Abstract
Tripartite motif-containing protein 44 (TRIM44) plays an important role in the development and progression of some human cancers; however, its role in skin squamous cell carcinoma (SCC) remains unknown. The aim of the present study was to investigate TRIM44 expression and clinicopathological significance of TRIM44 in SCC.
Immunohistochemistry (IHC) technique, reverse transcriptase-polymerase chain reaction (RT-PCR) and western blot were performed to evaluate differences in TRIM44 protein expression in SCC and normal skin tissues.
IHC showed that the positive rate of TRIM44 staining in SCC tissues 26.00% (9/30), while the positive rate of normal control group was 83.33% (25/30). The positive rate of TRIM44 staining in SCC tissues is significantly lower than normal skin tissue (P <.01). RT-PCR showed that the positive rates of TRIM44 mRNA expression in SCC tissues were 16.67% (5/30), but the positive rate of normal control group was 86.67% (26/30). TRIM44 mRNA expression in SCC group was significantly lower than that in the normal group (P <.01). Kaplan–Meier survival analysis showed that low expression was associated with poor overall survival in SCC patients (P =.004). Multi-factor survival analysis indicated that both low TRIM44 expression and tumor stage were independent factors affecting the overall survival of patients with SCC (P =.038 and P =.001, respectively). Low expression of TRIM44 in SCC was associated with staging (P =.009 and P =.008, respectively) and metastasis (P =.003 and P =.004, respectively).
The levels of TRIM44 protein and TRIM44 mRNA in SCC are both lowly expressed which is strongly associated with tumor staging, metastasis, and poor survival. And it also is an independent factor affecting the overall survival of patients with SCC.
Keywords: immunohistochemistry, prognosis, skin squamous cell carcinoma, tripartite motif-containing protein 44
1. Introduction
Skin squamous cell carcinoma (SCC) is the most common skin tumor in China,[1,2] which accounts for about 80% to 90% of the malignant tumors of the skin, and its incidence is increasing year by year, especially among the elderly. The 5-year survival rate of metastatic SCC is only 14%. This type of cancer has a high degree of malignancy, and there is no effective nonoperative treatment at present, even if the operation has a high recurrence rate. Since SCC is characterized by early metastasis and rapid progress, most patients are often diffused and metastasized before diagnosis, and SCC with metastatic and progressing are not sensitive to conventional treatment such as radiotherapy and chemotherapy.[3,4]
Tripartite motif (TRIM) protein family, also known as the RBCC family, has more than 70 protein molecules.[5] TRIM family proteins play E3 ubiquitin ligase function,[6] which play an important role in the physiological processes such as natural immune response, cell differentiation, transcription regulation, cytoskeleton remodeling, development, cell cycle, and apoptosis, DNA damage repair and other physiological processes. The abnormality of TRIM family proteins or genes plays a key role in the development of cancer by regulating gene expression, cell proliferation, DNA damage repair, and apoptosis.[7,8] Tripartite motif-containing protein 44 (TRIM44) has been identified as a member of TRIM protein family. Several studies showed that TRIM44 plays an important role in the development and progression of some human cancers, such as lung cancer, endometrial carcinoma, and hepatocelluar carcinoma.[9,10,11] However, so far, the correlation between TRIM44 expression and the prognosis of postoperative patients exhibiting SCC remain largely unknown.
Therefore, this study aimed to investigate the clinical significance of TRIM44 expression in SCC patients and to further determine whether TRIM44 could be used as a biomarker to predict metastasis and prognosis in SCC patients.
2. Patients and controls
Skin squamous carcinoma tissues were collected from 30 patients who received surgical resection in the First People's Hospital of Yancheng City, Yancheng, China from 2010 to 2015 and had been diagnosed by pathological confirmation.
Clinicopathological information of each case was recorded. None of them received preoperative chemotherapy or radiotherapy. Cancer patients included 17 males and 13 females (age 54.2–10.5 years, mean age of 59 years). According to Broders’ pathological grading criteria for SCC, 8 cases were grade I, 13 cases were grade II, and 9 cases were grade III. Normal tissue specimens were collected by surgical resection from 10 individuals to serve as a control group (age 48.5–10.6 years, mean age of 55 years). No statistically significant difference was detected in age or gender between the 2 groups.
This study was approved by the Ethical Review Committee of the First People's Hospital of Yancheng City and consent form was obtained from each patient (Identification No. HMU (Ethics) 20121103). Informed consent was obtained from all individual participants included in the study.
2.1. Immunohistochemical staining techniques
The immunohistochemical (IHC) staining method from Agilent Technologies, Inc. (Santa Clara, CA) was used to detect the distribution of TRIM44.
IHC procedures were performed in strict accordance with the manufacturer's instructions. The Envision and DAB chromogenic reagent kit (Agilent Technologies, Inc.) were used for IHC staining. All staining was performed under the same conditions; the tissue samples were sliced to a thickness of 2 to 3 μm, dehydrated in 80%, 90%, 95%, and 100% ethanol, dewaxed and antigen repair was performed using 0.01 mol/L citric acid (pH 6.0). Normal goat serum (Toyobo Co., Ltd., Osaka, China) was dropped onto the slide and incubation for 10 minutes at room temperature was performed. Subsequently, the corresponding specific antibody (anti-TRIM44 antibody (1:100; Novus Biologicals, Santa Cruz, CA) was added to the slide and incubated for 1.5 hours at room temperature. The slides were washed with phosphate-buffered saline (PBS) for 3 minutes 3 times. The secondary antibody (1:100; Novus Biologicals) was added and incubated for 30 minutes at room temperature. The slide was stained with DAB, and the nucleus was stained with hematoxylin, dehydrated using a gradient of ethanol, cleared with xylene, and sealed using natural gum. A known positive section reagent, which was supplied by Novus Biologicals, Santa Cruz. (TRIM44; 1:100), served as a positive control, and the specific antibodies were replaced with PBS to serve as the negative control.
The IHC results were determined by 3 pathologists, who observed the positive granule-stained cells in the SCC tissue samples and the adjacent healthy skin tissue using a BH-2 light microscope (Olympus Corporation, Tokyo, Japan). The staining score criteria were as follows: 0, 0% to 15%; 1, >15% to 30%; 2, > 30% to 45%; 3, >45%. According to the staining intensity for semi-quantitative determination, colorless was 0 and 3 (strong staining) was brown. The final staining score of a sample was determined as the product of the positive cell percentage score and the staining intensity score. Staining score <2, negative (−); staining score 2 to 4 points, weakly positive (+); staining score, 4 to 6 points, positive (+ +); staining score ≥6 points, strong positive (+ + +). For the convenience of statistical analysis of the data, the (−) group was defined as the negative expression group (−), and the (+), (+ +), and (+ + +) groups were designated as the positive expression group (+).
2.2. Detection the expression of TRIM44 mRNA by real-time PCR
According to the manufacturer's instructions,total RNA was extracted by using Trizol Reagent (Sangon Biotech Co., Ltd., Shanghai, China) and quantified using a Nandrop spectrophotometer. RNA (2 μg) was reverse transcribed to cDNA according to the Titanium One-Step reverse transcriptase-polymerase chain reaction (RT-PCR) kit (Takara Biotechnology Co., Ltd., Dalian, China), and was amplified by semi-quantitative PCR with β-actin serving as the reference. The primer sequences (Sangon Biotech Co., Ltd.) are presented in Table 1. The thermal cycling conditions were as follows: predenaturation at 94 °C for 4 minutes; 30 cycles of 94 °C for 10 seconds, 55 °C for 30 seconds, and 72 °C for 60 seconds.
Table 1.
Primer sequences for reverse transcription-polymerase chain reaction analysis.

Amplification of TRIM44 by PCR was examined by agarose gel electrophoresis and analyzed using Quantity One version 3 software (Bio-Rad Laboratories, Inc., Hercules, CA). The absorbance value of the belt and the reference were read, and the results were expressed as a ratio (sample value/reference value).
When the ratio of the SCC value and reference value was greater than the β-actin reference value, it was expressed positively. Otherwise, it was considered to be negative.
2.3. Detection of the expression of TRIM44 protein by using Western blot
The frozen specimens stored at 70 degrees were ground into liquid powder under liquid nitrogen. Cellular protein was measured by using BCA kit (Thermo). Fifteen microgram protein sample was sperated by 10% SDS-PAGE and electrotransferred to PVDF membrane (Millipore). The membrane was blocked with 5% non-fat milk at room temperature for 1hour. Then it was washed in TBST for 5 times (5 min/time). The first antibody of TRIM44 antibody (1: 300; Novus Biologicals)) and β-actin (1:1000) (Cell Signaling Technologies company) were added on the membrane, it was incubated at 4 °C overnight. It was washed by TBST for 3 times (5 min/time) again. The second antibody was added on the membrane, and shook for 1 hour at room temperature. Then it was washed by TBST for 3 times (5 min/time). It was imaged by ECL imaging system. The average gray value of TRIM44 protein band was analyzed by gel software. The β-actin protein as an internal standard and statistical analysis by the ratio of TRIM44 protein and β-actin respectively as the relative expression quantity.
2.4. Statistical methods
A statistical analysis was performed by using the SPSS 13.0 statistical software package. Data are given as mean and standard deviation (Mean ± SD). The χ2 test was used to compare the distribution of TRIM44 between SCC tissues and normal skin tissues. P <.05 was defined as statistically significant. Survival analyses were performed using the Kaplan–Meier method and log-rank test. Cox proportional-hazards regression was performed for multivariate analysis of prognostic predictors.
3. Results
3.1. Expression of TRIM44 protein and TRIM44 mRNA in SCC tissues and normal skin tissues
In order to investigate the expression of TRIM44 protein and mRNA in SCC tissues and normal skin tissues, IHC was performed and the result showed that the positive rate of TRIM44 staining in SCC tissues 26.00% (9/30), while the positive rate of normal control group was 83.33% (25/30). The positive rate of TRIM44 staining in SCC tissues is significantly lower than normal skin tissue (P <.01; Fig. 1).
Figure 1.

Expression of TRIM44 protein in SCC tissues and normal skin tissues. a: High expression of TRIM44 in normal tissues; b: Low expression of TRIM44 in SCC tissues. c: The positive rate of TRIM44 staining in SCC tissues is significantly lower than normal skin tissue. ∗P <.01. SCC = skin squamous cell carcinoma, TRIM44 = tripartite motif-containing protein 44.
RT-PCR showed that the positive rates of TRIM44 mRNA expression in SCC tissues were 16.67% (5/30), but the positive rate of normal control group was 86.67% (26/30). TRIM44 mRNA expression in SCC group were significantly lower than that in the normal group (P <.01; Fig. 2).
Figure 2.

Expression of TRIM44 mRNA in SCC tissues and normal skin tissues. a: High expression of TRIM44 mRNA in normal tissues and Low expression of TRIM44mRNA in SCC tissues; b:The positive rate of TRIM44 mRNA in SCC tissues is significantly lower than normal skin tissue. ∗P <.01. SCC = skin squamous cell carcinoma, TRIM44 = tripartite motif-containing protein 44.
3.2. Association between the expression level of TRIM44 and the overall survival of postoperative patients with SCC
The overall survival of patients with SCC who were high expression for the TRIM44 protein was 22.20 ± 2.45 months. The overall survival patients with SCC who were low expression for the TRIM44 protein was 12.25 ± 2.35 months. Based on Kaplan–Meier survival analysis, low expression was associated with poor overall survival in SCC patients (P =.004, Fig. 3; Table 2).
Figure 3.

Kaplan-Meier analysis of overall survival related to the expression of TRIM44. Patients with low expression of TRIM44 had a poorer prognosis than those of patients with high expression of TRIM44. P =.004. SCC = skin squamous cell carcinoma, TRIM44 = tripartite motif-containing protein 44.
Table 2.
Univariate survival analysis of overall survival in SCC.

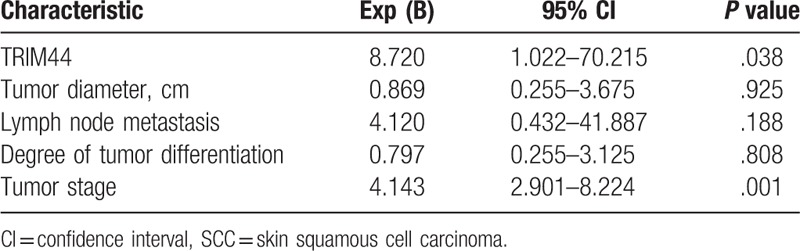
Multi-factor survival analysis indicated that both low TRIM44 expression and tumor stage were independent factors affecting the overall survival of patients with SCC (P =.038 and P =.001, respectively). Additionally, tumor invasion depth, lymph node metastasis and tumor differentiation (high, moderate, and poor) were not identified as independent factors affecting the overall survival of patients with SCC (all P >.05) (Table 3).
Table 3.
Multivariate survival analysis of overall survival in SCC.
3.3. Association between the expression levels of TRIM44 protein and TRIM44 mRNA in different pathological types of SCC
Compared with the control group, the expression levels of both TRIM44 protein and TRIM44 mRNA in SCC tissues were low. The expression tendency for TRIM44 protein and TRIM44 mRNA in SCC tissues is consistent. We further studied the association between the expression levels of TRIM44 protein and TRIM44 mRNA in different pathological types of SCC. The result for showed that low expression of TRIM44 in SCC was not associated with sex, age, tumor diameter, or degree of tumor differentiation (all P >.05), but was associated with staging (P =.009 and P =.008, respectively) and metastasis (P =.003 and P =.004, respectively) (Table 4).
Table 4.
Correlation of TRIM44 protein and mRNA expression levels with clinicopathological features in SCC.

4. Discussion
This is the first detailed report demonstrating that the levels of TRIM44 protein and TRIM44 mRNA in SCC are lowly expressed, which is strongly associated with tumor staging, metastasis and poor survival. And it also is an independent factor affecting the overall survival of patients with SCC.
Previous studies have demonstrated the particularly complicated processes involved in the occurrence and development of tumors,[12,13] which may be caused by the regulation of cell growth and proliferation.[14] In addition, abnormal expression of tumor-associated genes may be involved.[15,16] Cell growth and proliferation in the human body are affected and controlled by numerous factors.[17,18] Notably, cell signaling proteins, growth factors and their receptors, apoptotic proteins and transcription factors, and the changes of these factors are closely associated with the occurrence and development of tumors.[19] TRIM proteins are a large family of proteins that have been implicated in many biological processes including cell differentiation, apoptosis, transcriptional regulation, and signaling pathways.
Recent studies have demonstrated the expression levels changes and a multifaceted role of TRIM44 in cancer progression.[20] However, its correlation with the prognosis of postoperative patients exhibiting SCC remains unclear. In this study, IHC and RT-PCR revealed that both the TRIM44 protein and TRIM44 mRNA expression levels in SCC group were significantly lower than in normal skin tissue. This is not consistent with the result from Li P et al.[21] We think that the reason may be related to the different mechanisms of tumor in different parts and also the mechanisms of cancer are not the same.
Ong et al[22] showed that TRIM44 is associated with OS in patients with esophageal adenocarcinoma and can be an independent factor affecting the overall survival of patients. In our study, Kaplan–Meier survival analysis showed that low expression was associated with poor overall survival in SCC patients. Meanwhile, Multi factor survival indicated that both low TRIM44 expression and tumor stage were independent factors affecting the overall survival of patients with SCC, which is consistent with the result from Ong et al.
Zhu et al[23] demonstrated that high expression of TRIM44 can enhance proliferation, migration, invasion, and resistance to doxorubicin in hepatocellular carcinoma. In this study, we also studied the association between the expression levels of TRIM44 protein and TRIM44 mRNA in different pathological types of SCC. We found that low expression of TRIM44 in SCC was not associated with sex, age, tumor diameter, or degree of tumor differentiation, but was associated with staging and metastasis, which is also consistent with the result from Zhu et al.
A limitation of the present study was the relatively small sample size, lacked of further validation at animal level and cell level. The study was performed but all the patients are from 1 region only. However, studies in other regions have not yet been reported, and this is the first studies addressing TRIM44 expression in SCC.
In conclusion, the present study showed that low expression of TRIM44 is significantly correlated with poor prognosis in patients with SCC. It could be an independent factor affecting the overall survival of patients with SCC. TRIM44 may serve as a prognostic marker for predicting poor prognoses for patients with SCC.
However, further studies would provide a better test of this conclusion.
Author contributions
Conceptualization: Wei Wang, Jian Wu and Nai-Zhou Guo.
Data curation: Jian Wu, Zhou Nai Guo, Wei Wang, Lei Lei Cui, Yan Xue Zhang.
Formal analysis: Xue-yan Zhang, Lei Lei Cui.
Funding acquisition: Jian Wu.
Investigation: Lei-lei Cui, Wei Wang, Jian Wu.
Methodology: Cun-quan Xiong, Jian Wu, Zhou Nai Guo, Wei Wang, Lei Lei Cui, Quan Cun Xiong, Yan Xue Zhang.
Project administration: Jian Wu.
Software: Quan Cun Xiong, Yan Xue Zhang.
Validation: Jian Wu, Quan Cun Xiong.
Writing – original draft: Jian Wu, Cun-quan Xiong.
Writing – review & editing: Nai-Zhou Guo.
Footnotes
Abbreviations: IHC = immunohistochemistry, RT-PCR = reverse transcriptase-polymerase chain reaction, SCC = skin squamous cell carcinoma, TRIM44 = tripartite motif-containing protein 44.
JW and N-ZG equal contributors.
The present study was supported by the Youth Medical Talent of Jiangsu Province (grant no. QNRC2016475) to (JW) and the medical innovation team Research Fund of Yancheng City (2016).
The authors have no conflicts of interest to disclose.
References
- [1].Noori VJ, Trehan K, Savetamal A, et al. New onset squamous cell carcinoma in previous split-thickness skin graft donor site. Int J Surg 2018;52:16–9. [DOI] [PubMed] [Google Scholar]
- [2].Wu J, Zhang JR, Qin J. Clinical significance of methylation of E-cadherin and p14ARF gene promoters in skin squamous cell carcinoma tissues. Int J Clin Exp Med 2014;7:1808–12. [PMC free article] [PubMed] [Google Scholar]
- [3].Wu J, Lu WY, Cui LL. Inhibitory effect of curcumin on invasion of skin squamous cell carcinoma A431 cells. Asian Pac J Cancer Prev 2015;16:2813–8. [DOI] [PubMed] [Google Scholar]
- [4].Debaugnies M, Sánchez-Danés A, Rorive S, et al. YAP and TAZ are essential for basal and squamous cell carcinoma initiation. EMBO Rep 2018;6:e45809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reymond A, Meroni G, Fantozzi A, et al. The tripartite motif family identifies cell compartments. EMBO J 2001;20:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li Y, Wu H, Wu W, et al. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res 2014;24:762–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011;11:792–804. [DOI] [PubMed] [Google Scholar]
- [8].Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med 2011;3:513–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mcnab FW, Rajsbaum R, Stoy JP, et al. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol 2011;23:46–56. [DOI] [PubMed] [Google Scholar]
- [10].Yamada Y, Takayama Kl, Fujimura T, et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci 2017;108:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xing Y, Meng Q, Chen X, et al. TRIM44 promotes proliferation and metastasis in non-small cell lung cancer via mTOR signaling pathway. Oncotarget 2016;7:30479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mattina J, MacKinnon N, Henderson VC, et al. Design and reporting of targeted anticancer preclinical studies: a meta-analysis of animal studies investigating sorafenib antitumor efficacy. Cancer Res 2016;76:4627–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huo Y, Su T, Cai Q, et al. An in vivo gain-of-function screen identifies the williams-beuren syndrome gene gtf2ird1 as a mammary tumor promoter. Cell Rep 2016;15:2089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee J, Katzenmaier EM, Kopitz J, et al. Reconstitution of TGFBR2 in HCT116 colorectal cancer cells causes increased LFNG expression and enhanced N-acetyl-d-glucosamine incorporation into Notch1. Cell Signal 2016;28:1105–13. [DOI] [PubMed] [Google Scholar]
- [15].Gao R, Ma LQ, Du X, et al. Rnf25/AO7 positively regulates wnt signaling via disrupting Nkd1-axin inhibitory complex independent of its ubiquitin ligase activity. Oncotarget 2016;7:23850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fang M, Yuan J, Peng C, et al. Collagen as a double-edged sword in tumor progression. Tumor Biol 2014;35:2871–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bai J, Zhang X, Hu K, et al. Silencing DNA methyltransferase 1 (DNMT1) inhibits proliferation, metastasis and invasion in ESCC by suppressing methylation of RASSF1A and DAPK. Oncotarget 2016;7:44129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu JY, Lu JB, Xu Y. MicroRNA-153 inhibits the proliferation and invasion of human laryngeal squamous cell carcinoma by targeting KLF5. Exp Ther Med 2016;11:2503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin W, Zhong M, Yin H, et al. Emodin induces hepatocellular carcinoma cell apoptosis through MAPK and PI3K/AKT signaling pathways in vitro and in vivo. Oncol Rep 2016;36:961–7. [DOI] [PubMed] [Google Scholar]
- [20].Peng R, Zhang PF, Zhang C, et al. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing cell EMT via MAPK signaling. Cancer Med 2018;7:796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li P, Yin H, Meng F, et al. High TRIM44 expression in endometrial carcinoma is associated with a poorer patient outcome. Pathol Res Pract 2018;214:727–31. [DOI] [PubMed] [Google Scholar]
- [22].Ong CA, Shannon NB, Ross-Innes CS, et al. Amplification of TRIM44: pairing a prognostic target with potential therapeutic strategy. J Natl Cancer Inst 2014;106:2504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu X, Wu Y, Miao X, et al. High expression of TRIM44 is associated with enhanced cell proliferation, migration, invasion, and resistance to doxorubicin in hepatocellular carcinoma. Tumour Biol 2016;37:14615–28. [DOI] [PubMed] [Google Scholar]


