Abstract
Rationale: In lung cancer, upregulation of the PI3K (phosphoinositide 3-kinase) pathway is an early event that contributes to cell proliferation, survival, and tissue invasion. Upregulation of this pathway was recently described as associated with enrichment of the lower airways with bacteria identified as oral commensals.
Objectives: We hypothesize that host–microbe interactions in the lower airways of subjects with lung cancer affect known cancer pathways.
Methods: Airway brushings were collected prospectively from subjects with lung nodules at time of diagnostic bronchoscopy, including 39 subjects with final lung cancer diagnoses and 36 subjects with noncancer diagnoses. In addition, samples from 10 healthy control subjects were included. 16S ribosomal RNA gene amplicon sequencing and paired transcriptome sequencing were performed on all airway samples. In addition, an in vitro model with airway epithelial cells exposed to bacteria/bacterial products was performed.
Measurements and Main Results: The composition of the lower airway transcriptome in the patients with cancer was significantly different from the control subjects, which included up-regulation of ERK (extracellular signal–regulated kinase) and PI3K signaling pathways. The lower airways of patients with lung cancer were enriched for oral taxa (Streptococcus and Veillonella), which was associated with up-regulation of the ERK and PI3K signaling pathways. In vitro exposure of airway epithelial cells to Veillonella, Prevotella, and Streptococcus led to upregulation of these same signaling pathways.
Conclusions: The data presented here show that several transcriptomic signatures previously identified as relevant to lung cancer pathogenesis are associated with enrichment of the lower airway microbiota with oral commensals.
Keywords: microbiome, bronchoscopy, lung cancer
At a Glance Commentary
Scientific Knowledge on the Subject
Lung cancer prevalence is increasing among never smokers, suggesting that other environmental factors are relevant for lung cancer pathogenesis. Emerging evidence indicates that the lung microbiome plays an important role in lung disease. A detailed assessment of the lung microbiome in subjects with lung cancer may help us understand the contribution of the host–microbe interaction to the pathogenesis of this disease.
What This Study Adds to the Field
In this cross-sectional study, we identified transcriptomic and microbiomic signatures in the lower airways of subjects with lung cancer. Enrichment of the lower airway microbiota with oral commensals was associated with upregulation of the PI3K (phosphoinositide 3-kinase)-signaling pathway in this disease. These data highlight host–microbe interactions in patients with lung cancer and may provide a novel target for lung cancer prevention and/or treatment.
Lung cancer is the leading cause of cancer deaths worldwide and exceeds the combined total deaths for breast, colon, prostate, and kidney cancers (1). Although smoking is a well-established risk factor for lung cancer, only 15% of smokers will develop lung cancer (2). Similarly, chronic obstructive pulmonary disease (COPD)/emphysema, a risk factor for lung cancer independent of smoking, develops in only 15% of smokers (3). Both COPD and lung cancer development are characterized by chronic inflammation, with overlapping pathogenic pathways (4).
Evidence is mounting that the lung microbiome may play a role in cancer pathogenesis. Lung cancer is associated with diseases, such as COPD, HIV, and Chlamydia infections, where chronic airway infection is common (5–7). In experimentally challenged germ-free rats, lung cancer development is less frequent than in conventional control rats (8). Chronic administration of LPS in mice leads to lung tumorigenesis (9). Disruption of commensal bacterial growth with antibiotics in a mouse model affects the γδT17 cell response, leading to aggressive metastatic pulmonary tumor development (10).
The use of culture-independent techniques for sequencing bacterial 16S ribosomal RNA (rRNA) genes has led to an increased awareness that the lower airway microbial environment (collectively called the lung microbiome) may play an important role in the pathogenesis of lung diseases. In a small cohort study, Veillonella and Megasphaera were found to be enriched in the lower airways of subjects with lung adenocarcinoma (11). Distinct lung microbiota identified in the lower airways was found to have an impact on the host immune phenotype (12–15). We have also reported that the enrichment of the lower airway microbiome with oral anaerobic taxa, including Veillonella species, is associated with increased infiltration with inflammatory cells (Th17 cells) and upregulation of the ERK (extracellular signal–regulated kinase)/PI3K (phosphoinositide 3-kinase) pathway in bronchial epithelial cells (16). Importantly, PI3K pathway upregulation was previously shown to be an early pathogenetic event in non–small-cell lung carcinoma, regulating cell proliferation, survival, differentiation, and cell invasion (17). Current evidence suggests that a dysbiotic lower airway microbiota could affect lung carcinogenesis through different mechanisms, including induction of host inflammatory pathways, production of bacterial toxins that alters host genomic stability, and release of cancer-promoting microbial metabolites (18). In this study, we tested the hypothesis that disruption of the lower airway microbiota is associated with an altered airway transcriptome affecting signaling pathways, such as PI3K, that are related to lung cancer pathogenesis. Some of the results in this study have been previously reported in the form of an abstract (19).
Methods
Subjects
All subjects signed informed consent to participate in this study, which was approved by the institutional review board of New York University. Participants included patients who had suspicious nodules on chest imaging and underwent clinical bronchoscopy. The histopathological diagnosis separated these subjects into the “lung cancer” group (n = 39) or the “disease control” group (n = 36). In addition, a “healthy control” group (n = 10), which consisted of never smokers without respiratory symptoms, abnormalities on chest X-ray, or known lung disease, underwent research bronchoscopy. We excluded subjects with a prior history of cancer or recent (<1 mo) antibiotic use.
Bronchoscopic Procedure
Both background and supraglottic (buccal) samples were obtained before the procedure (see Figure E1A in the online supplement). The background sample was obtained by passing sterile saline through the suctioning channel of the bronchoscope before the procedure. In cases with suspicious nodules, lower airway samples were collected via cytology brush of: 1) the “involved” airway leading to the segment containing the lung nodule, and 2) the “uninvolved” airway leading to a segment that was spared of disease, usually in the lobe contralateral to the suspicious nodule (Figure E1B). In healthy control subjects, research bronchoscopy obtained only one lower airway brushing, labeled as “uninvolved” segment. See the online supplement for details.
Transcriptome of Bronchial Epithelial Cells
RNA sequencing was performed on bronchial epithelial cells obtained by airway brushing, as described (20–22), using the Hi-seq/Illumina platform at the New York University Genomic Technology Center (data available at Sequence Read Archive: #PRJNA412846). Kyoto Encyclopedia of Genes and Genomes annotation was summarized at levels 1 to 3. Genes with an false discovery rate (FDR)-corrected adjusted P value < 0.15 were considered significantly differentiated, unless otherwise specified. Pathway analysis using differentially regulated genes (FDR < 0.15) was done using Ingenuity Pathway Analysis (IPA; QIAGEN Inc.) (23). Gene set enrichment analysis (GSEA) was performed with differential genes (FDR < 0.1) for dataset comparison (R package fgsea v1.4.1) (24).
Bacterial 16S rRNA-Encoding Genes Sequencing
High-throughput sequencing of bacterial 16S rRNA-encoding gene amplicons (V4 region) (25) was performed (data available at Sequence Read Archive: #PRJNA397867). Reagent control samples and mock mixed microbial DNA were sequenced and analyzed in parallel. The obtained 16S rRNA gene sequences were analyzed with the Quantitative Insights into Microbial Ecology (QIIME) 1.9.1 package (26). Operational taxonomic units were not removed from upstream analysis. Permutational multivariate analysis of variance (PERMANOVA) testing was used to compare the compositional differences of groups.
Sample clustering of metacommunities was based on Dirichlet multinomial mixtures modeling (27). Sparse Correlations for Compositional data (SparCC) was used to evaluate cooccurrence between taxa (28). Genera cooccurring significantly (P < 0.05), with rho greater than 0.7 or rho less than −0.7, were included in network analyses. Random forest supervised learning (QIIME) was used to determine genera that were most discriminant between cancer and noncancer (29, 30).
In Vitro Epithelial Cell Line Exposure
A549 cell lines, and cells of the bacterial strains Prevotella melaninogenica (ATCC #25845), Streptococcus mitis (ATCC #49456), and Veillonella parvula (ATCC #10790) were used to assess interactions. Each well with airway epithelial cells (1 × 106 in 2 ml of media) was exposed to each of the following conditions for a total of 4 hours: BAL 100 μl, LPS 10 ng/ml, 50 μl bacterial supernatant, and 10 μl heat-killed bacteria, or dilution buffer alone (control). Cigarette smoke condensate (40 μg/ml) (Murty Pharmaceuticals) was added with or without bacterial products. A detailed description of the in vitro experiments can be found in the online supplement.
Statistical Analysis
Analysis was performed comparing samples from three different groups: lung cancer, disease control, and healthy control. For association with discrete factors, we used either the Mann-Whitney test or the Kruskal-Wallis ANOVA. Paired statistics (Wilcoxon signed rank test) were used for paired comparison of continuous parameters. To evaluate transcriptome and 16S rRNA gene sequencing data differences between cancer, disease control, and healthy control groups, we used linear discriminant analysis effect size (LEfSe) (31). To examine associations between specific operational taxonomic units and the differential expression of host genes in the context of lung cancer, we used the compositionally robust sparse partial least squares (compPLS) framework, as described (32, 33).
Results
Subjects and Clinical Characteristics
Eighty-five subjects participated in this study, and 39 subjects were eventually diagnosed with lung cancer. Subjects without lung cancer were separated into two groups: 1) a disease control group (n = 36), with a benign pulmonary nodule; and 2) a healthy control group (n = 10), which included volunteer subjects with no lung disease. The mean age of the total cohort was 61.2 years, with 66% men, 52.9% white, and 88.9% smokers, with a mean history of 30.9 pack-years (Table 1). The lung cancer group had a higher pack-year history (41.8 vs. 29.3 pack-years) than the disease control group, a difference that did not reach statistical significance. The healthy control group consisted of never smokers. Adenocarcinoma occurred in 22 of 39 (56.4%) subjects with lung cancer.
Table 1.
Demographic and Clinical Characteristics of the Cohort
| All Subjects (n = 85) | Healthy (n = 10) | Disease (n = 36) | Lung Cancer (n = 39) | P Value* | |
|---|---|---|---|---|---|
| Age, yr | 61.2 ± 13.6 | 42.1 ± 13.6 | 60.7 ± 12.2 | 66.2 ± 9.2 | ns† |
| Sex, male | 63 (48) | 90 (9) | 56 (20) | 69.2 (27) | ns |
| Race | |||||
| White | 52.9 (45) | 50 (5) | 56 (20) | 51.3 (20) | ns |
| Black | 17.6 (15) | 30 (3) | 5.6 (2) | 25.6 (10) | 0.024† |
| Asian | 10.6 (9) | 0 | 8.1 (3) | 15.4 (6) | ns† |
| Hispanic | 11.8 (11) | 10 (1) | 22.2 (8) | 5.1 (2) | 0.024† |
| Other | 5.9 (5) | 10 (1) | 8.1 (3) | 2.6 (1) | ns† |
| Smoking status | |||||
| Current smoker | 28.9 (22) | 0 | 30.6 (11) | 30.8 (12) | ns |
| Past smoker | 56.6 (43) | 0 | 50.0 (18) | 66.7 (26) | ns† |
| Never smoker | 14.5 (11) | 100 (10) | 19.4 (7) | 2.6 (1) | 0.019† |
| Pack-years | 30.9 ± 30.0 | 0 | 29.3 ± 25.2 | 41.83 ± 32.1 | ns† |
| Diagnosis | |||||
| Non–lung cancer | 54.1 (46) | ||||
| Healthy | 100 (10) | ||||
| Benign NOS‡ | 66.7 (24) | ||||
| Infection | 6.5 (3) | ||||
| Lymphoma | 4.3 (2) | ||||
| Organizing PNA | 2.2 (1) | ||||
| Sarcoidosis | 2.2 (1) | ||||
| TB | 6.5 (2) | ||||
| Other | 2.2 (1) | ||||
| Lung cancer | 45.9 (39) | ||||
| Adeno | 56.4 (22) | ||||
| Squamous | 25.6 (10) | ||||
| Small cell | 12.8 (5) | ||||
| Carcinoma NOS | 5.1 (2) |
Definition of abbreviations: NOS = not otherwise specified; ns = not significant; PNA = pneumonia; TB = tuberculosis.
Data are presented as % (n) or mean ± SD.
P values based on Mann-Whitney U or chi-square (continuous or categorical variables, respectively) comparing lung cancer versus disease control.
Significant (P value < 0.05) differences on the basis of Kruskal-Wallis test comparing all three groups.
NOS indicates clinically diagnosed as non–lung cancer; however, no specific diagnosis made.
Evaluation of the Lower Airway Transcriptome
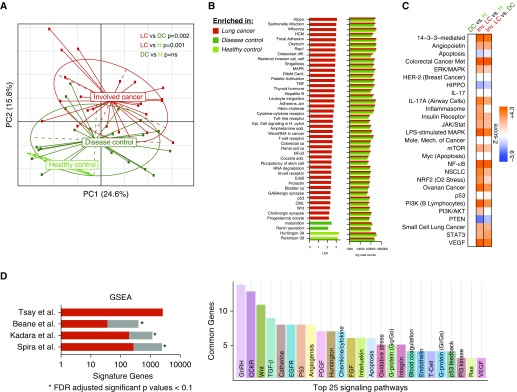
After quality control, RNAseq data were obtained on 94 lower airway samples. A principal coordinate analysis (PCoA) on the basis of the Bray-Curtis dissimilarity index showed that the global patterns of gene expression were significantly different between involved and uninvolved airway samples (Figure E2; PERMANOVA P = 0.001). We therefore examined for differences between cancer and the two control groups in each of these two different types of samples. Because of the diversity of diagnoses in the disease group, we used the noninvolved segments as the control samples (Figure E1B). PCoA analysis showed significant differences between involved-cancer airway samples, disease control samples, and healthy control samples (PERMANOVA P < 0.05; Figure 1A). Level 3 summarized Kyoto Encyclopedia of Genes and Genomes pathway evaluated for differences in functional pathways in involved-cancer, disease control, and healthy control samples (Figure 1B). Compared with both control groups, involved-cancer samples had significantly upregulated signaling pathways, including Hippo, MAPK (mitogen-activated protein kinase), TNF (tumor necrosis factor), toll-like receptor, T-cell receptor, B-cell receptor, bacterial invasion of epithelial cells, colorectal cancer, and epithelial cell signaling with Helicobacter pylori pathways.
Figure 1.
Differences in airway transcriptome between cancer, disease control, and healthy control. (A) Transcriptomic differences between involved airway of subjects with cancer (red, LC), disease control (dark green, DC), and healthy control (light green, H) were explored using principal coordinate analysis on the basis of Bray-Curtis dissimilarity index. Disease control samples were defined as airway samples from segments without disease from subjects with benign lung nodules. Significant differences were identified in β-diversity of transcriptome data in involved airways of subjects with cancer compared with both control subjects (permutational multivariate analysis of variance P value < 0.05). (B) Linear discriminant analysis (LDA > 2) detected significant differences in transcriptome (summarized L3 data) between involved cancer airway and both control groups. (C) Ingenuity pathway analysis was used to identify canonical pathways dysregulated in lung cancer when compared with the two control groups (differential gene FDR < 0.15). ERK and PI3K pathways were among those identified as upregulated in lung cancer airways when compared with control groups. (D) Gene set enrichment analysis of transcriptomic differences identified between lung cancer versus disease control (false discovery rate [FDR] < 0.15) was compared with transcriptomic differences previously published in three studies (34–36). Histogram on the left shows the number of transcriptomic signature genes identified in each study (34–36), with the genes in common with our current investigation highlighted in red. Histogram on the right shows signaling pathways shared between the current investigation and the other three prior studies. AKT = protein kinase B; CCKR = cholecystokinin receptor; CML = chronic myeloid leukemia; EGFR = epidermal growth factor receptor; ERK = extracellular signal–regulated kinase; FGF = fibroblast growth factor; HCM = hypertrophic cardiomyopathy; HER-2 = human epidermal growth factor receptor 2; GnRH = gonadotropin-releasing hormone; GSEA = gene set enrichment analysis; Inv. = involved; JAK/Stat = Janus kinase/signal transducer and activator of transcription protein; MAPK = mitogen-activated protein kinase; mTOR = mammalian target of rapamycin; NF-κB = nuclear factor-κB; NRF2 = nuclear factor (erythroid-derived-2)–like 2; NSCLC = non–small-cell lung cancer; PC = principal component; PDGF = platelet-derived growth factor; PI3K = phosphoinositide 3-kinase; PTEN = phosphatase and tensin homolog; STAT3 = signal transducer and activator of transcription 3; TGF-β = transforming growth factor-β; TNF = tumor necrosis factor; VEGF = vascular endothelial growth factor; Wnt = wingless/integrated.
We then evaluated functional enrichment of transcriptomic data between cancer, disease control, and healthy control samples, focusing on differentially expressed genes (FDR < 0.15). Compared with disease controls and healthy controls, the cancer group showed strong induction of the PI3K/AKT (protein kinase B) and ERK/MAPK signaling pathways (Figures 1C and E4, Table E1). Detailed analyses with uninvolved-cancer samples and involved disease samples are shown in Figure E3 and Figure E5, respectively.
Using GSEA with FDR less than 0.1, we compared the transcriptomic signature (lung cancer vs. disease control) to prior published datasets (34–36), which showed significant overlap in gene expressions, reproducing prior observations that p53, PI3K, EGF (epidermal growth factor) receptor, and Ras signaling pathways are induced in lower airways of subjects with lung cancer (Figure 1D).
16S rRNA Gene Sequence Data
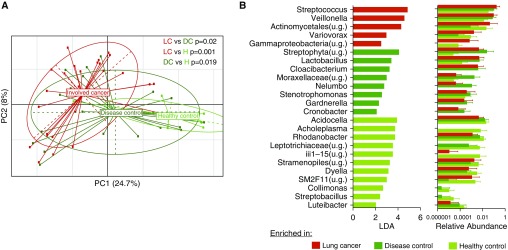
A total of 190 airway samples (buccal and lower airway) were used for analysis of 16S rRNA gene sequences. Of the 120 lower airway samples, 62 were obtained from airways leading to the involved segments, and 58 were from airways spared of disease. Unsupervised hierarchical clustering for all 190 airway samples and 72 background samples (Figure E6) showed some airway samples were enriched with oral anaerobes Prevotella, Streptococcus, and Veillonella, whereas others were enriched with taxa commonly found in background samples; this unsupervised hierarchical clustering based on relative abundance of most abundant taxa did not correlate with cancer status in either lower airway or buccal samples. PCoA did show significant differences in β-diversity by sample location type (P < 0.001, PERMANOVA) (Figure E7A).
Next, we analyzed data on the basis of cancer status. There were no differences (α- and β-diversity) seen in the buccal samples between cancer and noncancer (Figure E8A). However, in the lower airway samples, cancer (involved) and noncancer (disease and healthy) samples differed significantly in β-diversity (PERMANOVA P < 0.05) (Figure 2A). LEfSe identified several taxonomic differences between cancer versus the two control groups (Figure 2B). Compared with disease and healthy controls, involved cancer airway samples were enriched with Streptococcus and Veillonella. Disease control samples were enriched with Streptophyta, Moraxellaceae, and Stenotrophomonas, whereas healthy control samples were enriched with Acholeplasma and Acidocella. Analyses of uninvolved cancer airway samples shared many similar findings compared with the involved cancer airway samples (Figure E9).
Figure 2.
Differences in airway microbiota between cancer, disease control, and healthy control. (A) β-diversity comparisons of microbiota composition were explored using principal coordinate analysis on the basis of Bray-Curtis dissimilarity index between cancer (involved lung segments) and noncancer of airways. The microbiome of involved airways of subjects with cancer (red, LC) was significantly different when compared with disease control (dark green, DC) and healthy control (light green, H) (permutational multivariate analysis of variance P value < 0.05 for all comparisons). (B) Linear discriminant analysis (LDA > 2) detected differential taxonomic enrichment in involved airways from subjects with cancer when compared with both control groups. PC = principal component; u.g. = undetermined genus.
Multi-omic Analysis
To better characterize host–microbe interaction in this cohort, we used a multi-omic analytical approach combining microbiome and host epithelial transcriptomic data. First, using a Dirichlet multinomial model, we established that two clusters were optimal for the 16S rRNA gene sequence data (Figures E11A and E11B). LEfSe identified taxa most differentially enriched in these two distinct clusters (Figure E12A). In one cluster, oral taxa such as Streptococcus, Prevotella, Veillonella, and Rothia were enriched (Figure E12B), and we called it “cluster SPT” (for supraglottic-predominant taxa, which is in agreement with our prior publication) (16). In the other cluster, Xanthomondaceae, Staphylococcus, Corynebacterium, Methylobacterium, and Granulicatella (taxa found most abundantly in background samples) were enriched, and this was called “cluster BPT” (for background-predominant taxa). The lower airway samples from subjects with lung cancer were more commonly characterized as SPT-type microbiota than BPT-type microbiota (chi-square P = 0.026; Figure E11C). We then examined differences in the transcriptome associated with these two distinct microbiota clusters. Of the 2,458 differentially expressed genes (FDR < 0.15) between SPT and BPT, 1,605 (65.3%) were upregulated in SPT. Functional enrichment analysis using IPA showed that the most differential networks generated were the ERK1/2 and PI3K signaling pathways (Figure E12C and Table E3).
Next, we used SparCC to generate a cooccurrence network of taxa found in the lower airways (at the genus level). This analysis demonstrated two distinct taxa clusters: one dominated by oral commensals and the other dominated by taxa commonly found in background environmental controls (Figure E13). A random forests classifier identified that among the taxa predictive for lung cancer, the most abundant genera were Prevotella, Veillonella, and Streptococcus.
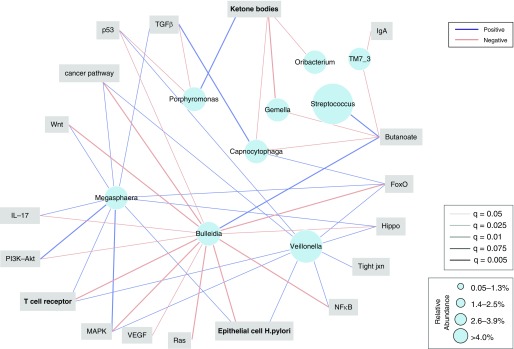
compPLS was used to identify associations between abundant genera from cluster SPT and signaling pathways related to lung cancer. This analysis was adjusted by smoking status and for paired samples obtained in subjects. Significant (adjusted P value [FDR < 0.1], on the basis of empirical P value calculated over 5,000 bootstraps) positive associations were identified between several oral commensals and ERK/MAPK, PI3K/AKT, transforming growth factor-β, p53, nuclear factor-κB, and other cancer-related signaling pathways (Figures 3, E14, and E15). Variance decomposition to adjust for the effects of smoking status did not change the taxa–pathway associations in the model. The most abundant taxa identified as significantly associated with cancer-relevant pathways were Megasphaera and Veillonella, which we have described as characteristic oral commensals and found in pneumotypeSPT (12, 16).
Figure 3.
Association network between microbiome and transcriptome. Compositionally robust sparse partial least squares (compPLS) was used to evaluate for associations between airway microbiota and transcriptome considering samples from lung cancer, disease control, and healthy control. Analysis was adjusted by smoking status and for paired samples obtained in subjects. Edge colors are representative of negative associations in red and positive associations in blue, and edge weight is scaled to represent the degree of the confidence for the associations. Only taxa identified as having significant correlations are displayed, and size of nodes denotes median relative abundance of taxa at a genus level. The most abundant taxon were: Veillonella, which was positively correlated with ERK/MAPK, T-cell receptor, p53, and cancer pathways; Megasphaera, which was positively correlated with ERK/MAPK, PI3K/Akt, IL-17, and TGF-β; and Bulleidia, which was negatively correlated with similar pathways. ERK = extracellular signal–regulated kinase; FoxO = forkhead box O; MAPK = mitogen-activated protein kinase; NF-κB = nuclear factor-κB; TGF-β = transforming growth factor-β; VEGF = vascular endothelial growth factor; Wnt = wingless/integrated.
To further explore the relationship between Veillonella and lung cancer, we performed 16S rRNA gene sequencing of surgically obtained lung tissue on a separate cohort of five subjects with adenocarcinoma. This analysis showed that Veillonella was more highly enriched in the involved lung tumor tissue than in the uninvolved lung (P = 0.04) (Figure E16), suggesting that a relationship may exist with lung cancer.
In vitro Coculture of Airway Epithelial Cells with Bacterial Products
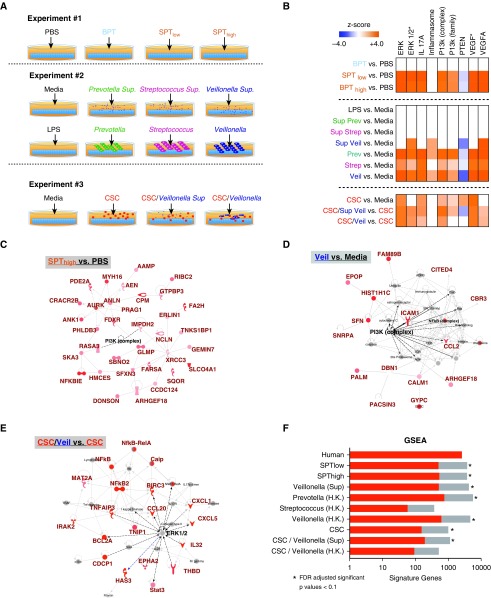
To explore possible causal relationships between exposure to bacteria and airway epithelial cell transcriptomic changes identified in the human cohort, we performed three in vitro experiments using cultured A549 cells exposed to microbial products (Figure 4A). For in vitro experiment 1, we used microbiota present in whole BAL available from a subgroup of control subjects (n = 8, uninvolved segments). These BAL samples were characterized as 1) samples enriched with BPT, 2) samples enriched with SPT with low bacterial load (SPTlow), and 3) samples enriched with SPT with high bacterial load (SPThigh) (Figure 17C; see online supplement for more details). Then, A549 cells were exposed in triplicate to the above-mentioned conditions (BPT, SPTlow, and SPThigh, as compared with PBS control) for 4 hours, followed by harvest for RNA. Compared with PBS, BPT did not alter the transcriptome. However, in SPT (low and high), IPA showed upregulation of upstream networks in the ERK, IL17, PI3K, and VEGF (vascular endothelial growth factor) signaling pathways and downregulation of the PTEN (phosphatase and tensin homolog) signaling pathway (FDR < 0.15), which is a known negative regulator of PI3K signaling (Figures 4B and 4C). This suggested that the transcriptomic changes were predominantly driven by microbial products found in a microbiota enriched with supraglottic taxa.
Figure 4.
In vitro transcript activation of epithelial cells by exposure to bacterial products. (A) Schematic experimental design for three in vitro experiments. For all conditions, A549 cells were exposed for 4 hours and then harvested for RNA isolation. Experiment 1: exposure to whole BAL from a subject whose lower airways were enriched with background-predominant taxa (BPT), supraglottic-predominant taxa (SPT) with high bacterial load (SPThigh), or SPT with low bacterial load (SPTlow). Experiment 2: exposure to media alone, LPS, or bacteria: Veillonella parvula, Prevotella melaninogenica, and Streptococcus mitis (both culture supernatant, heat-killed bacteria). Experiment 3: exposure to cigarette smoke condensate (CSC) and V. parvula (heat-killed and supernatant). Each experiment was done in triplicate or quadruplicates. (B) Ingenuity Pathway Analysis (IPA) was used to identify canonical pathways dysregulated in each in vitro experiment (false discovery rate [FDR] < 0.15). (C–E) IPA network analysis for transcriptomic changes (FDR < 0.15) annotated to ERK (extracellular signal–regulated kinase) or PI3K (phosphoinositide 3-kinase) pathways from one representative condition from each in vitro experiment. (F) Gene set enrichment analysis (GSEA) of transcriptomic signatures (FDR < 0.15) comparing in vitro experiment to in vivo human data. Bar chart compares transcriptomic changes for conditions identified in vitro with transcriptomic changes identified as associated with lung cancer on the basis of our human data. Overlapping signature genes are shown in red. PBS = phosphate-buffered saline.
We next exposed airway epithelial cells to supernatants from viable or heat-killed bacteria found in SPT (in vitro experiment 2). In addition, LPS was added as an experimental condition to explore TLR4 (Toll-like receptor 4) signaling. Each condition was performed in triplicate or quadruplicate. Exposure to supernatant from Veillonella or exposure to heat-killed bacteria led to upregulation of ERK, PI3K, and IL17A signaling pathways (Figures 4B and 4D). LPS and supernatant from Prevotella or Streptococcus did not upregulate PI3K, suggesting differences in pathways of activation and that TLR4 signaling is not responsible for the observed transcriptional changes. Interestingly, Veillonella supernatant, heat-killed Veillonella, and heat-killed Prevotella upregulated IL-1β, IL-18, CASP1 (caspase-1), and NLRP1 (NACHT, LRR, and PYD domains–containing protein 1), suggesting that the inflammasome might be recognizing some immune-active microbial product in these conditions (Figure 4B).
Finally, we explored the effects of microbes in conjunction with cigarette smoke condensate (CSC, in vitro experiment 3). First, addition of CSC alone led to upregulation of the aryl hydrocarbon receptor signaling (37, 38) and ERK1/2 and PI3K signaling pathways (Table E3). The addition of bacterial products to CSC-exposed A549 epithelial cells led to further upregulation in genes related to the ERK1/2 and PI3K signaling pathways (Figures 4B and 4E). Taken together, these findings suggest that exposure to oral commensals may further augment transcriptomic signaling changes beyond the effects of cigarette smoke. GSEA analysis showed the degree of overlap between the cancer transcriptome signature identified in the human cohort and in vitro transcriptome data. Several of the in vitro experimental conditions shared regulation of more than 100 genes with human transcriptome changes seen in lung cancer (FDR < 0.1) (Figure 4F). These changes included p53, apoptosis, p38 MAPK, and EGFR (epidermal growth factor receptor) signaling pathways (Figure E18).
Discussion
This investigation uncovered significant associations between the lower airway microbiota and transcriptional changes in epithelial cells that may be relevant for lung cancer pathogenesis. The results presented here extend our prior report of the association between the lower airway microbiota and the host immune endotype in healthy subjects (16) by identifying enrichment of the lower airways with several taxa commonly recognized as part of the oral commensal microbiota in lung cancer. Importantly, the transcriptional changes (identified using RNA sequence) associated with the lower airway microbiota and adjusted by multiple covariates occurred in several previously reported cancer pathways (as demonstrated by our GSEA analysis). Some of these associations occurred within the airway mucosae of lung segments proximal to the cancerous lesions as well as in uninvolved lung segments, which were previously described as the field of cancerization (39). Although multiple taxa were identified as enriched in lung cancer, the genus Veillonella was among the most abundant and was most strongly associated with transcriptomic changes. Using a multi-omic approach adjusted for relevant covariates (e.g., smoking status and paired samples), this genus was significantly associated with transcriptomic pathways known to participate in the development or progression of lung cancer. Using an in vitro model, we further validated that the PI3K signaling pathway is activated in airway epithelial cells exposed to Veillonella. The ERK and PI3K signaling pathways, previously shown to be upregulated in the lung field of cancerization (17, 35, 40, 41), consist of kinase cascades, which participate in regulating cell proliferation, survival, and differentiation (42). Specifically, PI3K/Akt activation in bronchial airway epithelium is an early event in lung tumor development (17), its deregulation is associated with disease progression (43), and, hence, it may be a therapeutic target (44). New evidence has shown that manipulation of the gut microbiome augments the antitumor immunity of checkpoint inhibitors, a class of drugs recently approved for treatment of lung cancer (45, 46). In light of these observations, the data presented here support a potential role for novel therapeutic strategies in lung cancer that target the lung microbial–host interface.
Using a multi-omic analysis, we noted interesting bacterial cooccurrence and identified specific taxa associated with lung cancer. Using cooccurrence network analysis (SparCC) we identified several members of the oral microbiota that, when present in the lower airways, were predictive for lung cancer. The most abundant of these oral commensals were Streptococcus, Prevotella, and Veillonella. Using compPLS analysis, Veillonella and Megasphaera were associated with upregulating cancer signaling pathways. These two organisms were previously identified as present in high relative abundance in BAL of patients with lung cancer (11, 47). Our experiments confirm and extend these observations.
To explore the directionality of the significant associations identified by this cross-sectional human study, we exposed malignant bronchial epithelial cell line to Streptococcus, Prevotella, and Veillonella. The observed upregulation in the PI3K and ERK1/2 signaling pathways in the in vitro model suggests a possible pathogenic mechanism of these common oral microbiota constituents. These bacteria may affect the host by shedding different microbial bioactive molecules. Because LPS did not upregulate the PI3K and ERK1/2 signaling pathways of airway epithelial cells in vitro, other immune-active microbial products are likely interacting with host epithelial cells through different pattern recognition receptors (such as the inflammasome) and are responsible for host responses. Interestingly, a novel therapy targeting specific components of the inflammasome pathway has recently been reported for non–small-cell lung cancer (48).
In addition, it is also possible that microbes such as Veillonella support the growth of other taxa with well-recognized pathogenic potential. For example, in a murine model to evaluate the tumor environment, Veillonella significantly increased the cell numbers of Pseudomonas aeruginosa in the tumor tissue (49). Cigarette exposure, along with bacterial exposure, on airway epithelial cells in the in vitro experiment showed further upregulation of the PI3K, ERK1/2, and VEGF signaling pathways, suggesting a synergistic combination that may drive carcinogenesis. Although transcriptome signature in the CSC experiment (experiment 3) showed large overlap between CSC alone and human data (GSEA), analysis of CSC/Veillonella exposure to the in vivo data showed less transcriptome overlap and nonsignificant enrichment analysis. It is possible that CSC exposure may have overshadowed the effect of the bacteria on a global level. However, through analysis with IPA at a targeted level for known relevant cancer-specific pathways, we were able to show Veillonella/CSC had an additional effect on upregulation of ERK, PI3K, and VEGF when compared with CSC alone. Overall, future investigations will need to identify relevant microbial molecules and host intermediate pathways responsible for the PI3K and ERK1/2 signaling activation, as well as to optimize the CSC concentration to reflect true in vivo conditions.
The current investigation has several limitations. Although we observed several statistically significant differences in transcriptomic and microbiomic analyses associated with cancer, the clinical significance of these findings cannot be assessed with the current investigation. We consider these findings as exploratory and hypothesis generating. The cross-sectional design of the human cohort prevents us from determining the directionality of the associations between microbes and host endotype. For example, changes in the airway transcriptome reflective of a proinflammatory host endotype related to lung cancer may exert distinct pressure on the airway microbiota. Conversely, the observed association could support the hypothesis that enrichment of the lower airway microbiome with oral commensals, probably through microaspiration, leads to upregulation of airway epithelial pathways that promote inflammation and affect cell apoptosis, dysplasia, and the development of nascent malignant cells. Although the in vitro model presented here supports the plausibility of the latter, the association between microbes and host is likely bidirectional. Further studies using longitudinal human cohorts and in vivo experimentation are needed.
The differences noted in both the airway transcriptome and airway microbiota in subjects with cancer were less clear when involved-cancer airway samples were compared with involved-diseased (non–lung cancer) airway samples (Figures E5 and E10). Given the multiple diagnoses in the control group, it is possible that other lung diseases might also affect both the transcriptome and the microbiome, confounding these measurements. The host–microbe interaction evaluated in involved and uninvolved segments also may reflect temporal differences of oncogenic field-of-injury development (50). Another limitation of the study is the lack of direct sampling of the lung tumor/tumor microenvironment. However, the Veillonella enrichment in the small cohort of surgically obtained cancer tissue samples suggests that the tumor microbiota may share features found in the airway microbiota. In addition, multiple lines of investigation have found anaerobic conditions within the tumor microenvironment, which favors colonization with anaerobes such as Veillonella (51). It is important to note that although we focused on the enrichment of the lower airway microbiota with Veillonella, other oral commensals, such as Streptococcus, tend to cooccur and were also enriched in the lower airways of patients with lung cancer. Varying bacteria may differentially contribute to the host microbial interactions; future investigations will be needed to dissect the nature of each of these associations. Much less certain is the role of the bacteria found negatively associated with lung cancer. Although a “protective” role is possible, it is important to highlight that, for the most part, the taxa identified as negatively associated with cancer are commonly found in background samples (e.g., bronchoscope) and may represent bacterial DNA from nonviable bacteria. We used immortalized airway epithelial cells to explore whether exposure to microbial products could elicit the transcriptomic changes observed in vivo. Although our data showed that some of the bacteria associated with lung cancer could induce pathways identified as upregulated in lung cancer, we found variability between different bacterial products. We also acknowledge that we cannot establish whether or not this is specific to the bacteria studied or related to the presence of viable bacteria in the lower airways, because microbial products present in BAL samples with BPT did not lead to similar changes. Evaluating bacteria viability is also challenging, given the lack of well-defined culture conditions for many of the organisms. Future investigations should consider alternative approaches in evaluating the activation of these pathways. Using a broad range of cell lines as well as ex vivo bronchial epithelial cells obtained from patients, such approaches will increase the confidence in the observed associations.
In conclusion, we determined that enrichment of the lower airway microbiome with oral commensals such as Veillonella was associated with transcriptomic changes of airway epithelial cells, including the ERK/PI3K pathways relevant to lung cancer. These findings shape our understanding of how microbes present in the lower airways may affect initial events in the malignant transformation of airway epithelial cells, the immune surveillance needed to control nascent malignant cells, and the tumor’s ability to proliferate and metastasize. Our work complements recent findings that antibiotic use affects lung tumor size and survival and that changing the gut microbiome affects immunotherapy outcomes (10, 45, 46). This study improves our understanding of the effects of the lower airway microbiota on the host endotype and provides a framework for future investigations that may uncover novel therapeutic microbial targets for lung cancer.
Acknowledgments
Acknowledgment
The authors thank Dr. Robert Smith, Dr. Kevin Felner, and Dr. Harald Sauthoff from the New York Harbor Manhattan VA Hospital and Dr. Jamie Bessich and Dr. Samaan Rafeq from the New York University (NYU) Langone Medical Center for their invaluable contributions to the NYU Lung Cancer Biomarker Center; Dr. Vivian Hayashi at the New York Harbor Manhattan VA Hospital for her review; and the participating subjects for their time and effort in this study.
Footnotes
Supported by NIH National Institute of Allergy and Infectious Diseases grant K23 AI102970 (L.N.S.), Early Detection Research Network grant 5 U01CA086137-13 (W.N.R.), Department of Defense grant W81XWH-16-1-0324 (J.-C.J.T.), the A Breath of Hope Lung Foundation (J.-C.J.T.), the Simons Foundation (R.B.), and Clinical and Translational Science Institute grant UL1 TR000038 (L.N.S.). The Genome Technology Center is partially supported by the Cancer Center Support grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center (A.H.), NIH National Cancer Institute grant T32 CA193111 (B.G.W.), and NIH National Center for Advancing Translational Sciences grant UL1TR001445 (B.G.W.).
Author Contributions: Conception and design: J.-C.J.T., A.H., H.P., W.N.R., M.J.B., and L.N.S. Acquisition of data: J.-C.J.T., B.G.W., P.M., Y.L., T.-A.Y., E.O., V.M., G.M., and L.N.S. Analysis and interpretation of data: J.-C.J.T., B.G.W., M.H.B., J.C.C., N.S., T.L., E.O., I.S., R.B., and L.N.S. Drafting or revising of article: J.-C.J.T., M.H.B., J.C.C., M.D.W., R.B., M.J.B., and L.N.S. Final approval of the manuscript: J.-C.J.T., J.C.C., G.M., A.T., A.H., H.P., M.D.W., W.N.R., D.H.S., R.B., M.J.B., and L.N.S.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201710-2118OC on June 4, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Samet JM, Avila-Tang E, Boffetta P, Hannan LM, Olivo-Marston S, Thun MJ, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15:5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terzikhan N, Verhamme KMC, Hofman A, Stricker BH, Brusselle GG, Lahousse L. Prevalence and incidence of COPD in smokers and non-smokers: the Rotterdam Study. Eur J Epidemiol. 2016;31:785–792. doi: 10.1007/s10654-016-0132-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer. 2015;90:121–127. doi: 10.1016/j.lungcan.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer. 2013;13:233–245. doi: 10.1038/nrc3477. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi AK, Gaydos CA, Agreda P, Holden JP, Chatterjee N, Goedert JJ, et al. Chlamydia pneumoniae infection and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1498–1505. doi: 10.1158/1055-9965.EPI-09-1261. [DOI] [PubMed] [Google Scholar]
- 7.Mani D, Haigentz M, Jr, Aboulafia DM. Lung cancer in HIV infection. Clin Lung Cancer. 2012;13:6–13. doi: 10.1016/j.cllc.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber H, Nettesheim P, Lijinsky W, Richter CB, Walburg HE., Jr Induction of lung cancer in germfree, specific-pathogen-free, and infected rats by N-nitrosoheptamethyleneimine: enhancement by respiratory infection. J Natl Cancer Inst. 1972;49:1107–1114. [PubMed] [Google Scholar]
- 9.Melkamu T, Qian X, Upadhyaya P, O’Sullivan MG, Kassie F. Lipopolysaccharide enhances mouse lung tumorigenesis: a model for inflammation-driven lung cancer. Vet Pathol. 2013;50:895–902. doi: 10.1177/0300985813476061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, et al. Microbiota modulate tumoral immune surveillance in lung through a γδT17 immune cell-dependent mechanism. Cancer Res. 2014;74:4030–4041. doi: 10.1158/0008-5472.CAN-13-2462. [DOI] [PubMed] [Google Scholar]
- 11.Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sze MA, Dimitriu PA, Suzuki M, McDonough JE, Campbell JD, Brothers JF, et al. Host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;192:438–445. doi: 10.1164/rccm.201502-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsay J, Clemente J, Lhakhang T, Li YH, Yie TA, Wu B, et al. Lung cancer and lung microbiome [abstract] Am J Respir Crit Care Med. 2017;195:A1001. [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 22.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 23.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLOS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 30.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 31.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Lesko M, Badri MH, Kapoor BC, Wu BG, Li Y, et al. Lung microbiome and host immune tone in subjects with idiopathic pulmonary fibrosis treated with inhaled interferon-γ. ERJ Open Res. 2017;3:00008-2017. doi: 10.1183/23120541.00008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res (Phila) 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadara H, Fujimoto J, Yoo SY, Maki Y, Gower AC, Kabbout M, et al. Transcriptomic architecture of the adjacent airway field cancerization in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:dju004. doi: 10.1093/jnci/dju004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 37.Tsay JJ, Tchou-Wong KM, Greenberg AK, Pass H, Rom WN. Aryl hydrocarbon receptor and lung cancer. Anticancer Res. 2013;33:1247–1256. [PMC free article] [PubMed] [Google Scholar]
- 38.Dertinger SD, Nazarenko DA, Silverstone AE, Gasiewicz TA. Aryl hydrocarbon receptor signaling plays a significant role in mediating benzo[a]pyrene- and cigarette smoke condensate-induced cytogenetic damage in vivo. Carcinogenesis. 2001;22:171–177. doi: 10.1093/carcin/22.1.171. [DOI] [PubMed] [Google Scholar]
- 39.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium: clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 40.Kadara H, Shen L, Fujimoto J, Saintigny P, Chow CW, Lang W, et al. Characterizing the molecular spatial and temporal field of injury in early-stage smoker non-small cell lung cancer patients after definitive surgery by expression profiling. Cancer Prev Res (Phila) 2013;6:8–17. doi: 10.1158/1940-6207.CAPR-12-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsay JC, Li Z, Yie TA, Wu F, Segal L, Greenberg AK, et al. Molecular characterization of the peripheral airway field of cancerization in lung adenocarcinoma. PLoS One. 2015;10:e0118132. doi: 10.1371/journal.pone.0118132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scrima M, De Marco C, Fabiani F, Franco R, Pirozzi G, Rocco G, et al. Signaling networks associated with AKT activation in non-small cell lung cancer (NSCLC): new insights on the role of phosphatydil-inositol-3 kinase. PLoS One. 2012;7:e30427. doi: 10.1371/journal.pone.0030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam S, Mandrekar SJ, Gesthalter Y, Allen Ziegler KL, Seisler DK, Midthun DE, et al. Cancer Prevention Network. A randomized phase IIb trial of myo-inositol in smokers with bronchial dysplasia. Cancer Prev Res (Phila) 2016;9:906–914. doi: 10.1158/1940-6207.CAPR-15-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111–3122. [PMC free article] [PubMed] [Google Scholar]
- 48.Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ CANTOS Trial Group. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390:1833–1842. doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
- 49.Pustelny C, Komor U, Pawar V, Lorenz A, Bielecka A, Moter A, et al. Contribution of Veillonella parvula to Pseudomonas aeruginosa-mediated pathogenicity in a murine tumor model system. Infect Immun. 2015;83:417–429. doi: 10.1128/IAI.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karnes WE, Jr, Samloff IM, Siurala M, Kekki M, Sipponen P, Kim SW, et al. Positive serum antibody and negative tissue staining for Helicobacter pylori in subjects with atrophic body gastritis. Gastroenterology. 1991;101:167–174. doi: 10.1016/0016-5085(91)90474-y. [DOI] [PubMed] [Google Scholar]
- 51.Graves EE, Maity A, Le QT. The tumor microenvironment in non-small-cell lung cancer. Semin Radiat Oncol. 2010;20:156–163. doi: 10.1016/j.semradonc.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]