, combinations of suboptimal concentrations of anti-Ig antibodies with type I or type II IFN resulted in further enhancement of VLRB N8 binding (Fig. 4D). In contrast, treatment of BJAB cells with a Toll-like receptor ligand (CpG) failed to induce VLRB N8 reactivity (Fig. 4C, bottom). These results demonstrate that B cell activation by antigen encounter or cytokine stimulation induces a unique epitope on HLA-I to allow binding of the VLRB N8 antibody to B lineage cells.
Fig. 4. Induction of VLRB N8 binding following B cell activation.
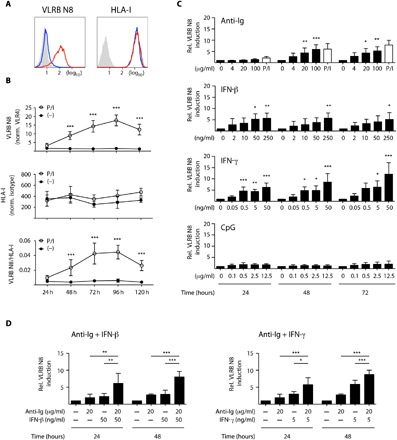
(A) BJAB cells were stimulated with PMA and ionomycin (P/I) for 1 hour and analyzed for VLRB N8 reactivity and HLA-I expression levels after 72 hours (red open histograms). Antibody binding obtained without stimulation is depicted in blue open histograms, and filled gray histograms represent the VLR4 and isotype control experiments. (B) Time course of BJAB response after stimulation with PMA and ionomycin. VLRB N8 binding normalized to negative control VLR4 (top), HLA-I expression normalized to isotype-matched control antibodies (middle), or the ratio of VLRB N8 relative to HLA-I (bottom) is shown. Statistical significance was determined using a multiple t test with Holm-Sidak post test. (C) BJAB cells were treated with the indicated stimuli, and VLRB N8 binding and HLA-I expression levels were assessed as in (A). Induction of VLRB N8 was determined by normalizing VLRB N8/HLA-I ratios to the corresponding unstimulated controls. Bars indicate means ± SD (n = 4). Statistical significance was determined using one-way analysis of variance (ANOVA) with Dunnett’s post test. For comparison, the VLRB N8 signals following PMA and ionomycin treatment are included in the graphic for anti-Ig responses (open bars). (D) Induction of VLRB N8 binding to BJAB cells following costimulation with anti-Ig and IFN. Induction of VLRB N8 reactivity was assessed as in (C). Statistical significance was determined using one-way ANOVA with Tukey’s post test. Statistically significant differences of P < 0.05 are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
VLRB N8 recognizes a tyrosine sulfation–dependent epitope on HLA-I
Recognition of HLA-I by VLRB N8 independently of HLA-I cell surface expression levels suggested that the epitope recognized by VLRB N8 could be formed by a posttranslational modification of HLA-I. No alternative glycosylation of HLA-I on VLRB N8–reactive cells could be determined. In addition to glycosylation, cell surface receptors are frequently sulfated on tyrosine residues, a posttranslational modification mediated by the TPST1 and TPST2 enzymes using 3′-phosphoadenosine-5′-phosphosulfate (PAPS) as a universal sulfate donor (35). Treatment of VLRB N8–reactive KMS-11 cells with NaClO3, an inhibitor of PAPS biosynthesis (36), resulted in a strong reduction of VLRB N8 reactivity without modulation of HLA-I cell surface expression levels (Fig. 5A). Similarly, incubation of BJAB cells with NaClO3 following PMA/ionomycin treatment also reduced the induction of VLRB N8 reactivity (Fig. 5B). The PAPS biosynthesis inhibition affects both sulfation of carbohydrate moieties and tyrosine residues. To discriminate between these sulfation reactions, we used short hairpin RNA (shRNA) to target the TPST1 and TPST2 tyrosine sulfotransferases in BJAB cells (Fig. 5C). HLA-I recognition of VLRB N8 following antigen receptor ligation of BJAB cells was strongly reduced in BJAB cells expressing TPST2-targeting shRNA, but not in cells expressing TPST1-targeting shRNA or negative control GFP shRNA (Fig. 5D).
Fig. 5. VLRB N8 recognizes a tyrosine sulfation–dependent antigen on HLA-I.

(A) KMS-11 cells were cultured in the presence of the indicated concentrations of NaClO3 for 48 hours followed by flow cytometric assessment of VLRB N8 and HLA-I reactivity. A representative experiment is depicted in the top panel, and VLRB N8/HLA-I ratios from five independent experiments are shown in the bottom bar diagram, depicted as means ± SD. Statistical significance was determined using one-way ANOVA with Dunnett’s post test (n = 5). (B) Inhibition of VLRB N8 recognition of HLA-I on BJAB cells following PMA and ionomycin stimulation. Cells were stimulated for 1 hour with PMA and ionomycin, and VLRB N8 and HLA-I binding were assessed following a 36-hour culture with the indicated concentrations of NaClO3. Means ± SD of VLRB N8 signals normalized to HLA-I are shown. Statistical significance was determined using two-way ANOVA test with Dunnett’s post test (n = 4). (C) shRNA-mediated down-regulation of transduced BJAB cells was verified by qRT-PCR. Transcript levels of TPST1 (left) and TPST2 (right) of the indicated cell populations are depicted as means ± SD (n = 3). Statistical significance was determined using a Student’s t test. (D) shRNA-transduced BJAB cells were stimulated with anti-Ig (20 μg/ml), followed by the assessment of VLRB N8 and anti–HLA-I recognition. Numbers indicate the mean fold induction of HLA-I normalized VLRB N8. Statistical significance for induced VLRB N8 binding was determined using a one-way ANOVA test with Tukey’s post test (n = 9). (E) Tyrosine sulfation of HLA-I following antigen receptor engagement. A representative autoradiogram (left) of anti–HLA-I immunoprecipitates of unstimulated and stimulated BJAB cells and the quantitation (right) of six independent experiments are shown. 35SO4 incorporation is shown with arbitrary units (AU). Statistical significance was determined using paired Student’s t test (n = 6). (F) TPST1 and TPST2 transcript analysis of tonsillar B cell populations. Means ± SD of qRT-PCR of TPST1 or TPST2 normalized to HPRT from five independent tonsil specimens are shown. Statistically significant differences of P < 0.05 are indicated by *P < 0.05, **P < 0.01, ***P < 0.001; n.s., P > 0.05.
In subsequent experiments, we used a metabolic labeling approach to directly demonstrate tyrosine sulfation of HLA-I in response to antigen receptor engagement (Fig. 5E). These experiments showed low levels of sulfate incorporation in HLA-I in our BJAB model system in the absence of B cell stimulation. Antigen receptor engagement resulted in significant increases in HLA-I tyrosine sulfation, which was inhibited by shRNA targeting TPST2 but not TPST1 or negative control shRNA targeting GFP. Last, to validate that our TPST2-dependent in vitro stimulation system could reflect tyrosine sulfation in primary B lineage cells, we determined the presence of TPST transcript levels in tonsillar B cell populations. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) demonstrated that TPST1 and TPST2 transcripts were detected in all tonsillar B cell populations with strong increases of TPST2 mRNA in the plasma cell compartment (Fig. 5F). Combined, these experiments indicate that the unique recognition of Bmem and PC by VLRB N8 is dependent on cell type–specific tyrosine sulfation modifications of HLA-I.
DISCUSSION
In the present study, we used the nonconventional VLR antibody platform of the evolutionarily distant sea lamprey for the discovery of a biomarker on Bmem and PC. The unexpected discovery of HLA-I as the antigen recognized by VLRB N8 on Bmem and PC independently of HLA-I expression levels suggested that the recognized epitope is likely formed by a posttranslational modification of HLA-I. While the only described posttranslational modification of HLA-I occurs at a conserved N-linked glycosylation site at position N110 of the α1 domain (37), our results are in accordance with tyrosine sulfation of HLA-I following B cell activation. The extracellular domain of HLA-I contains several conserved tyrosine residues, including tyrosine Y83, located within a tyrosine sulfation consensus sequence of the α1 domain. While we demonstrated the low-level baseline tyrosine sulfation of HLA-I in VLRB N8 nonreactive cells that is greatly increased following B cell activation in a TPST2-dependent manner and is accompanied by acquisition of VLRB N8 reactivity, the identity of the sulfotyrosine residue(s) under steady-state conditions and following B cell activation remains to be determined. Similarly, analysis of the structure of VLRB N8 in complex with HLA-I will determine whether VLRB N8 directly recognizes a sulfotyrosine residue or whether VLRB N8 recognizes HLA-I in a tyrosine sulfation–dependent manner without directly engaging the sulfotyrosine.
The strong inhibitory effect on VLRB N8 recognition of HLA-I that we observed following the transduction of TPST2 shRNA is in agreement with microarray studies and our demonstration showing the expression of TPST2 transcripts in all peripheral B cell compartments and strongly increased TPST2 levels in the PC compartment (38). Although VLRB N8 does not recognize the vast majority of naïve B lineage cells, these cells display levels of TPST2 transcripts that are comparable to those observed for VLRB N8–reactive Bmem. This indicates that tyrosine sulfation events required for VLRB N8 recognition of HLA-I are likely to be regulated on the level of enzymatic activity and are not necessarily dependent on enzyme expression levels.
Sulfation of tyrosine residues has been demonstrated for several cell surface and secreted proteins (39, 40) and can alter the affinities of receptor/ligand interactions (41). Among the various HLA-I functions, the antigen presentation of peptide antigens to the antigen receptor of CD8+ T cells represents arguably one of the most significant protein/protein interactions in adaptive immunity. It will be important to compare antigen presentation by VLRB N8–reactive HLA-I peptide complexes versus complexes not recognized by the VLR antibody. A potential Bmem/PC-specific posttranslational modification raises the possibility that T cell recognition of a peptide presented on HLA-I by these cells may be subject to an as yet unexplored level of immune regulation.
Our observation of strongly increased VLRB N8 recognition of HLA-I in SLE and MS indicates the potentially dysregulated tyrosine sulfation of Bmem in these autoimmune disorders. B cells are increasingly recognized as contributors to the pathophysiology of these disorders (42, 43), including in antibody-independent B cell effector functions such as generation of proinflammatory cytokines by Bmem in multiple sclerosis (44). It will be important to expand on our initial observation of increased VLRB N8 reactivity in Bmem found in PBMCs of patients with MS and SLE and to correlate the level of Bmem recognition by VLRB N8 with clinicopathological parameters. Similarly, it will be of interest to investigate potentially altered tyrosine sulfation of proteins distinct from HLA-I.
Cell surface receptors provide the interface where cues of the extracellular microenvironment are detected and integrated into appropriate cellular responses. Posttranslational modifications of intracellular proteins are extensively studied and represent a central mechanism controlling protein expression levels and function in response to a diverse set of stimuli. In contrast, protein modifications of the extracellular domains of cell surface receptors, such as glycosylation, the highly complex covalent attachment of carbohydrates to asparagine, serine, or threonine residues (45), or sulfation of tyrosine residues, are less well understood. Investigations into the complexities and functional consequences of posttranslational modifications of the extracellular domains of cell surface receptors are limited by a paucity of suitable detection reagents. Our study highlights the unique characteristics of VLR antibodies as a novel class of highly specific, broadly applicable research reagents to detect previously unrecognized epitopes on cell surfaces of otherwise only functionally defined cells. This is of particular interest for epitopes generated by enzymatic activities that may not be revealed in transcriptomic approaches. While the precise nature and the functional properties of the epitope recognized by VLRB N8 remains to be elucidated, our data demonstrate that HLA-I on Bmem/PC is structurally distinct from other lymphocytes and may reveal induced tyrosine sulfation as a novel mechanism in the regulation of HLA-I–dependent immune responses.
MATERIALS AND METHODS
Study design
The goal of this study was to harness the adaptive VLR-based immune system of the sea lamprey to interrogate the cell surface of Bmem, to isolate monoclonal VLR antibodies with binding patterns distinct from those of conventional antibodies, and to use these VLR antibodies as novel affinity reagents for antigen identification. We used cell line systems for biochemical experiments aimed at identification and characterization of the recognized epitope and primary human samples from healthy donors and autoimmune patients to validate specificity of the reagent. All experiments were performed at least three times.
Cells and reagents
Hematopoietic cell lines were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), glutamine, 50 μM 2-mercaptoethanol, and penicillin/streptomycin (100 U/ml). Human embryonic kidney (HEK) 293T cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 5% FBS, glutamine, and penicillin/streptomycin (100 U/ml). Cells were maintained in a humidified atmosphere at 37°C and 5% CO2. Antibodies to the human cell surface antigens CD19 (clone HIB-19), CD20 [clone H1(FB1)], CD27 (clone M-T271), IgD (clone IA6-2), HLA-I (clone G46-2.6), CD38 (clone HIT-2), and CD138 (clone MI15) were obtained from BD Biosciences (San Jose, CA, USA). Antibodies to CD3 (clone HIT-3a) were obtained from BioLegend (San Diego, CA, USA), and anti–β2-microglobulin antibody BM-63 was obtained from MilliporeSigma (St. Louis, MO, USA). Anti–HLA-I antibodies HC-10 and w6/32 and anti–β2-microglobulin antibody BBM.1 were gifts from D. Williams (University of Toronto, Canada). Phycoerythrin (PE)–labeled anti-HLA w6/32 was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Anti–6×His-PE antibodies (clone AD1.1.10) were obtained from Novus Biologicals (Littleton, CO, USA), and anti-human Ig was obtained from Jackson ImmunoResearch (West Grove, PA, USA). Recombinant IFN-β and IFN-γ were purchased from PeproTech (Rocky Hill, NJ, USA), and CpG was purchased from InvivoGen (San Diego, CA, USA).
Tonsil samples were obtained from the Hospital for Sick Children (Toronto, Ontario, Canada). Peripheral blood was obtained from healthy donors and heparinized. Primary cell sample collections were approved by the ethical review boards of the Hospital for Sick Children, Toronto, and the University of Toronto in accordance with the Declaration of Helsinki. All participants gave written informed consent.
Immunization of sea lamprey larvae and generation of monoclonal VLR antibodies
Sea lamprey (P. marinus) larvae were immunized with a cocktail of human plasmacytoma cell lines (NCI-H929, U266, and RPMI 8226) in 0.66× phosphate-buffered saline (PBS) and boosted twice in 2-week intervals before harvesting of lamprey lymphocytes from the blood. VLRB expression libraries were generated as described previously (20). Briefly, total RNA was prepared from the lamprey lymphocytes using RNeasy columns (Qiagen, Hilden, Germany), followed by complementary DNA (cDNA) generation using the SuperScript II system (Invitrogen, Carlsbad, CA, USA). A VLRB expression library was generated by PCR amplification of cDNA using oligonucleotides specific to the signal peptide (5′-ATATGCTAGCCACCATGTGGATCAAGTGGATCGCCACGC-3′) and C-terminal stalk region (5′-ATATACCGGTTCAACGTTTCCTGCAGAGGGCG-3′) of VLRB transcripts and cloning of the PCR products into the eukaryotic expression vector pIRESpuro2 (Invitrogen, Carlsbad, CA, USA). To generate monoclonal VLRB antibodies, plasmids encoding VLRB sequences were transfected into HEK293T cells using the polyethylenimine (PEI) method as described previously (20). Lamprey immunizations were approved by the animal care committees of the University of Toronto and Emory University.
Generation of recombinant monoclonal VLRB antibodies
VLRB sequences were subcloned into the vector pIRESpuro2-367HH, which contains sequences encoding the invariant VLRB stalk region as well as the 6×His and hemagglutinin (HA) epitope tags. The plasmids were transfected into HEK293T cells using the PEI method (46). Culture supernatants were analyzed 3 days after transfection for the presence of VLRB antibodies by Western blotting and used for staining of primary cells and cell lines. Transfectants were selected on puromycin (1 μg/ml), and purified VLRB antibodies were obtained using Ni-NTA (nitrilotriacetic acid) affinity chromatography (20).
Flow cytometric analysis of VLRB N8 binding to cell lines and primary lymphocytes
Cell lines were stained with purified monoclonal VLRB antibodies [500 ng/ml in PBS/1% bovine serum albumin (BSA)] for 25 min on ice as described (21). Bound VLRB antibodies were detected by subsequent incubation with anti-VLRB antibody 4C4 followed by fluorescently labeled goat anti-mouse antibodies. Alternatively, bound VLRB was detected by incubation with fluorescently labeled anti-VLRB antibody 5-8A3 or with anti-HA epitope tag antibodies. PBMCs and tonsillar lymphocytes were isolated by density gradient centrifugation over lymphocyte separation medium for 20 min at 750g. VLRB-stained primary cells were blocked extensively with 5% normal mouse serum for 15 min on ice before addition of a cocktail of lineage-specific, fluorescently labeled mouse monoclonal antibodies. Dead cells were excluded by propidium iodide (1 μg/ml) or an Aqua dead cell staining reagent (Life Technologies, Burlington, Ontario, Canada). Flow data were acquired using BD FACSCanto II or Millipore Guava easyCyte HT instruments and analyzed using the FlowJo software package (Ashland, OR, USA). Gating strategies for blood and tonsillar lymphocyte cell populations are shown in fig. S5.
Affinity purification and identification of antigens detected by VLRB N8
KMS-11 plasmacytoma cells (1 × 108) were surface-biotinylated and incubated with VLRB N8 followed by cross-linkage using the amine-reactive, membrane nonpermeable thiol-sensitive DTSSP cross-linker for 30 min at room temperature. The reaction was quenched by addition of 100 mM tris (pH7.5) before lysis of the cells in 1 ml of Nonidet P-40 lysis buffer [1% Nonidet P-40, 150 mM NaCl, 5 mM EDTA, and 50 mM tris (pH 7.5)] in the presence of protease inhibitors [leupeptin (5 μg/ml), pepstatin (1 μg/ml), aprotinin (5 μg/ml), soybean trypsin inhibitor (10 μg/ml), and phenylmethylsulfonyl fluoride (40 μg/ml)] for 10 min on ice. Cell lysates were centrifuged at 20,000g for 10 min at 4°C, and the supernatants were subjected to immunoprecipitation using 5 μg of anti-HA antibodies and 30 μl of a 50% slurry of protein G Sepharose (GE Healthcare, Pittsburgh, PA, USA). Five percent of the resulting immunoprecipitates was subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE), under reducing conditions and analyzed by Western blotting using horseradish peroxidase–labeled streptavidin and enhanced chemiluminesence reagent. The remaining 95% of the immunoprecipitates were eluted in 8 M urea/100 mM ammonium bicarbonate at 95°C. Eluates were reduced with 10 mM dithiothreitol for 20 min at 60°C, allowed to cool at room temperature, and alkylated with 10 mM iodoacetamide for 15 min at room temperature in the dark. Samples were diluted fourfold in 100 mM ammonium bicarbonate to reach a concentration of ≤2 mM urea before overnight proteolytic digest with trypsin (10 μg/ml) at room temperature. The resulting tryptic peptide samples were acidified with trifluoroacetic acid at a final concentration of 1% before desalting and purification using offline C18 reverse-phase chromatography. Samples were then dried in a vacuum centrifuge and redissolved in 0.1% formic acid for liquid chromatography (LC)–tandem mass spectrometry analysis. Inline C18 reverse-phase chromatography was performed over a 120-min gradient using an integrated nano-LC system (Easy-nLC, Thermo Fisher Scientific, San Jose, CA, USA), coupled to a linear ion trap Orbitrap hybrid mass spectrometer instrument (Orbitrap Elite, Thermo Fisher Scientific, San Jose, CA, USA). Profile mode mass spectrometry spectra were acquired at a 60,000 full width at half maximum resolution in the Orbitrap, whereas tandem mass spectrometry spectra were acquired in the linear ion trap.
Peptide and protein identification
Samples were analyzed with the Sequest (version 1.4.0.288, Thermo Fisher Scientific, San Jose, CA, USA) and X! Tandem [The GPM, version CYCLONE (2010.12.01.1); https://thegpm.org/)] search engines. For both search engines, the human Uniprot database was mined for tryptic peptides. Parent ion tolerance was set to 10 parts per million, and fragment ion mass tolerance was set at 0.60 Da. Variable modifications included in the search were asparagine and glutamine deamidation, methionine oxidation, cysteine carbamidomethylation, and N-terminal Glu->pyro-Glu, Gln>pyro-Glu and loss of ammonia. Scaffold (version Scaffold_4.3.4, Proteome Software Inc., Portland, OR, USA) was used to visualize and validate peptide and protein identifications. Peptide identifications required a >95% probability based on the PeptideProphet algorithm (47). Protein identifications required a 95.0% probability based on the Protein Prophet algorithm with at least two identified peptides matching the peptide identification criteria. Proteins that could not be differentiated on the basis of tandem mass spectrometry analysis alone were grouped to satisfy the principles of parsimony.
HLA-I ablation by siRNA targeting of β2-microglobulin and shRNA-targeting of TPST1/2
Ablation of β2-microglobulin expression was performed by transiently transfecting KMS-11 cells or primary PBMCs with 10 pmol of β2-microglobulin–specific siRNA or scrambled control siRNA (Silencer Select, Ambion, Grand Island, NY, USA) using the Amaxa Nucleofection T system (Lonza, Allendale, NJ, USA) and setting O-20. Cells were analyzed for VLR antibody binding and HLA-I expression levels 48 hours after transfection using a BD FACSCanto II instrument and Aqua LIVE/DEAD for dead cell exclusion. TPST expression was down-regulated using RNA interference consortium shRNA sequences, TRCN0000330250 targeting TPST1, and TRCN0000035732 targeting TPST2, cloned into the pLKO vector. HEK293T cells were cotransfected with TPST shRNA vector, packaging plasmid psPAX2, and the envelope vector pMD2.G. BJAB cells were incubated with viral supernatants 2 days after transfection and selected with puromycin (0.5 μg/ml) for 5 days followed by antigen receptor ligation and flow cytometric assessment of HLA-I recognition by VLRB N8 and anti–HLA-I antibody w6/32. The reduction of TPST1/2 transcripts was determined by qRT-PCR using oligonucleotides 5′-CTGGAACGGTGAAGGTGACA-3′ and 5′-GCTCCCCATGCTTAACGATAAT-3′ targeting TPST1, and 5′-CAGCTCGGCTATGACCCTTA-3′ and 5′-GCTGGTGTTTTATAGTCCCCTTTC-3′ targeting TPST2, respectively. qRT-PCR experiments were performed using the KAPA SYBR FAST qPCR system (Kapa Biosystems, Wilmington, MA, USA) on a CFX96 Touch Real-Time PCR Detection instrument (Bio-Rad, Hercules, CA, USA), and values were normalized to hypoxanthine-guanine phosphoribosyltransferase expression levels.
Blocking of VLRB N8 binding to HLA-I with conventional antibodies
KMS-11 cells were preincubated with anti–HLA-I antibodies w6/32 or HC-10, anti–β2-microglobulin antibodies BBM.1 or BM-63, or anti-CD138 antibodies in PBS (2 μg/ml) supplemented with 0.5% BSA for 10 min on ice. Subsequently, VLRB N8 or VLR4 antibodies were added for an additional 20 min. The cells were washed, bound VLRB antibodies were detected with anti–His-PE–labeled secondary antibodies, and bound blocking antibodies were detected with PE-labeled anti-mouse secondary reagents followed by detection with a Millipore Guava HT instrument. Primary tonsillar lymphocytes were incubated with the previously indicated antibody panels following preincubation with the blocking antibodies. VLRB N8 binding was detected with anti–His-PE antibodies.
Modulation of VLRB N8 binding to cell lines
BJAB cells were stimulated with the indicated concentrations of anti-Ig, CpG, IFN-β, or IFN-γ followed by flow cytometric assessment of VLRB N8 and anti–HLA-I binding. For PMA (50 ng/ml)/ionomycin (2 μg/ml) stimulations, cells were washed once after 1-hour incubation time before continued culture in RPMI 1640 medium supplemented with 10% FBS. Anti-Ig stimulations on shRNA-transduced BJAB cells were performed as three independent stimulations each on three independently performed shRNA transductions. Modulation of VLRB N8 recognition of KMS-11 cells following inhibition of sulfation was assessed by culture of the KMS-11 cells in the indicated concentrations of NaClO3. Culture medium was replaced every 12 hours, and VLRB N8 and anti–HLA-I binding were assessed by flow cytometry.
Metabolic labeling of BJAB cells
Radiolabeling of BJAB cells was conducted on the basis of a modified protocol for PSGL1 tyrosine sulfation detection (48). BJAB cells (1 × 106) were stimulated for 24 hours with anti-Ig (50 μg/ml) antibodies in RPMI supplemented with 10% FBS, glutamine, 50 μM 2-mercaptoethanol, and penicillin/streptomycin (100 U/ml). Subsequently, the cells were incubated for an additional 24 hours in Eagle’s minimum essential medium (Joklik modification for suspension cultures) supplemented with 23.8 mM NaHCO3, 0.4 mM CaCl2, nonessential amino acids, 20 mM Hepes, and 100 μCi/ml [35S]Na2SO4. Cells were lysed as described, followed by immunoprecipitation with anti–HLA-I clone w6/32 antibodies and the immunoprecipitates resolved by SDS-PAGE on 4 to 15% gradient gels. The gels were dried for 2 hours at 80°C, and [35S] signals were detected using a Typhoon FLA 9500 instrument (GE Healthcare).
Statistical analysis
Statistical analysis was performed using the GraphPad Prism6 software package. Statistical significance was determined using Student’s t test, repeated-measures ANOVA with Holm-Sidak post hoc test, one-way ANOVA with Dunnett’s post test, two-way ANOVA with Dunnett’s post test, Wilcoxon signed-rank test, and Friedman test with Dunn’s multiple comparison test as indicated.
Supplementary Material
Acknowledgments
We are grateful to D. White and J. Warzyszynska for assistance with flow cytometric analysis of primary cell preparations. Funding: This study was supported by Canadian Cancer Society grant 2012-701054 to G.R.A.E., NIH grant 5U19AI096187 to G.R.A.E. and M.D.C., and NIH grant AI072435 to M.D.C. Author contributions: J.T.H.C., Y.L., and S.K. designed, conducted, and analyzed experiments. J.R.S.-G., C.Z., L.Y.T.L., J.Y., and M.S. conducted and analyzed experiments. E.G., P.C., E.J.P., T.H., A.B.-O., and J.E.W. provided specimens and critically appraised the manuscript. C.W.C., M.F.M., A.F.P., and M.D.C. designed and analyzed experiments. G.R.A.E. designed, conducted, and analyzed experiments and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Reagent requests should be addressed to G.R.A.E. and M.D.C. The VLRB N8 antibody can be provided by G.R.A.E. pending scientific review and a completed material transfer agreement. Requests for the VLRB N8 antibody should be submitted to goetz.ehrhardt@utoronto.ca.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaar7653/DC1
Fig. S1. VLRB N8 binding to cell lines does not correlate with HLA-I cell surface expression levels.
Fig. S2. VLRB N8 immunoprecipitates a prominent 42-kDa protein antigen.
Fig. S3. Immunoprecipitation of HLA-I with VLRB N8.
Fig. S4. Recognition of HLA-I by VLRB N8 on Bmem and PCs is independent of HLA-I expression levels.
Fig. S5. Gating strategies for evaluation of lymphocyte populations from blood and tonsil.
REFERENCES AND NOTES
- 1.Hammarlund E., Lewis M. W., Hansen S. G., Strelow L. I., Nelson J. A., Sexton G. J., Hanifin J. M., Slifka M. K., Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9, 1131–1137 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Yu X., Tsibane T., McGraw P. A., House F. S., Keefer C. J., Hicar M. D., Tumpey T. M., Pappas C., Perrone L. A., Martinez O., Stevens J., Wilson I. A., Aguilar P. V., Altschuler E. L., Basler C. F., Crowe J. E. Jr., Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455, 532–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein U., Rajewsky K., Küppers R., Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 188, 1679–1689 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tangye S. G., Liu Y.-J., Aversa G., Phillips J. H., de Vries J. E., Identification of functional human splenic memory B cells by expression of CD148 and CD27. J. Exp. Med. 188, 1691–1703 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohnhorst J. Ø., Bjørgan M. B., Thoen J. E., Natvig J. B., Thompson K. M., Bm1–Bm5 classification of peripheral blood B cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the B cell subpopulations in patients with primary Sjögren’s syndrome. J. Immunol. 167, 3610–3618 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Pascual V., Liu Y. J., Magalski A., de Bouteiller O., Banchereau J., Capra J. D., Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 180, 329–339 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung J., Choe J., Li L., Choi Y. S., Regulation of CD27 expression in the course of germinal center B cell differentiation: The pivotal role of IL-10. Eur. J. Immunol. 30, 2437–2443 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Denoeud J., Moser M., Role of CD27/CD70 pathway of activation in immunity and tolerance. J. Leukoc. Biol. 89, 195–203 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Wei C., Anolik J., Cappione A., Zheng B., Pugh-Bernard A., Brooks J., Lee E.-H., Milner E. C. B., Sanz I., A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J. Immunol. 178, 6624–6633 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Ehrhardt G. R. A., Hsu J. T., Gartland L., Leu C.-M., Zhang S., Davis R. S., Cooper M. D., Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med. 202, 783–791 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fecteau J. F., Côté G., Néron S., A new memory CD27–IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J. Immunol. 177, 3728–3736 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Linthicum D. S., Hildemann W. H., Immunologic responses of Pacific hagfish. III. Serum antibodies to cellular antigens. J. Immunol. 105, 912–918 (1970). [PubMed] [Google Scholar]
- 13.Finstad J., Good R. A., The evolution of the immune response. III. Immunologic responses in the lamprey. J. Exp. Med. 120, 1151–1168 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pancer Z., Amemiya C. T., Ehrhardt G. R. A., Ceitlin J., Gartland G. L., Cooper M. D., Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Rogozin I. B., Iyer L. M., Liang L., Glazko G. V., Liston V. G., Pavlov Y. I., Aravind L., Pancer Z., Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 8, 647–656 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Alder M. N., Rogozin I. B., Iyer L. M., Glazko G. V., Cooper M. D., Pancer Z., Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310, 1970–1973 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Han B. W., Herrin B. R., Cooper M. D., Wilson I. A., Antigen recognition by variable lymphocyte receptors. Science 321, 1834–1837 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velikovsky C. A., Deng L., Tasumi S., Iyer L. M., Kerzic M. C., Aravind L., Pancer Z., Mariuzza R. A., Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat. Struct. Mol. Biol. 16, 725–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchdoerfer R. N., Herrin B. R., Han B. W., Turnbough C. L. Jr., Cooper M. D., Wilson I. A., Variable lymphocyte receptor recognition of the immunodominant glycoprotein of Bacillus anthracis spores. Structure 20, 479–486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C., Ali S., St-Germain J., Liu Y., Yu X., Jaye D. L., Moran M. F., Cooper M. D., Ehrhardt G. R. A., Purification and identification of cell surface antigens using lamprey monoclonal antibodies. J. Immunol. Methods 386, 43–49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu C., Liu Y., Chan J. T. H., Tong J., Li Z., Shi M., Davani D., Parsons M., Khan S., Zhan W., Kyu S., Grunebaum E., Campisi P., Propst E. J., Jaye D. L., Trudel S., Moran M. F., Ostrowski M., Herrin B. R., Lee F. E.-H., Sanz I., Cooper M. D., Ehrhardt G. R. A., Identification of human plasma cells with a lamprey monoclonal antibody. JCI Insight 1, e84738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn-Walters D. K., Isaacson P. G., Spencer J., Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) B cells suggests that the MGZ of human spleen is a reservoir of memory B cells. J. Exp. Med. 182, 559–566 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weller S., Braun M. C., Tan B. K., Rosenwald A., Cordier C., Conley M. E., Plebani A., Kumararatne D. S., Bonnet D., Tournilhac O., Tchernia G., Steiniger B., Staudt L. M., Casanova J.-L., Reynaud C.-A., Weill J.-C., Human blood IgM "memory" B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood 104, 3647–3654 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Das S., Herrin B. R., Hirano M., Cooper M. D., Definition of a third VLR gene in hagfish. Proc. Natl. Acad. Sci. U.S.A. 110, 15013–15018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehrhardt G. R. A., Hijikata A., Kitamura H., Ohara O., Wang J.-Y., Cooper M. D., Discriminating gene expression profiles of memory B cell subpopulations. J. Exp. Med. 205, 1807–1817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebrón J. A., Bjorkman P. J., The transferrin receptor binding site on HFE, the class I MHC-related protein mutated in hereditary hemochromatosis. J. Mol. Biol. 289, 1109–1118 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Jenei A., Varga S., Bene L., Mátyus L., Bodnár A., Bacsó Z., Pieri C., Gáspár R. Jr., Farkas T., Damjanovich S., HLA class I and II antigens are partially co-clustered in the plasma membrane of human lymphoblastoid cells. Proc. Natl. Acad. Sci. U.S.A. 94, 7269–7274 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ploegh H. L., Cannon L. E., Strominger J. L., Cell-free translation of the mRNAs for the heavy and light chains of HLA-A and HLA-B antigens. Proc. Natl. Acad. Sci. U.S.A. 76, 2273–2277 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perosa F., Luccarelli G., Prete M., Favoino E., Ferrone S., Dammacco F., β2-microglobulin-free HLA class I heavy chain epitope mimicry by monoclonal antibody HC-10-specific peptide. J. Immunol. 171, 1918–1926 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Palanichamy A., Apeltsin L., Kuo T. C., Sirota M., Wang S., Pitts S. J., Sundar P. D., Telman D., Zhao L. Z., Derstine M., Abounasr A., Hauser S. L., von Büdingen H.-C., Immunoglobulin class-switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci. Transl. Med. 6, 248ra106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi K. D., Ruderman D. L., Croze E., Wagner T. C., Velichko S., Reder A. T., Salamon H., IFN-β-regulated genes show abnormal expression in therapy-naïve relapsing–remitting MS mononuclear cells: Gene expression analysis employing all reported protein–protein interactions. J. Neuroimmunol. 195, 116–120 (2008). [DOI] [PubMed] [Google Scholar]
- 32.van Baarsen L. G. M., van der Pouw Kraan T. C. T. M., Kragt J. J., Baggen J. M. C., Rustenburg F., Hooper T., Meilof J. F., Fero M. J., Dijkstra C. D., Polman C. H., Verweij C. L., A subtype of multiple sclerosis defined by an activated immune defense program. Genes Immun. 7, 522–531 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Baechler E. C., Batliwalla F. M., Karypis G., Gaffney P. M., Ortmann W. A., Espe K. J., Shark K. B., Grande W. J., Hughes K. M., Kapur V., Gregersen P. K., Behrens T. W., Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. U.S.A. 100, 2610–2615 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett L., Palucka A. K., Arce E., Cantrell V., Borvak J., Banchereau J., Pascual V., Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197, 711–723 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone M. J., Chuang S., Hou X., Shoham M., Zhu J. Z., Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. N. Biotechnol. 25, 299–317 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Baeuerle P. A., Huttner W. B., Chlorate — A potent inhibitor of protein sulfation in intact cells. Biochem. Biophys. Res. Commun. 141, 870–877 (1986). [DOI] [PubMed] [Google Scholar]
- 37.Ploegh H. L., Orr H. T., Stominger J. L., Biosynthesis and cell surface localization of nonglycosylated human histocompatibility antigens. J. Immunol. 126, 270–275 (1981). [PubMed] [Google Scholar]
- 38.Jourdan M., Caraux A., Caron G., Robert N., Fiol G., Rème T., Bolloré K., Vendrell J.-P., Le Gallou S., Mourcin F., De Vos J., Kassambara A., Duperray C., Hose D., Fest T., Tarte K., Klein B., Characterization of a transitional preplasmablast population in the process of human B cell to plasma cell differentiation. J. Immunol. 187, 3931–3941 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Pouyani T., Seed B., PSGL-1 recognition of P-selectin is controlled by a tyrosine sulfation consensus at the PSGL-1 amino terminus. Cell 83, 333–343 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Farzan M., Mirzabekov T., Kolchinsky P., Wyatt R., Cayabyab M., Gerard N. P., Gerard C., Sodroski J., Choe H., Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Ludeman J. P., Stone M. J., The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 171, 1167–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinzel S., Weber M. S., B cell-directed therapeutics in multiple sclerosis: Rationale and clinical evidence. CNS Drugs 30, 1137–1148 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Kamal A., Khamashta M., The efficacy of novel B cell biologics as the future of SLE treatment: A review. Autoimmun. Rev. 13, 1094–1101 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Duddy M., Niino M., Adatia F., Hebert S., Freedman M., Atkins H., Kim H. J., Bar-Or A., Distinct effector cytokine profiles of memory and naive human B cell subsets and implication in multiple sclerosis. J. Immunol. 178, 6092–6099 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Spiro R. G., Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R (2002). [DOI] [PubMed] [Google Scholar]
- 46.Godbey W. T., Wu K. K., Mikos A. G., Poly(ethylenimine) and its role in gene delivery. J. Control. Release 60, 149–160 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R., Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Wilkins P. P., Moore K. L., McEver R. P., Cummings R. D., Tyrosine sulfation of P-selectin glycoprotein ligand-1 is required for high affinity binding to P-selectin. J. Biol. Chem. 270, 22677–22680 (1995). [DOI] [PubMed] [Google Scholar]
- 49.McGuffin L. J., Atkins J. D., Salehe B. R., Shuid A. N., Roche D. B., IntFOLD: An integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res. 43, W169–W173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaar7653/DC1
Fig. S1. VLRB N8 binding to cell lines does not correlate with HLA-I cell surface expression levels.
Fig. S2. VLRB N8 immunoprecipitates a prominent 42-kDa protein antigen.
Fig. S3. Immunoprecipitation of HLA-I with VLRB N8.
Fig. S4. Recognition of HLA-I by VLRB N8 on Bmem and PCs is independent of HLA-I expression levels.
Fig. S5. Gating strategies for evaluation of lymphocyte populations from blood and tonsil.


