Protumor factor FACT is removed from transcribed genes by anticancer drugs curaxins.
Abstract
Human FACT (facilitates chromatin transcription) is a multifunctional protein complex that has histone chaperone activity and facilitates nucleosome survival and transcription through chromatin. Anticancer drugs curaxins induce FACT trapping on chromatin of cancer cells (c-trapping), but the mechanism of c-trapping is not fully understood. Here, we show that in cancer cells, FACT is highly enriched within the bodies of actively transcribed genes. Curaxin-dependent c-trapping results in redistribution of FACT from the transcribed chromatin regions to other genomic loci. Using a combination of biochemical and biophysical approaches, we have demonstrated that FACT is bound to and unfolds nucleosomes in the presence of curaxins. This tight binding to the nucleosome results in inhibition of FACT-dependent transcription in vitro in the presence of both curaxins and competitor chromatin, suggesting a mechanism of FACT trapping on bulk nucleosomes (n-trapping).
INTRODUCTION
Histone chaperone FACT (facilitates chromatin transcription) is involved in DNA transcription (1–6), replication (7–10) and repair (11–15), cell differentiation, and cancer development (16–19) [reviewed in (20)]. Human FACT (hFACT) is a heterodimer composed of two proteins: SPT16 (suppressor of Ty16) and SSRP1 (structure-specific recognition protein 1) (21). FACT preferentially interacts with the histone H2A/H2B dimer and also binds the H3/H4 tetramer and DNA (21–26). By interacting with different targets in the nucleosome, yeast FACT has the ability to reorganize the nucleosomal structure (24, 27, 28); this activity is likely important for transcription initiation (24, 28–30). hFACT facilitates transcription through chromatin by RNA polymerase II (Pol II) in vitro and nucleosome survival during this process (1, 2, 5) by transiently interacting with the DNA binding surface of the H2A/H2B dimers (2, 5, 6); it also facilitates nucleosome survival during transcription in cancer cells (31). In cancer and stem cells, the expression of FACT is up-regulated so that it correlates with the malignancy of tumor cells (16–19, 32–34).
Recently, FACT has been reported as the potential target for anticancer compounds curaxins (17, 33–35) that intercalate into DNA without causing DNA damage (17, 36, 37). After treatment of cancer cells with curaxins, FACT is redistributed within the cell nuclei and becomes tightly bound to chromatin [c-trapped (17)]; there is a correlation between the ability to cause c-trapping and cytotoxicity of various curaxins (37). Treatment of cancer cells with curaxins is also accompanied by activation of p53, inhibition of nuclear factor κB– (NF-κB) and hypoxia-inducible factor 1α (HIF1α)–dependent transcription, and cancer cell death (17, 34). However, the mechanism of c-trapping upon treatment by curaxins is not fully understood.
In this work, the mechanism of c-trapping was analyzed using a combination of single-particle Förster resonance energy transfer (spFRET), biochemical, and genomic approaches. Genomic data suggest that in cancer cells, curaxins induce redistribution of FACT from transcribed chromatin to other genomic loci. Our in vitro studies indicate that curaxins induce FACT binding to nucleosomes that, in turn, results in nucleosome unfolding. Together, our data suggest that curaxins create multiple “false targets” for FACT, thus inducing FACT redistribution from transcribed genes to other chromatin regions.
RESULTS
Experimental rationale
FACT facilitates transcription in vitro by interacting with the histone surfaces transiently exposed during Pol II transcription (5). Because curaxins are DNA intercalators (36) and induce FACT relocation (c-trapping), we hypothesized that curaxins could induce changes in nucleosome structure and thus create high-affinity sites for FACT binding and induce genome-wide redistribution of FACT. This hypothesis makes the following predictions: (i) Curaxins create multiple sites for FACT binding genome-wide and therefore deplete FACT from highly transcribed genes. (ii) FACT is more tightly associated with bulk nucleosomes in the presence of curaxins. (iii) Curaxins inhibit FACT action during transcription through a nucleosome only in the presence of competitor chromatin. Below, we systematically evaluate these predictions. Throughout the study, we used curaxin CBL0137. This curaxin has been selected as our primary experimental model because, in preclinical studies, it can strongly inhibit growth of various, including drug-resistant, cancers (35, 38). Some in vitro data were confirmed with a structurally similar curaxin, CBL0100.
Curaxins induce redistribution of FACT from transcribed chromatin to other genomic loci
hFACT facilitates Pol II transcription through chromatin in vitro (1, 2, 5), raising the possibility that curaxins affect association of FACT with Pol II–transcribed chromatin in cancer cells. To obtain initial clues on colocalization of FACT and transcribing Pol II, we have analyzed immunofluorescence of SSRP1 and transcribing Pol II using anti-SSRP1 and anti–Pol II S2-P antibodies, respectively, before and after curaxin treatment (fig. S1). Because FACT is an abundant protein localized in the nuclei, it was difficult to detect colocalization of the small fraction of FACT bound to chromatin in the absence of curaxins with transcribing Pol II using an immunofluorescence approach (fig. S1). However, after curaxin treatment, Pol II and FACT were localized within different chromatin regions (fig. S1).
To evaluate the effect of curaxins on FACT association with transcribed genes in more detail, the distribution of FACT in the genome of HT1080 cancer cells was studied using chromatin immunoprecipitation with anti-SSRP1 antibodies followed by next-generation sequencing (ChIP-seq) before or after treatment with curaxin CBL0137 (19). FACT association with transcribed and nontranscribed genomic regions was notably reduced or increased, respectively, upon treatment with CBL0137 (Fig. 1, A and B). The total amount of FACT in CBL0137-treated cells was not changed [fig. S2 and (17)]. The total amount of chromatin-bound FACT was substantially increased (figs. S2 and S3). The SSRP1 ChIP-seq data were compared with nascent RNA sequencing (RNA-seq) data (37) to determine FACT occupancy in the transcribed regions of the genome before and after curaxin treatment. FACT is highly enriched on genes actively transcribed in untreated cells, especially on the 5% of most actively transcribed genes (n = 653; Fig. 1C and fig. S4). Consistently, there is a strong correlation between the extent of FACT enrichment and levels of transcription for the 500 highly transcribed genes in untreated cells (Fig. 1, D and E, and figs. S5 and S6). The depletion of SSRP1 upon CBL0137 treatment preferentially occurs at actively transcribed genes (Fig. 1 and figs. S4 to S6). Thus, general redistribution of FACT from transcribed to nontranscribed chromatin regions occurs upon treatment of the cells with curaxin CBL0137. It would be expected that global changes in the chromatin structure occur soon after curaxin treatment, and this is indeed the case (36).
Fig. 1. FACT subunit SSRP1 is preferentially enriched on highly transcribed genes, and curaxins remove SSRP1 from the gene bodies.
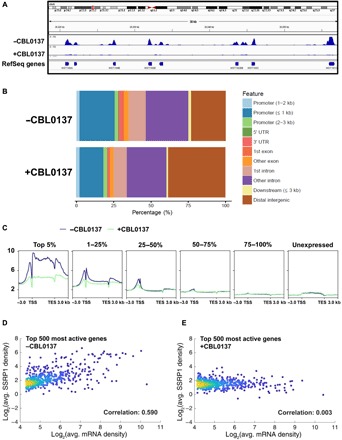
(A) Integrated genome views of the SSRP1 distributions at selected regions of chromosome 6 of the human genome. (B) Distribution of SSRP1 peaks in control and CBL0137-treated cells in relation to genome annotation features. (C) SSRP1 protein is preferentially enriched on highly transcribed genes. Average SSRP1 occupancy near the transcription start sites (TSSs), in the transcribed regions, and in transcription end sites (TESs) of the genes in HT1080 cells (incubated in the absence or presence of 3 μM curaxin CBL0137) was determined using ChIP-seq. The genes are grouped on the basis of the RNA-seq data. The corresponding heatmaps are shown in fig. S4. (D and E) Curaxins preferentially remove SSRP1 from actively transcribed genes. Density scatter plots representing average SSRP1 densities over gene bodies against the level of transcription of the corresponding genes, quantified by the average level of nascent transcripts determined in HT1080 cells by NET-seq (native elongating transcript sequencing). Analysis of all (fig. S5) or top 500 most actively transcribed genes (D and E) in the absence (D) or presence (E) of the curaxin. For the top 500 most active genes, the Pearson correlation coefficient is markedly decreased (from 0.590 to 0.003) upon treatment with curaxin.
FACT and curaxins strongly, synergistically, and reversibly uncoil nucleosomal DNA
The effect of curaxins on the nucleosome structure and FACT-nucleosome interaction was studied using the spFRET in solution in vitro. Precisely positioned nucleosomes assembled on a DNA template containing the 603 nucleosome positioning sequence (NPS) (39, 40) and fluorescent labels Cy3 and Cy5 at the +13 and +91 positions on nucleosomal DNA (from the 5′ end of nontemplate DNA strand of nucleosomal DNA; Fig. 2A), respectively, were gel purified (fig. S7). The donor fluorophore (Cy3) was excited by a 514.5-nm wavelength laser (28). The nucleosome complexes were diffused in solution, and only one particle of the nucleosome complex traversed the focal volume of the microscope at any given time. The fluorescence intensities of both donor (Cy3) and acceptor (Cy5) dyes were then measured, and the proximity ratios (EPR) that reveal the changes of distances between labeled DNA sites through changes in FRET efficiency were calculated (28). The data obtained using the fluorescently labeled nucleosome are typically described by two Gaussian peaks: a minor peak that corresponds to histone-free DNA in the solution and a major peak representing the intact nucleosome (Fig. 2, B and C). The signals obtained from control nucleosomes are stable during the experimental time periods (fig. S8).
Fig. 2. FACT and curaxins strongly and synergistically affect the nucleosomal structure: Analysis by spFRET.
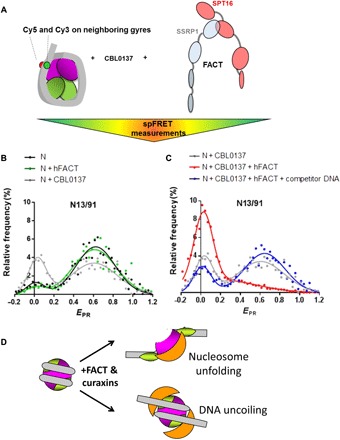
(A) Experimental approach. The mononucleosomes contained the single pair of Cy3 and Cy5 dyes on the nucleosomal DNA (the positions of Cy3 and Cy5 are shown by green and red circles, respectively). spFRET from nucleosomes was measured in the absence or presence of curaxin CBL0137, FACT, and/or competitor DNA. (B and C) Typical frequency distributions of FRET efficiencies (EPR). Analysis by spFRET microscopy. Sample sizes and other numerical parameters are shown in table S1. (B) Only minor changes in nucleosome structure are detected in the presence of either CBL0137 or FACT. (C) FACT and CBL0137 together induce marked and partially reversible uncoiling of the nucleosomal DNA. The uncoiling is partially reversed by subsequent addition of an excess of competitor DNA, resulting in removal of FACT from the complex. (D) Possible changes in the nucleosome structure in the presence of FACT and curaxins: nucleosome unfolding or uncoiling of nucleosomal DNA from the histone octamer.
FACT used in our studies has been purified from insect cells and therefore is likely phosphorylated and unable to bind nucleosomes (22, 25, 26, 41, 42); this is the state of bulk FACT complexes in cancer cells (6, 22, 26). Adding either CBL0137 or FACT alone to nucleosomes results in only minimal changes of EPR (Fig. 2B). In contrast, adding CBL0137 and FACT together results in a marked increase of the peak corresponding to the histone-free DNA and a corresponding decrease of the peak corresponding to the intact nucleosome (Fig. 2C). The data indicate that in the presence of CBL0137, FACT binds to nucleosomes, resulting in FACT trapping on nucleosomes (n-trapping). Thus, the data suggest that FACT binds to nucleosomes only in the presence of curaxins and induces a strong, partially reversible nucleosome unfolding or uncoiling of the nucleosomal DNA (Fig. 2D). Because FACT alone does not bind nucleosomes, the affinity of FACT to nucleosomes is strongly increased in the presence of curaxins. It should be noted that the term n-trapping was used previously to describe the interaction of FACT with the hexasomes (a nucleosome missing one H2A/H2B dimer) (36).
The changes in EPR are partially reversed by adding an excess of unlabeled histone-free DNA after the formation of FACT-nucleosome complex unfolded in the presence of curaxins to remove FACT (and likely some curaxins bound to nucleosomal DNA) from the complexes (28). The reversibility indicates that in the presence of CBL0137 and FACT, the nucleosome is either unfolded or the nucleosomal DNA is uncoiled without notable accompanying dissociation of the majority of core histones from the nucleosomal DNA. Any histone loss from the nucleosomes after FACT-dependent uncoiling is likely to be irreversible (43), especially in the presence of an excess competitor DNA. Even if nucleosomes would reform after the reversal of FACT binding, they will preferably form on the competitor DNA. Consistently, our previous experiments have shown that nucleosome reorganization by yeast FACT is accompanied by only minimal (typically 5 to 10%) loss of the dimers (28). The reversibility is incomplete, and the low EPR peak is slightly increased as compared to the control nucleosomes (Fig. 2C), likely because some nucleosomes are displaced by FACT in the presence of CBL0137 (see below) and some nucleosomes contain DNA that is partially uncoiled from the octamer in the presence of curaxins.
Next, the effect of curaxins on FACT-nucleosome interaction was analyzed in vitro using a gel shift assay. The nucleosome cores were assembled on the 32P-labeled 147–base pair (bp) DNA fragment containing the 603 NPS. Gel-purified nucleosomes contain less than 5% histone-free DNA (fig. S7). After incubation in the presence of FACT and/or curaxins CBL0137 or CBL0100, the samples were incubated either with or without an excess of unlabeled DNA competitor and analyzed by native polyacrylamide gel electrophoresis (PAGE) (Fig. 3A). FACT, competitor histone-free DNA, or curaxins alone minimally affect the nucleosomes (Fig. 3, B and C). Nucleosomes are slightly more efficiently disrupted after incubation with CBL0100 than after incubation with CBL0137 (Fig. 3, B and C), probably because CBL0100 is a more potent DNA intercalator than CBL0137 (17, 36).
Fig. 3. Nucleosomes are reversibly destabilized in the presence of curaxins and FACT.
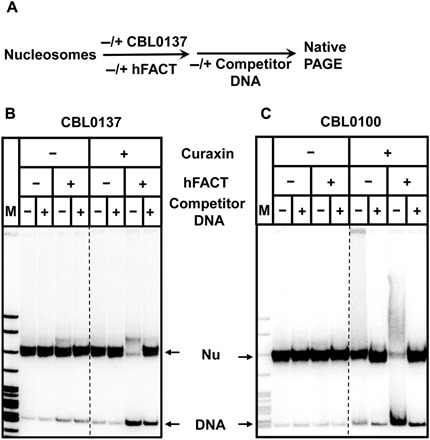
(A) Experimental approach. DNA-labeled nucleosomes were incubated in the presence of FACT, unlabeled competitor DNA, CBL0137 (B), or CBL0100 (C) and analyzed by native PAGE. Note that in the presence of both FACT and curaxins, nucleosomes are unstable (as reflected in release of histone-free DNA); this effect is reversed in the presence of DNA competitor that partially removes FACT from the nucleosomes (see Fig. 2).
In contrast, nucleosomes incubated in the presence of both FACT and curaxins are very unstable, with approximately 70% of the templates disrupted during gel electrophoresis in the absence of competitor DNA. This effect is largely reversed by adding the excess of competitor DNA after the reaction (Fig. 3, B and C), indicating that nucleosomes incubated in the presence of FACT and curaxins are stable in solution, but unstable during electrophoresis. Apparently, removal of FACT from the nucleosomes in the presence of competitor DNA in solution results in recovery of nearly intact nucleosomes that are stable during electrophoresis. Thus, consistent with the results of the spFRET studies (Fig. 2), biochemical studies suggest that nucleosomes are destabilized in the presence of FACT and curaxins, but the majority of these nucleosomes are recovered in the presence of an excess of unlabeled DNA competitor that removes FACT from the FACT-nucleosome complexes (Fig. 3).
Together, the data suggest that in the presence of curaxins, FACT binds to nucleosomes (n-trapped), causing a considerable change in the nucleosome structure that is partially reversible and is not accompanied by the loss of core histones (Figs. 2 and 3). The FACT-nucleosome complex formed in the presence of curaxins can be disrupted in the presence of competitor DNA or during electrophoresis.
Curaxins and competitor nucleosomes synergistically inhibit FACT-dependent transcription in vitro
To evaluate the effect of n-trapping on FACT-dependent transcription, a well-established in vitro Pol II transcription system containing the purified yeast Pol II and nucleosomes or corresponding DNA templates containing the 603 NPS was used [Fig. 4A (39, 40, 44)]. Authentic Pol II elongation complexes (ECs) were assembled and immobilized on Ni-NTA beads and ligated to DNA or nucleosomal templates (43, 45). The transcription was continued in the presence of incomplete combination of nucleotide triphosphates (NTPs), RNA was pulse labeled in the presence of α-32P–labeled guanosine 5′-triphosphate (GTP), and the ECs were stalled. The Pol II ECs were then washed, eluted from the beads, and further transcribed in the presence of all unlabeled NTPs, FACT, curaxin (CBL0137 or CBL0100), and/or competitor core nucleosomes. The pulse-labeled RNA transcripts were purified and analyzed by denaturing PAGE (Fig. 4, B and C).
Fig. 4. Curaxins and competitor nucleosomes synergistically inhibit FACT-dependent transcription in vitro.
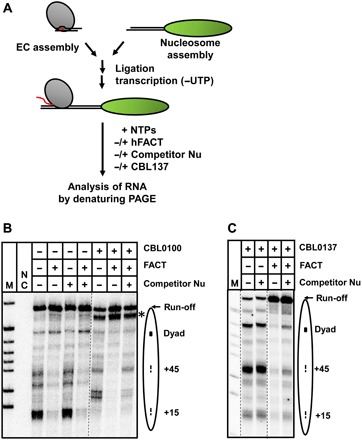
(A) Experimental approach. The assembled Pol II EC-119 was ligated to the 603 DNA or nucleosome (43, 58). The RNA was pulse labeled in the presence of a subset of NTPs and [α-32P]GTP, and Pol II was stalled at position −83 (43, 45, 58). Then, transcription was resumed by adding all unlabeled NTPs, FACT (to 0.2 μM), unlabeled competitor nucleosomes, and CBL0100 (B) or CBL0137 (C). Note that without competitor nucleosomes, curaxins moderately stimulate transcription through chromatin but do not inhibit FACT action. UTP, uridine 5′-triphosphate.
In the presence of the curaxins, the pattern of Pol II pausing is altered (Fig. 4, B and C), likely due to curaxin intercalation in DNA that affected the DNA structure in the nucleosome (36). The differences between CBL0137- and CBL0100-induced Pol II pausing patterns (Fig. 4, B and C) likely occur due to the different structures of the curaxins. In particular, CBL0100 is a more potent DNA intercalator than CBL0137 (17, 36); consistently, it has a stronger effect on the pausing. The +15 nucleosome-specific pausing is partially relieved in the presence of the curaxins, suggesting that they destabilize some DNA-histone interactions in the nucleosome. However, curaxins do not notably affect the catalytic activity of Pol II (fig. S9). In agreement with the previously published results (5), FACT facilitates Pol II transcription through the nucleosome, relieving nearly all nucleosomal pausing (Fig. 4, B and C). Curaxins do not considerably inhibit FACT action during transcription through the nucleosome (Fig. 4, B and C), suggesting that FACT redistribution upon curaxin treatment in vivo does not occur because of direct inhibition of Pol II or FACT by curaxins. Therefore, we next studied the effect of curaxin-induced n-trapping on FACT action during Pol II transcription.
In the control experiments in the absence of curaxins and FACT, free nontranscribed mononucleosomes present in the reaction minimally affect the nucleosomal pausing pattern and FACT activity; the curaxins themselves do not inhibit FACT action (Fig. 4, B and C). However, the action of FACT during Pol II transcription through the nucleosome is inhibited in the presence of both curaxins and excess of competitor mononucleosomes added together (Fig. 4, B and C).
Curaxin treatment results in FACT-induced nucleosome unfolding and n-trapping under the same experimental conditions (Figs. 2C and 3), suggesting that FACT-induced unfolding of competitor mononucleosomes and FACT n-trapping cause the inhibition of FACT-dependent transcription, likely after redistribution of FACT to competitor nucleosomes present in excess. To further evaluate this possibility, transcription was conducted in the presence of an excess of biotinylated competitor nucleosomes, the nucleosomes were immobilized on magnetic beads, and the amount of FACT remaining in supernatant was quantified (fig. S10). In the presence of CBL0137, less than 30% of FACT remained in the solution, confirming that FACT n-trapping on the competitor nucleosomes causes the inhibition of FACT-dependent transcription.
Mechanism of curaxin action in cancer cells: N-trapping
Together, our data suggest that FACT is preferentially associated with transcribed genes in cancer cells, most likely interacting with nucleosomes that are already destabilized by transcribing Pol II complexes. The association of FACT with bodies of transcriptionally active genes is considerably decreased upon curaxin treatment, preferentially on the highly transcribed genes (Fig. 1). FACT interacts with and unfolds nucleosomes in the presence of curaxins (n-trapping; Figs. 2 and 3). Destabilization of nucleosomes, caused by genome-wide CBL0137 binding, makes possible redistribution of FACT to multiple chromatin loci (Fig. 1), which thus compete with transcribed regions for FACT and deplete FACT from transcribed genes. Our in vitro data suggest that curaxins likely uncoil the nucleosomal DNA and thus affect the Pol II transcription through chromatin (Fig. 4). Curaxins inhibit FACT action during Pol II transcription through the nucleosome only in the presence of competitor mononucleosomes, suggesting that the FACT complexes are n-trapped on the excess of transcriptionally silent nucleosomes destabilized by the drug (Fig. 4).
The data suggest a model describing the effect of curaxins on FACT-dependent transcribed genes (Fig. 5). In cancer cells, FACT is associated with gene bodies, especially with the highly transcribed genes, and participates in transcription through chromatin and nucleosome survival by transiently interacting with histone H2A/H2B dimers within the transcribed nucleosomes (2, 5). Curaxins intercalate into DNA of nucleosomes present in various genomic locations, destabilize the nucleosomes, and, together with FACT, cause reversible, partial uncoiling of nucleosomal DNA. FACT tightly binds to the unfolded nucleosomes and thus becomes n-trapped on the vast excess of nontranscribed chromatin that is present in cancer cells; as a result, FACT is depleted from the transcribed regions of the genes.
Fig. 5. Mechanism of curaxin action in cancer cells: N-trapping.

In the cells without curaxin treatment (−Curaxins), FACT loosely interacts with the transiently exposed DNA binding surface of the H2A/H2B dimer and thus facilitates transcription through the nucleosome barrier. In the presence of curaxins (+Curaxins), FACT causes genome-wide nucleosome unfolding and is tightly trapped on the unfolded nucleosomes (n-trapping). Because the nontranscribed chromatin is present in vast excess, FACT is trapped primarily within inactive chromatin regions. Thus, curaxin-induced n-trapping reduces density of FACT on transcribed regions, resulting in loss of FACT from the transcribed genes and likely affects chromatin dynamics and/or transcript elongation process.
DISCUSSION
We observed a strong decrease of FACT association with transcribed genes after curaxin treatment. This should result in inhibition of FACT-dependent Pol II transcription. Indeed, n-trapping of FACT results in the inhibition of FACT-dependent Pol II transcription in vitro, suggesting a plausible mechanism for curaxin action in cancer cells through their effect on transcription. Curaxins also alter the structure of nucleosomes, suggesting that, together with FACT, they could act by perturbing global chromatin structure in cancer cells. Last, a recent study has demonstrated that curaxins can induce accumulation of Z-DNA and FACT trapping on the Z-DNA (z-trapping) in cancer cells (36); this mechanism could additionally contribute to the curaxin action.
The process of nucleosome unfolding by FACT in the presence of curaxins is adenosine 5′-triphosphate (ATP) independent and is partially reversed after removal of FACT with an excess of competitor DNA. Only nonphosphorylated (bacterially expressed) hFACT can interact with nucleosomes in the absence of curaxins (the structural state of the nucleosomes in the complex is unknown) (22, 41). In contrast, phosphorylated hFACT purified from insect cells [and likely representing the majority of hFACT complexes in cancer cells (6, 25, 26)] does not interact with intact nucleosomes (6, 22, 25, 26, 41, 42). In contrast, yeast FACT can induce reversible nucleosome unfolding even in the absence of curaxins, but only in the presence of Nhp6 protein (28); this activity is likely important for DNA replication and transcription initiation (7, 10, 46). In the complexes of nucleosomes with yeast or human FACT in the absence of curaxins, the interactions occur through multiple binding surfaces on the histone octamer (including on H2A/H2B dimers and H3/H4 tetramers) and FACT (including C-terminal tails of Spt16 and SSRP1 subunits, and mid-AID domain of Spt16) (22–26, 28). It is possible that this multipoint FACT-nucleosome interaction is a prerequisite for nucleosome unfolding that occurs in the presence of curaxins.
The difference in the nucleosome unfolding activities of yeast and human FACT are likely explained by the requirement for an additional protein that is involved in nucleosome unfolding in the case of yeast FACT. FACT subunits Spt16 and SSRP1 (Pob3 in yeast) are highly conserved between yeast and human; additional yeast Nhp6 protein is required for nucleosome unfolding by yeast FACT (7, 21, 46) and by phosphorylated hFACT (47). Yeast Nhp6 is an HMGB-like DNA binding protein that supports various functions of yeast FACT (7, 46). Similar to phosphorylated hFACT, yeast FACT does not induce nucleosome unfolding in the absence of Nhp6 protein (24, 28, 48), suggesting that the lack of Nhp6-like protein in hFACT results in additional requirements for curaxins to induce nucleosome unfolding. The ability of hFACT to unfold nucleosomes in the presence of curaxins suggests that hFACT has an intrinsic, conserved nucleosome unfolding activity. However, hFACT can unfold nucleosomes only if they are destabilized, e.g., by curaxins or by DNA damage (25). Alternatively, there could be an unknown Nhp6-like human protein that facilitates interaction of hFACT with nucleosomes.
Previously, the c-trapping of FACT was positively correlated with the curaxin toxicity for types of cancer that involved several signaling pathways related to cell death (17, 34). Transcriptions of cell survival–related genes such as NF-κB– and HIF1α-dependent genes were inhibited in the curaxin-treated cancer cells (17). During the c-trapping, curaxins also activate the CK2-p53 pathway involved in cell death (17), suggesting that curaxin-induced FACT-nucleosome complex could serve as a signal inducing the cell death signaling cascade. Because the c-trapping of FACT occurs during a very short time period (within 1 min) after adding curaxins to cancer cells (36), c-trapping therefore is a very early step, acting upstream of and possibly inducing the cell death signaling pathways.
In summary, we have shown that hFACT interacts with and unfolds bulk nucleosomes in the presence of curaxins (n-trapping). This, in turn, results in redistribution of FACT from the bodies of transcribed genes to other genomic regions, strongly affecting the function of FACT in cancer cells.
MATERIALS AND METHODS
Cells
HT1080 cells were obtained from the laboratory of A. Gudkov (Roswell Park Cancer Institute) and authenticated using short tandem repeat polymerase chain reaction (PCR) (100% match with American Type Culture Collection cell line). Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum and antibiotic/antimycotic solution. HT1080 cells constantly expressing green fluorescent protein–tagged SSRP1 or mCherry-tagged H2B were described previously (36).
Analysis of colocalization of FACT and transcribing Pol II using immunofluorescence
Cells were plated in 35-mm plastic dished with no 1.5 glass inserts. Cells were fixed in 4% paraformaldehyde for 10 min, washed three times with phosphate-buffered saline, and blocked in 3% bovine serum albumin (BSA) and 0.1% Triton X-100. Antibody staining was done in 0.5% BSA and 0.1% Triton X-100. Antibody against phosphorylated RNA polymerase (RBP1) at Ser2 was purchased from Abcam (clone H5, catalog no. ab24758) and diluted 1:200. Antibody against SSRP1 was purchased from BioLegend (clone 10D1, catalog no. 609702) and diluted 1:500. Images were taken using Zeiss Axio Observer A1 inverted microscope with N-Achroplan 100×/1.25 oil lens, Zeiss MRC5 camera, and AxioVision Rel.4.8 software.
Immunoblotting
Cells were lysed in 1× cell culture lysis reagent buffer (Promega) supplemented with protease inhibitor cocktail (Roche). After 10 min of incubation on ice, lysates were spun down at 13,000 rpm for 10 min at +4°C. Supernatant was collected and used as soluble protein extract. Pellet was suspended in the same buffer and sonicated for 10 cycles, 30 s on/30 s off on ice. Protein concentrations were measured using Bradford reagent (Bio-Rad). Electrophoresis and blotting were done using Bio-Rad Criterion running and blotting cameras and polyvinylidene difluoride (PVDF) membrane (Invitrogen). The following antibodies were used for the detection of SSRP1 (clone 10D1, catalog no. 609702, BioLegend), SPT16 (clone 8D2, catalog no. 607001, BioLegend), and β-actin (clone AC-74, catalog no. A2228, Sigma-Aldrich).
ChIP-seq and nascent RNA-seq
SSRP1 ChIP-seq data in the form of bed files are available at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45393 (19). Nascent RNA-seq data done in HT1080 cells are available at GSE107595 (37). Raw reads that passed quality filter from Illumina RTA were mapped to human reference genome (hg19) after quality control check using FastQC. For both ChIP-seq and nascent strand RNA-seq data, the raw counts in gene body were generated using featureCounts from Subsread R package with RefSeq gene annotation database (49, 50). The quantification data were normalized using DESeq2 R package. To remove impact of overlapping genes on both data, genes with overlapping were removed from analysis. The big wiggle files, heatmaps, and profiles of SSRP1 binding around genes were generated using deepTools software (51, 52).
Alignments of densities of ChIP-seq against the levels of nascent RNA-seq
Single-end SSRP1 ChIP-seq reads (19) were aligned to the human reference genome hg19 using Bowtie2 with parameters -X 1000 (to map sequences up to 1 kb with maximum accuracy). The aligned single-end reads were first analyzed using the SPP package (available at https://github.com/hms-dbmi/spp/) to estimate the average fragment length that resulted in the sonication process. This average length of 85 bp was used to extend the fragments from the 5′ end that was sequenced toward the 3′ end. The raw occupancy profiles were computed in MATLAB by stacking the extended (85 bp) reads and counting the number of reads that overlapped with each base pair. The raw occupancy profile for each chromosome was normalized by the chromosome average. For each RefSeq annotated gene, the average SSRP1 density was computed by averaging the normalized SSRP1 occupancy over the whole gene body. For counting the levels of transcriptions, single-end nascent RNA-seq reads (37) were aligned to the human transcriptome using TopHat2 with the default parameters. The raw nascent RNA-seq counts were computed in MATLAB by stacking all 5′ ends of the aligned reads, which correspond to the 3′ ends of the nascent transcripts. For each chromosome, the corresponding NET-seq profiles were normalized by the chromosome average. For each RefSeq annotated gene, the normalized nascent RNA-seq counts were averaged over the whole gene body.
Analysis of SSRP1 occupancy of various genomic regions in control and CBL0137-treated samples
MACS 2.0 (53) with default parameters for pair-end BAM files was used to identify peaks from ChIP-seq of SSRP1 (19). Heatmaps and profiles under all conditions were generated using deepTools on RPKM (reads per kilobase million) normalized coverage data using merged bam files of biological replicates. Tag densities for RefGenes body with 3 kb up- and downstream were plotted. Heatmaps of the unexpressed and top-expressed (5%) genes and quantiles of all genes were prepared using nascent RNA-seq data for control untreated HT1080 cells (37).
DNA templates
All DNA templates were amplified by PCR and purified from gel electrophoresis using a gel extraction kit (Omega Bio-Tek), as described (39). For spFRET, DNA templates were assembled from fluorescence oligos [Cy3/Cy5-labeled pairs at +13 and +91 bp relative to 603 NPS boundary], as described (28). All sequence of primers and templates will be provided per request.
Protein purification
Yeast Pol II was purified as described (45, 54). -H1 chicken erythrocyte chromatin, histone H2A/H2B dimer, and histone H3/H4 tetramer were purified as described (44, 55, 56). hFACT was purified as described (2).
Nucleosome assembly and purification
Nucleosomes were assembled as described (44). In short, NPS templates were mixed with purified chicken erythrocyte H2A/H2B dimers and H3/H4 tetramers or with -H1 chicken erythrocyte chromatin in the presence of salmon testes DNA as competitors in the buffer containing 2 M NaCl, 10 mM tris-HCl (pH 7.4), 0.1% NP-40, and 0.2 mM EDTA (pH 8). The samples were then dialyzed against buffers with progressively decreasing (2 M, 1.5 M, 1 M, 0.75 M, 0.5 M, and 10 mM) NaCl at 4°C for 2 hours at each step. For gel shift analysis of FACT binding and spFRET, nucleosomes were gel purified after assembly and analyzed as described (57).
Gel shift analysis of FACT binding
Core nucleosomes (final concentration, 15 nM) were incubated with hFACT (final concentration, 400 nM) in the transcription buffer [TB; 20 mM tris-HCl (pH 8.0), 5 mM MgCl2, and 2 mM β-mercaptoethanol] containing 40 mM KCl and then mixed with CBL0137 (final concentration, 1 μM) or CBL0100 (final concentration, 2.5 μM) for 1 min. Curaxins were provided by Incuron Inc. The samples were analyzed by native PAGE as described (57). To reverse FACT binding, DNA competitor [salmon testes DNA (250 ng/μl)] was added to the loading buffer.
Single-particle Förster resonance energy transfer
Nucleosomes reconstituted in the presence of donor chromatin from chicken erythrocytes and fluorescently labeled DNA templates (containing 603 positioning sequence) were gel purified and used for spFRET measurements at a concentration of 0.5 nM, as described (28). Nucleosomes were incubated in the presence of hFACT (0.1 μM) and/or CBL0137 (5 μM) in the TB containing 150 mM KCl for 5 min at 25°C. spFRET measurements and raw data analysis were conducted as described (28).
In vitro transcription assay
The in vitro transcription assay with yeast Pol II was performed as described (5, 58). In short, the ECs were assembled using purified yeast Pol II and DNA/RNA oligonucleotides. The assembled Pol II ECs were immobilized on Ni-NTA (nickel–nitrilotriacetic acid) resins (Qiagen), washed, and eluted from the beads. ECs and nucleosomal templates (or corresponding DNA fragments) were ligated by T4 ligase (Promega). Pol II was then progressed to position −83 using limited mixture of NTPs and α-32P–labeled GTP. ECs were washed from the unincorporated NTPs and eluted, and transcription was resumed in the presence of unlabeled NTPs, hFACT (final concentration, 0.1 μM), CBL0137 (1 μM) or CBL0100 (5 μM), and mononucleosomes (~0.18 μM) in the TB containing 150 mM KCl for 10 min. Transcription was terminated using phenol/chloroform extraction. RNA transcripts were purified and analyzed by denaturing PAGE.
Biotinylated nucleosome pull-down assay
The competitor core nucleosomes were assembled using 5′ end-biotinylated 147-bp DNA template. Transcription by Pol II in the presence or absence of curaxins, FACT, and competitor core nucleosomes (containing 50% of end-biotinylated DNA) was conducted as described above. After transcription, hydrophilic streptavidin magnetic beads (NEB) were added to the reaction mixture for 10 min at room temperature, and the supernatant was collected. The beads were resuspended in 1× TB40 buffer. The SDS loading buffer [1× SDS loading buffer containing 50 mM tris-HCl (pH 6.8), 2% SDS, 70 mM β-mercaptoethanol, and 10% glycerol] was added to the samples and heated at 99°C for 10 min. Electrophoresis and blotting were done using NuPAGE (4 to 12% bis-tris) (Invitrogen), NuPAGE MOPS SDS Running Buffer (Invitrogen), PVDF membrane (Invitrogen), and film developing solutions (SuperSignal West Femto Maximum Sensitivity Substrate, Thermo Fisher Scientific). The Flag-tagged SPT16 protein was detected using the DYKDDDDK Tag Monoclonal Antibody (FG4R) (Invitrogen). The data were quantified using OptiQuant software and normalized for total protein loading.
Supplementary Material
Acknowledgments
We thank K. Kudriashova for help with collecting spFRET data. Funding: This work was supported by NIH grant R01GM119398 to V.M.S., NIH grant HG004708 to A.V.M., Incuron LLC and NIH grant R01CA197967 to K.V.G., and NIH grant P30CA016056 to Roswell Park Comprehensive Cancer Center. spFRET experiments were supported by the Russian Science Foundation (grant 14-24-00031). R.V.C. was supported by the Intramural Research Program of the NIH. This study used the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov). Author contributions: Formulation or evolution of overarching research goals and aims: H.-W.C., M.E.V., M.P.K., A.V.M., K.V.G., and V.M.S.; development or design of methodology and creation of models: H.-W.C., M.E.V., A.S., O.I.K., A.V.M., A.V.F., K.V.G., and V.M.S.; programming and software development: R.V.C. and J.W.; verification of the overall replication/reproducibility of results/experiments: H.-W.C., M.E.V., A.S., R.V.C., O.I.K., A.V.F., K.V.G., and V.M.S.; application of statistical, mathematical, and computational techniques to analyze the data: H.-W.C., M.E.V., R.V.C., J.W, and A.V.M.; performing the experiments or data/evidence collection: H.-W.C., M.E.V., A.S., J.W., A.V.F., and K.V.G.; provision of reagents, materials, or other analysis tools: A.S., R.V.C., J.W., O.I.K., and A.V.F.; writing the initial draft: H.-W.C., A.S., A.V.F., K.V.G., and V.M.S.; preparation, creation, and/or presentation of the published work: H.-W.C., M.E.V., A.S., R.V.C., M.P.K., A.V.F., K.V.G., and V.M.S.; visualization/data presentation: H.-W.C., J.W., M.P.K., K.V.G., and V.M.S.; oversight and leadership responsibility for the research activity planning: H.-W.C., O.I.K., K.V.G., and V.M.S.; management and coordination responsibility for the research activity planning and execution: H.-W.C., O.I.K., M.P.K., K.V.G., and V.M.S.; acquisition of the financial support: M.P.K., K.V.G., and V.M.S. Competing interests: K.V.G. performs consulting services for Incuron Inc. and is an inventor on a patent application related to this work (no. WO 2010/ 042445 A1, filed on 15 April 2010). The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaav2131/DC1
Fig. S1. Analysis of colocalization of FACT and transcribing Pol II using immunofluorescence.
Fig. S2. Redistribution of FACT in HT1080 cells from nucleoplasm to chromatin.
Fig. S3. Redistribution of FACT in nucleus of HT1080-treated cells from nucleoplasm to chromatin.
Fig. S4. Heatmaps of SSRP1 occupancy in the vicinity of TSSs and TES of the genes in HT1080 cells.
Fig. S5. Analysis of the average SSRP1 densities over gene bodies against the levels of transcription of the corresponding genes.
Fig. S6. Curaxins preferentially remove SSRP1 from gene bodies of highly transcribed genes.
Fig. S7. Analysis of gel-purified nucleosomes by native PAGE.
Fig. S8. Typical frequency distributions of FRET efficiencies of the N13/91 nucleosomes.
Fig. S9. The catalytic activity of Pol II is minimally affected by curaxins.
Fig. S10. Trapping of FACT on immobilized competitor nucleosomes after Pol II transcription in the presence of CBL0137.
Table S1. Statistical data for spFRET analysis.
REFERENCES AND NOTES
- 1.Orphanides G., LeRoy G., Chang C.-H., Luse D. S., Reinberg D., FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., Reinberg D., FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Mason P. B., Struhl K., The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23, 8323–8333 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders A., Werner J., Andrulis E. D., Nakayama T., Hirose S., Reinberg D., Lis J. T., Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301, 1094–1096 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Hsieh F.-K., Kulaeva O. I., Patel S. S., Dyer P. N., Luger K., Reinberg D., Studitsky V. M., Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 110, 7654–7659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P., Dong L., Hu M., Wang Y.-Z., Xiao X., Zhao Z., Yan J., Wang P.-Y., Reinberg D., Li M., Li W., Li G., Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol. Cell 71, 284–293.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wittmeyer J., Formosa T., The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17, 4178–4190 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan B.C.-M., Chien C.-T., Hirose S., Lee S.-C., Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 25, 3975–3985 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abe T., Sugimura K., Hosono Y., Takami Y., Akita M., Yoshimura A., Tada S., Nakayama T., Murofushi H., Okumura K., Takeda S., Horikoshi M., Seki M., Enomoto T., The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J. Biol. Chem. 286, 30504–30512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurat C. F., Yeeles J. T. P., Patel H., Early A., Diffley J. F. X., Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65, 117–130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller D. M., Zeng X., Wang Y., Zhang Q. H., Kapoor M., Shu H., Goodman R., Lozano G., Zhao Y., Lu H., A DNA damage–induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7, 283–292 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Keller D. M., Lu H., p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2 hSPT16 SSRP1 complex. J. Biol. Chem. 277, 50206–50213 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Krohn N. M., Stemmer C., Fojan P., Grimm R., Grasser K. D., Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J. Biol. Chem. 278, 12710–12715 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Heo K., Kim H., Choi S. H., Choi J., Kim K., Gu J., Lieber M. R., Yang A. S., An W., FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell 30, 86–97 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Charles Richard J. L., Shukla M. S., Menoni H., Ouararhni K., Lone I. N., Roulland Y., Papin C., Ben Simon E., Kundu T., Hamiche A., Angelov D., Dimitrov S., FACT assists base excision repair by boosting the remodeling activity of RSC. PLOS Genet. 12, e1006221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia H., Fleyshman D., Kolesnikova K., Safina A., Commane M., Paszkiewicz G., Omelian A., Morrison C., Gurova K., Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget 2, 783–796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasparian A. V., Burkhart C. A., Purmal A. A., Brodsky L., Pal M., Saranadasa M., Bosykh D. A., Commane M., Guryanova O. A., Pal S., Safina A., Sviridov S., Koman I. E., Veith J., Komar A. A., Gudkov A. V., Gurova K. V., Curaxins: Anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci. Transl. Med. 3, 95ra74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh F.-K., Kulaeva O. I., Orlovsky I. V., Studitsky V. M., FACT in cell differentiation and carcinogenesis. Oncotarget 2, 830–832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia H., Miecznikowski J. C., Safina A., Commane M., Ruusulehto A., Kilpinen S., Leach R. W., Attwood K., Li Y., Degan S., Omilian A. R., Guryanova O., Papantonopoulou O., Wang J., Buck M., Liu S., Morrison C., Gurova K. V., Facilitates chromatin transcription complex is an "accelerator" of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 4, 159–173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurova K., Chang H.-W., Valieva M. E., Sandlesh P., Studitsky V. M., Structure and function of the histone chaperone FACT—Resolving FACTual issues. Biochim. Biophys. Acta 1861, 892–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orphanides G., Wu W.-H., Lane W. S., Hampsey M., Reinberg D., The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Winkler D. D., Muthurajan U. M., Hieb A. R., Luger K., Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., Zhang E. T., Nijmeijer B., Kotthoff C., Rybin V., Amlacher S., Hurt E., Ladurner A. G., Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499, 111–114 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kemble D. J., McCullough L. L., Whitby F. G., Formosa T., Hill C. P., FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsunaka Y., Fujiwara Y., Oyama T., Hirose S., Morikawa K., Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T., Liu Y., Edwards G., Krzizike D., Scherman H., Luger K., The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci. Alliance 1, e201800107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Formosa T., The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255 (2013). [PubMed] [Google Scholar]
- 28.Valieva M. E., Armeev G. A., Kudryashova K. S., Gerasimova N. S., Shaytan A. K., Kulaeva O. I., McCullough L. L., Formosa T., Georgiev P. G., Kirpichnikov M. P., Studitsky V. M., Feofanov A. V., Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 23, 1111–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahata S., Yu Y., Stillman D. J., FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34, 405–415 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erkina T. Y., Erkine A., ASF1 and the SWI/SNF complex interact functionally during nucleosome displacement, while FACT is required for nucleosome reassembly at yeast heat shock gene promoters during sustained stress. Cell Stress Chaperones 20, 355–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvalho S., Raposo A. C., Martins F. B., Grosso A. R., Sridhara S. C., Rino J., Carmo-Fonseca M., de Almeida S. F., Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41, 2881–2893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koman I. E., Commane M., Paszkiewicz G., Hoonjan B., Pal S., Safina A., Toshkov I., Purmal A. A., Wang D., Liu S., Morrison C., Gudkov A. V., Gurova K. V., Targeting FACT complex suppresses mammary tumorigenesis in Her2/neu transgenic mice. Cancer Prev. Res. 5, 1025–1035 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Safina A., Garcia H., Commane M., Guryanova O., Degan S., Kolesnikova K., Gurova K. V., Complex mutual regulation of facilitates chromatin transcription (FACT) subunits on both mRNA and protein levels in human cells. Cell Cycle 12, 2423–2434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maluchenko N. V., Chang H. W., Kozinova M. T., Valieva M. E., Gerasimova N. S., Kitashov A. V., Kirpichnikov M. P., Georgiev P. G., Studitsky V. M., Inhibiting the pro-tumor and transcription factor FACT: Mechanisms. Mol. Biol. 50, 599–610 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Burkhart C., Fleyshman D., Kohrn R., Commane M., Garrigan J., Kurbatov V., Toshkov I., Ramachandran R., Martello L. and Gurova K. V., Curaxin CBL0137 eradicates drug resistant cancer stem cells and potentiates efficacy of gemcitabine in preclinical models of pancreatic cancer. Oncotarget 5, 11038–11053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safina A., Cheney P., Pal M., Brodsky L., Ivanov A., Kirsanov K., Lesovaya E., Naberezhnov D., Nesher E., Koman I., Wang D., Wang J., Yakubovskaya M., Winkler D., Gurova K., FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res. 45, 1925–1945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesher E., Safina A., Aljahdali I., Portwood S., Wang E. S., Koman I., Wang J., Gurova K. V., Role of chromatin damage and chromatin trapping of FACT in mediating the anticancer cytotoxicity of DNA-binding small molecule drugs. Cancer Res. 78, 1431–1443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barone T. A., Burkhart C. A., Safina A., Haderski G., Gurova K. V., Purmal A. A., Gudkov A. V., Plunkett R. J., Anticancer drug candidate CBL0137, which inhibits histone chaperone FACT, is efficacious in preclinical orthotopic models of temozolomide-responsive and -resistant glioblastoma. Neuro Oncol. 19, 186–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bondarenko V. A., Steele L. M., Ujvari A., Gaykalova D. A., Kulaeva O. I., Polikanov Y. S., Luse D. S., Studitsky V. M., Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24, 469–479 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Thåström A., Lowary P. T., Widlund H. R., Cao H., Kubista M., Widom J., Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288, 213–229 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Tsunaka Y., Toga J., Yamaguchi H., Tate S.-i., Hirose S., Morikawa K., Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J. Biol. Chem. 284, 24610–24621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valieva M. E., Gerasimova N. S., Kudryashova K. S., Kozlova A. L., Kirpichnikov M. P., Hu Q., Botuyan M. V., Mer G., Feofanov A. V., Studitsky V. M., Stabilization of nucleosomes by histone tails and by FACT revealed by spFRET microscopy. Cancers 9, pii: E3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kireeva M. L., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V. M., Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Gaykalova D. A., Kulaeva O. I., Bondarenko V. A., Studitsky V. M., Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 523, 109–123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter W., Kireeva M. L., Tchernajenko V., Kashlev M., Studitsky V. M., Assay of the fate of the nucleosome during transcription by RNA polymerase II. Methods Enzymol. 371, 564–577 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Brewster N. K., Johnston G. C., Singer R. A., A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21, 3491–3502 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCullough L. L., Connell Z., Xin H., Studitsky V. M., Feofanov A. V., Valieva M. E., Formosa T., Functional roles of the DNA-binding HMGB domain in the histone chaperone FACT in nucleosome reorganization. J. Biol. Chem. 293, 6121–6133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., Formosa T., yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35, 365–376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Durbin R., Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kireeva M. L., Komissarova N., Waugh D. S., Kashlev M., The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 275, 6530–6536 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Simon R. H., Felsenfeld G., A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6, 689–696 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Holt C., Brandt W. F., Greyling H. J., Lindsey G. G., Retief J. D., Rodrigues J. d. A., Schwager S., Sewell B. T., Isolation and characterization of histones. Methods Enzymol. 170, 431–523 (1989). [DOI] [PubMed] [Google Scholar]
- 57.Studitsky V. M., Preparation and analysis of positioned nucleosomes. Methods Mol. Biol. 119, 17–26 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Kulaeva O. I., Gaykalova D. A., Pestov N. A., Golovastov V. V., Vassylyev D. G., Artsimovitch I., Studitsky V. M., Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 16, 1272–1278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaav2131/DC1
Fig. S1. Analysis of colocalization of FACT and transcribing Pol II using immunofluorescence.
Fig. S2. Redistribution of FACT in HT1080 cells from nucleoplasm to chromatin.
Fig. S3. Redistribution of FACT in nucleus of HT1080-treated cells from nucleoplasm to chromatin.
Fig. S4. Heatmaps of SSRP1 occupancy in the vicinity of TSSs and TES of the genes in HT1080 cells.
Fig. S5. Analysis of the average SSRP1 densities over gene bodies against the levels of transcription of the corresponding genes.
Fig. S6. Curaxins preferentially remove SSRP1 from gene bodies of highly transcribed genes.
Fig. S7. Analysis of gel-purified nucleosomes by native PAGE.
Fig. S8. Typical frequency distributions of FRET efficiencies of the N13/91 nucleosomes.
Fig. S9. The catalytic activity of Pol II is minimally affected by curaxins.
Fig. S10. Trapping of FACT on immobilized competitor nucleosomes after Pol II transcription in the presence of CBL0137.
Table S1. Statistical data for spFRET analysis.


