Abstract
The global production of cereal straw as an agricultural by-product presents a significant source of biomass, which could be used as feedstock for the production of second generation biofuels by fermentation. The production of sugars for fermentation is an important measure of straw quality and in its suitability for biofuel production. In this paper, we present a characterization of straw digestibility from a wide range of cereal. Our main objective is to evaluate the variability of fermentable sugars released from different species including wheat (Triticum durum L., Triticum aestivum L.), barley (Hordeum vulgare L.) and triticale (X Triticosecale Wittmack). To this end, we adapted a saccharification method (IAS Method) capable of detecting significant differences of released sugars between cultivars and species, while using separately another method that would serve as a control and with which we could contrast our results (CNAP method). ANOVA analyses revealed that barley has a higher saccharification potential than wheat and triticale and shows more variation between genotypes. Thus, populations derived from crosses among them such as Steptoe × Morex and OWB Dominant × OWB Recessive hold potential for the identification of genetic basis for saccharification-related traits. The correlation of glucose released between the two methods was moderate (R2 = 0.57). An evaluation of the inter- and intra- specific correlation between a number of chemical and agronomical parameters and saccharification suggests that the cell wall thickness and lignin content in straw could be used in breeding programs for the improvement of the saccharification potential. Finally, the lack of correlation between grain yield and saccharification suggests that it would be possible to make a selection of genotypes for dual purpose, low recalcitrance and grain yield.
Introduction
Widespread burning of fossil fuels produces approximately 81% of the world's energy, of which 41% comes from oil, mostly destined (92%) to the transport sector [1]. The environmental consequences of burning fossil fuels, and the threat of a shortage of energy due to finite oil reserves are well documented [2]. In response, the use of bioethanol as a liquid fuel has triggered a fivefold increase in ethanol production since 2000 [3]. Current commercial biofuel supply relies on first-generation biofuel production, which, although efficient, requires food and feed commodities as a feedstock and as such, poses a potential threat to food security. Although first generation biofuels can be produced efficiently, they use food and feed commodities as a feedstock posing a potential threat to food security. In addition, the cultivation of such feedstocks requires high agrochemical inputs that increase the carbon footprint of biofuels [4]. The development of second-generation biofuels from agricultural waste presents a valuable alternative as it can be obtained as a by-product from food crops [5]. At present, cereal straw is treated as a residue and is usually burnt or incorporated into the soil, but these by-products (including wheat, barley, rice, corn, oat, cotton straw, and bagasse from sugar cane, and totaling approximately 3 billion metric tons annually) present a great potential energy source [6]. Second generation bioethanol production from lignocellulosic biomass requires the conversion of lignocellulose into simple sugars, in three stages [7]: size reduction, thermochemical pretreatment and hydrolysis. The ease with which a biomass is hydrolyzed, also known as saccharification potential, can be used to evaluate recalcitrance of biomass in breeding programs. In this paper, two saccharification methods have been used, one developed by the Instituto de Agricultura Sostenible, (IAS hereinafter) and another procedure, which is widely used, [8] used as a control (Centre for Novel Agricultural Products, CNAP hereinafter).
Pretreatment serves to improve the accessibility of the hydrolysing enzymes to the lignocellulose feedstock. Each pretreatment process is optimised to the biomass to be hydrolyzed since this has a specific effect on the cellulose, hemicellulose and lignin fraction [9]. Due to the great variability in the composition of lignocellulosic materials, it is necessary to adapt the saccharification method to the properties of the biomass. The pretreatment conditions should be chosen in accordance with the configuration of the process selected for the subsequent hydrolysis and fermentation steps. This process, besides being crucial in the conversion of biomass to bioethanol, is considered as the second most expensive after the feedstock cost [9].
The variability in the cell wall degradability of lignocellulosic material can be affected by many factors such as genetic [10,11], morphological [12,13], environmental [14,15], experimental technique for releasing sugars [16], and crop harvesting [17,18]. To fully evaluate all sources of variability, it is advisable to take a multi-phase and multi-environment approach [19] with different experimental methods [16].
The deliberate modification of cell-wall properties is challenging considering the high number of genes involved. Indeed, recent findings in Arabidopsis thaliana estimate that 10–15% of plant genes are related to cell-wall biology [20]. This is not surprising since cell walls are essential to plants, contributing to pest and disease resistance and providing mechanical support to plant tissues. Consequently, breeding programs for bioethanol production should aim for a balance between saccharification potential and agronomic performance.
A number of biofuel research initiatives have developed high throughput methods for pretreatment and enzymatic hydrolysis (HTPH) to evaluate the saccharification properties of large collections of germplasm with high potential for the production of second generation biofuels [8,21–23].
The aims of this work are to evaluate the variation in sugar yield from straw obtained from wheat, barley and triticale cultivars under rain-fed environments and to select parental genotypes to develop mapping populations to detect QTL for saccharification.
Material and methods
Plant material
Four cereal species were studied: Hordeum vulgare L., Triticum aestivum L., Triticum durum L. and X Triticosecale Wittmack (Table 1). Triticale, barley and wheat lines were obtained from either the National Small Grains Collection (NSGC) of the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) (https://www.ars.usda.gov/pacific-west-area/aberdeen-id/small-grains-and-potato-germplasm-research/docs/national-small-grains-collection/) or from the Barley and Wild Plant Resource Center, Okayama University (http://earth.lab.nig.ac.jp). When available, accessions used as parental lines in mapping populations were selected with a dual purpose: Firstly, to allow for the identification of mapping populations suitable for studying the genetic bases of saccharification, and secondly to give a fair representation to the variability available in each species, as parental lines are normally selected to be as divergent as possible.
Table 1. Plant material used in this work.
More information on the genotypes can be found at https://npgsweb.ars-grin.gov/gringlobal/search.aspx.
| Accession name(a) | Species | Accession number |
|---|---|---|
| Apex** | H. vulgare | PI600966 |
| Azumamugi*** | H. vulgare | J698 |
| Cebada Capa** | H. vulgare | PI539113 |
| Clipper** | H. vulgare | PI349366 |
| Dicktoo** | H. vulgare | CIho 5529 |
| Franka** | H. vulgare | PI574293 |
| Franklin** | H. vulgare | PI373729 |
| Fredrickson** | H. vulgare | CIho 13647 |
| Golden Promise** | H. vulgare | PI467829 |
| Igri** | H. vulgare | PI406263 |
| Kanto Nakate Gold*** | H. vulgare | J518 |
| Ko A** | H. vulgare | PI383935 |
| L94** | H. vulgare | CIho 11797 |
| Lina** | H. vulgare | PI584808 |
| Mokusekko 3** | H. vulgare | PI420938 |
| Morex** | H. vulgare | Ciho 15773 |
| OWB dominant** | H. vulgare | GSHO3450 |
| OWB recessive** | H. vulgare | GSHO3451 |
| Stander** | H. vulgare | PI564743 |
| Steptoe** | H. vulgare | CIho 15229 |
| Vada** | H. vulgare | PI280422 |
| Anza* | T. aestivum | NA |
| Avocet** | T. aestivum | PI464644 |
| BobWhite* | T. aestivum | NA |
| Caledonia** | T. aestivum | PI610188 |
| Cayuga** | T. aestivum | PI595848 |
| CIGM90.248** | T. aestivum | PI610750 |
| Excalibur** | T. aestivum | PI572701 |
| JAYPEE** | T. aestivum | PI592760 |
| Kanqueen** | T. aestivum | PI401539 |
| M6** | T. aestivum | PI83534 |
| McNeal** | T. aestivum | PI574642 |
| Opata85** | T. aestivum | PI591776 |
| OS9A** | T. aestivum | PI658243 |
| P91193** | T. aestivum | GSTR 10001 |
| P92201** | T. aestivum | GSTR 10002 |
| Penawawa** | T. aestivum | PI495916 |
| Perico* | T. aestivum | NA |
| QCB36** | T. aestivum | PI658244 |
| Renan** | T. aestivum | PI564569 |
| SS550** | T. aestivum | GSTR 12501 |
| TAM107-R7** | T. aestivum | GSTR 11601 |
| Thatcher** | T. aestivum | CItr 10003 |
| UC1110** | T. aestivum | GSTR 13501 |
| USG 3209** | T. aestivum | PI617055 |
| Amadina** | T. durum | GSTR 12701 |
| Avalon** | T. durum | PI446910 |
| CO940610** | T. durum | GSTR 10702 |
| Grandin*5/ND614-A** | T. durum | GSTR 10401 |
| IDO444** | T. durum | GSTR 12902 |
| Jupateco 73S** | T. durum | GSTR 10501 |
| NY18/Clark's Cream 40–1** | T. durum | GSTR 10402 |
| Rio Blanco** | T. durum | PI531244 |
| Rugby** | T. durum | CItr 17284 |
| UC1113 Yr36 Gpc-B1** | T. durum | PI638741 |
| Weebill 1** | T. durum | GSTR 10502 |
| Armadillo 130** | X Triticosecale | PI583701 |
| Currency** | X Triticosecale | PI483066 |
| Drira** | X Triticosecale | PI520478 |
| Juanillo 95** | X Triticosecale | PI520488 |
| Kramer** | X Triticosecale | PI476216 |
| Navojoa** | X Triticosecale | PI520421 |
| Rahum** | X Triticosecale | PI422269 |
| Wapiti** | X Triticosecale | PI511870 |
| Yoreme Tehuacan 75** | X Triticosecale | PI519876 |
| Zebra** | X Triticosecale | PI429031 |
(a) Plant material availability
* IAS-CSIC
** USDA-ARS, National Small Grains Germplasm Research Facility, Aberdeen, ID 83210, USA
*** Barley and Wild Plant Resource Center. Institute of Plant Science and Resources. Okayama University, Kurashiki, 710–0046, Japan.
Field trials and sample processing
Three field trials, which were designed in three completely randomized blocks, were conducted in Córdoba (37.85981, -4.796895). Each field trial included sixty-six accessions belonging to four different species: barley (Hordeum vulgare L.) common wheat (T. aestivum), durum wheat (T. durum) and triticale (Triticosecale) (Table 1). Each plot consisted of four plants separated by 15cm with an inter-plot distance of 30cm and an inter-furrow distance of 50cm. The straw harvested included leaves and stems; and it was harvested at maturity for each genotype. Samples were chopped using a grinder before processing was performed using a cyclonic mill (Cyclotec 1093, Foss-Tekator) with a 1mm sieve.
Phenotyping
The all genotypes were scored for: plant height at different stages of growth, total plant biomass, grain yield, biomass yield and stem wall thickness at several internodes. All determinations but plant height was taken at harvest.
Theoretical ethanol yield calculation
The theoretical ethanol yield was calculated considering the total biomass conversion per surface area unit (ha), according to the National Renewable Energy Laboratory Standards (NREL) [24]. Theoretical ethanol was conducted through the following formula:
Where Glu: Glucose released (μl · mg−1DW), Biomass: Theoretical biomass (Kg · ha−1), produced by genotype from the quantity of straw in plots of 0.3 square meters, 0.511: theoretical ethanol yield conversion.
Saccharification systems
Both systems to determine saccharification were calibrated based on previous knowledge for near optimal hydrothermal pretreatment of straw and optimal enzyme loading [25,26].
IAS method
Assays to determine saccharification involved three main steps: pretreatment, hydrolysis and sugar detection. The conditions established by Gomez et al. [8] and Santoro et al. [23] were adapted for sample processing in a single 2 mL tube as used by Santoro et al [23]. Briefly, 20 mg of ground straw were loaded into 2 mL screw-cap tubes. A pretreatment solution (6.25 mM) NaOH was used as described by Santoro et al. [23] using 1.5 mL of pretreatment solution and incubated at 90°C for 3 h in a water bath, then cooled on ice. Enzymatic hydrolysis was performed using an enzyme cocktail with a 4:1 ratio of Celluclast: Novozyme 188 (Novozymes, Bagsvaerd, Denmark) [8]. Hydrolysis was performed during 20 h with constant shaking, at 50°C in a 0.5 M sodium citrate buffer at pH 4.5. Different enzyme concentrations were assayed to optimize the digestion in a single tube (S1 Fig), the concentration selected as optimal to determine differences between genotypes was 0.05 μL/mg DW. Nine serial dilutions were established from a maximum enzyme concentration of 2 μL/mg of dry weight (DW). The determination of sugars released after hydrolysis was carried out using the glucose oxidase/peroxidase (GOPOD) kit (K-Gluc, Megazyme, Ireland). The assay volumes were reduced to allow the procedure to be performed in 96-well ELISA plates. Glucose determination was performed using 8 μL of the digestion reaction mixture and 240 μL of the GOPOD assay reagent followed by incubation at 50°C during 20 min. The yield of glucose was analyzed using 96 well plates. Absorbance readings were determined at 490 nm in a BioTek ELx800 Absorbance Microplate Reader (BioTek Instruments, Inc.). The adapted protocol used at IAS was validated with the saccharification protocol described by Gomez et al. [8].
CNAP method
96-well plates containing biomass underwent saccharification analysis using a liquid handling platform (Tecan Evo 200; Tecan Group Ltd.) which pretreated the samples with 0.5N NaOH at 90°C for 20 min, followed by enzymatic hydrolysis 50°C for 8 hours. The enzyme cocktail contained commercially available Celluclast and Novozyme 188 (Novozymes A/S, Bagsvaerd, Denmark) at a ratio of 4:1 at an enzyme loading of 22.5 Filter Paper Units (FPU)/g. The reducing sugars released during hydrolysis were determined using a colorimetric assay involving 3-methyl-2-benzothiazolinone hydrozone (MTBH) [8]. Each plate contained standard reactions of 50 nmol, 100 nmol, and 150 nmol of glucose. Change in color was read with a Tecan Sunrise microplate absorbance reader at 620 nm.
Lignin determination
Lignin content was quantified using the acetyl bromide method according to Foster et al. [27]. Briefly, 3mg of biomass alcohol insoluble residue (AIR) were weighed into a 5 mL volumetric flask, and 250 μL of freshly prepared acetyl bromide solution (25% v/v acetyl bromide in glacial acetic acid) was added. Samples were incubated at 50°C for 2h, followed by a further 1h, mixing every 15min. Samples were taken to 5 mL with glacial acetic acid and mixed. The absorption was read using a Shimadzu UV 1800 spectrophotometer (http://www.shimadzu.com) at 280nm. Lignin content was (μg x mg-1 cell wall) determined using the following formula: Lignin Content = Absorbance / (Coefficient x Path length) x (Total volume/Biomass weight) x 100. The coefficient used for grasses was 17.75.
Statistical analyses
All statistical analyses were conducted with the software R version 3.2.3 [28]. Data was adjusted to a linear model with the function lm and the significance was established using analysis of the variance (ANOVA) (function aov, package agricolae [29]. Differences between species or genotypes were determined by Tuckey HSD test (P ≤ 0.05) (function LSD.test, agricolae package). Pearson correlations were calculated with cor function (stats package) and all boxplot and art-graph were depicted with boxplot function (ggplot2 package [30]). The main assumptions of linear model were confirmed using the Shapiro-Wilk test for normal distribution (function shapiro.test, stats package [28] and by the Levene test for homogeneity of variances (function leveneTest, package car [31]) and variables were transformed if required.
Results and discussion
Variation of the saccharification potential in a range of cereal cultivars
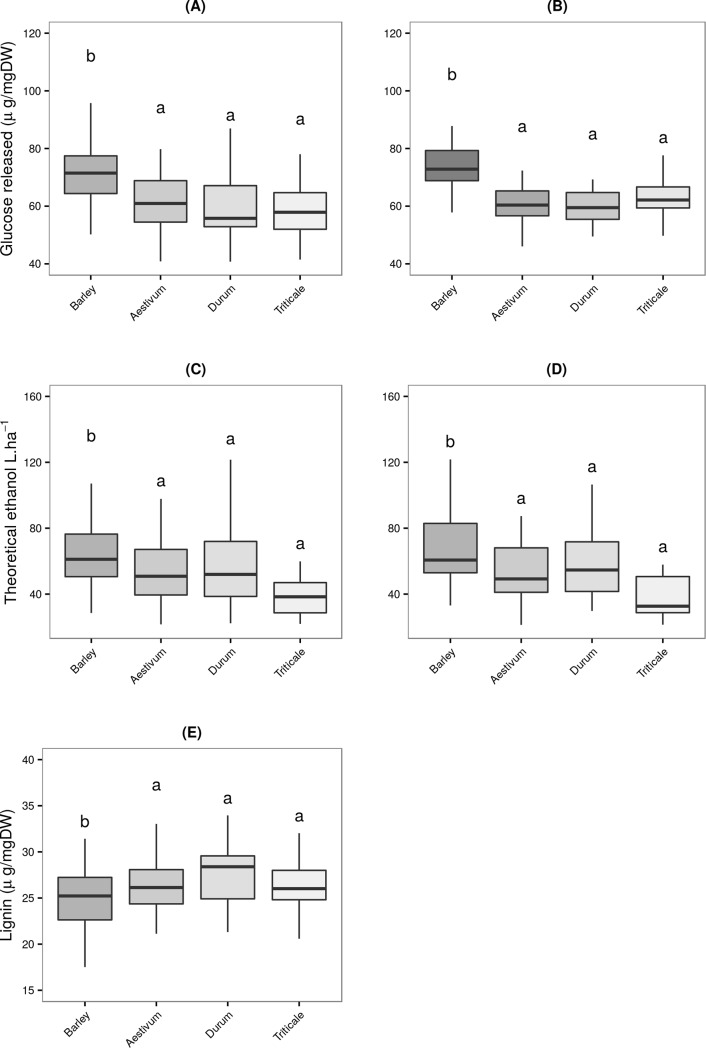
To assess the differences in recalcitrance among species and cultivars of triticale, wheat (T. durum and T. aestivum) and barley, all samples were analysed for saccharification potential using the IAS and CNAP methods described above. Glucose yields were standardized using inter-plate checks to control inter-plate variance. ANOVA analysis revealed significant differences among species, barley being the species with the highest saccharification potential (Fig 1A).
Fig 1. Comparative yield of glucose released in barley, wheat and triticale under different saccharification conditions.
Boxplot of glucose’s quantification released for wheat, barley and triticale under different saccharification conditions. (a) IAS; (b) CNAP. Mean (line), 25th-75th percentile (box) and 10th-90th percentile (whiskers) of glucose released for each genus. For each saccharification method, bars with different letters are significantly different (ANOVA, Tuckey HSD test, P≤0.05).
To validate the results obtained through IAS method, a biological replicate of each sample was analysed using the CNAP method, as control. The correlation of standardized glucose yields between IAS and CNAP methods was moderate (R2 = 0.5688) but it is significantly higher than the values reported by Lindedam et al.[16] for two high throughput systems (R2 = 0.2139), differences between IAS and CNAP could be due to the different methods used for the quantification of the sugars released. IAS determines only glucose, while CNAP determines all sugars as reducing sugars. Lindedam et al. [16] analysed three different methods, but only reported their best correlation which implies that the other correlations were lower. Both methods used here show that barley presents the highest saccharification potential (Fig 1B). Further analyses were conducted to evaluate the relative recalcitrance among genotypes for each genus/species (Table 2).
Table 2. Mean values of total sugar released (μg/mgDW) for sixty-six accessions under IAS-CSIC saccharification conditions.
Post-hoc test independently for all genotypes in each. The Study in Wheat was made with LSD test (p ≤ 0.05) with Benjamini-Yekutieli p-values adjust. Values with same letter are not significantly different at level 0.05.
| T.aestivum Genotype | Glucose Yield | T.durum genotype | Glucose yield | Barley genotype | Glucose Yield | Triticale genotype | Glucose Yield |
|---|---|---|---|---|---|---|---|
| Caledonia | 75.11 a | Avalon | 78.4 a | OWB recessive | 98.00 a | Juanillo 95 | 68.32 a |
| Kanqueen | 68.94 ab | NY18/Clark's Cream 40–1 | 69.55 ab | Steptoe | 89.62 ab | Currency | 65.5 ab |
| Excalibur | 68.6 abc | IDO444 | 66.05 abc | Apex | 83.24 abc | Yoreme Tehuacan 75 | 63.38 abc |
| SS550 | 67.93 abc | Rugby | 63.59 abc | Golden Promise | 79.56 abc | Armadillo 130 | 59.53 abcd |
| USG3209 | 67.41 abcd | UC1113 YR36 Gpc-B1 | 63.52 abc | Capa | 76.95 bcd | Drira | 58.69 abcd |
| Avocet | 66.94 abcd | Jupateco | 58.15 bcd | Lina | 75.57 bcd | Rahum | 56.48 abcd |
| Cayuga | 66.45 abcd | GRA614A | 56.04 bcd | Fredrickson | 73.10 bcd | Zebra | 55.07 abcd |
| McNeal | 65.37 abcd | Amadina | 53.45 bcd | Clipper | 73.07 bcd | Navajoa | 54.05 bcd |
| QCB36 | 64.68 abcd | Weebill_1 | 53.27 bcd | Azumamugi | 72.07 bcd | Wapiti | 51.36 cd |
| P92201 | 64.58 abcd | CO940610 | 50.06 cd | Dicktoo | 71.60 bcd | Kramer | 47.76 d |
| Renan | 63.59 abcd | Rio Blanco | 47.1 d | Igri | 71.55 bcd | ||
| P91193 | 63.57 abcd | Mokusekko | 69.71 bcd | ||||
| CIGM90.248 | 62.71 abcd | Koa | 67.44 bcd | ||||
| OS9A | 62.33 abcd | OWB dominant | 66.49 cd | ||||
| Penawawa | 59.83 bcd | Stander | 65.84 cd | ||||
| M6 | 59.79 bcd | Franka | 65.44 cd | ||||
| UC1110 | 57.44 bcd | Morex | 65.34 cd | ||||
| Thatcher | 57.23 bcd | Kanto Nakte Gold | 64.88 cd | ||||
| Jaypee | 56.07 bcd | Vada | 64.33 cd | ||||
| TAM107 R7 | 56 bcd | Franklin | 62.06 cd | ||||
| Anza | 55 bcd | L94 | 52.41 d | ||||
| Opata85 | 53.61 cd | ||||||
| Bobwhite | 51.1 d | ||||||
| Perico | 50.72 d |
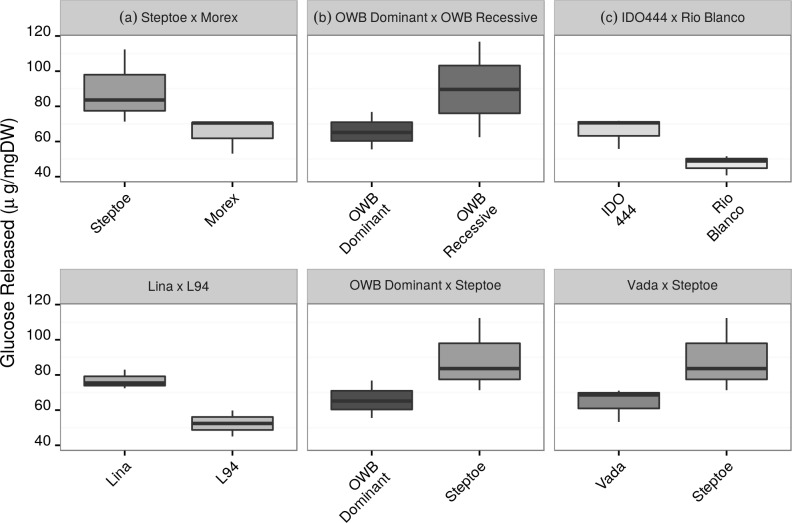
Significant differences were detected among barley, wheat and triticale genotypes. In the screening of the 66 cultivars of wheat, barley and triticale, we are able to identify a large variability in the enzymatic hydrolysis of the cell walls of straw. The variability for saccharification among cultivars of different species ranged between 47.09 μg/mg DW and 89.62 μg/mg DW (mean values) using the IAS method, and from 51.59 to 85.07 μg/mg DW using the CNAP method. The variance coefficients (CV) between all genotypes in this trial were 14.7% and 12.2% for the IAS and CNAP methods, respectively. H. Chilense had a coefficient of variation of 11.6% and 8.54%, T. Aestivum of 9.6% and 7.6%, T. Durum of 15.1% and 7.82%, and Triticosecale of 9.8% and 6.2%, respectively. The differences between methods for CVs between cultivars of each species are always higher for the IAS method. This could be explained to a large extent because in the IAS method only one 96-well plate could be assayed each time, whereas in the high-throughput method of CNAP a larger number of plates per assay (usually six). In terms of variability in cell wall saccharification, similar results have been previously reported in other collections of different cultivars [11,32,33]. The block factor was also significant in the ANOVA analysis, but it is likely related to a short flooding period during the growing season. A significant block effect was also reported by [19] due to a short period of drought stress. Taken together, these results suggest that the water balance during the crop cycle could marginally affect the release of glucose. In the present work we do not have the possibility of separating the environmental effect of experimental error, but environmental interactions on the degradability of the cell wall have been previously investigated [10,32]. However, several genotypes differing in biomass recalcitrance to enzymatic hydrolysis have been used as parental lines in mapping populations for different traits. These mapping populations constitute a valuable resource for barley genetic studies. Indeed, since the development of the Steptoe x Morex and OWB populations [34] they have been successfully used for genetic mapping, including regulatory genes [35] or resistance to leaf stripe [36]. Furthermore, both mapping populations were used to develop a consensus SNP genetic linkage map in barley [37]. Given the contrasting saccharification potential of the parental lines, the barley mapping populations Steptoe × Morex, Vada × Steptoe, OWB Dominant × Steptoe, OWB Dominant × OWB Recessive, and Lina × L94, could be used to identify the genetic factors underlying differential recalcitrance (Fig 2A). Similarly, the IDO444 × Rio Blanco mapping population [38] could be used in wheat (Fig 2C). However, only the OWB populations and Steptoe × Morex should be considered for mapping purposes since the CNAP method did not detect significant differences at p < 0.05 between the other parental listed above.
Fig 2. Yield of glucose released in selected barley and wheat lines.
Boxplot of glucose’s quantification. Mean (line), 25th-75th percentile (box) and 10th-90th percentile (whiskers) of glucose released for each genotype. Each graph (a to f) shows significant differences at significance level of 0.05 (using IAS-CSIC saccharification conditions) between parental lines used for the development of mapping populations in the literature. Differences shown in graphs a and b were also significant using the CNAP saccharification conditions, and differences shown in graph c was the only one significant different for wheat.
Determinants of sugar yield
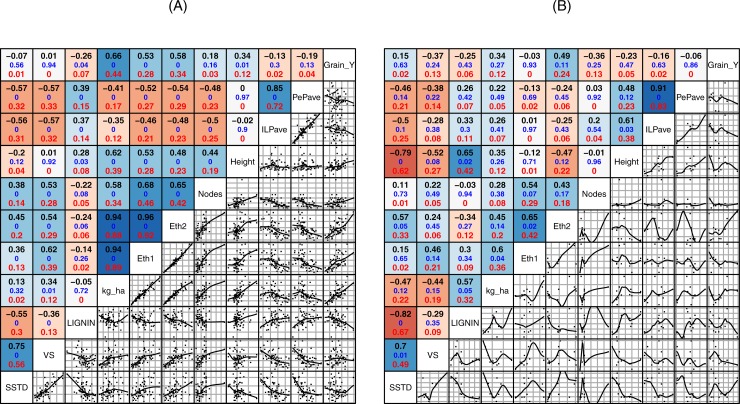
Fig 3 shows the degree of correlation between a number of phenotypic characters and saccharification in all genotypes. Lignin content presents a significant negative correlation with sugar yield (r = -0.55) for all genotypes (Fig 3A), which is in agreement with previous results by Lindedam et al. [39]. When we compared the top 10 genotypes for biomass yield (Fig 3B), we found a stronger Pearson correlation (r = -0.82) and a better relationship between saccharification and lignin content. These results are comparable to previous findings in Solanum pennellii by Caruso et al. [40], transgenic alfalfa lines by Chen et al. [41] and Arabidopsis thaliana by Van Acker et al [42]. Collectively these results suggest that lignin content should be considered in breeding for saccharification potential. In the current study we observed a negative correlation (r = -0.79) between plant height and saccharification using the CNAP method and a positive correlation between plant height and lignin content (r = 0.65) (Fig 3B), both correlations for high biomass yield selected lines. This relationship between plant height and plant cell wall recalcitrance could be due to the requirement of increased lignin for mechanical stiffness with the consequent reduction in saccharification. Similar results were showed by [11] and [43]. The negative correlation between plant height and degradability could also partly be explained by higher plants having relatively smaller leaf fraction. For correlation analysis with all samples, we could not see correlation between height and degradability; this fact could be explained because breeding programs for semi-dwarf cultivars may in fact have affected the degradability of modern cultivar [44]. ILPave (Average for straw wall thickness for largest internode) and PePave (Average for straw wall thickness for peduncle) showed a significant negative correlation with degradability, theoretical ethanol and number of nodes, and also showed a positive correlation between thickness and lignin content (Fig 3A). Generally, barley has a higher number of nodes and low wall thickness, which is consistent with high saccharification and low lignin content results showed in Fig 1. Differences in lignin content in cell wall of one genotype of wheat, one barley, and one triticale straw, have been reported previously, showing that barley contains less lignin than wheat [45,46]. Our results obtained from many genotypes for each species are in agreement with previous reports and extend the observation across genotypes. Plants with the same height and stems with low wall thickness will have more short internodes, implying more numbers of nodes, and consequently are less susceptible to lodging. Correlations between lodging resistance, thickness and number of nodes were shown by Jezowski et al. [47], Tandon et al. [48], and Brady et al. [49] On the other hand, as reported Saint Pierre et al. [50] thickness is an ideal factor to maximize water soluble carbohydrate reserves, and it appears to be important under water limited conditions, where these could be mobilized for grain filling. ILPave and PePave were uncorrelated with plant height and grain yield, hence allowing breeding for that character without compromising high grain yield. Moreover, we could assess that grain yield and saccharification are not correlated (Fig 3A), establishing a degree of independence between these two traits.
Fig 3. Scatter Plot and Pearson’s correlation coefficient matrix for comparison among phenotyping, saccharification and theoretical ethanol data.
Pairwise correlation analyses were performed for all assayed genotypes (a) and the 10 best genotypes for biomass yield (b). The upper panel above the diagonal shows Pearson’s correlation coefficients, p-value and regression coefficient. The lower panel below the diagonal gives their scatter plot. (SSTD = Saccharification standardized values under CNAP conditions, VS = Saccharification standardized values under IAS-CSIC conditions, Kg_ha = estimated weight of straw by hectare, Eth1 = Theoretical ethanol calculated with CNAP’s saccharification values and estimated biomass, Eth2 = Theoretical ethanol calculated with IAS-CSIC’s saccharification values and estimated biomass, ILPave = Average for straw wall thickness for largest internode, PePave = Average for straw wall thickness for peduncle, and Grain_Y = grain yield).
Final remarks
In the current work we analysed a collection of wheat (T. durum and T. aestivum), barley and triticale genotypes in order to investigate interspecific and intraspecific differences. The methodology adapted at IAS could be useful for genotype selection in biomass quality since it shows a good degree of concordance with previous methodologies. Thus, it would be useful for the identification of improved varieties with good saccharification potential in a breeding program. Collectively, our results indicate that barley is a better source of lignocellulosic material for bioethanol production than wheat and triticale. The ranking of genotypes was slightly different with IAS and CNAP methods, but the most contrasting genotypes were picked up by both methodologies. Interestingly, some of the most dissimilar genotypes have been used to develop mapping populations in barley. For instance, both Steptoe × Morex and OWB Dominant × OWB Recessive barley mapping populations would be good tools for the identification of the genetic basis of saccharification-related traits. Finally, correlation analyses suggest that sugars released, lignin content and its correlation with straw wall thickness would be good predictors of biomass degradability in breeding programs. Furthermore, the lack of correlation between grain yield and saccharification suggests that it would be possible to select genotypes with low recalcitrance and high grain yield for dual use (grain and energy).
Supporting information
Glucose released in wheat genotypes (Anza, Bobwhite and Perico) with different concentrations of enzyme cocktail. R2 values correspond to different wheat genotypes and enzyme concentrations between 2 and 0.0078 μL/mg DW.
(TIFF)
Acknowledgments
We would like to thank Tadeo Bellot for his work growing these plants. We are also grateful for the help we have received in laboratory procedures from Rachael Simister.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was supported by AGL2011-22596 financed by the Spanish Ministry of Economy and Competitiveness (http://www.mineco.gob.es/) and co-financed with FEDER. FJO was supported by the funding of FPI grant BES-2012-052455 from the Spanish Ministry of Economy and Competitiveness. A short-term stay in CNAP by FJO was funded by EEBB-I-15-10360. Research at CNAP was funded by BBSRC projects BB/L001926/1, BB/P02372, and BB/N023269.
References
- 1.IEA I. World energy outlook 2011 Int Energy Agency; 2011; 666. [Google Scholar]
- 2.IEA. World energy outlook 2010. Paris: organisation for economic cooperation and development; 2010. Available: http://www.oecd-ilibrary.org/content/book/weo-2010-en
- 3.From IEA. 1st to 2nd generation biofuel technologies; An overview of current industry and R&D activities Paris, France: Int Energy Agency; 2008; [Google Scholar]
- 4.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. PNAS. 2006; 103:11206–11210. 10.1073/pnas.0604600103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ercolano MR, Gomez LD, Andolfi A, Simister R, Troise C, Angelino G, et al. Residual biomass saccharification in processing tomato is affected by cultivar and nitrogen fertilization. Biomass and Bioenergy. 2015; 72:242–250. 10.1016/j.biombioe.2014.10.030 [DOI] [Google Scholar]
- 6.Yuan T-Q, Sun R-C. Chapter 1—Introduction. Cereal straw as a resource for sustainable biomaterials and biofuels Amsterdam: Elsevier; 2010. pp. 1–7. [DOI] [Google Scholar]
- 7.Carroll A, Somerville C. Cellulosic Biofuels. Annual review of plant biology. 2009; 60:165–182. 10.1146/annurev.arplant.043008.092125 [DOI] [PubMed] [Google Scholar]
- 8.Gomez LD, Whitehead C, Barakate A, Halpin C, McQueen-Mason SJ. Automated saccharification assay for determination of digestibility in plant materials. Biotechnology for Biofuels. 2010; 3:23 10.1186/1754-6834-3-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, et al. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresource Technology. 2005; 96:673–686. 10.1016/j.biortech.2004.06.025 [DOI] [PubMed] [Google Scholar]
- 10.Capper BS. Genetic variation in the feeding value of cereal straw. Animal feed science and technology. 1988; 21:127–140. 10.1016/0377-8401(88)90095-8 [DOI] [Google Scholar]
- 11.Jensen JW, Magid J, Hansen-Møller J, Andersen SB, Bruun S. Genetic variation in degradability of wheat straw and potential for improvement through plant breeding. Biomass and Bioenergy. 2011; 35:1114–1120. 10.1016/j.biombioe.2010.11.036 [DOI] [Google Scholar]
- 12.Duguid KB, Montross MD, Radtke CW, Crofcheck CL, Shearer SA, Hoskinson RL. Screening for sugar and ethanol processing characteristics from anatomical fractions of wheat stover. Biomass and Bioenergy. 2007; 31:585–592. 10.1016/j.biombioe.2007.03.002 [DOI] [Google Scholar]
- 13.Tolera A, Tsegaye B, Berg T. Effects of variety, cropping year, location and fertilizer application on nutritive value of durum wheat straw. Journal of Animal Physiology and Animal Nutrition. 2008; 92:121–130. 10.1111/j.1439-0396.2007.00717.x [DOI] [PubMed] [Google Scholar]
- 14.Orskov ER (Rowett RI. Consistency of differences in nutritive value of straw from different varieties in different seasons ILCA; 1988. Available: http://agris.fao.org/agris-search/search.do?recordID=QM9000090 [Google Scholar]
- 15.Rao SC. Regional environment and cultivar effects on the quality of wheat straw. Agronomy journal. 1989; Available: http://agris.fao.org/agris-search/search.do?recordID=US201302680656 [Google Scholar]
- 16.Lindedam J, Bruun S, Jørgensen H, Decker SR, Turner GB, De Martini JD, et al. Evaluation of high throughput screening methods in picking up differences between cultivars of lignocellulosic biomass for ethanol production. Biomass and Bioenergy. 2014; 66:261–267. 10.1016/j.biombioe.2014.03.006 [DOI] [Google Scholar]
- 17.Ohlde GW, Becker K, Akin DE, Rigsby LL, Lyon CE. Differences in rumen bacterial degradation of morphological fractions in eight cereal straws and the effect of digestion on different types of tissues and mechanical properties of straw stalks. Animal Feed Science and Technology. 1992; 36:173–186. 10.1016/0377-8401(92)90055-B [DOI] [Google Scholar]
- 18.Tan ZL, Chen HP, He LH, Fang RJ, Xing TX. Variation in the nutritional characteristics of wheat straw. Animal Feed Science and Technology. 1995; 53:337–344. 10.1016/0377-8401(94)00721-K [DOI] [Google Scholar]
- 19.Oakey H, Shafiei R, Comadran J, Uzrek N, Cullis B, Gomez LD, et al. Identification of crop cultivars with consistently high lignocellulosic sugar release requires the use of appropriate statistical design and modelling. Biotechnology for Biofuels. 2013; 6:185 10.1186/1754-6834-6-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mewalal R, Mizrachi E, Mansfield SD, Myburg AA. Cell wall-related proteins of unknown function: missing links in plant cell wall development. Plant Cell Physiol. 2014; 55:1031–1043. 10.1093/pcp/pcu050 [DOI] [PubMed] [Google Scholar]
- 21.Decker SR, Brunecky R, Tucker MP, Himmel ME, Selig MJ. High-Throughput screening techniques for biomass conversion. Bioenerg Res. 2009; 2:179 10.1007/s12155-009-9051-0 [DOI] [Google Scholar]
- 22.Studer MH, DeMartini JD, Brethauer S, McKenzie HL, Wyman CE. Engineering of a high-throughput screening system to identify cellulosic biomass, pretreatments, and enzyme formulations that enhance sugar release. Biotechnol Bioeng. 2010; 105:231–238. 10.1002/bit.22527 [DOI] [PubMed] [Google Scholar]
- 23.Santoro N, Cantu SL, Tornqvist C-E, Falbel TG, Bolivar JL, Patterson SE, et al. A High-Throughput platform for screening milligram quantities of plant biomass for lignocellulose digestibility. Bioenerg Res. 2010; 3:93–102. 10.1007/s12155-009-9074-6 [DOI] [Google Scholar]
- 24.Dowe N, McMillan J. SSF Experimental Protocols—Lignocellulosic biomass hydrolysis and fermentation. 2008; Available: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.693.5856&rep=rep1&type=pdf [Google Scholar]
- 25.Wyman CE, Dale BE, Elander RT, Holtzapple M, Ladisch MR, Lee YY. Comparative sugar recovery data from laboratory scale application of leading pretreatment technologies to corn stover. Bioresource Technology. 2005; 96:2026–2032. 10.1016/j.biortech.2005.01.018 [DOI] [PubMed] [Google Scholar]
- 26.Wyman CE, Dale BE, Elander RT, Holtzapple M, Ladisch MR, Lee YY. Coordinated development of leading biomass pretreatment technologies. Bioresource Technology. 2005; 96:1959–1966. 10.1016/j.biortech.2005.01.010 [DOI] [PubMed] [Google Scholar]
- 27.Foster CE, Martin TM, Pauly M. Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) Part I: Lignin. J Vis Exp. 2010; 10.3791/1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. R: The R Stats Package. 2015 [cited 18 Mar 2016]. Available: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/stats-package.html
- 29.Felipe de Mendiburu. CRAN—Package agricolae. 2015 [cited 18 Mar 2016]. Available: http://cran.fyxm.net/web/packages/agricolae/
- 30.Wickham Hadley. ggplot2: Elegant graphics for data analysis Springer-Verlag; New York; 2009. Available: http://ggplot2.org [Google Scholar]
- 31.Fox J, Weisberg S, Adler D, Bates D, Baud-Bovy G, Ellison S, et al. car: Companion to applied regression. 2015. Available: https://cran.r-project.org/web/packages/car/index.html [Google Scholar]
- 32.Ørskov ER, Shand WJ, Tedesco D, Morrice L a. F. Rumen degradation of straw. 10. Consistency of differences in nutritive value between varieties of cereal straws. Animal Science. 1990; 51:155–162. 10.1017/S0003356100005250 [DOI] [Google Scholar]
- 33.White LM, Hartman GP, Bergman JW. In vitro digestibility, crude protein, and phosphorus content of straw of winter wheat, spring wheat, barley, and oat cultivars in eastern Montana. Agronomy Journal. 1981; 73:117–121. 10.2134/agronj1981.00021962007300010026x [DOI] [Google Scholar]
- 34.Kleinhofs A, Kilian A, Maroof MAS, Biyashev RM, Hayes P, Chen FQ, et al. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theoret Appl Genetics. 1993; 86:705–712. 10.1007/BF00222660 [DOI] [PubMed] [Google Scholar]
- 35.Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, et al. Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet. 2005; 112:445–454. 10.1007/s00122-005-0144-7 [DOI] [PubMed] [Google Scholar]
- 36.Arru L, Francia E, Pecchioni N. Isolate-specific QTLs of resistance to leaf stripe (Pyrenophora graminea) in the “Steptoe” x “Morex” spring barley cross. Theoretical and applied genetics. 2003; 106:668–675. 10.1007/s00122-002-1115-x [DOI] [PubMed] [Google Scholar]
- 37.Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009; 10:582 10.1186/1471-2164-10-582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chao S, Zhang W, Dubcovsky J, Sorrells M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (L.) germplasm representing different market classes. Crop Science. 2007; 47:1018 10.2135/cropsci2006.06.0434 [DOI] [Google Scholar]
- 39.Lindedam J, Andersen SB, DeMartini J, Bruun S, Jørgensen H, Felby C, et al. Cultivar variation and selection potential relevant to the production of cellulosic ethanol from wheat straw. Biomass and Bioenergy. 2012; 37:221–228. 10.1016/j.biombioe.2011.12.009 [DOI] [Google Scholar]
- 40.Caruso G, Gomez LD, Ferriello F, Andolfi A, Borgonuovo C, Evidente A, et al. Exploring tomato Solanum pennellii introgression lines for residual biomass and enzymatic digestibility traits. BMC Genetics. 2016; 17:56 10.1186/s12863-016-0362-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotech. 2007; 25:759–761. 10.1038/nbt1316 [DOI] [PubMed] [Google Scholar]
- 42.Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnology for Biofuels. 2013; 6:46 10.1186/1754-6834-6-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellucci A, Torp AM, Bruun S, Magid J, Andersen SB, Rasmussen SK. Association mapping in scandinavian winter wheat for yield, plant height, and traits important for second-generation bioethanol production. Front Plant Sci. 2015; 6. 10.3389/fpls.2015.01046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Travis AJ, Murison SD, Hirst DJ, Walker KC, Chesson A. Comparison of the anatomy and degradability of straw from varieties of wheat and barley that differ in susceptibility to lodging. The Journal of Agricultural Science. 1996; 127:1–10. 10.1017/S0021859600077327 [DOI] [Google Scholar]
- 45.Chen Y, Sharma-Shivappa RR, Keshwani D, Chen C. Potential of agricultural residues and hay for bioethanol production. Appl Biochem Biotechnol. 2007; 142:276–290. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Sharma-Shivappa RR, Chen C. Ensiling agricultural residues for bioethanol production. Appl Biochem Biotechnol. 2007; 143:80–92. [DOI] [PubMed] [Google Scholar]
- 47.Jezowski S (Polska AN. Variation correlation and heritability of characters determining lodging of spring barley (Hordeum vulgare L.). 2. Analysis of relationship between lodging grade and some morphological characters of spring barley varieties. Polish Journal of Theoretical and Applied Genetics. 1981; Available: http://agris.fao.org/agris-search/search.do?recordID=PL8200169 [Google Scholar]
- 48.Tandon JP, Jain KBL. Relationship between lodging resistance and some morphological characters in barley. 1973. [cited 2 Jun 2016]. Available: http://www.indianjournals.com/ijor.aspx?target=ijor:ijgpb&volume=33&issue=3&article=012 [Google Scholar]
- 49.Brady J. Some factors influencing lodging in cereals. The Journal of Agricultural Science. 1934; 24:209–232. 10.1017/S0021859600006602 [DOI] [Google Scholar]
- 50.Saint Pierre C, Trethowan R, Reynolds M. Stem solidness and its relationship to water-soluble carbohydrates: association with wheat yield under water deficit. Funct Plant Biol. 2010; 37:166–174. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Glucose released in wheat genotypes (Anza, Bobwhite and Perico) with different concentrations of enzyme cocktail. R2 values correspond to different wheat genotypes and enzyme concentrations between 2 and 0.0078 μL/mg DW.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.