Abstract
Introduction
An Elecsys® Amyloid β (Aβ [1–42]) immunoassay cutoff for classification of patients with Alzheimer's disease was investigated.
Methods
Cerebrospinal fluid samples collected from patients with mild-to-moderate Alzheimer's disease were analyzed by Elecsys® immunoassays: (1) Aβ (1–42), (2) total tau, and (3) phosphorylated tau. Cutoffs (Aβ [1–42] and ratios with tau) were estimated by method comparison between AlzBio3 (n = 206), mixture modeling (n = 216), and concordance with florbetapir F 18 imaging-based classification (n = 75).
Results
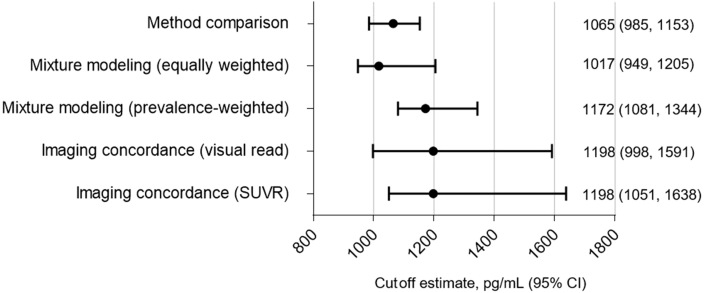
A 1065-pg/mL (95% confidence interval: 985–1153) Elecsys® Aβ (1–42) cutoff provided 94% overall percentage agreement with AlzBio3. Comparable cutoff estimates (95% confidence interval) were derived from mixture modeling (equally weighted: 1017 [949–1205] pg/mL; prevalence weighted: 1172 [1081–1344] pg/mL) and concordance with florbetapir F 18 imaging (visual read: 1198 [998–1591] pg/mL; automated: 1198 [1051–1638] pg/mL).
Discussion
Based on three approaches, a 1100-pg/mL Elecsys® Aβ (1–42) cutoff is suitable for clinical trials with similar populations and preanalytical handling.
Keywords: Alzheimer's disease, Biomarkers, Immunoassay, Amyloid β, Cerebrospinal fluid, Method comparison, Florbetapir F 18 imaging, Cutoff determination, Patient selection
Highlights
-
•
Biomarkers can facilitate appropriate patient recruitment into clinical trials.
-
•
Amyloid beta measurement can aid the identification of amyloid-positive patients.
-
•
Similar cutoff estimates were derived by three different approaches.
1. Background
Alzheimer's disease (AD) is becoming increasingly prevalent, in part due to the aging population [1]. In the United States, the number of death certificates stating AD as the cause of death increased by 89% between 2000 and 2014, and the number of individuals (aged ≥65 years) with AD is expected to increase from 5.3 million in 2017 to 13.8 million in 2050 [1], [2]. At present, the diagnosis of AD is based principally on an assessment of clinical symptoms, with tests and imaging techniques used to rule out other possible causes of symptoms. Histopathological hallmarks of AD include the deposition of extracellular amyloid plaques (primarily composed of amyloid β [Aβ] peptides), tau protein neurofibrillary tangles, and subsequent neuronal degeneration [3]. Therefore, establishing biomarkers that can accurately identify amyloid and tau pathology could be beneficial in aiding the earlier diagnosis of AD compared with the use of clinical symptoms alone. The use of biomarkers may also serve to identify individuals who could experience a greater benefit from disease-modifying AD therapies [4], [5]; for example, therapies that aim to reduce Aβ and phosphorylated tau levels in the brain may alleviate cognitive dysfunction and prevent further synaptic loss, axon degeneration, and neuronal cell death [6].
In the recent EXPEDITION (NCT00905372) and EXPEDITION2 (NCT00904683) studies, 26% of patients with clinically diagnosed mild-to-moderate AD lacked evidence of amyloid pathology established by positron emission tomography (PET) [7], [8]. A subsequent trial, EXPEDITION3 (NCT01900665), was enriched for patients with objective, biomarker-based evidence of amyloid pathology, assessed by amyloid-PET imaging or cerebrospinal fluid (CSF) Aβ (1–42) analysis using the INNO-BIA AlzBio3 Aβ (1–42) immunoassay (cutoff value of 249 pg/mL) [9]. Compared with the AlzBio3 Aβ (1–42) immunoassay, the fully automated Elecsys® Aβ (1–42) immunoassay has an improved analytical performance and is the first assay to be standardized to a candidate reference measurement procedure [10], [11]. Therefore, the Elecsys® Aβ (1–42) assay may improve the precision of Aβ (1–42) quantification in CSF compared with the AlzBio3 Aβ (1–42) assay.
This exploratory study aimed to establish a cutoff estimate for the Elecsys® Aβ (1–42) immunoassay (Roche Diagnostics) for future clinical trials with similar population and preanalytical procedures, based on the analysis of CSF samples from the EXPEDITION and EXPEDITION2 studies. Cutoff estimates for the ratios between measurements derived with the Elecsys® Total-Tau (tTau) and Elecsys® Aβ (1–42) immunoassays and between the Elecsys® Phospho-Tau (181P) (pTau) and Elecsys® Aβ (1–42) immunoassays were also evaluated.
2. Methods
2.1. Study design
EXPEDITION and EXPEDITION2 were phase 3, double-blind, placebo-controlled international trials of solanezumab in patients aged 55–94 years with mild-to-moderate AD [7], [8]. In both the trials, mild AD was defined as a Mini-Mental Status Examination (MMSE) score of 20–26, and moderate AD was defined as a MMSE score of 16–19. Patients were required to provide written informed consent for the collection of CSF samples and data, and the studies were approved by the relevant institutional ethics committees and conducted in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki).
In the present analysis, frozen archived aliquots of CSF samples from a subset of patients in the EXPEDITION and EXPEDITION2 studies, with mild or moderate AD and for whom AlzBio3 Aβ (1–42) data were available, were analyzed using the Elecsys® Aβ (1–42), pTau, and tTau immunoassays on a cobas e601 analyzer. All measurements were performed at the University of Pennsylvania [9]. Three different statistical approaches (outlined in the following section) were used to estimate a cutoff for Elecsys® Aβ (1–42), the Elecsys® tTau/Aβ (1–42) ratio, and the Elecsys® pTau/Aβ (1–42) ratio: (1) method comparison versus AlzBio3, mixture modeling, and concordance with florbetapir F 18 imaging.
2.2. Assays
The Elecsys® Aβ (1–42), Elecsys® tTau, and Elecsys® pTau CSF immunoassays are currently under development and are for investigational use only. The Elecsys® Aβ (1–42) assay has a measuring range of 200–1700 pg/mL. Values above the upper limit of the measuring range are provided based on an extrapolation of the calibration curve; they are restricted to exploratory research use only and are excluded from clinical decision-making and the derivation of medical decision points (as performance of the assay outside the measuring range has not been formally validated). The Elecsys® tTau and Elecsys® pTau immunoassays have a measuring range of 80–1300 pg/mL and 8–120 pg/mL, respectively. Ratios between the tTau or pTau assays and the Aβ (1–42) assay were calculated from single-marker concentrations. Elecsys® assays were run on a fully automated cobas e analyzer (Roche Diagnostics).
The AlzBio3 Aβ (1–42) assay (Fujirebio; research use only) has a measuring range of 16–1600 pg/mL for the measurement of Aβ (1–42) and was run on a Luminex 200 xMAP platform (Luminex Corp).
2.3. Imaging
A subset of patients enrolled in the EXPEDITION and EXPEDITION2 studies underwent a florbetapir F 18 PET scan to determine amyloid burden at baseline. A PET scan was performed with data acquired for 15 minutes, beginning 50 minutes after intravenous administration of approximately 370-MBq florbetapir F 18. Images were reconstructed via an iterative algorithm with a 5-mm full-width half-maximum Gaussian filter in a 128 × 128 matrix. A central reader visually interpreted each image as amyloid positive (likely associated with moderate-to-frequent neuritic plaques) or amyloid negative (likely associated with zero-to-sparse neuritic plaques), in accordance with previous published methods and the manufacturers' label instructions [12], [13]. For quantitative analysis, images were fitted to a PET template, and a composite standard uptake value ratio (SUVR) was also derived from the average estimate of florbetapir F 18 retention in six cortical volumes of interest (frontal, temporal and parietal cortex, anterior and posterior cingulate, and precuneus) relative to a whole cerebellum reference region [14]. An SUVR cutoff of 1.1 was used, whereby patients with SUVR ≥ 1.1 were classified as PET positive. This cutoff estimate was initially proposed based on the upper confidence limits of a group of clinically normal middle-aged subjects [15] and subsequently demonstrated to be able to distinguish autopsy-confirmed amyloid-positive subjects (moderate-to-frequent neuritic plaque) from amyloid-negative subjects with high sensitivity and specificity [12], [14]. All quantitative image analyses and visual interpretation were performed blind to clinical data and treatment assignment.
2.4. Statistical analysis
Method comparison is a commonly used approach for bridging cutoffs between two nonstandardized biomarkers to guarantee backward comparability. A method comparison was performed between the INNO-BIA AlzBio3 assay β-Amyloid (1–42) (Fujirebio) and the Elecsys® Aβ (1–42) immunoassay, Elecsys® tTau/Aβ (1–42), and Elecsys® pTau/Aβ (1–42) ratios. Method comparison was performed using a weighted Deming (Aβ [1–42]) or Deming (tTau/Aβ [1–42] and pTau/Aβ [1–42]) regression after log transformation of both AlzBio3 and Elecsys biomarkers (to achieve linearity) and after removal of Elecsys® Aβ (1–42) values above the upper limit of the measuring range. A Spearman's correlation coefficient was also calculated. Using the regression, a cutoff estimation corresponding to the previously validated AlzBio3 Aβ (1–42) cutoff of 249 pg/mL was determined for the Elecsys® Aβ (1–42) assay. Confidence intervals (CIs; 95%) for the Elecsys® cutoff estimate were calculated by jackknife estimation [16]. Agreement measures were obtained between AlzBio3 (using cutoff 249 pg/mL) and Elecsys®-based classification (using cutoff transferred via the regression line). Overall percentage agreement (OPA) was calculated as the proportion of subjects with the same classification according to both assays; negative percentage agreements (“specificity”), positive percentage agreements (“sensitivity”), negative predictive values, and positive predictive values were also calculated.
Mixture modeling was used to obtain two cutoff estimates each for the Elecsys® Aβ (1–42) immunoassay and the Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42) ratios after log transformation; one estimate was based on equally weighted densities of both the groups (see the study by De Meyer et al. [17]), and the second was based on prevalence-weighted densities. The two estimates vary with respect to their dependence on prevalence within the data set (in this case, the greater proportion of “positive” patients); the former approach based on equally weighted densities is less dependent on prevalence and thus less specific for the population of interest. The distribution of Aβ (1–42) concentrations was expected to follow a mixture of two normal distributions with means μG1 and μG2 and standard deviations σG1 and σG2, respectively, where G1 refers to the group with lower Aβ (1–42) concentrations, “AD like,” and G2 refers to the group with higher Aβ (1–42) concentrations, “non-AD like”. Parameters of the two normal distributions dG1 and dG2 were estimated using an expectation-maximization algorithm, which maximizes the expected complete-data log likelihood. Biomarker values above the measuring range estimated from the extrapolated calibration curve were used in these analyses.
Single biomarker data (Elecsys® Aβ [1–42] immunoassay and AlzBio3 Aβ [1–42] immunoassay) and combination biomarker data (Elecsys® tTau/Aβ [1–42] and Elecsys® pTau/Aβ [1–42] ratios) were assessed for concordance with the florbetapir F 18 imaging classifications according to visual read and SUVR outcomes. Area under the receiver operating characteristic (ROC) curve analysis was determined, and cutoffs were chosen as the value maximizing the Youden Index for concordance with outcome amyloid PET. Values outside the measuring range in this analysis were handled as follows. For single-marker analyses, values outside the measuring range were set to the respective assay's limit of the measuring range and included in the analysis; for biomarker ratios pTau/Aβ (1–42) and tTau/Aβ (1–42), Aβ (1–42), pTau, and tTau concentrations outside the measuring range were set to the respective limit. As an exception, the handling of Aβ (1–42) concentrations above the upper limit of the measuring range was included in rule defining CSF biomarker status: (1) if tau/Aβ (1–42) ratio was greater than the cutoff value and Aβ (1–42) ≤ 1700 pg/mL, CSF biomarker status was defined as positive and (2) if tau/Aβ (1–42) ratio was less than or equal to the cutoff or Aβ (1–42) > 1700 pg/mL, CSF biomarker status was defined as negative. It has been shown previously that this way of handling Aβ (1–42) values above the measuring range yields almost identical results for concordance with PET imaging as an analysis using Aβ (1–42) values estimated from the extrapolated calibration curve [18].
3. Results
3.1. Baseline characteristics
Baseline characteristics of patients who provided the CSF samples are outlined in Table 1. In the overall cohort, mean (standard deviation) age was 71.9 (7.9) years, 102 of 217 (47%) patients were male, and proportions of patients with no, one, or two apolipoproteins E (APOE) 4 alleles were 40.2%, 39.2%, and 20.6%, respectively. The mean (standard deviation) MMSE total score was 21.54 (3.64) in the overall cohort, 22.89 (2.88) in the cohort of 161 patients with mild AD, and 17.65 (2.72) in the cohort of 54 patients with moderate AD. Using the Elecsys® Aβ (1–42) assay (cutoff, 1065 pg/mL), 180 of 206 (87.4%) samples were determined to be amyloid positive and 26 (12.6%) amyloid negative. Using the AlzBio3 assay (cutoff, 249 pg/mL), 177 (85.9%) samples were determined to be amyloid positive and 29 (14.1%) amyloid negative.
Table 1.
Baseline characteristics of patients who provided CSF samples
| Variable | Mild AD (n = 161) | Moderate AD (n = 54) | All (N = 217)∗ |
|---|---|---|---|
| Mean age (SD), years | 71.76 (7.49) | 72.41 (9.19) | 71.91 (7.93) |
| Sex, n (%) | |||
| Female | 83 (51.6) | 31 (57.4) | 115 (53.0) |
| Male | 78 (48.4) | 23 (42.6) | 102 (47.0) |
| APOE genotype, n (%) | |||
| N | 151 | 51 | 204 |
| E2/E3 | 4 (2.6) | 2 (3.9) | 6 (2.9) |
| E2/E4 | 5 (3.3) | 0 (0) | 5 (2.5) |
| E3/E3 | 56 (37.1) | 20 (39.2) | 76 (37.3) |
| E3/E4 | 54 (35.8) | 20 (39.2) | 75 (36.8) |
| E4/E4 | 32 (21.2) | 9 (17.6) | 42 (20.6) |
| APOE genotype grouped, n (%) | |||
| N | 151 | 51 | 204 |
| 0 E4 | 60 (39.7) | 22 (43.1) | 82 (40.2) |
| 1 E4 | 59 (39.1) | 20 (39.2) | 80 (39.2) |
| 2 E4 | 32 (21.2) | 9 (17.6) | 42 (20.6) |
| MMSE total score, mean (SD) | 22.89 (2.88) | 17.65 (2.72) | 21.54 (3.64) |
| ADAS, mean (SD) | 28.87 (8.41) | 41.50 (10.66) | 32.15 (10.56) |
| CDR-SB, mean (SD) | 4.23 (2.16) | 5.71 (2.16) | 4.62 (2.26) |
| IADL, mean (SD) | 44.14 (10.27) | 36.61 (9.76) | 42.17 (10.64) |
| SUVR | |||
| n | 55 | 20 | 77 |
| Mean (SD) | 1.34 (0.28) | 1.47 (0.23) | 1.37 (0.27) |
Abbreviations: AD, Alzheimer's disease; ADAS, Alzheimer's Disease Assessment Scale; APOE, apolipoproteins E; CDR-SB, Clinical Dementia Rating sum of boxes; CSF, cerebrospinal fluid; IADL, Instrumental Activities of Daily Living; MMSE, Mini-Mental State Examination; SD, standard deviation; SUVR, standard uptake value ratio.
n = 2 missing AD status.
3.2. Elecsys® Aβ (1–42) cutoff estimation
Each approach of Elecsys® Aβ (1–42) cutoff estimation produced similar numerical values, with considerable overlap in the 95% CI range (Fig. 1).
Fig. 1.
Summary of Elecsys® amyloid β (1–42) cutoff estimates with 95% CIs, as derived by the different approaches. Abbreviations: CI, confidence interval; SUVR, standard uptake value ratio.
3.2.1. Method comparison
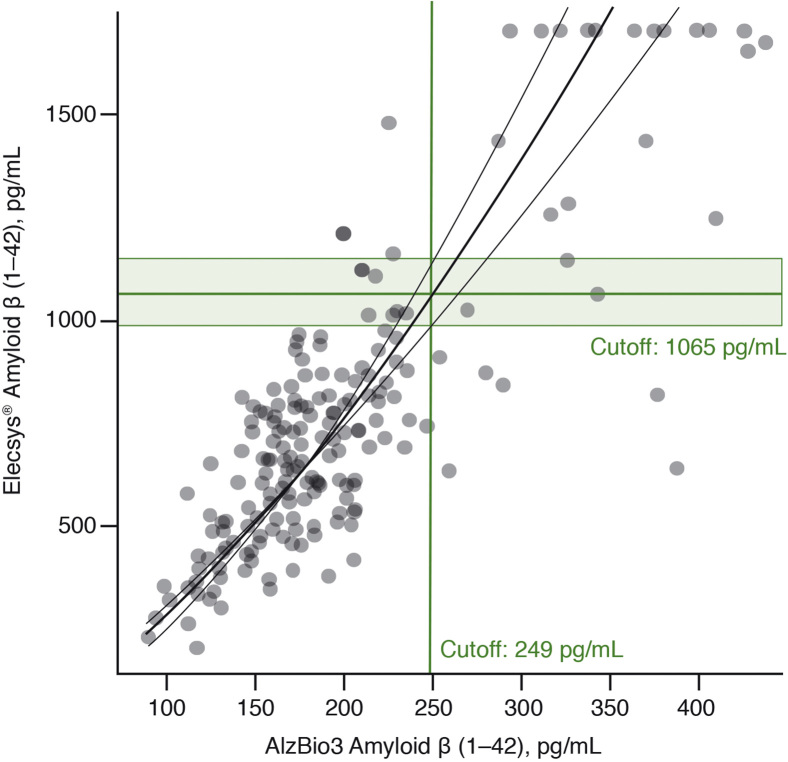
A cutoff estimate of 1065 pg/mL (95% CI: 985–1153; n = 206) was derived for the Elecsys® Aβ (1–42) immunoassay, based on the transfer of the AlzBio3 Aβ (1–42) cutoff (249 pg/mL) via the regression curve (Fig. 2). The Elecsys® Aβ (1–42) immunoassay showed high-percentage agreements with AlzBio3 Aβ (1–42) assay (Table 2), with an OPA of 94% (95% CI: 90–97).
Fig. 2.
Method comparison of the Elecsys® amyloid β (1–42) with AlzBio3 amyloid β (1–42) using weighted Deming regression. The curve represents a linear fit on the log-transformed values of both assays.
Table 2.
Agreements between amyloid β (1–42) concentrations from different assays and between amyloid β (1–42) concentrations and florbetapir F 18 imaging outcomes
| Comparator | Assay | OPA | PPA | NPA | PPV | NPV |
|---|---|---|---|---|---|---|
| AlzBio3 amyloid β (1–42) (n = 206) | Elecsys® amyloid β (1–42) | 94 (90–97) | 97 (94–99) | 72 (53–87) | 96 (91–98) | 81 (61–93) |
| Florbetapir F 18 imaging concordance: visual read (n = 75) | AlzBio3 amyloid β (1–42), % (95% CI) | 97 (91, 100) | 98 (91, 100) | 93 (66, 100) | 98 (91, 100) | 93 (66, 100) |
| Elecsys® amyloid β (1–42), % (95% CI) | 96 (89, 99) | 98 (91, 100) | 86 (57, 98) | 97 (89, 100) | 92 (64, 100) | |
| Florbetapir F 18 imaging concordance: SUVR (n = 75) | AlzBio3 amyloid β (1–42), % (95% CI) | 95 (87, 99) | 98 (91, 100) | 81 (54, 96) | 95 (87, 99) | 93 (66, 100) |
| Elecsys® amyloid β (1–42), % (95% CI) | 96 (89, 99) | 100 (94, 100) | 81 (54, 96) | 95 (87, 99) | 100 (75, 100) |
Abbreviations: CI, confidence interval; NPA, negative percentage agreement; NPV, negative predictive value; OPA, overall percentage agreement; PPA, positive percentage agreement; PPV, positive predictive value; SUVR, standard uptake value ratio.
3.2.2. Mixture modeling
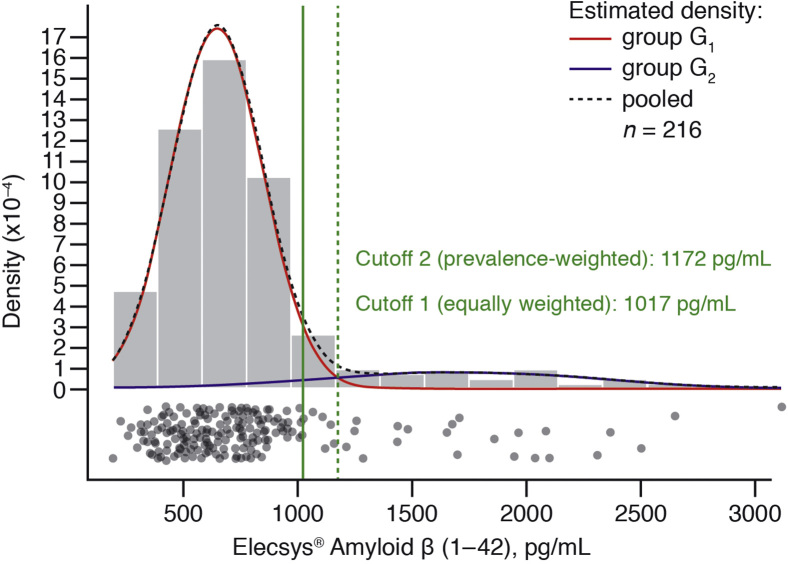
Based on equally weighted densities, a cutoff estimate of 1017 pg/mL (95% CI: 949–1205; n = 216) was derived (Fig. 3). When the densities were prevalence weighted, the cutoff estimate was 1172 pg/mL (95% CI: 1081–1344; n = 216; Fig. 3).
Fig. 3.
Mixture modeling of Elecsys® amyloid β (1–42). The estimated probability densities (solid lines) and the joint density (dashed line) are plotted; group G1 (“AD-like” group); group G2 (“non–AD-like” group). Cutoffs are estimated as the crossing point of the equally weighted densities (cutoff 1, solid line) and the prevalence-weighted densities (cutoff 2, dashed line). Abbreviation: AD, Alzheimer's disease.
3.2.3. Concordance with florbetapir F 18 imaging
The Elecsys® Aβ (1–42) immunoassay demonstrated good concordance with florbetapir F 18 imaging. Scatter plots of SUVR versus Elecsys® Aβ (1–42) are presented in Supplementary Fig. 1A. The cutoff estimate optimized relative to florbetapir F 18 visual interpretation was 1198 pg/mL (95% CI: 998–1591; n = 75), and the area under the ROC curve was 93% (95% CI: 81–100; Supplementary Fig. 2A). OPA was 96% (95% CI: 89–99) for the Elecsys® Aβ (1–42) immunoassay, highly similar to the OPA of AlzBio3 with florbetapir F 18 (OPA, 97% [95% CI: 91–100]), based on the optimized cutoff 254 pg/mL (95% CI: 213–275). Using SUVR, the cutoff estimate was 1198 pg/mL (95% CI: 1051–1638; n = 75), and the area under the ROC curve was 93% (83%–100%; Supplementary Fig. 2B). OPA was 96% (95% CI: 89–99) for the Elecsys® Aβ (1–42) immunoassay, highly similar to OPA of AlzBio3 with florbetapir F 18 (OPA, 95% [95% CI: 87–99]), based on the optimized cutoff 227 pg/mL (95% CI: 160–257). Percentage agreements and predictive values are presented in Table 2.
3.3. Analysis of marker combinations
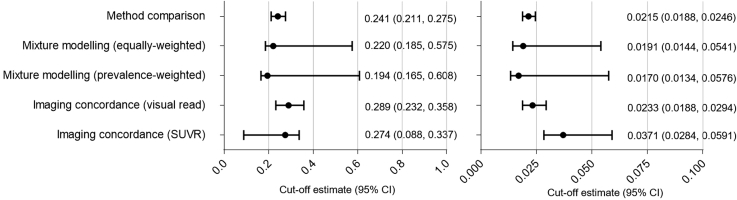
When the analyses were repeated for the biomarker ratios Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42), relatively similar cutoff estimates were derived by each approach, with considerable overlap in the 95% CI (Fig. 4).
Fig. 4.
Summary of cutoff estimates with 95% CIs for the Elecsys® tTau/amyloid β (1–42) ratio (left panel) and the Elecsys® pTau/amyloid β (1–42) ratio (right panel), as derived by the different approaches. Abbreviations: CI, confidence interval; SUVR, standard uptake value ratio.
3.3.1. Method comparison
The cutoff estimates for the Elecsys® tTau/Aβ (1–42) ratio and Elecsys® pTau/Aβ (1–42) ratio were 0.241 (95% CI: 0.211–0.275; n = 214) and 0.0215 (95% CI: 0.0188–0.0246; n = 211), respectively (Supplementary Fig. 3). Classification based on the Elecsys® tTau/Aβ ratio and the Elecsys® pTau/Aβ ratio showed good concordance with that based on AlzBio3 Aβ (1–42) (Supplementary Table 1). The OPA of the Elecsys® tTau/Aβ (1–42) ratio with AlzBio3 was 94% (95% CI: 89–97), and the OPA of Elecsys® pTau/Aβ (1–42) ratio with AlzBio3 was 96% (95% CI: 92–98).
3.3.2. Mixture modeling
The cutoff estimates for the Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42) ratios derived using equally weighted densities were 0.220 (95% CI: 0.185–0.575; n = 215) and 0.0191 (95% CI: 0.0144–0.0541; n = 212), respectively (Supplementary Fig. 4). The cutoff estimates for the Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42) ratios derived using prevalence-weighted densities were 0.194 (95% CI: 0.165–0.608; n = 215) and 0.0170 (95% CI: 0.0132–0.0576; n = 212), respectively (Supplementary Fig. 4).
3.3.3. Concordance with florbetapir 18 F PET imaging
Scatter plots of Elecsys® tTau and pTau against Aβ (1–42) are presented in Supplementary Fig. 5.
Classification based on the Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42) ratios demonstrated good concordance with classification based on florbetapir imaging (visual read and SUVR).
Cutoff estimates for the Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42) ratios optimized relative to florbetapir F 18 visual interpretation were 0.289 (95% CI: 0.232–0.358; n = 68) and 0.0233 (95% CI: 0.0188–0.0294; n = 68), respectively; corresponding areas under the ROC curves were 100% (95% CI: 99–100) and 100% (95% CI: 100–100; Supplementary Fig. 6A and C). The OPA of both ratios versus the visual read was 97% (95% CI: 91–100). Percentage agreements and predictive values are presented in Supplementary Table 1.
Using SUVR-based classification (with cutoff 1.1), the cutoff estimates for the Elecsys® tTau/Aβ (1–42) and Elecsys® pTau/Aβ (1–42) ratios were 0.274 (95% CI: 0.0875–0.337; n = 68) and 0.0371 (95% CI: 0.0284–0.0591; n = 68), respectively; corresponding areas under the ROC curves were 98% (95% CI: 96–100) and 99% (95% CI: 97–100; Supplementary Fig. 6B and D). The OPA of the Elecsys® tTau/Aβ (1–42) ratio versus SUVR was 96% (95% CI: 89–99) and OPA of Elecsys® pTau/Aβ (1–42) ratio versus SUVR was 92% (95% CI: 83–97). Percentage agreements and predictive values are presented in Supplementary Table 1. Scatter plots of SUVR versus Elecsys® tTau/Aβ (1–42) ratio and Elecsys® tTau/Aβ (1–42) ratio are presented in Supplementary Fig. 1B and C.
4. Discussion
The Elecsys® Aβ (1–42) immunoassay has previously demonstrated good analytical performance [10] and may provide advantages (e.g., increased precision, linearity over the desired measuring range, no high-dose hook effect) compared with currently available commercial and “research-use-only” assays [10], [11]. Importantly, the Elecsys® Aβ (1–42) immunoassay was the first CSF Aβ (1–42) immunoassay to be standardized to a Joint Committee for Traceability in Laboratory Medicine–approved reference measurement procedure [10]. Similarly, the Elecsys® tTau and pTau assays provide quantification of total tau and phosphorylated tau, which may serve as later biomarkers of disease progression [4], [18], [19]. The new evidence provided herein confirms that the Elecsys® Aβ (1–42), Elecsys® tTau, and Elecsys® pTau immunoassays are suitable for the diagnostic quantification of their respective analytes in CSF.
In the present analysis, the Elecsys® Aβ (1–42) immunoassay–based patient classification showed good concordance with the classification based on CSF Aβ (1–42) determined by the AlzBio3 immunoassay and based on florbetapir F 18 imaging and was confirmed by mixture modeling. Each statistical approach used to derive cutoff estimates for the Elecsys® Aβ (1–42) immunoassay produced similar numerical values with considerable overlap in the 95% CIs. Cutoff estimates for the Elecsys® tTau/Aβ (1–42) and the Elecsys® pTau/Aβ (1–42) ratios were also comparable across the three statistical approaches. Differences in the cutoff estimates derived using each of the applied approaches are most likely due to inherent differences of the three statistical methods and variation in the size of the data sets available for use within each approach.
Based on the multiple cutoff estimates for the Elecsys® Aβ (1–42) assay derived herein and the comparable cutoffs derived from the BioFINDER study [20], an Elecsys® Aβ (1–42) cutoff point of 1100 pg/mL is proposed; this cutoff point was contained within the 95% CIs arising from all three approaches of estimation evaluated in the present study. This cutoff is higher than the value of 1000 pg/mL for the CE-marked Elecsys® Aβ (1–42) immunoassay, which was established for use with a different preanalytical protocol. Notably, data used to determine the cutoff of 1100 pg/mL in the present study were derived using samples obtained after highly standardized preanalytical procedures for CSF specimen collection and handling specific to the Eli Lilly and Company–sponsored EXPEDITION and EXPEDITION2 trials. Thus, this Elecsys® Aβ (1–42) immunoassay cutoff point of 1100 pg/mL may be specific for use in clinical trials that use identical preanalytic procedures. Preanalytic conditions have been shown to affect the consistency of CSF biomarker measurements, including variables such as plastic tube type, the number of tube transfers, and the number of freeze-thaw cycles [21], [22]. Use of other CSF collection and handling procedures may necessitate the determination of alternate cutoffs. Where this proves necessary, we recommend determination of cutoff estimates by multiple statistical approaches, as reported herein. We believe this approach provides greater confidence in cutoff values ultimately adopted and is especially valuable when sponsors are compelled to select study-specific cutoffs based on limited data, as was the case in this study.
Previously, it was likely that participants with clinical AD but without AD pathology were enrolled in trials, potentially contributing to the failure of such trials to meet their primary endpoints [23]. Moreover, as highlighted by the third renewal of the AD Neuroimaging Initiative, the ability to enroll patients with earlier predementia or even at presymptomatic stages could facilitate the development of disease-modifying agents [24]. Currently, the only FDA-approved method for assessing the presence of amyloid plaques in the brain is amyloid-PET imaging, which has a number of disadvantages including high cost and resource use [25]. The use of the biomarkers investigated in this study could potentially aid the diagnosis of AD and enable intervention at an earlier stage of disease. The use of the ratio of Aβ (1–42) with tTau or pTau as an AD biomarker may also improve selection of patients into AD clinical trials, particularly in earlier disease stages (Hansson O et al. Alz & Dement 2017 [in revision/submission]).
Patient inclusion and exclusion in the EXPEDITION and EXPEDITION2 trials were determined by clinical characteristics (e.g. MMSE score) rather than by biomarkers. An interesting analysis would be to compare biomarker results in patients who were included (randomized) versus those who were excluded. Unfortunately, few if any excluded patients underwent biomarker testing, obviating this type of analysis.
A limitation of the study is that the evaluation of the benefit of the single biomarker and combined biomarker ratios in terms of PET concordance was constrained by the small sample size. In addition to the limited number of patients in the “non–AD-like” distribution (with higher Aβ [1–42] concentrations) in the EXPEDITION and EXPEDITION2 population (mild-to-moderate AD), these patients were most likely clinically misdiagnosed, explaining the presence of biomarker-negative patients in the clinically mild-to-moderate AD population [24]. The small sample size of biomarker-negative patients reduces certainty around the cutoff estimates for the two densities in the mixture modeling analysis. Although the classifications of biomarker positive and biomarker negative have been used, in reality, all biomarker measurements are continuous variables. Even with regard to the amyloid-PET SUVR cutoff of 1.1, ongoing discussions suggest that this value may be too high for the study of persons with preclinical (i.e., presymptomatic) AD. The need to dichotomize diagnostic biomarkers is clear; however, in clinical trials, the exact cutoff point could vary depending on the population being studied. The EXPEDITION and EXPEDITION2 studies were performed in 16 countries including over 200 sites. Some, but not all, of the sites were university based; thus, although a quantitative comparison of the patients enrolled in EXPEDITION and EXPEDITION2 studies to patients enrolled to a typical memory clinic is not possible, a broad selection of patients was a characteristic of the EXPEDITION and EXPEDITION2 studies.
A key strength of the study, as noted previously, was the fact that all samples analyzed from EXPEDITION and EXPEDITION2 studies underwent the same preanalytical handling procedure and that multiple statistical approaches were used to determine the cutoff estimates, each of which have different theoretical bases, providing confidence in the robustness of the derived cutoffs. The availability of standardized CSF biomarker analytical methods, such as that provided by the Elecsys® Aβ (1–42) immunoassay, and future adoption of standardized procedures for CSF specimen collection and handling are expected to move the field closer to universal cutoffs for use in clinical research and clinical practice. At that time, large studies to establish cutoffs based on conventional, age- and other demographic-appropriate reference intervals will be warranted.
In summary, using the cutoffs estimated in this study, the Elecsys® Aβ (1–42), Elecsys® tTau, and Elecsys® pTau immunoassays are suitable for use in future clinical trials using identical preanalytical collection methods to determine biomarker levels and identify evidence of amyloid pathology in patients with AD who may achieve greater benefit from disease-modifying treatments.
Research in Context.
-
1.
Systematic review: The authors reviewed the literature using PubMed and database searches of conference abstracts. Data on the use of the Elecsys® Amyloid β (Aβ) (1–42) immunoassay for the establishment of a cutoff to identify individuals with Alzheimer's disease have been reviewed appropriately and cited.
-
2.
Interpretation: This study confirms that the Elecsys® Aβ (1–42) immunoassay is suitable for identifying individuals with evidence of amyloid pathology and could facilitate appropriate patient selection for future clinical trials aimed at reducing Aβ peptide accumulation into neuritic plaques. An Elecsys® Aβ (1–42) cutoff of 1100 pg/mL appears to be suitable for this purpose in the present study.
-
3.
Future directions: Comparison of the derived cutoff-point estimates against estimates obtained from different clinical cohorts, with varying preanalytical procedures and populations, using the Elecsys® immunoassays, would be of interest.
Acknowledgments
Authors' contributions: All authors made substantial contributions to the conception or design of the work, or the acquisition, analysis, or interpretation of data for the work; drafted or revised the manuscript critically for important intellectual content; approved the version to be published; and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding: The EXPEDITION clinical trials were sponsored by Eli Lilly and Company. The present analyses (analysis and interpretation of data) were sponsored by Roche Diagnostics and supported by Eli Lilly and Company. Third-party medical writing assistance, under the direction of the authors, was provided by David Evans, PhD, and Louise Kelly, BSc (Gardiner-Caldwell Communications) and was funded by Roche Diagnostics.
Footnotes
Disclosure and competing interests: L.M.S. provides quality control oversight for the Roche Elecsys CSF AD biomarkers measured by immunoassay on the cobas e601 analyzer in the ADNI study and has received research support from Roche for studies using this analytical system and from Lilly for CSF AD biomarker studies. J.Q.T., L.F., M.F., M.K., and T.W. have nothing to disclose. M.J.P. is an employee of Avid Radiopharmaceuticals, a wholly-owned affiliate of Eli Lilly and Company, and is a stockholder in Eli Lilly and Company. U.E. and S.W. are employees of Roche Diagnostics GmbH. M.Q. is an employee of Roche Diagnostics. J.A.T. and S.W.A. are employees of Eli Lilly and Company. D.R.L., E.R.S., and R.A.D. are retired, former employees of Eli Lilly and Company and are stockholders in Eli Lilly and Company.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2018.07.002.
Supplementary data
References
- 1.Alzheimer's Association 2017 Alzheimer's disease facts and figures. Alzheimers Dement. 2017;13:325–373. [Google Scholar]
- 2.Kochanek K.D., Murphy S.L., Xu J.Q., Tejada-Vera B. Deaths: Final data for 2014. Natl Vital Stat Rep. 2016;65:1–122. [PubMed] [Google Scholar]
- 3.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 4.Buchhave P., Minthon L., Zetterberg H., Wallin A.K., Blennow K., Hansson O. Cerebrospinal fluid levels of beta-amyloid 1-42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch Gen Psychiatry. 2012;69:98–106. doi: 10.1001/archgenpsychiatry.2011.155. [DOI] [PubMed] [Google Scholar]
- 5.Hansson O., Zetterberg H., Buchhave P., Londos E., Blennow K., Minthon L. Association between CSF biomarkers and incipient Alzheimer's disease in patients with mild cognitive impairment: A follow-up study. Lancet Neurol. 2006;5:228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- 6.Blennow K., Zetterberg H. Cerebrospinal fluid biomarkers for Alzheimer's disease. J Alzheimers Dis. 2009;18:413–417. doi: 10.3233/JAD-2009-1177. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y.-F., Ma X., Sundell K., Alaka K., Schuh K., Raskin J. Use of quantile regression to characterize solanezumab effects across percentiles of disease progression in EXPEDITION Alzheimer's disease trials. Alzheimers Dement. 2014;10:448. doi: 10.1016/j.trci.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doody R.S., Thomas R.G., Farlow M., Iwatsubo T., Vellas B., Joffe S. Phase 3 trials of solanezumab for mild-to-moderate Alzheimer's disease. N Engl J Med. 2014;370:311–321. doi: 10.1056/NEJMoa1312889. [DOI] [PubMed] [Google Scholar]
- 9.Shaw L.M., Fields L., Korecka M., Waligorksa T., Trojanowski J.Q., Allegranza D. Method comparison of Aβ(1-42) measured in human cerebrospinal fluid samples by liquid chromatography-tandem mass spectrometry, the INNO-BIA AlzBio3 assay, and the Elecsys® β-Amyloid(1-42) assay. Alzheimers Dement. 2016;12:668. [Google Scholar]
- 10.Bittner T., Zetterberg H., Teunissen C.E., Ostlund R.E., Jr., Militello M., Andreasson U. Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of beta-amyloid (1-42) in human cerebrospinal fluid. Alzheimers Dement. 2016;12:517–526. doi: 10.1016/j.jalz.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 11.The Alzheimer's Association QC Program for CSF Biomarkers; Round 24 Results. 2017. http://neurophys.gu.se/sektioner/psykiatri_och_neurokemi/neurokem/TheAlzAssQCProgram/Resultat Available at: [Google Scholar]
- 12.Clark C.M., Pontecorvo M.J., Beach T.G., Bedell B.J., Coleman R.E., Doraiswamy P.M. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study. Lancet Neurol. 2012;11:669–678. doi: 10.1016/S1474-4422(12)70142-4. [DOI] [PubMed] [Google Scholar]
- 13.Amyvid [package insert] Lilly USA, LLC; Indianapolis, IN: 2013. [Google Scholar]
- 14.Joshi A.D., Pontecorvo M.J., Lu M., Skovronsky D.M., Mintun M.A., Devous M.D. A semiautomated method for quantification of F 18 florbetapir PET images. J Nucl Med. 2015;56:1736–1741. doi: 10.2967/jnumed.114.153494. [DOI] [PubMed] [Google Scholar]
- 15.Joshi A.D., Pontecorvo M.J., Clark C.M., Carpenter A.P., Jennings D.L., Sadowsky C.H. Performance characteristics of amyloid PET with florbetapir F 18 in patients with Alzheimer's disease and cognitively normal subjects. J Nucl Med. 2012;53:378–384. doi: 10.2967/jnumed.111.090340. [DOI] [PubMed] [Google Scholar]
- 16.Efron B., Stein C. The jacknife estimate of variance. Ann Stat. 1981;9:586–596. [Google Scholar]
- 17.De Meyer G., Shapiro F., Vanderstichele H., Vanmechelen E., Engelborghs S., De Deyn P.P. Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67:949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindler S.E., Gray J.D., Gordon B.A., Xiong C., Batrla-Utermann R., Quan M. Cerebrospinal fluid biomarkers measured by Elecsys assays compared to amyloid imaging. Alzheimers Dement. 2018 doi: 10.1016/j.jalz.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansson O., Seibyl J., Stomrud E., Zetterberg H., Trojanowski J.Q., Bittner T. CSF biomarkers of Alzheimer's disease concord with amyloid-β PET and predict clinical progression: A study of fully-automated immunoassays in BioFINDER and ADNI cohorts. Alzheimers Dement. 2018 doi: 10.1016/j.jalz.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lifke V., Manuilova E., Knop C., Selle T., Kraus W., Oelschlaegel T. Elecsys® Total-Tau CSF and Elecsys® Phospho-Tau (181P) CSF: Novel, fully automated immunoassays for rapid and accurate quantitation of CSF biomarkers for clinical use. J Prev Alzheimers Dis. 2017;4:359. [Google Scholar]
- 21.Janelidze S., Pannee J., Mikulskis A., Chiao P., Zetterberg H., Blennow K. Performance of different amyloid immunoassays in predicting outcome of visual assessment of amyloid PET imaging. JAMA Neurol. 2017;74:1492–1501. doi: 10.1001/jamaneurol.2017.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann R., Lelental N., Ganslandt O., Maler J.M., Kornhuber J., Lewczuk P. Preanalytical sample handling and sample stability testing for the neurochemical dementia diagnostics. J Alzheimers Dis. 2011;25:739–745. doi: 10.3233/JAD-2011-110212. [DOI] [PubMed] [Google Scholar]
- 23.Leitão M.J., Baldeiras I., Herukka S.K., Pikkarainen M., Leinonen V., Simonsen A.H. Chasing the effects of pre-analytical confounders – a multicenter study on CSF-AD biomarkers. Front Neurol. 2015;6:153. doi: 10.3389/fneur.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vellas B., Carrillo M.C., Sampaio C., Brashear H.R., Siemers E., Hampel H. Designing drug trials for Alzheimer's disease: What we have learned from the release of the phase III antibody trials: A report from the EU/US/CTAD Task Force. Alzheimers Dement. 2013;9:438–444. doi: 10.1016/j.jalz.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Green R.C. The Alzheimer's Disease Neuroimaging Initiative 3: Continued innovation for clinical trial improvement. Alzheimers Dement. 2017;13:561–571. doi: 10.1016/j.jalz.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.