Abstract
Rationale: Recurrent wheeze and asthma are thought to result from alterations in early life immune development following respiratory syncytial virus (RSV) infection. However, prior studies of the nasal immune response to infection have assessed only individual cytokines, which does not capture the whole spectrum of response to infection.
Objectives: To identify nasal immune phenotypes in response to RSV infection and their association with recurrent wheeze.
Methods: A birth cohort of term healthy infants born June to December were recruited and followed to capture the first infant RSV infection. Nasal wash samples were collected during acute respiratory infection, viruses were identified by RT-PCR, and immune-response analytes were assayed using a multianalyte bead-based panel. Immune-response clusters were identified using machine learning, and association with recurrent wheeze at age 1 and 2 years was assessed using logistic regression.
Measurements and Main Results: We identified two novel and distinct immune-response clusters to RSV and human rhinovirus. In RSV-infected infants, a nasal immune-response cluster characterized by lower non-IFN antiviral immune-response mediators, and higher type-2 and type-17 cytokines was significantly associated with first and second year recurrent wheeze. In comparison, we did not observe this in infants with human rhinovirus acute respiratory infection. Based on network analysis, type-2 and type-17 cytokines were central to the immune response to RSV, whereas growth factors and chemokines were central to the immune response to human rhinovirus.
Conclusions: Distinct immune-response clusters during infant RSV infection and their association with risk of recurrent wheeze provide insights into the risk factors for and mechanisms of asthma development.
Keywords: respiratory syncytial virus, human rhinovirus, immune response, wheeze, systems approach
At Glance Commentary
Scientific Knowledge on the Subject
Respiratory syncytial virus acute respiratory illness is strongly associated with childhood asthma. Our understanding of the mechanisms underlying the association of viral infection with asthma development is limited.
What This Study Adds to the Field
This study provides evidence that acute respiratory syncytial virus respiratory infection in infants results in two distinct immune-response phenotypes. The response profile characterized by type-2 and type-17 high and low non-IFN antiviral immune response was significantly associated with later recurrent wheezing. The newly identified infant viral–nasal immune-response cluster association with recurrent wheeze may provide insights into acute viral disease mechanisms and the established relationship of these viruses with later asthma development.
Acute respiratory infection (ARI) with respiratory syncytial virus (RSV) is one of the risk factors most strongly and consistently associated with asthma risk (1–3). Human rhinovirus (HRV) has also been strongly associated with subsequent asthma risk (4, 5). However, these viruses differ in their mode of entry into the cells and subsequent behavior (6–8), therein we hypothesized, potentially evoking distinct immune-responses (9). As the innate immune response is central to the initial defense against RSV (10–13), infection may contribute to early life reprogramming of an infant’s immune response (14–18). Prior studies have assessed only individual cytokines or immune cell lineage (19–22), which does not capture the whole spectrum of immune response to infection (23). Advances in technologies and computational approaches make it possible to examine the immune-response pattern in response to an environmental risk factor, such as RSV, through analysis of multiple components of the immune system simultaneously (24, 25). This integrative analytical approach identifies the central pathways and dynamic relationships among the myriad of immune-response mediators involved in the pathogenesis of early childhood wheezing.
Our objective was to understand how RSV may contribute to later childhood asthma by identifying infant immune-response patterns to RSV infection (which we term the viral–nasal immune-response pattern) at the site of infection, to compare it with another known common infant respiratory virus and asthma risk factor, HRV, and to establish pathways through which RSV immunomodulation may contribute to the development of childhood recurrent wheeze.
Methods
Population
The study was conducted on a subset of the ongoing INSPIRE (Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure) cohort (n = 193; 9.8%), for whom nasal immune-response mediators data were available during ARI with either RSV or HRV. The details of the cohort have been described previously (26). Infancy is defined as first-year of child life. Recurrent wheeze at age 1 and 2 years was assessed as a known precursor to later childhood asthma, and time points are the most current available for this ongoing longitudinal birth cohort study. The institutional review board of Vanderbilt University approved this study.
Acute Respiratory Illness Visit and Nasal Wash Sample Collection
Infants underwent biweekly surveillance for ARI during winter viral season (November through March). If infants met predefined criteria for ARI, a respiratory illness visit was conducted to assess disease severity and a nasal wash sample was collected (26). The nasal wash was performed using 5 ml of sterile saline and immediately placed at 4°C for transport and then snap frozen at −80°C for storage.
Viral Detection and Viral–Nasal Immune-Response Pattern Analytes Intensity Quantification
Nasal samples collected during ARI were profiled for respiratory viruses (RSV and HRV) using RT-PCR and assayed for a panel of analytes. Nasal samples from 193 infants (with a total of 232 samples including repeated infections) who were either RSV (108 first and 4 second infections) or HRV (102 first and 18 second infections) PCR positive were profiled using Luminex xMap multianalyte bead assays for 53 analytes (Milliplex Human Cytokine/Chemokine Panel II MAGNETIC Premixed 23 Plex Kit, EMD Millipore; and Cytokine 30-Plex Human Panel, Life Technologies Corporation). We used only the samples from the first infections for each virus (RSV and HRV). Table 1 provides sample demographic and population characteristics. Two replicates of each sample assay were performed on plates with 96 wells each. The multiplex bead assays included 53 immune-response analytes: VEGF (vascular endothelial growth factor), G-CSF (granulocyte colony–stimulating factor), GM-CSF (granulocyte-macrophage colony–stimulating factor), EGF (epidermal growth factor), FGF (fibroblast growth factor), eotaxin-1/CCL11, eotaxin-2/CCL24, eotaxin-3/CCL26, MCP-1 (monocyte chemotactic protein-1)/CCL2, MCP-2/CCL8, MCP-4/CCL13, I-309/CCL1, IL-16, TARC (thymus- and activation-regulated chemokine)/CCL17, 6CKine (chemokine with 6 cysteines)/CCL21, LIF (leukemia inhibitory factor), TPO (thrombopoietin), SCF (stem cell factor), TSLP (thymic stromal lymphopoietin), TRAIL (TNF [tumor necrosis factor]-related apoptosis-inducing ligand), BCA-1 (B cell–attracting chemokine-1), CTACK (cutaneous T cell-attracting chemokine)/CCL27, SDF-1ab (stromal cell–derived factor-1ab)/ CXCL12, ENA-78 (epithelial cell-derived neutrophil attractant with 78 amino acids)/CXCL5, IL-1β, IL-1RA (receptor antagonist), IL-2, IL-2R, IL-7, IL-10, IL-13, IL-4, IL-5, IL-6, IL-8, IL-12, IL-15, IL-17, IL-20, IL-21, IL-23, IL-28A, IL-33, IFN-γ, IFN-α, TNF-α, RANTES (regulated on activation, normal T cell expressed and secreted)/CCL5, MIP-1α (macrophage inflammatory protein-1α)/CCL3, MIP-1β/CCL4, MIP-1d/CCL15, IP-10/ CXCL10, MIG (monokine induced by IFN-γ)/CXCL9, and HGF (human growth factor). A blank well was used to estimate the background. HGF was below detectable levels in more than 50% of the infant nasal wash sample, and was therefore removed from further analysis.
Table 1.
Demographic, Parental History, and Illness Characteristics by Virus Type and Clusters of Immune Responses to RSV and HRV
| RSV |
HRV |
|||||
|---|---|---|---|---|---|---|
| Total (n = 108) | Cluster 1 (n = 47) | Cluster 2 (n = 61) | Total (n = 102) | Cluster 1 (n = 45) | Cluster 2 (n = 57) | |
| Sex, n (%) | ||||||
| Male | 61 (56.5) | 27 (57.5) | 34 (55.7) | 54 (52.9) | 26 (57.8) | 28 (49.1) |
| Female | 47 (43.5 | 20 (42.6) | 27 (44.3) | 48 (47.1) | 19 (42.2) | 29 (50.9) |
| Race, n (%) | ||||||
| White | 76 (70.4) | 33 (70.2) | 43 (70.5) | 75 (75.8) | 32 (74.4) | 43 (76.8) |
| Black | 13 (12.0) | 3 (6.4) | 10 (16.4) | 7 (7.1) | 2 (4.7) | 5 (8.9) |
| Hispanic | 11 (10.2) | 5 (10.6) | 6 (9.8) | 12 (12.1) | 7 (16.3) | 5 (8.9) |
| Multiple/other | 8 (7.4) | 6 (12.8) | 2 (3.3) | 5 (5.0) | 2 (4.6) | 3 (5.4) |
| Maternal smoking, n (%) | ||||||
| Never | 74 (68.5) | 37 (77.10) | 39 (63.9) | 67 (67.7) | 28 (65.1) | 39 (69.6) |
| Former | 14 (13.0) | 5 (10.4) | 8 (13.1) | 14 (14.1) | 7 (16.3) | 7 (12.5) |
| Current | 20 (18.5) | 6 (12.5) | 14 (23.0) | 18 (18.2) | 8 (18.6) | 10 (17.9) |
| Maternal asthma, n (%) | ||||||
| Yes | 17 (15.7) | 5 (10.6) | 12 (19.7) | 18 (18.1) | 8 (18.6) | 10 (17.9) |
| No | 91 (84.3) | 42 (89.4) | 49 (80.3) | 81 (81.8) | 35 (81.4) | 46 (82.1) |
| Maternal allergy, n (%) | ||||||
| Yes | 27 (25.0) | 9 (19.1) | 18 (29.5) | 27 (27.3) | 14 (32.6) | 13 (23.2) |
| No | 81 (75.0) | 38 (80.9) | 43 (70.5) | 72 (72.7) | 29 (67.4) | 43 (76.8) |
| Maternal education, n (%) | ||||||
| <High school | 9 (8.3) | 3 (6.4) | 6 (9.8) | 3 (3.0) | 3 (7.0) | 0 (0.0) |
| High school graduate | 19 (17.6) | 4 (8.5) | 15 (24.6) | 20 (20.2) | 9 (20.9) | 11 (19.6) |
| College | 80 (74.1) | 40 (85.1) | 40 (65.6) | 76 (76.8) | 31 (72.1) | 45 (80.4) |
| Health care received for ARI, n (%) | ||||||
| None | 14 (13.0) | 5 (14.6) | 9 (14.8) | 53 (52.0) | 24 (53.3) | 29 (51.0) |
| Outpatient | 64 (59.3) | 30 (63.8) | 34 (55.7) | 45 (44.1) | 19 (42.2) | 26 (45.6) |
| Emergency | 12 (11.1) | 6 (12.8) | 6 (9.8) | 3 (2.9) | 2 (4.4) | 1 (1.7) |
| Hospitalization | 18 (16.7) | 6 (12.8) | 12 (19.7) | 1 (1.0) | 0 (0.0) | 1 (1.7) |
| Age at RSV or HRV ARI, mo, mean (SD) | 4 (1.9) | 4.5 (1.9) | 3.6 (1.8) | 3.8 (1.8) | 3.4 (1.7) | 4.0 (1.8) |
| Days from symptom onset to the nasal sample, mean (SD) | 3.1 (1.5) | 3.1 (1.4) | 3.1 (1.4) | 2.7 (1.3) | 2.6 (1.1) | 2.8 (1.4) |
| Severity score of ARI illness, mean (SD)* | 3.1 (2.3) | 3.1 (2.2) | 3.1 (2.4) | 1.9 (1.3) | 2.1 (1.6) | 1.6 (0.9) |
Definition of abbreviations: ARI = acute respiratory infection; HRV = human rhinovirus; RSV = respiratory syncytial virus.
Respiratory severity score on scale to 0–12, with higher scores indicating more severe disease.
Infant Viral–Nasal Immune-Response Pattern Analytes Data Processing
Because our primary interest was to determine the pattern of immune-responses, we used the relative abundances of the analytes, which is fluorescence intensity (FI), in our analysis (27, 28). This is an approach first promoted by Won and colleagues to process the individual bead FI data and has been validated (28, 29) and used in a number of studies as alternative approach to overcome several issues with curve-based FI data (30–33). Won and coworkers’ (27) approach differs from the typical standard curve–based FI data-processing approach, which imposes range limits on the median FI of analytes, thus yielding excessive missing data for low FI analytes with median values below limits of detection. Assay wells with less than 50 beads were eliminated from further data processing because of unreliability. Subsequently, the pooled 5% trimmed mean of fluorescence values from blank/buffer samples was subtracted from the FI values of each analyte. Generalized log transformation was applied to stabilize variance of the FI values (34, 35).
The assays were originally processed using the standard curve methods; therefore, we used known standard replicates between plates. In addition, we used standard curve method derived concentration to test for statistically significant differences between plates and did not find any.
Statistical Analysis
We normalized the FI data using the median of all FI values to guard against any systematic variance introduced by technical factors, such as sample-to-sample cell count differences, suboptimal antibodies and fluorescence marker combinations, instrument variability over time, changes in reagent lots, and variability in sample handling (36, 37). Finally, we scaled the normalized and log transformed values using the Pareto scaling method (38). The multivariate kernel density graph of the raw data and the processed data is provided in the online supplement (see Figure E1 in the online supplement).
We first assessed the number of infant nasal immune-response pattern using principal component (PC) analysis and hierarchical clustering. We then assessed the statistically significant distance between immune-response patterns with Gaussian mixture model. Subsequently, we used the number of patterns observed in the PC analysis, hierarchical clustering, and Gaussian mixture model to cluster the infants with k-means clustering method. The immune-response patterns were visualized using heatmaps, where k-mean cluster labels were included. We then used partial least square discriminant analysis (PLS-DA) and orthogonal PLS-DA to obtain a ranked list of cytokines ordered by their variable importance in projection score based on their ability to differentiate between the identified clusters. This ranking was validated with five-fold cross-validation. Finally, we used multivariable logistic regression to assess the association of the clusters identified with recurrent wheezing during the first and second year of life, the most recent time point available for the birth cohort. The model was adjusted for infant age at ARI, sex, ARI severity score, maternal asthma, maternal education level, and number of days from symptom onset to collection of the nasal wash sample. Recurrent wheezing was defined as wheezing frequency greater than three or asthma medication use in the past 12 months for 1- and 2-year recurrent wheezing outcomes. ARI severity score was assessed for each infant, and is a composite score ranging from 0 to 12, with a higher score indicating more severe disease (1). In addition, we conducted discriminant analysis using orthogonal PLS-DA to differentiate between RSV and HRV infections and partial correlation-based network analysis to identify cytokines central to HRV and RSV immune-responses. Significance of the partial correlation between analytes was cutoff at P less than 0.05 for visualization. Multivariate computation was conducted in R version 3.3.3 (39) and Metaboanalyst 3.0 (40), and network analysis was performed in Cytoscape 3.1 (41).
Results
Variation in the Infant Viral–Nasal Immune-Response Data and Variation Attributable to the Viruses
To explore the variation in the infant viral–nasal immune-response mediator data we performed PC analysis of the immune-response mediators and evaluated the association of PCs with non-immune-response covariates (see the Results section of the online supplement and Figure E2). The first, second, and third PCs accounted for 45.1%, 10.7%, and 6.7% of the variation in the viral–nasal immune-response mediators data. The first and second PCs, which cumulatively account for 55.8% of the variation in the data (see Figure E2), were associated with age at ARI, where older age (in months) is negatively associated with PC1 (ordinary least square regression, β = −1.19; P < 0.001) and positively associated with PC2 (ordinary least square regression, β = 0.59; P = 0.002). The first PC, which explained 45.1% of the variation in the data, was significantly associated with recurrent wheezing in the first year (logistic regression, odds ratio [OR], 1.04; confidence interval [CI], 1.01–1.07). The third PC, which is responsible for 6.7% of the variation, is associated with maternal asthma (logistic regression, OR, 1.15; CI, 1.03–1.29). The fourth PC, which contributed 5.5% of the variation in the data, is associated with RSV (vs. HRV) (logistic regression, OR, 1.12; CI, 1.02–1.20). Analyses using orthogonal PLS-DA show that 5% of the variation was attributable to the virus type (see Figures E3A and Figure E3B). The variation in the immune-response mediators data caused by virus type was small, but biologically meaningful, characterizing the comparison of immune response to RSV and to that of HRV.
Infant Viral–Nasal Immune-Response Pattern
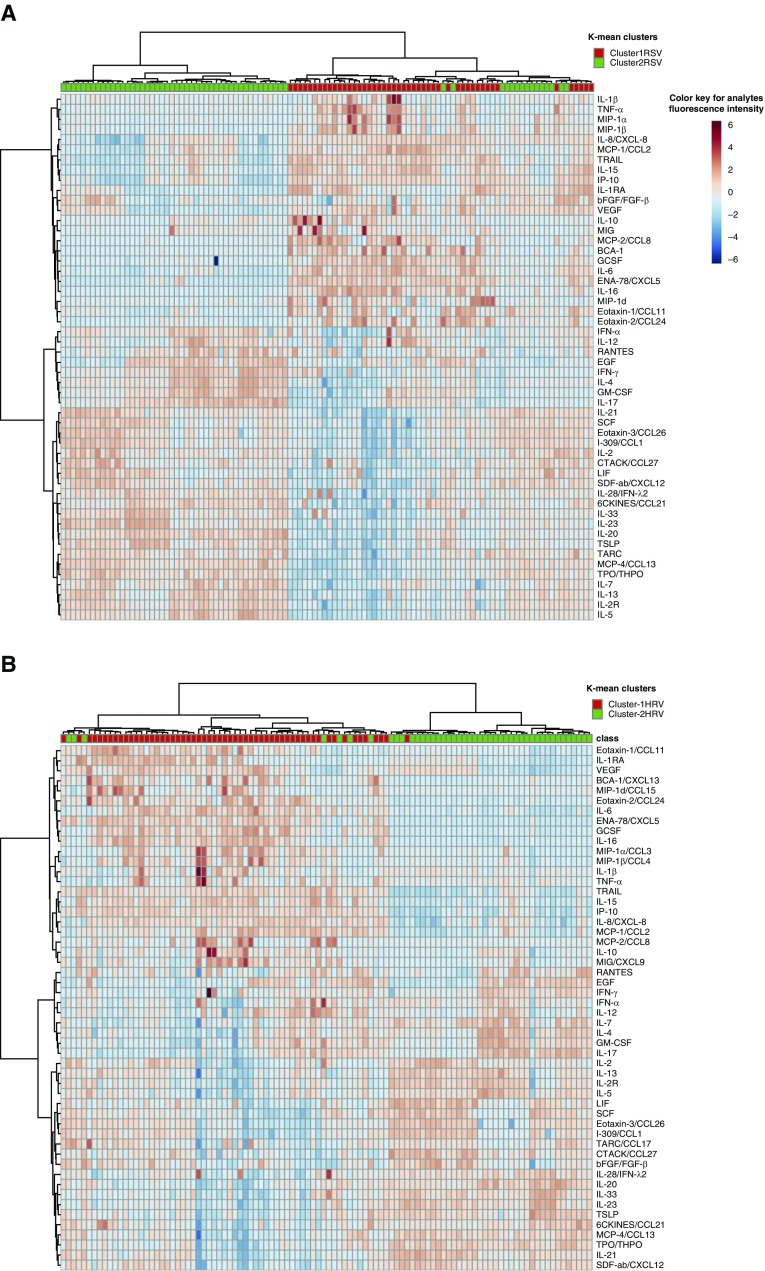
To identify immune-response mediator patterns to RSV and HRV acute respiratory illness, we next used a combination of clustering approaches on each separately. Hierarchical clustering and Gaussian mixture model or latent class clustering, a model-based soft clustering approach, followed by k-mean clustering identified two distinct immune-response clusters each for RSV and HRV ARI. We refer to the clusters as cluster-1RSV (n = 47) and cluster-2RSV (n = 61), and cluster-1HRV (n = 45) and cluster-2HRV (n = 57) hereafter. The intensity of type-2 and type-17 immune response to the viruses differed between the two identified clusters as visualized using k-mean clusters labeled heatmap (Figure 1).
Figure 1.
Heatmap of infant immune-response pattern using hierarchical clustering. (A) Heatmap of infant immune response using hierarchical clustering shows two distinct immune-response patterns to respiratory syncytial virus infection. (B) Heatmap of infant immune response using hierarchical clustering shows two distinct immune-response patterns to human rhinovirus infection. The infants are also labeled with the k-means cluster category shown at the top of the heatmap. The heatmap cell color represents the fluorescence intensity of immune-response mediators as shown in the color keys. HRV = human rhinovirus; RSV = respiratory syncytial virus.
Profile of Infant Viral–Nasal Immune-Response Clusters
Clusters 1RSV and 2RSV had distinctly different immune-response patterns from clusters 1HRV and 2HRV, respectively (Figure 1). Details are presented in the Results section of the online supplement. A nonparametric univariate test (Wilcoxon rank test) for the difference between the immune-response mediators in the two immune-response clusters for each virus indicates that 47 immune-response mediators were significantly different between RSV clusters and 46 immune-response mediators were significantly different between HRV clusters after false discovery rate correction for multiple testing (see Tables E4 and E5 and Figures E4A and E4B). In summary, elevated type-2 and type-17 immune-response mediators were characteristic of cluster-2RSV and cluster-2HRV. However, decreased levels of chemokines that are known to limit viral replication, and immune-response regulators were characteristic of cluster-2RSV and cluster-2HRV. Immune-response mediators including IL-1β, IL-7, IL-8/CXCL8, IL-15, IP-10/CXCL10, MIG/CXCL9, MCP-1/CCL2, TNF-α, VEGF, G-CSF, eotaxin-1/CCL11, FGF-Basic, and ENA-78/CXCL5 and immunoregulatory cytokines, such as IL-1RA, IL-6, IL-10, and IL-16 were decreased in cluster-2RSV. Moreover, both cluster-2RSV and cluster-2HRV were characterized by type-2 and type-17 cytokines: IL-4, IL-5, IL-13, IL-17, IL-20, IL-23, IL-33, and TSLP. In cluster-2RSV and cluster-2HRV, the level of IFN-γ was elevated and the level of IL-10 was decreased.
Association of Distinct Infant Viral–Nasal Immune-Response Clusters with Recurrent Wheeze
To understand the impact of the observed distinct infant immune-response cluster during RSV or HRV ARI on the development of recurrent wheezing, we performed adjusted and unadjusted logistic regression assessing the association between first and second year recurrent wheezing and the identified immune-response clusters (Table 2; see Tables E1 and Table E2). The RSV cluster characterized by type-2 and type-17 cytokine dominance (cluster-2RSV) was significantly associated with wheezing at 1 year (adjusted OR, 4.78; 95% CI, 1.73–13.24) and at 2 years (adjusted OR, 2.83; 95% CI, 1.08–7.44). The associations between HRV cluster (cluster-2HRV) and recurrent wheezing were not statistically significant at any time point (see the Results section of the online supplement and Table E2). The frequency of recurrent wheeze by virus, immune-response cluster, and year is provided in Table E3.
Table 2.
Association of RSV Clusters with 1- and 2-Year Recurrent Wheezing Outcomes in Unadjusted and Adjusted Multivariable Logistic Regression Analyses
| First-Year Recurrent Wheeze [OR (95% CI)]* | Second-Year Recurrent Wheeze [OR (95% CI)]* | |
|---|---|---|
| RSV | ||
| Cluster-2RSV:cluster-1RSV (unadjusted) | n = 101 | n = 102 |
| 2.82 (1.23– 6.48) | 2.35 (0.99–5.57) | |
| Cluster-2RSV:cluster-1RSV (adjusted†) | n = 96 | n = 96 |
| 4.78 (1.73–13.24) | 2.83 (1.08–7.44) | |
| HRV | ||
| Cluster-2HRV:cluster-1HRV (unadjusted) | n = 94 | n = 90 |
| 2.08 (0.76–5.69) | 1.46 (0.58–3.65) | |
| Cluster-2HRV:cluster-1HRV (adjusted†) | n = 88 | n = 86 |
| 2.05 (0.67–6.32) | 1.61 (0.55–4.64) | |
Definition of abbreviations: CI = confidence interval; HRV = human rhinovirus; OR = odds ratio; RSV = respiratory syncytial virus.
Recurrent wheezing was defined as wheezing frequency >3 episodes or asthma medication use in the past 12 months for every year.
Adjusted for: sex, age at acute respiratory infection, severity of acute respiratory infection, time from symptom onset to nasal wash sample (d), maternal asthma, maternal education level (see Tables E2 and E3 for details on covariates).
What Are the Most Important Immune-Response Mediators in Distinguishing the Infant Viral–Nasal Immune-Response Clusters?
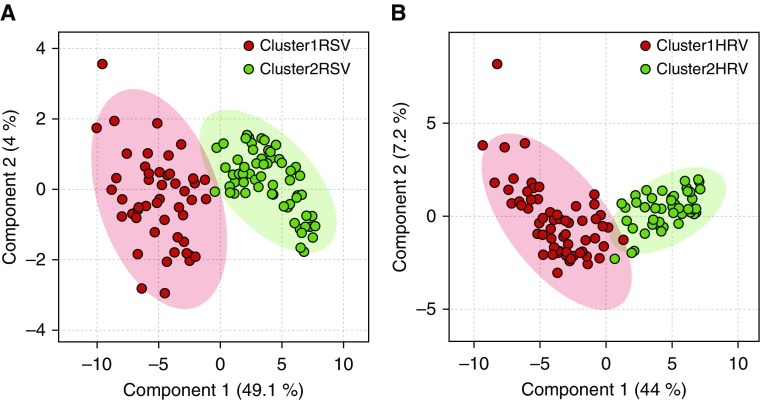
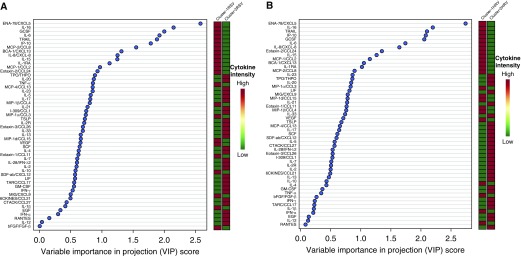
We used PLS-DA to establish whether the clusters were distinctly separated (Figure 2) and to identify important mediators that distinguish the two immune-response clusters for each viral agent (Figure 3). The PLS-DA variable importance in projection procedure ranked the mediators according to their importance in distinguishing the two clusters for each virus (Figure 3; see the Results section of the online supplement).
Figure 2.
Partial least square discriminant analysis to discriminate between immune-response clusters. (A) Partial least square discriminant analysis score plot shows discrimination between respiratory syncytial virus (RSV) immune-response clusters (cluster-1 RSV vs. cluster-2 RSV) that were identified using hierarchical and k-means clustering. (B) Partial least square discriminant analysis score plot shows discrimination between human rhinovirus (HRV) immune-response clusters (cluster-1 HRV vs. cluster-2 HRV) that were identified using hierarchical and k-means clustering.
Figure 3.
Immune-response mediators importance rank (variable importance in projection) in discriminating between immune-response clusters using partial least squares discriminant analysis. (A) Immune-response mediators’ importance rank in discriminating between respiratory syncytial virus (RSV) immune-response clusters (cluster-1 RSV vs. cluster-2 RSV). (B) Immune-response mediators importance rank in discriminating between human rhinovirus (HRV) immune-response clusters (cluster-1 HRV vs. cluster-2 HRV). The immune-response mediators ranking or variable importance in projection was internally validated with 10-fold cross-validation.
Infant Viral–Nasal Immune-Response Mediators Network and Topologic Analysis
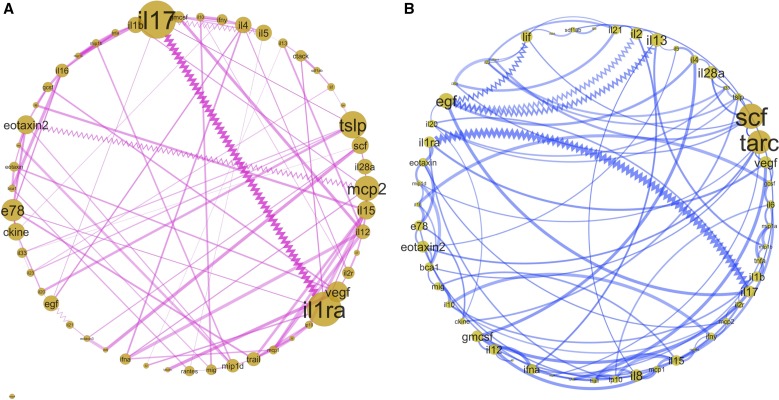
To further characterize the differential infant immune response to RSV and HRV, we used partial correlations with an alpha threshold of P less than 0.05 for constructing the network of immune responses during RSV and HRV ARIs (Figure 4; see the Results section of the online supplement).
Figure 4.
Infant viral–nasal immune-response mediators network analysis and topologic analysis. (A) Infant nasal immune-response mediators network in response to respiratory syncytial virus infection. (B) Infant nasal immune-response mediators network in response to human rhinovirus infection. The network is based on partial correlations between the immune-response mediators with P < 0.05. The zigzag lines and straight lines represent negative and positive correlation between nodes, respectively. The thickness of the line represents the strength of correlation between nodes/immune-response mediators. The size of the node represents betweenness-centrality of the node in the immune-response network; the larger the size of the nodes, the more central the cytokines are to the immune-response network. Betweenness-centrality measures the extent to which a node lies on the paths between other nodes.
Topologic analysis focusing on the centrality of the network revealed mediators that play central roles in the immune response to these viruses (Figure 4). IL-17 and IL-1RA were central to the immune response to RSV. MCP-2/CCL8, TSLP, eotaxin-2/CCL24, IL-15, ENA-78/CXCL5, and 6Ckine/CCL21 were also important hubs in response to RSV. The network analysis of HRV immune response showed that TARC/CCL17 and SCF were at the center of the immune response with the highest between-centrality score. EGF, VEGF, IL-13, IL-28α, eotaxin-2/CCL24, IL-1RA, and GM-CSF had high betweenness-centrality scores during HRV-associated ARI. As noted, TARC/CCL17 and SCF were elevated in the cluster with type-2 and type-17 characteristics (cluster-2HRV).
Discussion
Using a prospective birth cohort designed to identify the first RSV infection during infancy and track ARIs throughout RSV season, we identified two unique viral–nasal immune-response clusters during acute RSV infection, one of which identifies infants at increased risk of childhood recurrent wheeze. To determine if these findings were specific to RSV, we compared the viral–nasal immune-response patterns of infants with RSV ARI to those with HRV ARI, demonstrating some overlap in profiles, but differences between the immune-mediator responses to the two viruses.
One of immune-response clusters we identified both for RSV and HRV (cluster-2RSV and cluster-2HRV), and which was associated with recurrent wheeze in RSV ARI, was characterized by a profile of increased type-2 and type-17 immune-response mediators. The mediators that have been previously demonstrated to be instrumental in clearing RSV (42–46) and HRV (47–51) infections, including IL-1β, IL-7, IL-8/CXCL8, IL-15, IP-10/CXCL10, MIG/CXCL9, MCP-1/CCL2, TNF-α, VEGF, G-CSF, eotaxin-1/CCL11, FGF-Basic, and ENA-78/CXCL5 and immunoregulatory cytokines, such as IL-1RA, IL-6, IL-10, and IL-16 were decreased in cluster-2RSV. Moreover, both cluster-2RSV and cluster-2HRV were characterized by type-2 and type-17 cytokines: IL-4, IL-5, IL-13, IL-17, IL-20, IL-23, IL-33, and TSLP, a key regulator of asthma development (52–55). In cluster-2RSV and cluster-2HRV, the level of IFN-γ was elevated and the level of IL-10 was decreased, which is consistent with published experimental evidence showing that IL-10 deficiency enhances the inflammatory effects of type-2 cytokines and chemokines (56–58).
An infant immune-response cluster to RSV ARI (cluster-2RSV) that was significantly associated with recurrent wheeze was characterized by increased type-2 and type-17 immune mediators, and decreased immunoregulatory and non-IFN antiviral immune-response mediators (TNF-α and macrophages). In comparison, we did not observe this association in HRV-infected infants. Prior studies that have compared individual cytokine concentrations between wheezers and nonwheezers have demonstrated increased concentrations of IFN-γ, IL-2, MIP-1α, and MIP-1β in nasopharyngeal aspirates among recurrent wheezers (22), consistent with the expanded immune-response profile of the RSV cluster that was associated with recurrent wheezing in our study. Because there is a difference in severity of ARI caused by the two viruses, we not only adjusted for severity of ARI but we did a matched analysis, which did not yield different results.
Based on network analyses, we demonstrated differences in the immune mediators central to the immune responses to each of these viruses. Topologic analysis of the immune mediators in the network, and the relationship between individual mediators may shed light on potential synergy, or lack thereof, in immune response to each virus. Between centrality analyses of the networks revealed that type-2 and type-17 cytokines were central to the immune response to RSV, whereas growth factors and chemokines were central to the immune response to HRV.
The observed negative correlation between IL-17 and IL-1RA (receptor antagonist) is explained by animal experiments that demonstrated IL-1RA suppresses IL-1R expression and IL-1RA-deficient mice exhibit elevated IL-1R expression, which plays a critical role in inducing IL-17 production (59). It has been also shown that IL-17 can enhance the RSV-induced production of neutrophil-attractant chemokines (60, 61), thereby increasing neutrophil recruitment and contributing to heightened lung inflammation (62). It is important to note that high IL-17 and low IL-1RA were the hallmark of the RSV cluster that was associated with recurrent wheeze (cluster-2RSV). The negative correlation between EGF and type-2 immune-response mediators (IL-13 and IL-2) during HRV-associated ARI is interesting because EGF has a role in cell proliferation and tissue repair; it may protect the epithelial cell from inflammatory immune-response mediators or virus-mediated damage (63). However, EGF has a role in viral entry, delayed apoptosis during ARIs associated with both HRV and RSV, and promotes viral replication by suppressing antiviral-related immune mediators (63–67). In addition, our findings demonstrate a prominent role of EGF in the immune response to HRV. Supportive of the influence of EGF in HRV infection, several prior studies have shown that HRV-associated asthma exacerbations are characterized by major airway mucin production involving a novel EGFR-dependent pathway (68, 69). These findings are consistent with the literature that RSV, via the various immunomodulatory mechanisms, is acting at every level of defense (18, 70), including on the early life immune system that is inexperienced and vulnerable to modulation (71–73). This enhanced understanding of the changes in the immune system during first year RSV infections may help to explain the pathogenesis of RSV and its strong association with childhood wheeze and asthma.
This study is innovative in the use of a birth cohort specifically designed to capture the first infant RSV infection with ARI surveillance throughout winter virus season and use of high throughput infant nasal immune-response mediator data in combination with computational methods to discover and characterize two novel clusters and identify those mediators that are central in the immune response to RSV and HRV. Despite its novelty in linking changes in nasal immune response to specific viral infection and subsequent recurrent wheeze, this study has some limitations that should be considered. First, even though we ascertained the number of reported days from ARI symptom onset to sample collection, in human studies of natural infection, it is not possible to know the exact number of days from actual viral infection to the nasal sampling assessment of the immune response to ARI. To address this issue, we adjusted for the number of days from symptom onset to the nasal sample collection to account for differences in viral kinetics.
Second, our study of ARIs includes infections of varied severity level, including both lower and upper respiratory tract infections. Nevertheless, we note that our findings did not change when we adjusted the model for disease severity using a continuous respiratory severity score and a dichotomous measure of infection severity, lower respiratory tract infection versus upper respiratory tract infection. We additionally conducted the analyses excluding those who were hospitalized, and the results did not change. Racial/ethnic differences as a proxy for genetic and cultural differences, which may influence diet and other lifestyle factors, may influence immune response. However, race/ethnicity equally distributed between the immune-response clusters and adjusting for it did not change our result. Finally, we used the same sample for testing and validating the results in the discriminant analysis steps. This was done because the sample size was not large enough to split into training and validation sets; however, we used bootstrap resampling for validation of the separation between the clusters, which is standard for machine learning approaches. We also confirmed the existence and significant difference between the two clusters using Gaussian mixture model.
Conclusions
Our findings identify two distinct infant nasal immune-response clusters in response to infant RSV infection, one of which is associated with increased risk of subsequent wheeze. These findings support the concept that the immune-response pattern to RSV is an early insight into host response to environmental stimuli that is a known risk factor for subsequent wheeze, and that infant RSV infection may inhibit immunoregulatory and non-IFN antiviral response mediators and promote a type-2 and type-17 immune phenotype. The findings provide new insights into the distinct infant nasal immune response to ARI and its associations with subsequent childhood wheeze, which may enhance the understanding of the pathways through which RSV may be associated with the development of asthma.
Acknowledgments
Acknowledgment
The authors thank Dr. Jacek J. Hawiger for providing feedback on this manuscript.
Footnotes
Supported by the NIH (U19AI95227, K24 AI 77930, R21HD087864, and T32HL087738).
Author Contributions: K.N.T. conceived the study, performed the analyses, and drafted and edited the manuscript. J.S. and T.G. performed data processing, participated in data analysis, and edited the manuscript. T.V.H. designed the study, oversaw the clinical study conduct, obtained funding for the study, was involved in data analysis and interpretation of the results, and edited the manuscript. L.J.A., D.R., and K.G. assayed the nasal wash samples, participated in data processing and interpretation of the results, and edited the manuscript. S.R.D., C.S., E.K.L., C.R.-S., S.M.B., M.L.M., and R.S.P. participated in parent study design and interpretation of the results and edited the manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201711-2348OC on May 7, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Feldman AS, Hartert TV, Gebretsadik T, Carroll KN, Minton PA, Woodward KB, et al. Respiratory severity score separates upper versus lower respiratory tract infections and predicts measures of disease severity. Pediatr Allergy Immunol Pulmonol. 2015;28:117–120. doi: 10.1089/ped.2014.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sigurs N, Gustafsson PM, Bjarnason R, Lundberg F, Schmidt S, Sigurbergsson F, et al. Severe respiratory syncytial virus bronchiolitis in infancy and asthma and allergy at age 13. Am J Respir Crit Care Med. 2005;171:137–141. doi: 10.1164/rccm.200406-730OC. [DOI] [PubMed] [Google Scholar]
- 3.Wu P, Dupont WD, Griffin MR, Carroll KN, Mitchel EF, Gebretsadik T, et al. Evidence of a causal role of winter virus infection during infancy in early childhood asthma. Am J Respir Crit Care Med. 2008;178:1123–1129. doi: 10.1164/rccm.200804-579OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemanske RF, Jr, Jackson DJ, Gangnon RE, Evans MD, Li Z, Shult PA, et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–577. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Zheng LL, Li CM, Zhen SJ, Li YF, Huang CZ. A dynamic cell entry pathway of respiratory syncytial virus revealed by tracking the quantum dot-labeled single virus. Nanoscale. 2017;9:7880–7887. doi: 10.1039/c7nr02162c. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Kim K, Kwon T, Kim DW, Kim SS, Kim YJ. Secondary structure conservation of the stem-loop IV sub-domain of internal ribosomal entry sites in human rhinovirus clinical isolates. Int J Infect Dis. 2015;41:21–28. doi: 10.1016/j.ijid.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Kaminski A, Pöyry TA, Skene PJ, Jackson RJ. Mechanism of initiation site selection promoted by the human rhinovirus 2 internal ribosome entry site. J Virol. 2010;84:6578–6589. doi: 10.1128/JVI.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandini S, Calamelli E, Faldella G, Lanari M. Immune and inflammatory response in bronchiolitis due to respiratory syncytial virus and rhinovirus infections in infants. Paediatr Respir Rev. 2017;24:60–64. doi: 10.1016/j.prrv.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y, López CB. The innate immune response to RSV: advances in our understanding of critical viral and host factors. Vaccine. 2017;35:481–488. doi: 10.1016/j.vaccine.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee S, Lukacs NW. Innate immune responses to respiratory syncytial virus infection. Curr Top Microbiol Immunol. 2013;372:139–154. doi: 10.1007/978-3-642-38919-1_7. [DOI] [PubMed] [Google Scholar]
- 12.Mella C, Suarez-Arrabal MC, Lopez S, Stephens J, Fernandez S, Hall MW, et al. Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis. 2013;207:564–573. doi: 10.1093/infdis/jis721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García C, Soriano-Fallas A, Lozano J, Leos N, Gomez AM, Ramilo O, et al. Decreased innate immune cytokine responses correlate with disease severity in children with respiratory syncytial virus and human rhinovirus bronchiolitis. Pediatr Infect Dis J. 2012;31:86–89. doi: 10.1097/INF.0b013e31822dc8c1. [DOI] [PubMed] [Google Scholar]
- 14.Openshaw PJM, Chiu C, Culley FJ, Johansson C. Protective and harmful immunity to RSV infection. Annu Rev Immunol. 2017;35:501–532. doi: 10.1146/annurev-immunol-051116-052206. [DOI] [PubMed] [Google Scholar]
- 15.Lambert L, Sagfors AM, Openshaw PJ, Culley FJ. Immunity to RSV in early-life. Front Immunol. 2014;5:466. doi: 10.3389/fimmu.2014.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt ME, Varga SM. Modulation of the host immune response by respiratory syncytial virus proteins. J Microbiol. 2017;55:161–171. doi: 10.1007/s12275-017-7045-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhivaki D, Lemoine S, Lim A, Morva A, Vidalain PO, Schandene L, et al. Respiratory syncytial virus infects regulatory B cells in human neonates via chemokine receptor CX3CR1 and promotes lung disease severity. Immunity. 2017;46:301–314. doi: 10.1016/j.immuni.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Openshaw PJM. RSV takes control of neonatal Breg cells: two hands on the wheel. Immunity. 2017;46:171–173. doi: 10.1016/j.immuni.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chkhaidze I, Zirakishvili D, Shavshvishvili N, Barnabishvili N. Prognostic value of TH1/TH2 cytokines in infants with wheezing in a three year follow-up study. Pneumonol Alergol Pol. 2016;84:144–150. doi: 10.5603/PiAP.2016.0016. [DOI] [PubMed] [Google Scholar]
- 20.Kato M, Tsukagoshi H, Yoshizumi M, Saitoh M, Kozawa K, Yamada Y, et al. Different cytokine profile and eosinophil activation are involved in rhinovirus- and RS virus-induced acute exacerbation of childhood wheezing. Pediatr Allergy Immunol. 2011;22:e87–e94. doi: 10.1111/j.1399-3038.2010.01026.x. [DOI] [PubMed] [Google Scholar]
- 21.Pitrez PM, Machado DC, Jones MH, Andrade F, Camozzato C, Stein RT. Th-1 and Th-2 cytokine production in infants with virus-associated wheezing. Braz J Med Biol Res. 2005;38:51–54. doi: 10.1590/s0100-879x2005000100008. [DOI] [PubMed] [Google Scholar]
- 22.Sugai K, Kimura H, Miyaji Y, Tsukagoshi H, Yoshizumi M, Sasaki-Sakamoto T, et al. MIP-1α level in nasopharyngeal aspirates at the first wheezing episode predicts recurrent wheezing. J Allergy Clin Immunol. 2016;137:774–781. doi: 10.1016/j.jaci.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 23.Watson ML. Chemokines: linking receptors to response. Immunology. 2002;105:121–124. [PMC free article] [PubMed] [Google Scholar]
- 24.Gaze S, Driguez P, Pearson MS, Mendes T, Doolan DL, Trieu A, et al. An immunomics approach to schistosome antigen discovery: antibody signatures of naturally resistant and chronically infected individuals from endemic areas. PLoS Pathog. 2014;10:e1004033. doi: 10.1371/journal.ppat.1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furman D, Davis MM. New approaches to understanding the immune response to vaccination and infection. Vaccine. 2015;33:5271–5281. doi: 10.1016/j.vaccine.2015.06.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin EK, Gebretsadik T, Moore ML, Anderson LJ, Dupont WD, Chappell JD, et al. INSPIRE Study. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE) BMC Pulm Med. 2015;15:45. doi: 10.1186/s12890-015-0040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Won JH, Goldberger O, Shen-Orr SS, Davis MM, Olshen RA. Significance analysis of xMap cytokine bead arrays. Proc Natl Acad Sci USA. 2012;109:2848–2853. doi: 10.1073/pnas.1112599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breen EJ, Tan W, Khan A. The statistical value of raw fluorescence signal in Luminex xMAP based multiplex immunoassays. Sci Rep. 2016;6:26996. doi: 10.1038/srep26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breen EJ, Polaskova V, Khan A. Bead-based multiplex immuno-assays for cytokines, chemokines, growth factors and other analytes: median fluorescence intensities versus their derived absolute concentration values for statistical analysis. Cytokine. 2015;71:188–198. doi: 10.1016/j.cyto.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 30.Pepple KL, Rotkis L, Van Grol J, Wilson L, Sandt A, Lam DL, et al. Primed mycobacterial uveitis (PMU): histologic and cytokine characterization of a model of uveitis in rats. Invest Ophthalmol Vis Sci. 2015;56:8438–8448. doi: 10.1167/iovs.15-17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shankar J, Nguyen MH, Crespo MM, Kwak EJ, Lucas SK, McHugh KJ, et al. Looking beyond respiratory cultures: microbiome-cytokine signatures of bacterial pneumonia and tracheobronchitis in lung transplant recipients. Am J Transplant. 2016;16:1766–1778. doi: 10.1111/ajt.13676. [DOI] [PubMed] [Google Scholar]
- 32.Smith SE, Neier SC, Reed BK, Davis TR, Sinnwell JP, Eckel-Passow JE, et al. Multiplex matrix network analysis of protein complexes in the human TCR signalosome. Sci Signal. 2016;9:rs7. doi: 10.1126/scisignal.aad7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski JA, Muehling LM, Eccles JD, Capaldo BJ, Agrawal R, Shirley DA, et al. TH1 signatures are present in the lower airways of children with severe asthma, regardless of allergic status. J Allergy Clin Immunol. 2018;141:2048–2060.e13. doi: 10.1016/j.jaci.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18:S105–S110. doi: 10.1093/bioinformatics/18.suppl_1.s105. [DOI] [PubMed] [Google Scholar]
- 35.Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 36.Inoue M, Nishimura S, Hori G, Nakahara H, Saito M, Yoshihara Y, et al. Improved parameter estimation for variance-stabilizing transformation of gene-expression microarray data. J Bioinform Comput Biol. 2004;2:669–679. doi: 10.1142/s0219720004000806. [DOI] [PubMed] [Google Scholar]
- 37.Hendriks MM, Smit S, Akkermans WL, Reijmers TH, Eilers PH, Hoefsloot HC, et al. How to distinguish healthy from diseased? Classification strategy for mass spectrometry-based clinical proteomics. Proteomics. 2007;7:3672–3680. doi: 10.1002/pmic.200700046. [DOI] [PubMed] [Google Scholar]
- 38.Eriksson L, Byrne T, Johansson E, Trygg J, Vikström C. Multi-and megavariate data analysis basic principles and applications. Umetrics Academy; 2013. [Google Scholar]
- 39.Team RDCR. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 40.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foronjy RF, Dabo AJ, Cummins N, Geraghty P. Leukemia inhibitory factor protects the lung during respiratory syncytial viral infection. BMC Immunol. 2014;15:41. doi: 10.1186/s12865-014-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schijf MA, Lukens MV, Kruijsen D, van Uden NO, Garssen J, Coenjaerts FE, et al. Respiratory syncytial virus induced type I IFN production by pDC is regulated by RSV-infected airway epithelial cells, RSV-exposed monocytes and virus specific antibodies. PLoS One. 2013;8:e81695. doi: 10.1371/journal.pone.0081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrag MA, Almajhdi FN. Human respiratory syncytial virus: role of innate immunity in clearance and disease progression. Viral Immunol. 2016;29:11–26. doi: 10.1089/vim.2015.0098. [DOI] [PubMed] [Google Scholar]
- 45.Russell CD, Unger SA, Walton M, Schwarze J. The human immune response to respiratory syncytial virus infection. Clin Microbiol Rev. 2017;30:481–502. doi: 10.1128/CMR.00090-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viuff B, Tjørnehøj K, Larsen LE, Røntved CM, Uttenthal A, Rønsholt L, et al. Replication and clearance of respiratory syncytial virus: apoptosis is an important pathway of virus clearance after experimental infection with bovine respiratory syncytial virus. Am J Pathol. 2002;161:2195–2207. doi: 10.1016/S0002-9440(10)64496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajan D, McCracken CE, Kopleman HB, Kyu SY, Lee FE, Lu X, et al. Human rhinovirus induced cytokine/chemokine responses in human airway epithelial and immune cells. PLoS One. 2014;9:e114322. doi: 10.1371/journal.pone.0114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayaraman A, Jackson DJ, Message SD, Pearson RM, Aniscenko J, Caramori G, et al. IL-15 complexes induce NK- and T-cell responses independent of type I IFN signaling during rhinovirus infection. Mucosal Immunol. 2014;7:1151–1164. doi: 10.1038/mi.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reza Etemadi M, Ling KH, Zainal Abidin S, Chee HY, Sekawi Z. Gene expression patterns induced at different stages of rhinovirus infection in human alveolar epithelial cells. PLoS One. 2017;12:e0176947. doi: 10.1371/journal.pone.0176947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bondanese VP, Francisco-Garcia A, Bedke N, Davies DE, Sanchez-Elsner T. Identification of host miRNAs that may limit human rhinovirus replication. World J Biol Chem. 2014;5:437–456. doi: 10.4331/wjbc.v5.i4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherjee S, Lindell DM, Berlin AA, Morris SB, Shanley TP, Hershenson MB, et al. IL-17-induced pulmonary pathogenesis during respiratory viral infection and exacerbation of allergic disease. Am J Pathol. 2011;179:248–258. doi: 10.1016/j.ajpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JW, Taube C, Yang ES, Joetham A, Balhorn A, Takeda K, et al. Respiratory syncytial virus-induced airway hyperresponsiveness is independent of IL-13 compared with that induced by allergen. J Allergy Clin Immunol. 2003;112:1078–1087. doi: 10.1016/j.jaci.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 54.Schwarze J, Cieslewicz G, Hamelmann E, Joetham A, Shultz LD, Lamers MC, et al. IL-5 and eosinophils are essential for the development of airway hyperresponsiveness following acute respiratory syncytial virus infection. J Immunol. 1999;162:2997–3004. [PubMed] [Google Scholar]
- 55.Schwarze J, Cieslewicz G, Joetham A, Ikemura T, Mäkelä MJ, Dakhama A, et al. Critical roles for interleukin-4 and interleukin-5 during respiratory syncytial virus infection in the development of airway hyperresponsiveness after airway sensitization. Am J Respir Crit Care Med. 2000;162:380–386. doi: 10.1164/ajrccm.162.2.9903057. [DOI] [PubMed] [Google Scholar]
- 56.Loebbermann J, Schnoeller C, Thornton H, Durant L, Sweeney NP, Schuijs M, et al. IL-10 regulates viral lung immunopathology during acute respiratory syncytial virus infection in mice. PLoS One. 2012;7:e32371. doi: 10.1371/journal.pone.0032371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun L, Cornell TT, LeVine A, Berlin AA, Hinkovska-Galcheva V, Fleszar AJ, et al. Dual role of interleukin-10 in the regulation of respiratory syncitial virus (RSV)-induced lung inflammation. Clin Exp Immunol. 2013;172:263–279. doi: 10.1111/cei.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss KA, Christiaansen AF, Fulton RB, Meyerholz DK, Varga SM. Multiple CD4+ T cell subsets produce immunomodulatory IL-10 during respiratory syncytial virus infection. J Immunol. 2011;187:3145–3154. doi: 10.4049/jimmunol.1100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akitsu A, Ishigame H, Kakuta S, Chung SH, Ikeda S, Shimizu K, et al. IL-1 receptor antagonist-deficient mice develop autoimmune arthritis due to intrinsic activation of IL-17-producing CCR2(+)Vγ6(+)γδ T cells. Nat Commun. 2015;6:7464. doi: 10.1038/ncomms8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laan M, Cui ZH, Hoshino H, Lötvall J, Sjöstrand M, Gruenert DC, et al. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J Immunol. 1999;162:2347–2352. [PubMed] [Google Scholar]
- 62.Stoppelenburg AJ, Salimi V, Hennus M, Plantinga M, Huis in ’t Veld R, Walk J, et al. Local IL-17A potentiates early neutrophil recruitment to the respiratory tract during severe RSV infection. PLoS One. 2013;8:e78461. doi: 10.1371/journal.pone.0078461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu K, Gualano RC, Hibbs ML, Anderson GP, Bozinovski S. Epidermal growth factor receptor signaling to Erk1/2 and STATs control the intensity of the epithelial inflammatory responses to rhinovirus infection. J Biol Chem. 2008;283:9977–9985. doi: 10.1074/jbc.M710257200. [DOI] [PubMed] [Google Scholar]
- 64.Kalinowski A, Ueki I, Min-Oo G, Ballon-Landa E, Knoff D, Galen B, et al. EGFR activation suppresses respiratory virus-induced IRF1-dependent CXCL10 production. Am J Physiol Lung Cell Mol Physiol. 2014;307:L186–L196. doi: 10.1152/ajplung.00368.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueki IF, Min-Oo G, Kalinowski A, Ballon-Landa E, Lanier LL, Nadel JA, et al. Respiratory virus-induced EGFR activation suppresses IRF1-dependent interferon λ and antiviral defense in airway epithelium. J Exp Med. 2013;210:1929–1936. doi: 10.1084/jem.20121401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monick MM, Cameron K, Staber J, Powers LS, Yarovinsky TO, Koland JG, et al. Activation of the epidermal growth factor receptor by respiratory syncytial virus results in increased inflammation and delayed apoptosis. J Biol Chem. 2005;280:2147–2158. doi: 10.1074/jbc.M408745200. [DOI] [PubMed] [Google Scholar]
- 67.Zheng K, Kitazato K, Wang Y. Viruses exploit the function of epidermal growth factor receptor. Rev Med Virol. 2014;24:274–286. doi: 10.1002/rmv.1796. [DOI] [PubMed] [Google Scholar]
- 68.Zhu L, Lee PK, Lee WM, Zhao Y, Yu D, Chen Y. Rhinovirus-induced major airway mucin production involves a novel TLR3-EGFR-dependent pathway. Am J Respir Cell Mol Biol. 2009;40:610–619. doi: 10.1165/rcmb.2008-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hewson CA, Haas JJ, Bartlett NW, Message SD, Laza-Stanca V, Kebadze T, et al. Rhinovirus induces MUC5AC in a human infection model and in vitro via NF-κB and EGFR pathways. Eur Respir J. 2010;36:1425–1435. doi: 10.1183/09031936.00026910. [DOI] [PubMed] [Google Scholar]
- 70.Lo MS, Brazas RM, Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol. 2005;79:9315–9319. doi: 10.1128/JVI.79.14.9315-9319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Basha S, Surendran N, Pichichero M. Immune responses in neonates. Expert Rev Clin Immunol. 2014;10:1171–1184. doi: 10.1586/1744666X.2014.942288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malloy AM, Falsey AR, Ruckwardt TJ. Consequences of immature and senescent immune responses for infection with respiratory syncytial virus. Curr Top Microbiol Immunol. 2013;372:211–231. doi: 10.1007/978-3-642-38919-1_11. [DOI] [PubMed] [Google Scholar]
- 73.Ruckwardt TJ, Malloy AM, Morabito KM, Graham BS. Quantitative and qualitative deficits in neonatal lung-migratory dendritic cells impact the generation of the CD8+ T cell response. PLoS Pathog. 2014;10:e1003934. doi: 10.1371/journal.ppat.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]