Abstract
Determining the extent to which Symbiodinium communities in corals are inherited versus environmentally acquired is fundamental to understanding coral resilience and to predicting coral responses to stressors like warming oceans that disrupt this critical endosymbiosis. We examined the fidelity with which Symbiodinium communities in the brooding coral Seriatopora hystrix are vertically transmitted and the extent to which communities are genetically regulated, by genotyping the symbiont communities within 60 larvae and their parents (9 maternal and 45 paternal colonies) using high-throughput sequencing of the ITS2 locus. Unexpectedly, Symbiodinium communities associated with brooded larvae were distinct from those within parent colonies, including the presence of types not detected in adults. Bayesian heritability (h2) analysis revealed that 33% of variability in larval Symbiodinium communities was genetically controlled. Results highlight flexibility in the establishment of larval symbiont communities and demonstrate that symbiont transmission is not exclusively vertical in brooding corals. Instead, we show that Symbiodinium transmission in S. hystrix involves a mixed-mode strategy, similar to many terrestrial invertebrate symbioses. Also, variation in the abundances of common Symbiodinium types among adult corals suggests that microhabitat differences influence the structure of in hospite Symbiodinium communities. Partial genetic regulation coupled with flexibility in the environmentally acquired component of Symbiodinium communities implies that corals with vertical transmission, like S. hystrix, may be more resilient to environmental change than previously thought.
Introduction
Symbiosis is fundamental to life on Earth, underpinning the existence of numerous prokaryotic and eukaryotic species and shaping the physiology and health of many organisms (Moya et al. 2008; Gilbert et al. 2012; Lewis et al. 2015). Microbial symbionts also enable hosts to expand their niche breadth to survive in environments otherwise unsuited to their physiology (Goffredi et al. 2007). For example, symbiosis with photosynthetic dinoflagellates of the genus Symbiodinium has allowed corals to thrive in oligotrophic tropical seas through the utilization of symbiont photosynthates. Similar nutritional facilitation has been described for sap-sucking insects that rely on microbial partners to supplement their diets (Baumann 2005). Compared to these well-characterized systems, coral endosymbioses are poorly described at the Symbiodinium-type level during early ontogeny.
Nutritional symbioses can drive diversification of host and symbiont lineages (Douglas 1989; Brucker and Bordenstein 2012; Oliver et al. 2014), with eukaryotic symbionts like Symbiodinium that have gone through multiple cycles of diversification and expansion (Thornhill et al. 2014). This standing genetic variation provides new material upon which selection may operate (Moran et al. 2008; Russell et al. 2012), facilitating coevolution between hosts and symbionts or among symbionts (Moran and Dunbar 2006; Moran et al. 2008; Moya et al. 2008). Understanding the fidelity (here defined as the exactness of transfer of symbionts from parent to offspring) of Symbiodinium community inheritance is key to determining the degree to which endosymbiotic Symbiodinium communities have coevolved with their coral hosts and is central to coral nutrition and health. Despite this, little is known about genetic regulation underpinning this symbiosis.
Symbionts may be acquired from the environment (horizontal transmission) or passed maternally into eggs or larvae (vertical transmission), with the latter thought to be the most prevalent mode of transmission in brooding scleractinian corals (Baird et al. 2009). Maternally derived symbionts may involve the transmission of one or multiple symbionts (superinfections), at least in well-studied insect vertically transmitting symbioses. Transmission of bacterial symbionts in insects may be exclusively vertical or may occur initially as vertical transfer followed later by horizontal transmission (Fujishima and Fujita 1985; Sandström et al. 2001; Kaltz et al. 2003; Scheuring and Yu 2012; Andersen et al. 2013; Oliver et al. 2014). Although similar mixed-mode transmission has been hypothesized for corals (Byler et al. 2013), the absence of experimental data means that it is not yet clear if transmission is exclusively vertical in brooding corals or if mixed-mode transmission also occurs in this group. Given recent evidence of differences in the diversity of symbiont communities transmitted from parents to offspring in two broadcast-spawning species (Padilla-Gamiño et al. 2012; Quigley et al. 2017a, 2017b), Symbiodinium transmission dynamics in corals may be as complex as those observed in the Arthropoda.
In general, symbiont-host specificity is theorized to be much greater when symbionts are transmitted vertically compared to horizontally (Douglas 1998; Baker 2003). In corals, hosts may form strict associations with only one Symbiodinium type (and vice versa) or associate with multiple partners, and superinfections of multiple Symbiodinium types and subtypes of varying abundances are common among species (Little et al. 2004; Abrego et al. 2009a, 2009b; Fabina et al. 2012; Byler et al. 2013; Poland and Coffroth 2017). Although maternal transfer of Symbiodinium and bacteria is less well-characterized in corals than in terrestrial invertebrates (Apprill et al. 2009; Padilla-Gamiño et al. 2012; Sharp et al. 2012; Byler et al. 2013, 2017a, 2017b), the presence of superinfections raises the possibility that Symbiodinium dynamics are similar to the mixed-mode transmission dynamics characteristic of superinfections described in terrestrial invertebrates like aphids and sharpshooter cicada (Moran et al. 2008; Oliver et al. 2014). However, unlike studies of insect symbiont specificity, no studies have used high-throughput sequencing to examine maternally transmitted Symbiodinium communities in brooding corals or the diversity of low abundance Symbiodinium types in detail. Similarly, the genetic component of parental contributions to the maturation of coral-Symbiodinium symbioses remains unquantified.
It is clear that Symbiodinium types vary in their impact on holobiont physiology because of variation in their stress tolerance and ability to produce and transfer photosynthates to the coral host under differing light, temperature and nutrient regimes (Little et al. 2004; Berkelmans and van Oppen 2006; Reynolds et al. 2008; Cantin et al. 2009; Hume et al. 2015; LaJeunesse et al. 2015). Moreover, environmental variation and stress may bring about shifts in the dominance of Symbiodinium types, in some cases benefiting the host under the altered conditions (Jones et al. 2008; Cunning et al. 2015). The extent of a coral’s flexibility to acquire resilient types or shuffle symbionts may be genetically regulated, for example by heritable host immune responses, similar to those that shape symbiont diversity in Drosophila (Mateos et al. 2006). Complete inheritance results in complete fidelity of symbiont transmission, and, hence, little scope for flexibility in coral-Symbiodinium symbioses. However, the extent of such potential regulation of symbiont transmission and its underlying basis are unknown for corals.
It is increasingly revealed that the genetic architecture behind traits and pathologies can be complex (Cho 2015). For example, both the diversity and abundance of microbial symbionts in the human gut are complex traits under partial genetic control (Zoetendal et al. 2001; Ley et al. 2006; Benson et al. 2010; Turnbaugh et al. 2010; Campbell et al. 2012; Liu et al. 2015). Narrow-sense heritability (h2) is the parameter typically used to describe the degree to which variability in a trait is explained by genetic factors. Assuming that the Symbiodinium community associated with a coral can be represented as a complex trait, then an h2 value of 1 would imply that variability of the community is mostly due to host genetics. Conversely, an h2 value estimated at 0 would imply no genetic basis for variability in the community, thus the community would not be under selection and could not evolve (no evolvability; Lynch and Walsh 1998). Although an h2 estimate close to 1 does not necessarily guarantee absolute genetic determination as a result of gene segregation (Visscher et al. 2008), a large heritability estimate of the Symbiodinium community would imply that changes in host genotypes are required for shifts in symbiont communities. Conversely, changes in the environmental availability of Symbiodinium or in environmental conditions would have limited influence on in hospite communities. Understanding the relative contributions that host genetics versus environmental conditions make to the composition of Symbiodinium communities through estimations of h2 will improve the accuracy with which the potential, direction and speed of changes in Symbiodinium communities can be predicted.
To examine Symbiodinium community transfer between adults and their offspring in a brooding coral and quantify the narrow-sense heritability (h2) of this trait, we quantified the in hospite Symbiodinium communities of individual planula larvae and their parents across a spectrum of relatedness using high-throughput sequencing. Relatedness was based on a population genetic parentage analysis that assigned the likely paternal identity of each larva. In light of results on heritability and fidelity of symbiont transfer, we discuss the potential of larvae from brooding corals like S. hystrix to acclimate to novel environments.
Materials and methods
Study species and sampling design
The common, hermaphroditic coral Seriatopora hystrix broods sexually produced larvae following internal fertilization of eggs by sperm from surrounding colonies (Ayre and Resing 1986; Warner et al. 2016). DNA extracts of planula larvae for the present study were selected from samples that were collected in an earlier study to assess sperm dispersal distances and larval parentage of a cryptic species within the S. hystrix species complex, specified as S. hystrix (ShA) (Warner et al. 2015, 2016). In Warner et al.’s study, colonies were tagged and sampled for molecular analyses within a 16 m × 16 m sampling area, with additional colonies sampled from two adjacent transects (totaling 16 m × 40 m area) in the Lizard Island lagoon (S14°41.248, E145°26.606; Warner et al. 2015, 2016). Microsatellite genotypes and paternity assigned to individual larvae in this earlier study (Warner et al. 2016) enabled us to examine the effect of both maternal and paternal identity on larval Symbiodinium communities across a full pedigree of larval relatedness. Hence, our study included full-sib and half-sib larvae, and four individuals produced by selfing (further details in supporting information and Table S1).
Symbiodinium community genotyping
Symbiodinium communities of adults and larvae were quantified with amplicon sequencing of the ITS2 locus using the same DNA extractions that had been used to assign microsatellite genotypes and paternity in Warner et al. (2016). Maternal and paternal identities of each of the 60 larvae were estimated in (Warner et al. 2016). Specifically, 9 maternal and 45 assigned paternal colonies (which included the nine maternal colonies), plus all larvae whose paternity was designated with a confidence level of very high (95% posterior paternity probability + 95% confidence assignment), high (95 + 80%), or medium (95% only) by Warner et al. (2016) (n = 60 larvae) were sequenced with the primers ITS2alg-F and ITS2alg-R (Pochon et al. 2001) using paired-end Illumina Miseq technology. Library preparation and sequencing were performed at the University of Texas at Austin’s Genomics Sequencing and Analysis Facility (USA) using their standard protocols, including Bioanalyzer (Agilent)-based DNA standardization and pooled triplicate PCR before library preparation.
Raw reads (total = 6,875,177) were analyzed using the USEARCH and UPARSE pipelines (v.7; Edgar 2013), as outlined in Quigley et al. (2016) (further details in supporting information). Because there is currently no single copy marker for Symbiodinium genotyping (Pochon et al. 2012), the ITS2 marker was selected for the broadest comparisons to the vast literature that has used this marker to describe Symbiodinium diversity, including some using next generation sequencing (e.g., Arif et al. 2014; Green et al. 2014; Thomas et al. 2014; Quigley et al. 2016, 2017a, 2017b; Ziegler et al. 2017a). Additional steps were taken to assess the presence and impact of intragenomic variants (further explained below). Briefly, reads were filtered, clustered into OTUs at 97% similarity, annotated with NCBI nt database and Symbiodinium-specific searches (further details in Table S2). Using these methods, the majority of the OTUs were re-assigned to a clade/type level, leaving only 0.03% of cleaned reads (1459 reads, 78 OTUs) that could not be classified, and which may represent new Symbiodinium types (Table S3, Fig. S1, supporting information).
To account for variable read-depth across all samples, sample reads were normalized using “DESeq2” and “Phyloseq” implemented in R (R Core Team 2012; McMurdie and Holmes 2013; Love et al. 2014a, 2014b). Nonmetric multidimensional scaling (NMDS) was performed and plotted using the normalized counts matrix using “Phyloseq”, “vegan”, and “ggplot” (Wickham 2009; Schloerke et al. 2014). A permutational multivariate analysis of variance and permutation test for homogeneity of multivariate dispersions were used to determine whether significant differences in Symbiodinium community structure exist between broods using the “adonis” and “betadisper” functions in “vegan.” Genetic distances between OTUs were calculated in “Ape” (Paradis et al. 2004). Statistical testing of variation in OTU abundance was performed on raw reads in “DESeq2”, which incorporates variance normalization of OTU abundance, and interpreted using the Bejamini–Hochberg correction for multiple-inferences of p-adjusted alpha at 0.05. DESeq2 calculates p-values based on independent filtering criteria and Benjamini–Hochberg multiple testing adjustments to determine whether p-values are significant. “DESeq2” outputs are expressed as the mean of normalized counts of treatment groups (baseMeans) and multiplicative (log2 fold) terms between or among treatments (Love et al. 2014a, 2014b). Network analysis on planula larvae was performed using the “igraph” package (Csardi and Nepusz 2006) and custom scripts from (Cunning et al. 2017).
Estimating the diversity and heritability of Symbiodinium communities
To describe the Symbiodinium community in coral samples, we used a diversity measure (qDZij(p)) that incorporates OTU richness, evenness, and sequence similarity (Leinster and Cobbold 2012). This metric therefore combines a variance-normalized matrix of OTU abundances and diversity, rarity, and a matrix of pairwise similarities between sequences (sequence similarity). Sequence similarity was calculated using pairwise percent similarities between OTU sequences using the “Ape” package with a “raw” model of molecular evolution. Through this diversity metric (further described in Quigley et al. 2017a), the Symbiodinium community is presented as a continuous quantitative host trait. Heritability of Symbiodinium diversity associated with the 60 larvae was calculated using the package “MCMCglmm” (Hadfield 2010) utilizing the diversity metrics described above, where the coefficient of relatedness between individuals was set as a random effect. The coefficient of relatedness is the degree to which individuals share genetic material. For example, full siblings share 50% of their genetic material, whereas clones would share 100%. Maternal environmental effects and paternal effects were assessed and were not significant. Deviance Information Criterion was used to test if adding a maternal random effect had a statistically significant effect on heritability estimates. Models were run with 1.5 × 106 iterations, a thinning of 50, and burn-in of 10% of the total iterations. A non-informative flat prior specification was used following an inverse gamma distribution (see Wilson et al. 2010 for further analysis details and scripts). Assumptions of chain mixing, normality of posterior distributions, and autocorrelation were met. The posterior heritability was calculated by dividing the model variance attributed to relatedness by the sum of additive and residual variance.
Multiple ITS2 copies and intragenomic variation
Intragenomic variation within and between Symbiodinium types makes classifying type-level diversity in Symbiodinium based on sequence data difficult (Thornhill et al. 2007; Sampayo et al. 2009; Arif et al. 2014; Quigley et al. 2014). We addressed intragenomic variation by clustering across samples at 97% similarity and provide two additional analyses to test for their presence and potential impact on the heritability estimate; and both confirm the robust nature of our conclusions in regards to this issue (Supporting Methods and Results).
Colony size and spatial distribution of adult S. hystrix (ShA) colonies
To determine if Symbiodinium communities varied with colony size (as a proxy for colony age), adult colonies were divided into five size classes based on their mean diameter (Warner et al. 2016): <8 cm (n = 1 colony), 8 – <14 cm (n = 19), 14 – <20 cm (n = 13), 20 – <26 cm (n = 11), and 26–32 cm (n = 1). Differential abundance testing of Symbiodinium OTUs among size classes was performed as for larval communities.
Sitepainter (Gonzalez et al. 2012) and Inkscape (Bah 2009) were used to visualize spatial patterns in the distribution of Symbiodinium OTUs associated with the 45 adult colonies of S. hystrix (ShA) that were genotyped across the 16 m × 40 m sampling area. Gradient-boosted models and linear models were run in the package “gbm” (Ridgeway 2006) to examine spatial distributions of the ten most abundant OTUs. These types of Boosted Regression Trees uses machine learning and predictive modeling to construct models, thereby making them particularly suited to spatially patchy data of S. hystrix colonies in our sampling area (Leathwick et al. 2008). Linear models were checked for assumptions of linearity, normality, and homogeneity of variance. Square-root transformations were used to correct for issues of normality or heterogeneity. Latitude and longitude coordinates were centered before fitting models. The package “Spatstat” (Baddeley and Turner 2005) was used to visualize spatial variability in abundances of the three most significantly heterogeneous OTUs across the sampling area (OTUs: 1, 3, and 6). Spearman’s rho rank correlation coefficients were calculated to test for competitive exclusion amongst the three OTUs that varied significantly across the sampling area. Pairwise p-values were generated for all OTU comparisons using the base “stats” package in R.
Results
Symbiodinium communities differ between parents and brooded larvae
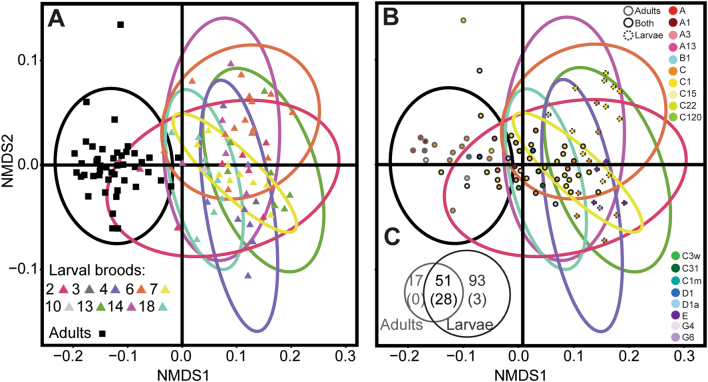
Symbiodinium communities differed between adults and their larvae in the brooding coral Seriatopora hystrix (ShA) (Fig. 1a, b). Overall, the composition of Symbiodinium communities was significantly different between adults and larvae, with higher similarity among adult corals, but more variable among larvae (Permutation test for homogeneity of multivariate dispersions, df = 1, p = 0.001). On average, adults contained 29.9 ± 0.6 (SE) OTUs and larvae had 22 ± 0.4 OTUs (Fig. 2). However, the number of unique OTUs recovered was more than five times greater from larvae than from adults (93 vs. 17 OTUs, respectively; Fig. 1c). Of the 17 unique adult OTUs, ten belonged to clade C (C1, C15, and other variants), three from clade A (A1 and variants), three from D (including D1 and D1a), and one was a putative C type (Fig. 1b). Unique to larvae were 17 OTUs from clade C (likely C1 variants), four from clade E, one from each of A3, B1, and G6, and 69 that were of putative Symbiodinium-type level identity (Fig. 1b). Of the 93 larval-specific OTUs, only the abundance of C1_OTU136 (type followed by OTU designation) and two putative clade D OTUs (OTU148 and OTU149) were significantly different from zero with the Bejamini–Hochberg correction (Fig. 1b). Although raw read counts were low, C1_OTU136 was present in larvae from every dam but dam 3.
Fig. 1.
Nonmetric multidimensional scaling (NMDS) Split Biplot, based on a Bray–Curtis distance matrix of variance-normalized OTU abundances and sequence similarity between OTUs (pairwise percent identities), illustrating differences between Symbiodinium communities associated with adult colonies and larvae of the brooding coral Seriatopora hystrix (ShA) based on parentage analysis. Ellipses encircling symbols of the corresponding color represent 95% probability regions for adults (black) and larval broods (colored), where each brood represents all larvae sharing the same dam (color-coded). Samples and OTUs have been separated to increase Biplot clarity whilst maintaining ellipse positions to facilitate comparisons of ordination space between samples and OTU positions. a Each point represents the Symbiodinium community associated with a unique coral adult or larval sample. b Each point represents an OTU colored by type level. Only 83 of the 161 OTUs are shown, representing those OTUs with the greatest taxonomic confidence (see Table S7 for full names and selection criteria). Outlining around each point represents the origin of the OTU, i.e., those found uniquely in adult (gray outline) or larval (broken gray outline) samples, or retrieved from both (black outline). Samples presented in a and OTUs presented in b share the same ordination space but were separated for clarity. c Venn diagram, illustrating the number of Symbiodinium OTU’s that were unique to larvae (dark gray text) versus adults (light gray text). The number of OTUs that were significant after p-adjustments are in parentheses. Ellipses corresponding to dams 3 and 10 are not represented, as only one larva per dam was collected and sequenced
Fig. 2.

Barplots of variance-normalized abundances of Symbiodinium diversity associated with a adults and b planula larvae of Seriatopora hystrix. Colors represent different Symbiodinium types. Numbers below planulae barplots represent the different broods
Fifty-one OTUs were shared by adult colonies and planula larvae (43 of known Symbiodinium, 8 of putative Symbiodinium taxonomy, Fig. 3a), and the abundance of 28 of these OTUs differed significantly between the two groups at the adjusted p-level, including both dominant (i.e., C1/C120_OTU1) and background OTUs (C15_OTU46) (Table S4). Of these 28 OTUs, 23 were from clade C (including C1, C3w, C120 among others), three from clade D (D1, D1a), and two from clade A (A1, A3). Adult Symbiodinium communities were characterized by up to 4–5.7 (2–2.5 log2 fold) times more D-types (D1_OTU3, D1_OTU597, D1a_OTU6), and A-types (A1_OTU10 and A3_OTU8) compared to larvae (Bejamini–Hochberg corrections, Table S4). Nine of the 23 C-types had up to 9.2 times (3.2 log2 fold) significantly higher abundances in adults (including multiple C1 types, C120/C120a_OTU1, C1m_OTU5/105, C1v6/C22_OTU228, C15_OTU46, C31_OTU733, and C3W_OTU165) and the remaining C1 types had between 0.02–0.13 (−5.4 to −2.9 log2 fold) (times significantly lower abundances in adults (Table S4). D1, D1a, A1, and A3 were found at high relative abundances in adult and larval communities (Fig. 2, Table S4). C1/C120 OTUs were detected at the greatest mean relative abundances. In contrast, some of the C1/C15 OTUs were detected at mean relative abundances at the background level (<0.16, Table S4).
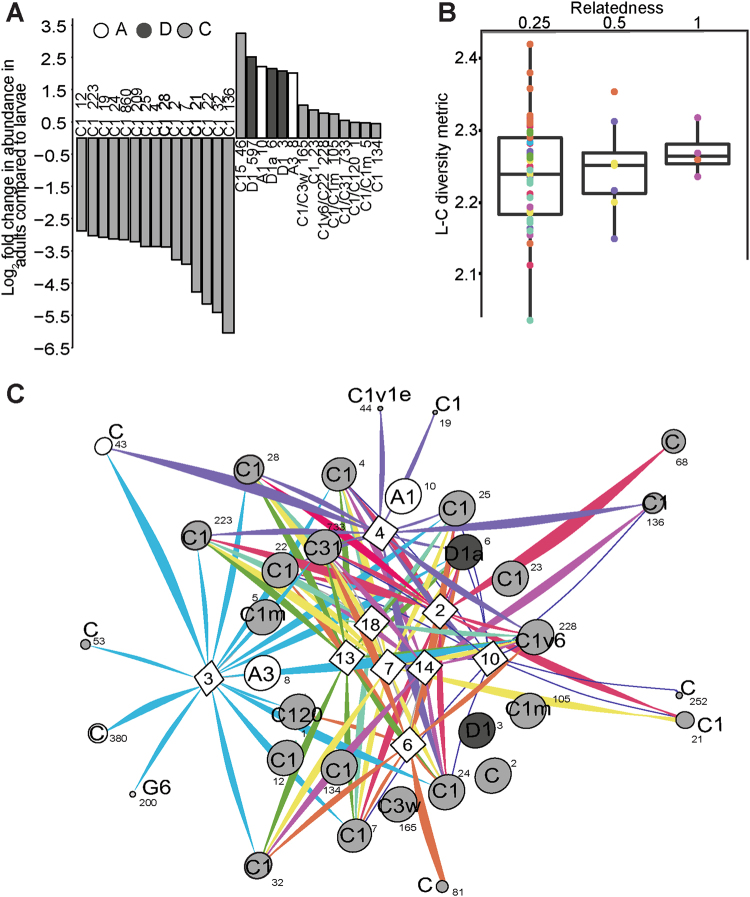
Fig. 3.
a Log2 fold change in abundances of Symbiodinium OTU’s that differed significantly between communities associated with adults versus larvae of Seriatopora hystrix (ShA). Gray-scale in the bar plot identify Symbiodinium clades. A positive change indicates the OTU is more abundant in adults. b Boxplots showing medians, quartiles, and minimum/maximum values of Symbiodinium community diversity (Leinster and Cobbold metric) in relation to individual larval relatedness. On the x-axis, 0.25 denotes half sibs, 0.5 full sibs, and 1.0 denotes larvae produced from selfing. Each larva is colored by its respective dam. c Network analysis of planula larvae showing OTUs present in 50% or more of larvae per brood. Edges have been removed for OTUs found in 100% of larvae from each brood (OTUs:1, 2, 3, 5, 8, 10, 12, 23, 105, 134, and 165). White diamonds correspond to maternal broods, where each brood sharing the same dam is color-coded. Small numbers next to each node indicate the OTU number for that Symbiodinium type. Line thickness denotes relative abundance of the Symbiodinium type per brood
Larval Symbiodinium communities vary among broods
Planula larvae that shared the same maternal parent generally grouped together when Symbiodinium OTU richness, abundance, and DNA distance between OTUs were incorporated into analyses. Maternal broods differed significantly in their Symbiodinium community structure (permutational analysis of variance by dam; R2 = 0.57, df = 44, p = 0.001) although 95% probability regions overlapped (Fig. 1a, b). Thirty-one OTUs (including multiple C1 variants, D1, D1a, A1, and A3) were found in >50% of larvae per brood and were generally present across all broods (Fig. 3c) and drive the structure among larval broods.
Eleven OTUs dominated larval communities and were found in all larvae from all broods, including C120, C1m, and D1 (see full OTU list in Fig. 3c). Other OTUs contributed to the unique structure of each brood and included OTUs that were highly prevalent within broods of highly related larvae (e.g. brood 3 and 4: G6 and C1v1e, respectively). At least 8 of these key OTUs were not shared between broods (i.e., OTUs 81, 252, 68), and highlight the distinctiveness of larval samples across broods and contributed to full-sib phenotypic correlations compared to those of half sibs. Alternatively, OTUs that are prevalent across all broods (e.g., C120, D1, A3, and A1) contributed to structuring between larval broods (overlap between OTUs shared between broods). Furthermore, OTUs also varied in their relative abundance across larval broods, with particular OTUs being numerically dominant in specific broods compared to others, such as C_68 in brood 2 and C_81 in brood 6 (Fig. 3c).
However, differences in the abundance of OTUs amongst larval broods were detected for symbiont types A1, A3, C1, D1, and D1a, amongst others. Briefly, larvae from dam 2 displayed higher abundances of A1 and A3. Larvae from dams 3, 7 and 10 had significantly less of C1_OTU2, whilst broods from dams 4, 6, 13, 14, and 18 had significantly different abundances of many C-types, including C120/C120a, C1, C1v1e, C1m, and C31. The abundances of D1_OTU3 and D1a_OTU6 also varied significantly among larval broods, particularly among those from dams 2, 4, and 18 vs. dam 13 (for a full description see supporting information and Table S5).
Heritability
Leinster and Cobbold estimates of Symbiodinium community diversity varied across the 60 larvae. Notably, more closely related larvae had more similar Symbiodinium communities (Fig. 3b). The posterior mean heritability of the Symbiodinium community in S. hystrix (ShA) larvae was 0.43 ± 0.21 SD, with a posterior mode of 0.33 (95% Bayesian credibility interval (BCI) 0.1–0.8; Fig. S2, supporting information). The effect of larvae sharing the same maternal environment was not significant (no model improvement; Deviance Information Criterion <2 units with the addition of maternal identity as a random effect) but decreased the posterior mean and mode of heritability slightly (mean = 0.37 ± 0.21 SD and mode = 0.19; BCI: 0.1–0.8).
Patterns in adult Symbiodinium communities of varying spatial distribution and colony size
The distributions of three of the ten most abundant OTUs in adult corals varied significantly across the sampling area (p > 0.05; Fig. 4), although not in a consistent manner with distance either along or down the sampling area. For example, although abundances of Symbiodinium C120/C120a were greatest in colonies that were closest to the lower left of the sampling area (corresponding to 55,737–29,885 non-normalized reads, the most abundant Symbiodinium OTU in S. hystrix; gradient-boosted model (GBM): p = 0.019), consistent with a gradual increase in distance down the reef slope, this pattern was not consistent along the reef slope (GBM: p = 0.00841). The abundance of D1 was significantly higher in the top-right and lower left side of the sampling area than in other aspects (x and y interaction, corresponding to 7802–800 non-normalized reads; GBM: p = 0.0393). D1a was least abundant in the top left and inner portion of the sampling area (corresponding to 876-0 non-normalized reads; GBM: p = 0.0405). Finally, although the variance normalized abundances of all three OTUs were significantly positively correlated overall (Spearman’s rank correlation ρ: 0.42–0.77, all p < 0.004), extremely low abundances of C120/C120a at x-coordinates >15 contrast markedly with high abundances of the two D-types in the same region (Fig. 3). Of the 68 Symbiodinium OTUs found in adults, the abundance of only C1_OTU4 differed significant among the five coral size classes, with colonies from the 8–14 cm size class hosting 1.7 times lower C1 abundances compared to corals in the 14–20 cm size class (p > 0.05; Table S6).
Fig. 4.
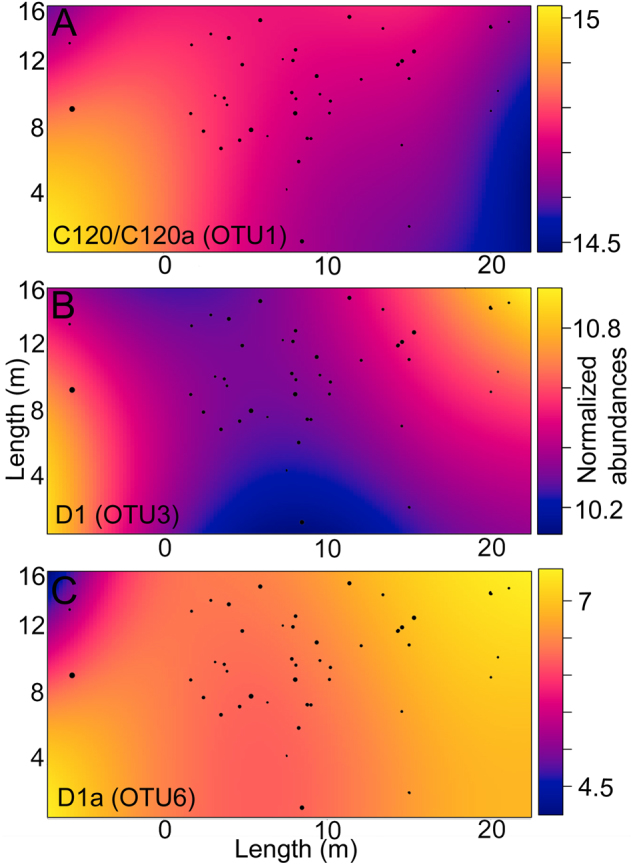
Spatial patterns in the normalized abundance of three Symbiodinium OTU’s associated with adult colonies of Seriatopora hystrix (ShA) that differed significantly in their abundances across a portion of the 16 m × 40 m sampling area at Lizard Island. Positions of the 45 genotyped adult colonies are denoted by black circles. a C120/C120a, b D1, and c D1a. Colors represent changes in the normalized abundance of each OTU across sampling site coordinates, with yellow representing the highest abundance and blue the lowest. Sizes of the black circles represent size classes of coral colonies in cm drawn to scale of the sampling area (smallest circles are 10 cm and the largest are 30 cm)
Discussion
Mixed-mode transmission structures larval Symbiodinium communities in a brooding coral
The availability of a full larval pedigree for Seriatopora hystrix (ShA) (Warner et al. 2016) provided a unique opportunity to evaluate the relative contributions of heritability (i.e., the degree to which variability in a trait is explained by genetic factors) versus maternal environmental effects (the effect of larvae sharing a common maternal environment) to the composition of larval Symbiodinium communities in a brooding coral. Here we show that Symbiodinium communities associated with larvae of S. hystrix (ShA) differ from those associated with their parents, providing experimental evidence that at least a portion of the Symbiodinium community is horizontally transmitted in a brooding coral. These results further demonstrate that coral-Symbiodinium symbioses align with well-characterized models of invertebrate symbioses. Results from previous studies suggesting exclusively vertical transmission in brooding corals may have been due to the lower detection capability of earlier methods and will no doubt change with the continued application of NGS technology to more brooding coral species. Overall, Symbiodinium communities were found to be moderately heritable, with only 33% of variability in larval symbiont communities under genetic regulation. Model selection also showed that, for larvae released from the same colony, sharing the same maternal environment did not significantly explain variability in Symbiodinium communities found among larvae from the same brood. This result, combined with the moderate heritability estimate, indicates that similarities in Symbiodinium communities among larvae of the same maternal brood were due to gene(s) inherited by these larvae.
Heritability estimates reveal important information about the evolvability of a trait, such as the capacity of brooding corals to vary their symbiont communities in response to changing environmental conditions. If levels of heritability and genetic variance are low, then responses to natural or artificial selection (evolvability) would be limited (Visscher et al. 2008). Conversely, high heritability and high genetic variance of a trait would enable greater responses to selection pressures. On the other hand, highly heritable symbiont communities with low genotypic variation could be problematic for vertically transmitting coral populations if adult communities are thermally sensitive (Matz et al. 2017). We found moderate heritability of Symbiodinium communities in S. hystrix (ShA). Much greater heritability of the Symbiodinium community was expected in this vertically transmitting coral, and in comparison with what is known of other important reproductive and fitness traits. For example, fertilization success, larval heat tolerance, protein content, settlement success, settlement substrate preferences, and juvenile growth and survivorship are all heritable traits (Meyer et al. 2009; Kenkel et al. 2011, 2015; Baums et al. 2013; Dixon et al. 2015). Although the distribution of posteriors was skewed towards values greater than that of our heritability estimate, it is unlikely that heritability (i.e., genetic regulation) for this trait will resolve to be much greater with increased sampling effort (~0.5–0.6, Fig. S2). The moderate levels of genetic regulation (i.e., heritability) found here suggest that S. hystrix (ShA) has some capacity to respond to changing environmental conditions. Thus, intervention efforts to facilitate such phenotypic change may be possible (Visscher et al. 2008). Given that assisted evolution efforts involving heat-selected Symbiodinium types show promise in horizontally transmitting corals (Levin et al. 2016), it may be that vertically transmitting, brooding species with moderate fidelity like S. hystrix (ShA) could also be candidates for assisted Symbiodinium uptake.
Combined maternal and environmental uptake produces locally adapted but flexible Symbiodinium communities
Detection of 93 larval-specific OTUs in this study demonstrates that brooding corals like S. hystrix (ShA) have a mixed-mode transmission strategy, in which dominant symbionts are transmitted vertically but additional background strains are acquired from environmental sources. Although adult diversity may have been under-sampled by only sequencing one branch of each parental colony, unique larval OTUs were not detected in any of the 45 adult colonies that were genotyped. Environmental uptake of novel Symbiodinium by larvae of this species is further supported by the appreciable amount of variation in the composition of larval Symbiodinium communities that was not under genetic control, according to our heritability model. These results validate the hypothesis of potential mixed-mode transmission initially raised by Byler et al. (2013) although they did not find differences in diversity between S. pistillata adults and larvae (Byler et al. 2013). Finally, although many of the OTUs unique to larvae or significantly differentially abundant between life stages were at background abundances, rare Symbiodinium have important functional roles in regulating host fitness (Quigley et al. 2017b; Ziegler et al. 2017b).
Evidence of mixed-mode transmission in S. hystrix (ShA) contradicts previous assumptions that maternally transmitted symbiont communities are transferred to offspring with high fidelity in corals (Douglas 1998; Baker 2003; Fabina et al. 2012). Our findings are consistent with transmission patterns documented in other symbiotic systems, such as wild Drosophilia hydei populations (Oliver et al. 2014), Acromyrmex ants (Scheuring and Yu 2012; Andersen et al. 2013), and paramecium (Fujishima and Fujita 1985; Kaltz et al. 2003), and aligns symbiotic transmission ecology in corals with terrestrial invertebrate symbioses. In addition, the novel diversity found in S. hystrix (ShA) larvae mirrors increased diversity of Symbiodinium communities detected in eggs of Montipora capitata and M. digitata compared to adults (Padilla-Gamiño et al. 2012; Quigley et al. 2017a). Similarly, novel, bacterial community diversity was detected in the larvae of the brooding coral Porites astreoides, and various bacterial communities associated with larvae of sponge species with supposed vertical transmission (Schmitt et al. 2008; Sharp et al. 2012).
Mounting evidence for mixed-mode transmission across phyla suggests that it may be evolutionarily advantageous to compromise between completely vertically and horizontally acquired symbiont communities, as both strategies provide distinct advantages and disadvantages (Baird et al. 2009; Byler et al. 2013). In S. hystrix (ShA), vertical transmission of Symbiodinium that are locally adapted to the parental environment is likely to provide benefits for a species that is able to self-fertilize (Sherman 2008; Warner et al. 2016) and has highly localized larval dispersal (e.g., Underwood et al. 2007; van Oppen et al. 2008; Noreen et al. 2009). However, a locally adapted community might become a liability if environmental conditions change or if larval dispersal distances are long. Negative effects include deregulation or disruption of symbiont abundances, which may have harmful physiological effects on the host (Xie et al. 2010; Oliver et al. 2014; Cunning et al. 2015). Thus, a mixed-mode strategy that results in superinfections of multiple symbionts can be beneficial (e.g., parasitoid protection in aphid hosts, Sandström et al. 2001; Oliver et al. 2014) and may provide more flexibility for adjusting to variable environmental conditions. Similarly, a mixed mating strategy of selfing and outcrossing in S. hystrix (ShA), combined with a functional, established symbiosis upon release, may facilitate both local and long-distance dispersal (Warner et al. 2016). Our findings confirm that diversity and flexibility of Symbiodinium transmission in a brooding coral are greater than previously thought, highlighting the potential for evolvability that may confer greater resilience over time than coral species with strict vertical transmission.
Additional to environmental uptake of Symbiodinium during early ontogeny and processes like competitive exclusion may contribute to differences between larval and adult communities in S. hystrix (ShA). Theory suggests that competition among symbionts may preclude transmission of an exact replica of the parental symbiont community because conditions promoting growth for some symbionts may differ between life stages (Moran et al. 2008). The novel symbiont diversity found in S. hystrix (ShA) larvae may provide benefits similar to those observed in insect symbioses, for example to provide larvae with the flexibility to host optimal symbiont types for the changing conditions through ontogeny (Abrego et al. 2009b; Byler et al. 2013). For example, Symbiodinium C1_OTU136, which was uniquely identified in larvae, may represent an adaptive advantage for this early life stage. Clade C-types are taxonomically and physiologically diverse (LaJeunesse 2005; Thornhill et al. 2014), and exhibit a range of tolerances for light and temperature, which are also reflected in their in hospite distributions across individual adult colonies and species (Sampayo et al. 2008). Larval settlement and early juvenile survival are generally highest in cryptic, low-light areas that offer protection from predation (Maida et al. 1994; Suzuki et al. 2013). Given that optimal settlement environments differ substantially from light environments experienced by adults, potentially by as much as 10-fold (Suzuki et al. 2013), it is possible that variation in Symbiodinium communities between larvae and adults observed here relates to different selective pressures associated with differing light environments (Rowan et al. 1997; Gómez-Cabrera et al. 2008; Kemp et al. 2008). Other potentially numerous, uncharacterized differences between larval and adult microhabitats may also contribute to differences in selective pressures between life stages. The potential ecological roles for the larval-specific OTUs recorded here are unknown. Indeed, it is possible that they represent non-symbiotic, free-living types (Lee et al. 2016) that may have attached to the exterior of the larvae following release or that may have entered brooded larvae without engaging in symbiosis. Further work is needed to determine how many of these OTUs represent physiologically important versus transient Symbiodinium.
Potential mechanisms shaping larval Symbiodinium communities
The immune system is an obvious mechanism by which the host could exert control over its symbiotic community by regulating the establishment of individual Symbiodinium types (Bay et al. 2011) or of either whole clades or functional units (i.e., clades or types with similar metabolic roles) (Ley et al. 2006). The symbioses of Wolbachia and Spiroplasma bacteria among Drosophila and lepidopteran genera, for example, are highly specific and exclude other bacterial lineages through a dynamic and mature immune response, to the extent that specific Drosophila species host novel and specific Wolbachia and/or Spiroplasma strains (Mateos et al. 2006; Russell et al. 2012). Mechanisms of immunity that could be transmitted through inheritance of parental genes include components of both the innate and adaptive immune response, including some that have been implicated in shaping invertebrate symbiont communities, such as T-cells, Nod2, defensins, and antimicrobial peptides (as reviewed in Franzenburg et al. 2013; Raina et al. 2016). These mechanisms have been documented during Symbiodinium establishment in corals (Wood-Charlson et al. 2006; Bay et al. 2011; Davy et al. 2012) and observed in the Hydra/bacteria symbiosis (Fraune et al. 2010; Franzenburg et al. 2013).
Conversely, the greater variation and diversity found in larval compared to adult Symbiodinium communities may be a function of an immature immune response that is not yet able to differentiate appropriate Symbiodinium types, rather than an adaptive response. As the coral immune system matures over time (Frank et al. 1997; Puill-Stephan et al. 2012), it is possible that a winnowing process eliminates symbionts that are not physiologically beneficial to the coral host (Abrego et al. 2009b; Byler et al. 2013). If true, then the ubiquitous presence of Symbiodinium C1_OTU136 in larvae may be a consequence of an opportunistic Symbiodinium type taking advantage of immature host immunity. Further work is needed to identify the role that the immune response has in shaping Symbiodinium communities; in particular what (if any) immune-related genes are being transmitted from parents to offspring and whether novel symbionts are a function of an under-developed immune response.
Winnowing and microhabitat variation may shape adult Symbiodinium communities
The disparate Symbiodinium communities in larvae versus adults found here further indicate that the re-shaping of the Symbiodinium community through ontogeny is an important developmental process in corals. Ontogenetic variability in microbial communities (both Symbiodinium and bacteria) is common in both vertically and horizontally transmitting cnidarian species (Coffroth et al. 2001; Nyholm and McFall-Ngai 2004; Abrego et al. 2009a, 2009b; Apprill et al. 2009; Littman et al. 2009; Sharp et al. 2012; Padilla-Gamiño et al. 2012; Poland et al. 2013; Byler et al. 2013; Lema et al. 2014; Poland and Coffroth 2017; Quigley et al. 2017b). The low level of variation in Symbiodinium communities associated with corals ranging in diameter from 8 cm to >30 cm (3–10 years Babcock 1991) suggests that the end of the winnowing process likely occurs earlier in the development of the brooding coral S. hystrix (ShA) (i.e., before 3 years) than in broadcast-spawning corals (~3.5 years; Abrego et al. 2009a, 2009b). Although evidence for switching of symbiont communities in adults corals exists (Boulotte et al. 2016), the pre-winnowing period may be the most flexible time for hosts to associate with a diversity of microbes. Microhabitat variation in the abundances of C120, D1, and D1a in adult corals at meter-level scales found in this study could be important for structuring in hospite Symbiodinium diversity, and may be partly responsible for the variability found at the level of individual larvae and broods. Therefore, identifying at what stage winnowing occurs in brooding corals and the influence of fine-scale environmental variables will provide crucial insights into when the flexibility to associate with environmentally acquired and potentially stress-tolerant types diminishes and specialization of the Symbiodinium community begins.
Conclusion
On the basis of novel heritability and paternity analyses, we show that Symbiodinium communities associated with the brooding coral S. hystrix (ShA) are only partially genetically regulated by their host and that larvae retain the flexibility to associate with novel symbionts across generations. Our results reveal a mixed-mode transmission strategy for establishing Symbiodinium communities in larvae of a brooding coral, based on demonstrations that a subset of novel and unique Symbiodinium types are found in brooded larvae but not in adults (although adults also had their own unique types). Importantly, this information aligns symbiosis transmission ecology in corals with well-known terrestrial invertebrate symbioses that typically exhibit mixed-mode transmission strategies. Advances in the understanding of heritable genetic mechanisms quantified here provide important insights into how Symbiodinium communities may be targeted for intervention strategies to increase reef resilience.
Data accessibility
DNA sequences: All sequencing data will be made available through NCBI SRA SRP082130.
Electronic supplementary material
Acknowledgements
This study was carried out with permission and in accordance with the recommendations from the Great Barrier Reef Marine Park Authority.
Author contributions
KMQ and BLW conceived of the experiment. KMQ and PAW designed the sampling scheme and performed the experiment, collected and analyzed the data. LKB provided reagents and materials. KMQ wrote and BLW, LKB, and PAW edited and critically reviewed the manuscript. All authors read and approved the final version.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0059-0) contains supplementary material, which is available to authorized users.
References
- Abrego D, van Oppen MJH, Willis BL. Highly infectious symbiont dominates initial uptake in coral juveniles. Mol Ecol. 2009;18:3518–3531. doi: 10.1111/j.1365-294X.2009.04275.x. [DOI] [PubMed] [Google Scholar]
- Abrego D, van Oppen MJH, Willis BL. Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny. Mol Ecol. 2009;18:3532–3543. doi: 10.1111/j.1365-294X.2009.04276.x. [DOI] [PubMed] [Google Scholar]
- Andersen SB, Hansen LH, Sapountzis P, Sørensen SJ, Boomsma JJ. Specificity and stability of the Acromyrmex–Pseudonocardia symbiosis. Mol Ecol. 2013;22:4307–4321. doi: 10.1111/mec.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill A, Marlow HQ, Martindale MQ, Rappé MS. The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 2009;3:685–699. doi: 10.1038/ismej.2009.3. [DOI] [PubMed] [Google Scholar]
- Arif C, Daniels C, Bayer T, Banguera Hinestroza E, Barbrook A, Howe CJ, et al. Assessing Symbiodinium diversity in scleractinian corals via next generation sequencing based genotyping of the ITS2 rDNA region. Mol Ecol. 2014;23:4418–4433. doi: 10.1111/mec.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre DJ, Resing JM. Sexual and asexual production of planulae in reef corals. Mar Biol. 1986;90:187–190. [Google Scholar]
- Babcock RC. Comparative demography of three species of scleractinian corals using age and size dependent classifications. Ecol Monogr. 1991;61:225–244. [Google Scholar]
- Baddeley A, Turner R. Spatstat: an R package for analyzing spatial point patterns. J Stat Softw. 2005;12:1–42. [Google Scholar]
- Baird AH, Guest JR, Willis BL. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu Rev Ecol Evol Syst. 2009;40:551–571. [Google Scholar]
- Bah T (2009) Inkscape: Guide to a Vector Drawing Program (Digital Short Cut). Pearson Education.
- Baker AC. Flexibility and specificity in coral–algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst. 2003;34:661–689. [Google Scholar]
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Baums IB, Devlin-Durante MK, Polato NR, Xu D, Giri S, Altman NS, et al. Genotypic variation influences reproductive success and thermal stress tolerance in the reef building coral, Acropora palmata. Coral Reefs. 2013;32:703–717. [Google Scholar]
- Bay LK, Cumbo VR, Abrego D, Kool JT, Ainsworth TD, Willis BL. Infection dynamics vary between Symbiodinium types and cell surface treatments during establishment of endosymbiosis with coral larvae. Diversity. 2011;3:356–374. [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans R, van Oppen MJH. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc R Soc B Biol Sci. 2006;273:2305–2312. doi: 10.1098/rspb.2006.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulotte NM, Dalton SJ, Carroll AG, Harrison PL, Putnam HM, Peplow LM et al. (2016) Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J 10:2693–2701 [DOI] [PMC free article] [PubMed]
- Brucker RM, Bordenstein SR. Speciation by symbiosis. Trends Ecol Evol. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Byler KA, Carmi-Veal M, Fine M, Goulet TL. Multiple symbiont acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata. PLoS ONE. 2013;8:e59596. doi: 10.1371/journal.pone.0059596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6:2033–2044. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin N, van Oppen M, Willis B, Mieog J, Negri A. Juvenile corals can acquire more carbon from high-performance algal symbionts. Coral Reefs. 2009;28:405–414. [Google Scholar]
- Cho J. The heritable immune system. Nat Biotechnol. 2015;33:608–609. doi: 10.1038/nbt.3256. [DOI] [PubMed] [Google Scholar]
- Coffroth MA, Santos S, Goulet T. Early ontogenetic expression of specificity in a cnidarian–algal symbiosis. Mar Ecol Prog Ser. 2001;222:85–96. [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. Inter Complex Syst. 2006;1695:1–9. [Google Scholar]
- Cunning R, Gates RD, Edmunds PJ (2017) Using high-throughput sequencing of ITS2 to describe Symbiodinium metacommunities in St. John, US Virgin Islands. PeerJ 5:e3472 [DOI] [PMC free article] [PubMed]
- Cunning R, Silverstein RN, Baker AC. Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc R Soc B. 2015;282:20141725. doi: 10.1098/rspb.2014.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev. 2012;76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon GB, Davies SW, Aglyamova GV, Meyer E, Bay LK, Matz MV. Genomic determinants of coral heat tolerance across latitudes. Science. 2015;348:1460–1462. doi: 10.1126/science.1261224. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Mycetocyte symbiosis in insects. Biol Rev. 1989;64:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Host benefit and the evolution of specialization in symbiosis. Heredity. 1998;81:599–603. [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Fabina NS, Putnam HM, Franklin EC, Stat M, Gates RD. Transmission mode predicts specificity and interaction patterns in coral-Symbiodinium networks. PLoS ONE. 2012;7:e44970. doi: 10.1371/journal.pone.0044970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank U, Oren U, Loya Y, Rinkevich B. Alloimmune maturation in the coral Stylophora pistillata is achieved through three distinctive stages, 4 months post–metamorphosis. Proc R Soc Lond B Biol Sci. 1997;264:99–104. [Google Scholar]
- Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci USA. 2013;110:E3730–E3738. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima M, Fujita M. Infection and maintenance of Holospora obtusa, a macronucleus-specific bacterium of the ciliate Paramecium caudatum. J Cell Sci. 1985;76:179–187. doi: 10.1242/jcs.76.1.179. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. Q Rev Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Johnson SB, Vrijenhoek RC. Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl Environ Microbiol. 2007;73:2314–2323. doi: 10.1128/AEM.01986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cabrera MdelC, Ortiz JC, Loh WKW, Ward S, Hoegh-Guldberg O. Acquisition of symbiotic dinoflagellates (Symbiodinium) by juveniles of the coral Acropora longicyathus. Coral Reefs. 2008;27:219–226. [Google Scholar]
- Gonzalez A, Stombaugh J, Lauber CL, Fierer N, Knight R. SitePainter: a tool for exploring biogeographical patterns. Bioinformatics. 2012;28:436–438. doi: 10.1093/bioinformatics/btr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EA, Davies SW, Matz MV, Medina M. Quantifying cryptic Symbiodinium diversity within Orbicella faveolata and Orbicella franksi at the Flower Garden Banks, Gulf of Mexico. PeerJ. 2014;2:e386. doi: 10.7717/peerj.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw. 2010;33:1–22. [Google Scholar]
- Hume BCC, D’Angelo C, Smith EG, Stevens JR, Burt J, Wiedenmann J. Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci Rep. 2015;5:1–8. doi: 10.1038/srep08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Berkelmans R, van Oppen MJH, Mieog JC, Sinclair W. A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc R Soc B Biol Sci. 2008;275:1359–1365. doi: 10.1098/rspb.2008.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltz O, Koella JC, Poulin R. Host growth conditions regulate the plasticity of horizontal and vertical transmission in Holospora undulata, a bacterial parasite of the protozoan Paramecium caudatum. Evolution. 2003;57:1535–1542. doi: 10.1111/j.0014-3820.2003.tb00361.x. [DOI] [PubMed] [Google Scholar]
- Kemp DW, Fitt WK, Schmidt GW. A microsampling method for genotyping coral symbionts. Coral Reefs. 2008;27:289–293. [Google Scholar]
- Kenkel CD, Setta SP, Matz MV. Heritable differences in fitness-related traits among populations of the mustard hill coral, Porites astreoides. Heredity. 2015;115:509–516. doi: 10.1038/hdy.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel CD, Traylor MR, Wiedenmann J, Salih A, Matz MV (2011) Fluorescence of coral larvae predicts their settlement response to crustose coralline algae and reflects stress. Proc R Soc B Biol Sci 278:2691–2697 [DOI] [PMC free article] [PubMed]
- LaJeunesse TC. ‘Species’ radiations of symbiotic dinoflagellates in the atlantic and Indo-Pacific since the Miocene-pliocene transition. Mol Biol Evol. 2005;22:570–581. doi: 10.1093/molbev/msi042. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Lee SY, Gil-Agudelo DL, Knowlton N, Jeong HJ. Symbiodinium necroappetens sp. nov. (Dinophyceae): an opportunist ‘zooxanthella’ found in bleached and diseased tissues of Caribbean reef corals. Eur J Phycol. 2015;50:223–238. [Google Scholar]
- Leathwick J, Moilanen A, Francis M, Elith J, Taylor P, Julian K, Hastie T, Duffy C. Novel methods for the design and evaluation of marine protected areas in offshore waters. Conserv Lett. 2008;1:91–102. [Google Scholar]
- Lee MJ, Jeong HJ, Jang SH, Lee SY, Kang NS, Lee KH et al. (2016) Most low-abundance “background’‘ Symbiodinium spp. are transitory and have minimal functional significance for symbiotic corals. Microb Ecol 71:771–783 [DOI] [PubMed]
- Leinster T, Cobbold CA. Measuring diversity: the importance of species similarity. Ecology. 2012;93:477–489. doi: 10.1890/10-2402.1. [DOI] [PubMed] [Google Scholar]
- Lema KA, Bourne DG, Willis BL. Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol Ecol. 2014;23:4682–4695. doi: 10.1111/mec.12899. [DOI] [PubMed] [Google Scholar]
- Levin RA, Beltran VH, Hill R, Kjelleberg S, McDougald D, Steinberg PD et al. (2016) Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol Biol Evol 33:3032 [DOI] [PMC free article] [PubMed]
- Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:1. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Little AF, van Oppen MJH, Willis BL. Flexibility in algal endosymbioses shapes growth in reef corals. Science. 2004;304:1492–1494. doi: 10.1126/science.1095733. [DOI] [PubMed] [Google Scholar]
- Littman RA, Willis BL, Bourne DG. Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades. Mar Ecol Prog Ser. 2009;389:45–59. [Google Scholar]
- Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 2015;1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M, Anders S, Huber W. Differential analysis of count data–the DESeq2 package. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates, Inc., Sunderland, MA
- Maida M, Coll JC, Sammarco PW. Shedding new light on scleractinian coral recruitment. J Exp Mar Bio Ecol. 1994;180:189–202. [Google Scholar]
- Mateos M, Castrezana SJ, Nankivell BJ, Estes AM, Markow TA, Moran NA. Heritable endosymbionts of Drosophila. Genetics. 2006;174:363–376. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz MV, Treml EA, Aglyamova GV, van Oppen MJH, Bay LK (2017) Adaptive pathways of coral populations on the Great Barrier Reef. bioRxiv 114173 10.1101/114173
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Davies S, Wang S, Willis BL, Abrego D, Juenger TE, et al. Genetic variation in responses to a settlement cue and elevated temperature in the reef-building coral Acropora millepora. Mar Ecol Prog Ser. 2009;392:81–92. [Google Scholar]
- Moran NA, Dunbar HE. Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci USA. 2006;103:12803–12806. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote–animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Noreen AME, Harrison PL, van Oppen MJH. Genetic diversity and connectivity in a brooding reef coral at the limit of its distribution. Proc R Soc Lond B Biol Sci. 2009;276:3927–3935. doi: 10.1098/rspb.2009.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai M. The winnowing: establishing the squid–Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Smith AH, Russell JA. Defensive symbiosis in the real world–advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol. 2014;28:341–355. [Google Scholar]
- van Oppen MJH, Lutz A, De’ath G, Peplow L, Kininmonth S. Genetic traces of recent long-distance dispersal in a predominantly self-recruiting coral. PLoS ONE. 2008;3:e3401. doi: 10.1371/journal.pone.0003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Gamiño JL, Pochon X, Bird C, Concepcion GT, Gates RD. From parent to gamete: vertical transmission of Symbiodinium (Dinophyceae) ITS2 sequence assemblages in the reef building coral Montipora capitata. PLoS ONE. 2012;7:e38440. doi: 10.1371/journal.pone.0038440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pochon X, Pawlowski J, Zaninetti L, Rowan R. High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar Biol. 2001;139:1069–1078. [Google Scholar]
- Pochon X, Putnam H, Burki F, Gates R. Identifying and characterizing alternative molecular markers for the symbiotic and free-living dinoflagellate genus Symbiodinium. PLoS ONE. 2012;7:e29816. doi: 10.1371/journal.pone.0029816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland DM, Coffroth MA (2017) Trans-generational specificity within a cnidarian–algal symbiosis. Coral Reefs 36:1–11
- Poland DM, Mansfield JM, Hannes AR, Lewis CLF, Shearer TL, Connelly SJ, et al. Variation in Symbiodinium communities in juvenile Briareum asbestinum (Cnidaria: Octocorallia) over temporal and spatial scales. Mar Ecol Prog Ser. 2013;476:23–37. [Google Scholar]
- Puill-Stephan E, Willis BL, Abrego D, Raina JB, van Oppen MJH. Allorecognition maturation in the broadcast-spawning coral Acropora millepora. Coral Reefs. 2012;31:1019–1028. [Google Scholar]
- Quigley K, Davies S, Kenkel C, Willis B, Matz M, Bay L. Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS ONE. 2014;9:e94297. doi: 10.1371/journal.pone.0094297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley K, Willis B, Bay L. Maternal effects and Symbiodinium community composition drive differential patterns in juvenile survival in the coral Acropora tenuis. R Soc Open Sci. 2016;3:1–17. doi: 10.1098/rsos.160471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley K, Willis B, Bay L. Heritability of the Symbiodinium community in vertically and horizontally transmitting broadcast-spawning corals. Sci Rep. 2017;7:8219. doi: 10.1038/s41598-017-08179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley K, Bay L, Willis B (2017b) Temperature and water quality-related patterns in sediment-associated Symbiodinium communities impact symbiont uptake and fitness of juveniles in the genus acropora. Front Mar Sci 4:401
- R Core Team (2012) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
- Raina JB, Tapiolas D, Motti CA, Foret S, Seemann T, Tebben J, et al. Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ. 2016;4:e2275. doi: 10.7717/peerj.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JM, Bruns BU, Fitt WK, Schmidt GW. Enhanced photoprotection pathways in symbiotic dinoflagellates of shallow-water corals and other cnidarians. Proc Natl Acad Sci USA. 2008;105:13674–13678. doi: 10.1073/pnas.0805187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgeway G (2006) gbm: Generalized boosted regression models. R Packag version 1.3: 55
- Rowan R, Knowlton N, Baker A, Jara J. Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature. 1997;388:265–269. doi: 10.1038/40843. [DOI] [PubMed] [Google Scholar]
- Russell JA, Funaro CF, Giraldo YM, Goldman-Huertas B, Suh D, Kronauer DJC, et al. A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: broad molecular surveys and a systematic review. PLoS ONE. 2012;7:e51027. doi: 10.1371/journal.pone.0051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampayo EM, Dove S, Lajeunesse TC. Cohesive molecular genetic data delineate species diversity in the dinoflagellate genus Symbiodinium. Mol Ecol. 2009;18:500–519. doi: 10.1111/j.1365-294X.2008.04037.x. [DOI] [PubMed] [Google Scholar]
- Sampayo EM, Ridgway T, Bongaerts P, Hoegh-Guldberg O. Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc Natl Acad Sci USA. 2008;105:10444–10449. doi: 10.1073/pnas.0708049105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström JP, Russell JA, White JP, Moran NA. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol. 2001;10:217–228. doi: 10.1046/j.1365-294x.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- Scheuring I, Yu DW. How to assemble a beneficial microbiome in three easy steps. Ecol Lett. 2012;15:1300–1307. doi: 10.1111/j.1461-0248.2012.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloerke B, Crowley J, Cook D, Hofmann H, Wickham H, Briatte F et al. (2014) Ggally: extension to ggplot2. R package version 1, no.0: 28
- Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol. 2008;74:7694–7708. doi: 10.1128/AEM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Distel D, Paul VJ. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 2012;6:790–801. doi: 10.1038/ismej.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman CDH. Mating system variation in the hermaphroditic brooding coral, Seriatopora hystrix. Hered. 2008;100:296–303. doi: 10.1038/sj.hdy.6801076. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Yamashita H, Kai S, Hayashibara T, Suzuki K, Iehisa Y, et al. Early uptake of specific symbionts enhances the post-settlement survival of Acropora corals. Mar Ecol Prog Ser. 2013;494:149–158. [Google Scholar]
- Thomas L, Kendrick GA, Kennington WJ, Richards ZT, Stat M. Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Mol Ecol. 2014;23:3113–3126. doi: 10.1111/mec.12801. [DOI] [PubMed] [Google Scholar]
- Thornhill DJ, Lajeunesse TC, Santos SR. Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol Ecol. 2007;16:5326–5340. doi: 10.1111/j.1365-294X.2007.03576.x. [DOI] [PubMed] [Google Scholar]
- Thornhill DJ, Lewis AM, Wham DC, LaJeunesse TC. Host-specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evol. 2014;68:352–367. doi: 10.1111/evo.12270. [DOI] [PubMed] [Google Scholar]
- Tonk L, Bongaerts P, Sampayo EM, Hoegh-Guldberg O. SymbioGBR: a web-based database of Symbiodinium associated with cnidarian hosts on the Great Barrier Reef. BMC Ecol. 2013;13:7. doi: 10.1186/1472-6785-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JN, Smith LD, van Oppen MJH, Gilmour JP. Multiple scales of genetic connectivity in a brooding coral on isolated reefs following catastrophic bleaching. Mol Ecol. 2007;16:771–784. doi: 10.1111/j.1365-294X.2006.03187.x. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Warner PA, Oppen MJH, Willis BL. Unexpected cryptic species diversity in the widespread coral Seriatopora hystrix masks spatial‐genetic patterns of connectivity. Mol Ecol. 2015;24:2993–3008. doi: 10.1111/mec.13225. [DOI] [PubMed] [Google Scholar]
- Warner PA, Willis BL, van Oppen MJH. Sperm dispersal distances estimated by parentage analysis in a brooding scleractinian coral. Mol Ecol. 2016;25:1398–1415. doi: 10.1111/mec.13553. [DOI] [PubMed] [Google Scholar]
- Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer Science & Business Media, New York
- Wilson AJ, Reale D, Clements MN, Morrissey MM, Postma E, Walling CA, et al. An ecologist’s guide to the animal model. J Anim Ecol. 2010;79:13–26. doi: 10.1111/j.1365-2656.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- Wood-Charlson EM, Hollingsworth LL, Krupp DA, Weis VM. Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cell Microbiol. 2006;8:1985–1993. doi: 10.1111/j.1462-5822.2006.00765.x. [DOI] [PubMed] [Google Scholar]
- Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE. 2010;5:e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler M, Arif C, Burt JA, Dobretsov S, Roder C, LaJeunesse TC et al. (2017a) Biogeography and molecular diversity of coral symbionts in the genus Symbiodinium around the Arabian Peninsula. J Biogeogr 44: 674–686 [DOI] [PMC free article] [PubMed]
- Ziegler M, Eguíluz VM, Duarte CM, Voolstra CR. Rare symbionts may contribute to the resilience of coral–algal assemblages. ISME. 2017;12:161–172. doi: 10.1038/ismej.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis. 2001;13:129–134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: All sequencing data will be made available through NCBI SRA SRP082130.