Abstract
Recent evidence has revealed that heterotrimeric G-proteins can be activated by cytoplasmic proteins that share an evolutionarily conserved sequence called the Gα-binding-and-activating (GBA) motif. This mechanism provides an alternative to canonical activation by G-protein–coupled receptors (GPCRs) and plays important roles in cell function, and its dysregulation is linked to diseases such as cancer. Here, we describe a discovery pipeline that uses biochemical and genetic approaches to validate GBA candidates identified by sequence similarity. First, putative GBA motifs discovered in bioinformatics searches were synthesized on peptide arrays and probed in batch for Gαi3 binding. Then, cDNAs encoding proteins with Gαi3-binding sequences were expressed in a genetically-modified yeast strain that reports mammalian G-protein activity in the absence of GPCRs. The resulting GBA motif candidates were characterized by comparison of their biochemical, structural, and signaling properties with those of all previously described GBA motifs in mammals (GIV/Girdin, DAPLE, Calnuc, and NUCB2). We found that the phospholipase Cδ4 (PLCδ4) GBA motif binds G-proteins with high affinity, has guanine nucleotide exchange factor activity in vitro, and activates G-protein signaling in cells, as indicated by bioluminescence resonance energy transfer (BRET)-based biosensors of G-protein activity. Interestingly, the PLCδ4 isoform b (PLCδ4b), which lacks the domains required for PLC activity, bound and activated G-proteins more efficiently than the full-length isoform a, suggesting that PLCδ4b functions as a G-protein regulator rather than as a PLC. In summary, we have identified PLCδ4 as a nonreceptor activator of G-proteins and established an experimental pipeline to discover and characterize GBA motif–containing proteins.
Keywords: G-protein–coupled receptor (GPCR), guanine nucleotide exchange factor (GEF), GTPase, bioluminescence resonance energy transfer (BRET), heterotrimeric G-protein, phospholipase C
Introduction
Heterotrimeric G-proteins are gatekeepers of signal transduction that regulate fundamental aspects of cell biology across the full spectrum of the eukaryota domain, from unicellular fungi to multicellular mammals (1, 2). They work as molecular switches that are turned on or off depending on their guanine nucleotide-binding status. Thus, the biological activity of heterotrimeric G-proteins is determined by their nucleotide-handling properties (such as GDP/GTP exchange and GTP hydrolysis), which control the amplitude and duration of downstream signaling. At the same time, nucleotide-binding and hydrolytic activity are modulated by a large network of regulators (2). The best characterized components of this network are membrane receptors of the GPCR3 superfamily. In the canonical paradigm, ligand-activated GPCRs act as Guanine-nucleotide Exchange Factors (GEFs) that promote the exchange of GDP for GTP in the Gα subunit of a resting Gαβγ heterotrimer. As a consequence of GTP binding, Gα and Gβγ dissociate from each other and subsequently activate downstream effectors that propagate signaling intracellularly. In turn, signaling is terminated upon hydrolysis of GTP by Gα, which returns it to the GDP-bound form that reassociates with Gβγ. These series of reactions constitute the so-called canonical G-protein cycle (1).
In addition to the components described above as core elements of the canonical regulatory cycle, G-proteins interact with a wide range of “accessory proteins” (3). These are cytoplasmic factors that can be classified based on their biochemical activity toward G-proteins. The best characterized of these accessory proteins are members of the regulators of G-protein signaling (RGS) family, which bind to active G-proteins and have GTPase Activating Protein (GAP) activity (4, 5), thereby facilitating the termination of G-protein signaling via acceleration of GTP hydrolysis. This family of proteins shares a signature domain of ∼120 aa called the RGS box, which confers the GAP activity (6–9). Another well-characterized family of accessory proteins is defined by a signature sequence of 30–40 aa called the GoLoco motif, which confers guanine nucleotide dissociation inhibitor (GDI) activity (10–12). Members of this family also bind to Gα subunits, but they do so when they are in the inactive, GDP-bound form to prevent nucleotide exchange and subsequent activation (13–15). Both RGS and GoLoco proteins have been shown to play crucial roles in cell biology and human disease (12, 16, 17).
Another group of accessory proteins can be broadly defined based on their biochemical activity as GEFs. These are cytoplasmic factors that mimic the action of GPCRs, i.e. they promote the exchange of GDP for GTP on Gα subunits. However, in contrast to RGS and GoLoco proteins, the initial discovery of nonreceptor GEFs was not accompanied by the identification of a shared domain or motif responsible for the biochemical activity, as this group was constituted by unrelated proteins, and the domains responsible for the G-protein regulatory activity were not mapped (18–22). The exception to this was GIV (also known as Girdin), for which a short sequence of ∼30 aa, named the Gα-binding-and-activating (GBA) motif, was identified (23, 24). Subsequently, it has been shown that the GBA motif is present in evolutionarily unrelated proteins and defines a mechanism of G-protein activation by nonreceptor proteins already existing in invertebrates (25–27). Thus, the GBA motif represents the defining structural element for a subfamily of nonreceptor G-protein activators with GEF activity (28). In mammals, this group currently consists of four proteins (GIV, DAPLE, Calnuc, and NUCB2), which bind and activate Gα subunits of the Gi subfamily (i.e. Gαi1, Gαi2, and Gαi3). The structural basis for the binding and activation of Gαi proteins by the GBA motif in vitro has been studied in detail (29–31), and it has also been shown that the GBA motif activates G-proteins in cells using approaches that monitor the formation of Gαi–GTP (cAMP or conformation-specific antibodies) or free Gβγ (live-cell biosensors or PI3K–Akt signaling) (23, 26, 32–36). Such signaling mechanisms have a profound impact on cellular processes such as cell migration, mitosis, polarity, or autophagy (24, 26, 37–39), which underlie the linkage between GBA protein dysregulation and human disorders such as cancer, hepatic fibrosis, renal dysfunction, insulin resistance, or pathological vascularization (24, 40).
Despite these advances in characterizing the structural basis, cell signaling and disease linkage properties of GBA proteins, a fundamental question that remains unanswered is how prevalent this mechanism of G-protein regulation is. For this, a critical first step is to identify which proteins, and how many, contain a functional GBA motif. In this regard, the amino acid sequence of the GBA motif of proteins identified to date is known, but this information is insufficient to systematically identify other GBA motifs based solely on sequence similarity. Although sequence similarity has been successfully used to identify members of other families of G-protein regulators (e.g. RGS GAPs), the short length of the GBA motif and the low number of validated GBA sequences limit the power of bioinformatics searches. For these reasons, bioinformatics searches for GBA motifs yield a large number of false positives. To overcome this limitation, here we describe a pipeline of biochemical and genetic assays that can be used to validate and characterize GBA candidates identified by sequence similarity to currently known GBA proteins. Using peptide arrays, functional assays in genetically-engineered yeast, and other biochemical assays, we identify PLCδ4 as a novel GBA motif–containing nonreceptor GEF and subsequently characterize its signaling properties by systematic comparison with all the other GBA motifs described to date.
Results
Identification of putative GBA motifs by bioinformatics and peptide arrays
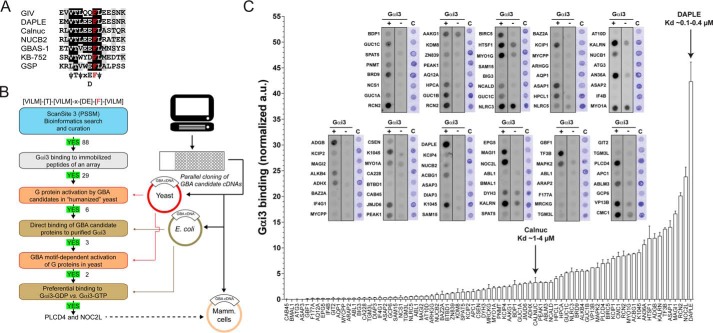
We envisioned a screening strategy to discover GBA motif–containing G-protein activators (Fig. 1, A and B). We started by using bioinformatics to identify putative GBA motifs by sequence similarity, followed by a high-throughput peptide array assay to identify Gαi3 binders as initial hits. The subsequent validation pipeline involved genetic assays in yeast and further biochemical characterization. The underlying logic of this approach is that previous evidence indicates that Gαi3 binding is an obligatory requirement for GBA-mediated G-protein activation and that the extent of binding and GEF activity correlate tightly (23, 25, 26, 29, 31). For this, we used the consensus sequence of the seven amino acids of the highly conserved core of the GBA motifs identified to date (23, 25, 36, 41–43) to search for similar sequences using ScanSite3 (44). This program uses a Position-specific Scoring Matrix (PSSM), which in our case was defined as (VLIM)(T)(VLIM)X(DE)F(VLIM) based on the consensus GBA motif ψTψX(D/E)Fψ, where ψ is an aliphatic residue, and X is any residue (Fig. 1A). Approximately 200 hits were obtained using this PSSM to search the human proteome based on the most stringent cutoff setting of ScanSite3 (0.2 score). Proteins that were annotated in UniProt (45) as secreted/extracellular or as transmembrane proteins in which the predicted GBA sequence was either in the membrane-spanning region or not facing the cytosol were eliminated. The 88 candidate GBA motifs that resulted from this curation were synthesized as 24-mer peptides (in which the invariable Phe of the GBA motif occupied the 12th position) immobilized on cellulose and probed in batch with purified His–Gαi3 (Fig. 1C). This set of peptides included sequences derived from DAPLE and Calnuc, two previously validated GBA motifs that served as internal controls. We found that DAPLE was the strongest binder in the peptide array (Fig. 1C), which is consistent with previous observations showing that DAPLE has the highest affinity for Gαi3 of all the GBA motifs described to date (Kd ∼0.1–0.4 μm) (26, 29). Calnuc binding was weaker (Fig. 1C), which is also consistent with the Kd value previously reported for its interaction with Gαi3 (Kd ∼1–4 μm) (25). We reasoned that peptides binding Gαi3 at least as much as the Calnuc GBA peptide might represent physiologically relevant interactors because Calnuc has been previously shown to interact directly with Gαi3 in a cellular context (25, 46). Based on this criterion, 29 GBA candidates were selected for further investigation.
Figure 1.
Identification of putative GBA motifs by bioinformatics and peptide array screening. A, alignment of known GBA sequences of GIV, DAPLE, Calnuc, NUCB2, and GBAS-1 proteins and the synthetic peptides KB-752 and GSP. The background was shaded black or gray if the residue was identical or similar, respectively, in ≥50% of the sequences. The invariable phenylalanine, required for G-protein binding, is in red. The consensus sequence for the 7 amino acids of the core of the GBA motif is shown below the alignment (ψ, hydrophobic; x, any). B, discovery pipeline for the identification and validation of G-protein activators with a GBA motif. Putative GBA motifs are identified by bioinformatics, tested for G-protein binding in a GBA peptide array, and the corresponding genes evaluated for G-protein activation in yeast-based signaling assays. Subsequent steps include validation of direct binding of GBA motif–containing proteins to GDP-bound G-proteins and assessment of GBA dependence for the activation of signaling in yeast. C, peptide array screening for binding of GBA motif candidates to Gαi3. The 24-mer peptides corresponding to each one of the indicated GBA motif candidates (full sequences of positive hits is shown in Fig. S1) were synthesized and immobilized on slides as one peptide per spot. The immobilized peptides were probed in batch with purified His–Gαi3 (Gαi3 +), and binding was determined after sequential incubation with primary and secondary antibodies coupled to fluorescent probes. Control reactions in which the slides were incubated with primary and secondary antibodies but without probing with purified His–Gαi3 (Gαi3 −) were performed in parallel. A third slide was stained with Coomassie Blue (C). The arbitrary units (a.u.) of Gαi3 binding were determined by subtracting the signal in the “Gαi3 −” condition from the signal in the “Gαi3 +” condition and normalizing to the Coomassie staining signal. Results are the average of four experiments (performed with two independent batches of peptide synthesis), and the error bars are the S.E. Two peptides corresponding to the characterized GBA proteins Calnuc and DAPLE were included as internal controls, and the previously determined Kd values for their interaction with Gαi3 are indicated.
Validation of GBA candidate hits as G-protein activators in yeast
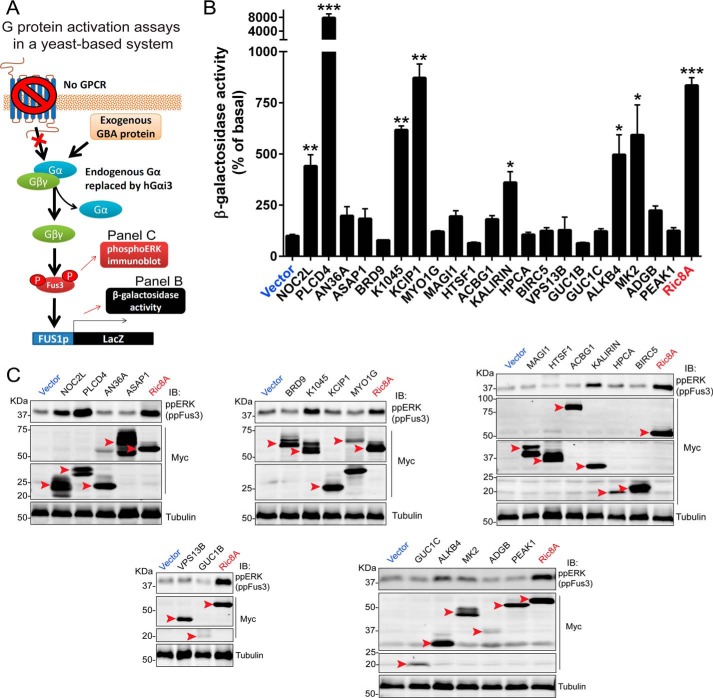
Although binding of Gαi3 to immobilized GBA peptides suggests that these sequences are capable of G-protein binding in isolation, this might not be the case when the sequence is placed in the context of a folded protein and/or might be insufficient for activation of G-protein signaling. To start validating the GBA candidates, we used a previously described genetic screening approach in yeast (Fig. 2A) (47, 48). Briefly, we expressed the cDNA of GBA candidates in a genetically-engineered yeast strain that lacks GPCRs and with the endogenous yeast Gα protein Gpa1 replaced by human Gαi3. In this “humanized” yeast system, only an exogenous G-protein activator can trigger a signaling pathway that is normally activated as a pheromone response that leads to an increase in transcriptional activation of the FUS1 gene (Fig. 2A). The cDNAs of GBA candidates were cloned as full-length proteins based on naturally occurring isoforms annotated in UniProt or as truncations corresponding to domains known or predicted to fold as independent units (see details in Fig. S1). These constructs were expressed in the genetically-engineered yeast strain described above, and G-protein activation was assessed by the activation of a FUS1 promoter β-gal reporter (Fig. 2B). As an internal control, we used Ric-8A, a protein that displays GEF activity toward Gαi proteins in vitro (19) and that we have previously validated as a G-protein activator in this yeast-based assay (27). From the 29 candidates identified as Gαi3 binders in the peptide array (Fig. 1), eight were toxic in yeast or excluded for other reasons (Fig. S1). From the remaining 21 candidate genes expressed in yeast, PLCD4 (which encodes for the phospholipase PLCδ4) stood out from the rest by inducing FUS1 activation ∼80-fold over basal levels. Another six candidates (NOC2L, K1045, KCIP1, KALIRIN, ALKB4, and MAPK2) induced FUS1 activation of much lower magnitude (∼3–8-fold) than PLCD4 but still comparable with the positive control Ric-8A (Fig. 2B). As a secondary validation of these results, we measured Fus3 phosphorylation in lysates of the same yeast cells used in the reporter assay. Because Fus3 phosphorylation is an event upstream of FUS1 transcriptional activation in the signaling cascade triggered by G-proteins (Fig. 2A), we reasoned that GBA candidates leading to increased β-gal activity should also have increased Fus3 phosphorylation. We found that this was the case except for KCIP1 (Fig. 2C), which failed to increase Fus3 phosphorylation despite leading to FUS1 activation based on β-gal activity measurements. We concluded that KCIP1's effect on β-gal activity is likely due to a G-protein–independent mechanism, whereas the GBA candidates PLCD4, NOC2L, K1045, KALIRIN, ALKB4, and MAPK2 might be G-protein activators.
Figure 2.
Assessment of GBA motif–containing gene candidates as G-protein activators in “humanized” yeast. A, schematic of humanized yeast reporter system to determine G-protein activation by candidate GBA proteins. The pheromone-response pathway in Saccharomyces cerevisiae is regulated by GPCR-mediated activation of heterotrimeric G-proteins, which induces Fus3 phosphorylation and transcriptional activation of the FUS1 promoter. A genetically engineered strain lacking endogenous pheromone-sensitive GPCRs, expressing human Gαi3 instead of the endogenous Gα Gpa1 and bearing a lacZ reporter under the control of the FUS1 promoter, was used to determine GPCR-independent G-protein activation by exogenously expressed GBA proteins or by Ric-8A (positive control). FUS1 promoter activity was determined by β-gal activity assays (B), and Fus3 activation was determined by phospho-ERK (ppERK, which recognizes yeast ppFus3) immunoblotting (C). B, β-gal activity of yeast cells expressing the cDNA of the indicated GBA candidates (see details for each construct in Fig. S1) was measured as described under “Experimental procedures” and normalized relative to the activity in cells expressing an empty vector. Results are the average of six independent experiments, and the error bars represent the S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with vector using the Student's t test. C, yeast cells expressing the cDNA of the indicated GBA candidates (see details for each construct in Fig. S1) were lysed and proteins analyzed by immunoblotting with the indicated antibodies as described under “Experimental procedures.” One representative experiment of at least three is shown. The red arrowheads indicate the expected position for the full-length protein corresponding to each construct.
Validation of GBA candidate hits as direct G-protein binders
As an independent validation of GBA candidates, we also investigated whether they bound directly to G-proteins. As mentioned above, it has been previously shown that binding of GBA proteins to Gαi3 correlates tightly with their ability to activate G-protein signaling (23, 25, 26, 29, 31); therefore, we established binding as a selection criterion to identify bona fide GBA motif–containing G-protein activators. For this, we cloned the exact same sequences of the 29 GBA candidate genes used for the yeast assays described above as GST fusions for expression in Escherichia coli (Fig. 1B). We could purify 15 of them in sufficient quantity and quality for protein–protein binding experiments. This set included one protein that could not be tested in the yeast-based assays due to toxicity (ABLIM3). RCN2 and TF3B were toxic in yeast and could not be purified from E. coli, but subsequent pulldown experiments with lysates of mammalian cells expressing these candidates ruled out binding to Gαi3 (Fig. S2).
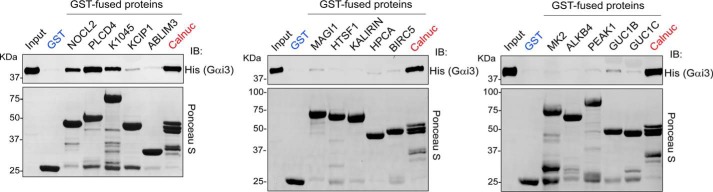
Binding of purified His–Gαi3 to the 15 GST-fused GBA candidates purified from E. coli was tested in pulldown assays using permissive conditions (i.e. relatively high concentrations of proteins) to facilitate the identification of binders. As a positive control, we used GST–Calnuc. As indicated above, Calnuc is a validated GBA protein that binds with relatively weak affinity compared with other GBA motif–containing proteins (25, 46), so we considered positive for binding only those GBA candidates that displayed binding similar to that detected with Calnuc. Based on this criterion, KCIP1, KALIRIN, ALKB4, and MAPK2 did not bind to His–Gαi3 (Fig. 3). Thus, it is likely that the activity of these candidates in the yeast-based system (Fig. 2, B and C) is exerted via G-protein–independent mechanisms, and we excluded them from further characterization as a possible GBA motif containing G-protein activators. The other three GBA candidates that were active in the yeast assays (Fig. 2, B and C), i.e. PLCD4, NOC2L, and K1045, did bind directly to His–Gαi3 (Fig. 3).
Figure 3.
PLCD4, NOC2L, and K1045 GBA candidates bind directly to Gαi3. Approximately 20 μg of the indicated purified GST-fused constructs (see details for each construct in Fig. S1) were immobilized on GSH-agarose beads and incubated with 5 μg of purified His–Gαi3 in the presence of GDP. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. Input = 5% of the total amount of His–Gαi3 added in each binding reaction. One representative experiment of at least three is shown.
PLCD4 and NOC2L, but not K1045, activate G-protein signaling in yeast via their GBA motif
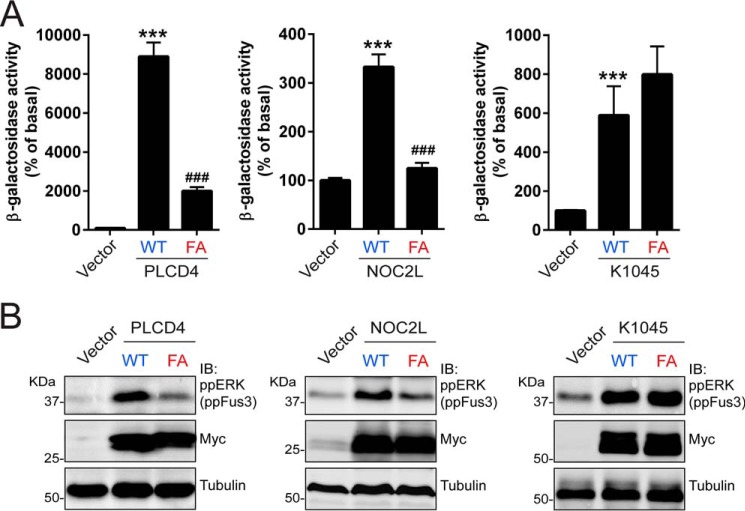
Next, we investigated whether the GBA motif identified in PLCD4, NOC2L, and K1045 in our bioinformatics searches (Fig. 1) is responsible for the ability of these proteins to activate G-protein signaling in yeast (Fig. 2). For this, we mutated the conserved phenylalanine in the GBA motif (Fig. 1A) of each one of these candidates to alanine and assessed the effect of these mutants on β-gal FUS1 reporter activity and Fus3 phosphorylation in the humanized yeast strain described above (Fig. 2A). It has been previously shown that equivalent “FA” mutations in any of the previously validated GBA motif–containing proteins (GIV, DAPLE, Calnuc, NUCB2, and GBAS-1) abolish G-protein binding and GEF activity almost completely (23, 25–27, 29). Consistent with these previous observations, we found that the FA mutants of PLCD4 and NOC2L failed to recapitulate the activation exerted by their wildtype (WT) counterparts in either β-gal FUS1 reporter activity assays (Fig. 4A) or Fus3 phosphorylation immunoblots (Fig. 4B). In contrast, the equivalent mutation in K1045 did not have an effect in the same assays (Fig. 4, A and B), suggesting that the GBA motif of this protein is not responsible for its G-protein regulatory activity. Taken together, these results indicate that PLCD4 and NOC2L activate G-protein signaling via their GBA motifs.
Figure 4.
PLCD4 and NOC2L activate G-protein signaling in yeast via their GBA motif. A, β-gal activity of yeast cells expressing wild-type (WT) or “FA” mutant versions of the indicated GBA candidates (PLCD4, left; NOC2L, center; K1045, right) was measured as described under “Experimental procedures” and normalized relative to the activity in cells expressing an empty vector. Results are the average of six independent experiments, and the error bars represent the S.E. ***, p < 0.001 compared with the vector control, or ###, p < 0.001 compared with WT using the Student's t test. B, yeast cells expressing the same constructs as in A were lysed and proteins analyzed by immunoblotting (IB) with the indicated antibodies as described under “Experimental procedures.” One representative experiment of at least three is shown.
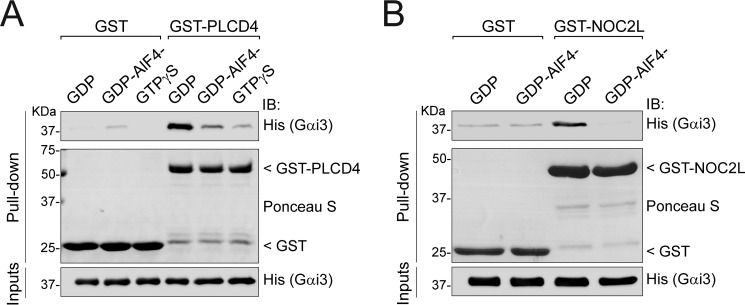
PLCD4 and NOC2L bind preferentially to GDP-bound Gαi3 over the GTP-bound form
Much like other proteins with GEF activity, all previously characterized GBA proteins (GIV, DAPLE, Calnuc, NUCB2, and GBAS-1) bind preferentially to GDP-bound, inactive Gαi3 over GTP-bound Gαi3 (23, 25–27, 29). Next, we investigated whether this was the case for PLCD4 and NOC2L. Purified His–Gαi3 was preloaded with GDP, GDP/AlF4− (which mimics the GTP-bound transition state (49, 50)), or GTPγS (which is a nonhydrolyzable GTP analog) by incubation at 30 °C for 3 h and used as the soluble ligand in pulldowns with resin-immobilized GST–PLCD4. We found that PLCD4 binding to Gαi3 loaded with either of the two GTP mimetics (GDP/AlF4− or GTPγS) was much weaker than to Gαi3–GDP (Fig. 5A), suggesting that PLCD4 has G-protein state–dependent binding properties analogous to those previously described for other proteins with GBA motifs. The same nucleotide preincubation conditions of 3 h at 30 °C diminished the already weak binding of NOC2L to Gαi3–GDP to undetectable levels, which precluded any comparison with GTP mimetics. To overcome this limitation, we omitted the GTPγS condition and performed the preloading step at 4 °C for 20–30 min, which is sufficient to load the G-protein with AlF4−. Using these modified preincubation conditions, we found that NOC2L binds preferentially to inactive (Gαi3–GDP) versus active (Gαi3–GDP/AlF4−) (Fig. 5B). Interestingly, K1045 showed the opposite pattern of binding, i.e. it bound preferentially to GTP mimetic-bound Gαi3 over Gαi3–GDP (data not shown), which along with the lack of effect of the FA mutant in G-protein activity assays in yeast (Fig. 4) supports that this protein does not behave like other GBA proteins. Studies on the significance of the K1045–Gαi3 interaction are being pursued out of the scope of this work.
Figure 5.
PLCD4 and NOC2L bind preferentially to GDP-bound Gαi3 over GTP-bound Gαi3. Approximately 20 μg of purified GST–PLCD4 (left) or GST-NOC2L (right) (or GST, as a negative control) immobilized on GSH-agarose beads were incubated with 5 μg of purified His–Gαi3 preloaded with GDP (inactive), GDP + AlF4− (mimics GTP-bound transition state), or GTPγS (nonhydrolyzable GTP analog) as indicated. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. Inputs = 5% of the total amount of His–Gαi3 added in each binding reaction. One representative experiment of at least three is shown.
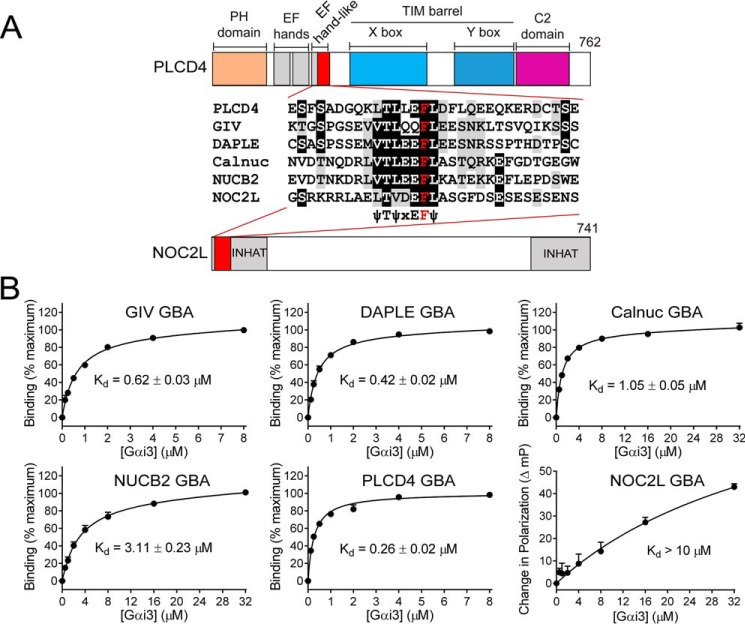
PLCD4 GBA motif, but not NOC2L GBA motif, binds with high affinity to Gαi3
Next, we set out to characterize the binding properties of the GBA motifs of PLCD4 and NOC2L. We have recently shown that a 31-mer peptide corresponding to the GBA motif of GIV is sufficient to recapitulate the high-affinity binding (Kd ∼0.5–0.6 μm) of the protein (30). Using NMR spectroscopy, we also found that the G-protein binding contacts of a GIV GBA peptide are the same as for a larger 200-aa fragment of GIV(1660–1870) (29) that is necessary and sufficient to activate G-proteins (31, 42). Consistently, the NMR spectroscopy studies also showed that the conformational changes in Gαi3 associated with G-protein activation by GIV were the same for the GBA peptide and the larger 200-aa construct (36). Thus, peptides corresponding to the GBA motif can recapitulate well the G-protein–binding properties of the proteins in which they are embedded. To quantify the Gαi3-binding affinity of the GBA motifs of PLCD4 and NOC2L compared with previously described GBA motifs in mammalian proteins (GIV, DAPLE, Calnuc, and NUCB2), we synthesized fluorescently-labeled peptides and used them in fluorescence polarization (FP) assays to calculate the Kd values. Following the design of the GIV GBA motif peptide, we generated analogous 31-mer peptides corresponding to the GBA motifs of DAPLE, Calnuc, NUCB2, PLCD4, and NOC2L (Fig. 6A), which would be expected to yield an increase in FP upon binding to Gαi3 (∼40 kDa). Consistent with previously published data, the GIV GBA motif bound Gαi3 with a Kd ∼0.6 μm (Fig. 6B). DAPLE, Calnuc, and NUCB2 GBA motifs also bound to Gαi3 with Kd values of ∼0.4, ∼1, and ∼3 μm, respectively, in this assay (Fig. 6B), which are in agreement with the binding affinities of larger fragments of the corresponding proteins using other assays (25, 26). These results further support that isolated GBA motifs recapitulate well the G-protein–binding properties of the proteins in which they are embedded. As for PLCD4 and NOC2L, we found that the former binds with high affinity to Gαi3, although the latter does not (Fig. 6B). PLCD4 GBA motif binds Gαi3 with a Kd ∼0.25 μm (Fig. 6B), which is even lower than that of the GBA motif of GIV or of DAPLE. Because GIV and DAPLE have been shown to regulate G-protein signaling in cells via GBA-dependent binding (23, 26, 31, 36, 37, 51), this suggests that the GBA motif of PLCD4 might also be bioactive. In contrast, NOC2L binding was so weak that the FP signal did not reach saturation even at the highest concentration of G-protein tested (32 μm) (Fig. 6B), thereby precluding Kd calculation. We estimate that the Kd value for the NOC2L GBA–Gαi3 interaction is at least 1 order of magnitude larger than the Kd value for the weakest GBA motif (NUCB2). This low affinity of the isolated GBA peptide likely reflects the G-protein–binding properties of the NOC2L protein because G-protein activation in yeast assays and direct Gαi3 binding were also weak (compared with PLCD4) in previous experiments using a larger domain of NOC2L (Figs. 2 and 3). Although we cannot rule out that the weak affinity interaction of NOC2L with Gαi3 is biologically relevant, we focused our subsequent efforts on characterizing the GBA motif of PLCD4.
Figure 6.
GBA motif of PLCD4 binds Gαi3 with high affinity. A, alignment of GBA 31-mer peptide sequences of PLCD4, NOC2L, GIV, DAPLE, Calnuc, and NUCB2 used in fluorescence polarization (FP) assays. The background was shaded black or gray if the residue was identical or similar, respectively, in ≥50% of the sequences. The invariable phenylalanine, required for G-protein binding, is in red. A bar diagram of the domains of PLCD4 and NOC2L is presented to show the position of the GBA motif in the context of the full proteins. B, indicated fluorescently-labeled GBA peptides were incubated with increasing amounts of purified rat His–Gαi3 in the presence of GDP and binding determined by FP. Data were normalized to maximal binding (except for NOC2L, for which data are presented as increase in millipolarization (mP) units) and fitted to a one-site binding model (solid lines) to determine the equilibrium dissociation constant (Kd). Results expressed as average ± S.E. of n = 3.
Identification of critical residues involved in forming the Gαi3/PLCD4 interface
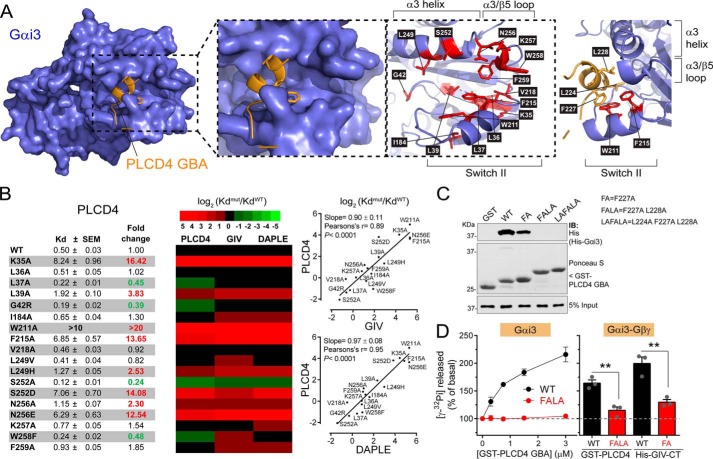
To start characterizing the structural properties of the interface formed between PLCD4 and Gαi3, we generated a homology-based model of the complex. For this, we used the previously described crystal structure of a GBA-like synthetic peptide called KB-752 bound to Gαi1 as a template (41). This model resolves a 13-amino acid stretch of PLCD4 containing the core of the GBA motif. As expected, this model predicts that the GBA motif of PLCD4 binds to Gαi3 similar to other GBA motifs, i.e. it docks onto the cleft formed by the Switch II (SwII) region and α3 helix of Gαi3 (Fig. 7A). Consistent with this mode of binding, we found that myristoylation of the N terminus of Gαi, which is far from the SwII/α3 pocket in crystal structures, does not affect binding to the GBA motif of PLCD4 (Fig. S3). To validate more extensively the predicted conservation between the mode of binding of PLCD4 GBA motif and that of other GBA motifs, we tested its binding to a battery of 18 Gαi3 mutants previously used to characterize the binding of GIV and DAPLE GBA motifs (29). This set includes mutants within or near the SwII/α3 region previously shown to inhibit, enhance, or have no effect on binding to the GBA motif of GIV or DAPLE (29). We reasoned that this systematic approach would allow us to define a “fingerprint” of PLCD4 GBA motif–binding properties based on the effect of different mutants and that this fingerprint could be compared with that of previously characterized GBA motifs (29). For this, we performed FP experiments with a fluorescently labeled PLCD4 GBA peptide to quantify the Kd for each mutant and then calculated the fold change in affinity compared with Gαi3 WT (Fig. 7B). This dataset for PLCD4 GBA motif was compared with previously obtained data (29) for GIV GBA motif and DAPLE GBA motif using the same experimental conditions. As shown in the heat map and correlation plots in Fig. 7B, there was good agreement between the effects of Gαi3 mutants on the binding of PLCD4 GBA motif and on GIV or DAPLE GBA motif. The Pearson's correlation coefficient was 0.89 for PLCD4/GIV and 0.95 for PLCD4/DAPLE. In general, mutants that diminished affinity for GIV or DAPLE also had diminished affinity for PLCD4, and mutants that increased affinity, like S252A, also did so for all the GBA motifs (Fig. 7B). A notable exception was the effect of the W258F mutation. Although this mutation diminishes the affinity for GIV (∼3.5-fold) and DAPLE (∼2-fold), it increases affinity for PLCD4 (∼2-fold). Although our homology model does not include the predicted contacts made by Trp-258 with the GBA motif (Fig. 7A), it is reasonable to think that the difference in binding upon the Trp-258 mutation arises from the divergence of the sequence in the C-terminal region (i.e. downstream of the conserved Phe) of the GBA motifs of PLCD4, GIV, and DAPLE (Fig. 6A). Overall, these results indicate that, barring some nuanced differences, the GBA motif of PLCD4 physically engages G-proteins in a manner similar to previously characterized GBA motifs.
Figure 7.
PLCD4 GBA motif binds to the SwII/α3 helix region of Gαi3 and has GEF activity in vitro. A, different views of a structure homology model of the complex formed between Gαi3 and the GBA motif of PLCD4 with details of the residues within or near the predicted protein–protein interface. Left panel, overview of the G-protein (blue), GBA motif (orange) complex. PLCD4 GBA motif is predicted to bind to the groove formed between the SwII and α3 helix. Middle panels, two representations of the same view of the Gαi3/PLCD4 interface. On the view on the right, side chains of amino acids in the SwII/α3 helix pocket of Gαi3 mutated in the experiments shown in B and displayed in red, and the PLCD4 motif is omitted for clarity. Right panel, view of conserved hydrophobic amino acid side chains in PLCD4 GBA motif predicted to make contact with hydrophobic residues in the SwII/α3 helix pocket of Gαi3. B, effect of Gαi3 mutations on PLCD4 binding is highly correlated with their effect on GIV and DAPLE binding. Left panel, equilibrium dissociation constants (Kd) for the binding of purified human His–Gαi3 mutants to the GBA motif of PLCD4 was determined from FP binding curves as described under “Experimental procedures.” Results are expressed as average ± S.E. of n = 3–5. Fold changes in the Kd compared with WT that were larger than two or smaller than 0.5 are indicated in red or green, respectively. Middle panel, heat map (green to red scale shown on top) of log2(KdMUT/KdWT) for each Gαi3 mutant relative to WT comparing PLCD4 GBA binding to previously obtained data (29) for GIV or DAPLE GBA binding. Right panels, correlation plots of log2(KdMUT/KdWT) values for PLCD4 and GIV (top) or PLCD4 and DAPLE (bottom). C, mutation of hydrophobic residues in PLCD4 GBA motif impairs Gαi3 binding. Approximately 10 μg of purified GST or GST-fused PLCD4 GBA motif (aa 213–243, WT, or bearing the indicating mutations) were immobilized on GSH-agarose beads and incubated with 1 μg of purified rat His–Gαi3 in the presence of GDP. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. Input = 5% of the total amount of His–Gαi3 added in each binding reaction. One representative experiment of at least three is shown. D, PLCD4 GBA motif increases Gαi3 (left) or Gαi3–Gβγ heterotrimer (right) activity in vitro. The steady-state GTPase activity of purified rat His–Gαi3 or His–Gαi3–Gβγ was determined in the presence of GST–PLCD4 GBA motif WT (black) or FALA mutant (red) by measuring the production of [32P]Pi at 15 min as described under “Experimental procedures.” The effect of His–GIV–CT (aa 1660–1870, containing the GBA motif) WT (black) or F1685A mutant (red) on the activity of the Gαi3–Gβγ heterotrimer was also tested. Results are the average ± S.E. of n = 3. **, p < 0.01 using the Student's t test.
Consistent with the homology model (Fig. 7A), some of the most marked defects in GBA motif binding were observed upon mutation of the aromatic residues Trp-211 and Phe-215 in Gαi3 (Fig. 7B), which are predicted to form extensive hydrophobic contacts with PLCD4 GBA motif. More specifically, residues Leu-224, Phe-227, and Leu-228 of PLCD4 are located on one side of the α-helical region of the GBA motif that docks onto the Gαi3 hydrophobic groove dominated by Trp-211 and Phe-215 (Fig. 7A, right). Again, this is a feature conserved with previously described GBA motifs, as all of them have hydrophobic residues in these positions (Fig. 1A), and mutation of the invariable phenylalanine almost completely abolishes binding for GIV, DAPLE, Calnuc, and NUCB2 (23, 25, 26). When we tested whether this was also the case for PLCD4, we found that mutation of Phe-227 to alanine (“FA”) caused a marked but incomplete decrease in Gαi3 binding compared with PLCD4 GBA WT (Fig. 7C). This incomplete reduction in binding is also in agreement with the incomplete ablation of activity in yeast-based assays using the same mutant (Fig. 4, A and B). We reasoned that PLCD4 binding to Gαi3 might be stabilized by contacts established by the other conserved hydrophobic residues in the GBA motif (i.e. Leu-224 and Leu-228), so we tested whether multiposition mutants F227A/L228A (“FALA”) and L224A/F227A/L228A (“LAFALA”) were more efficient in disrupting the interaction. We found that this is the case because both FALA and LAFALA displayed undetectable binding to Gαi3 (Fig. 7C). These results not only indicate that this GBA/G-protein interface has properties slightly different from those described for other GBA motifs, as suggested by the Gαi3 mutants profiling results (Fig. 7A), but also identify the double mutant F227A/L228A as a useful tool to completely blunt the physical coupling of PLCD4 GBA motif to Gαi3.
PLCD4 GBA motif activates Gαi3 in vitro
Having identified a powerful tool to assess the specificity of PLCD4 GBA motif action (i.e. the FALA mutant), we set out to determine whether this motif had GEF activity like previously characterized GBA proteins and related synthetic peptides (23, 25–27, 41, 43). We have previously validated that steady-state GTPase assays accurately reflect the effect of the GBA proteins GIV, DAPLE, Calnuc, and NUCB2 on Gαi3 nucleotide exchange, providing results that are indistinguishable from those obtained from GTPγS binding experiments done in parallel (25, 26, 29, 42). This is explained well by the much faster rate of GTP hydrolysis versus nucleotide exchange (2 orders of magnitude for Gαi) (52), which makes GDP/GTP exchange the rate-limiting step under steady-state GTPase conditions. Using this assay, we found that increasing amounts of the GST-fused PLCD4 GBA motif increased the rate of nucleotide exchange on purified His–Gαi3 more than 2-fold (Fig. 7D), which is comparable with levels previously observed for other regulators of the same family (23, 25–27, 41, 43). This GEF activity was specific because parallel reactions with the G-protein binding–deficient PLCD4 FALA mutant revealed no activity (Fig. 7D). When analogous experiments were performed with Gαi3–Gβγ heterotrimers instead of with free Gαi3, we found that PLCD4 WT, but not the FALA mutant, also leads to increased G-protein activity, although somewhat less efficiently (∼1.7-fold for the trimer versus ∼2-fold for free Gαi3) (Fig. 7D). Similar results were obtained for His–GIV-CT (aa 1660–1870, containing the GBA motif), in that it also increased activity with Gαi3–Gβγ heterotrimers but somewhat less efficiently than with free Gαi3 (∼2-fold for the trimer (Fig. 7D) versus ∼2.5–3-fold for Gαi3) (23, 31, 42). The lower GEF efficacy on heterotrimers compared with monomeric Gαi is consistent with previously reported competition between GBA motifs and Gβγ for binding to Gαi (23, 26). These results indicate that the GBA motif of PLCD4 has GEF activity in vitro.
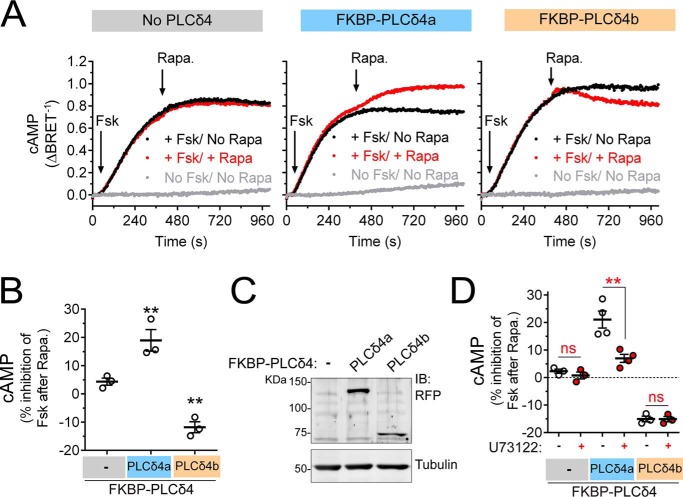
PLCD4 GBA motif activates G-protein signaling in mammalian cells
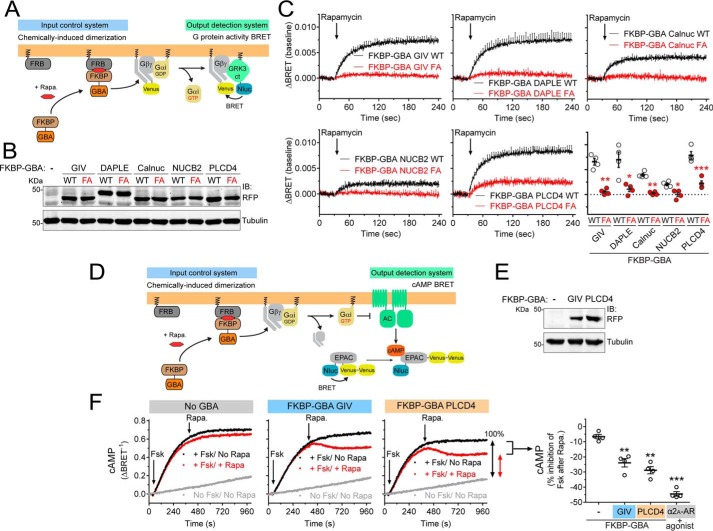
Next, we investigated whether the GBA motif of PLCD4 works as an activator of G-proteins in cells. For this, we took advantage of a recently developed experimental system used to demonstrate that membrane recruitment of GIV is sufficient to trigger G-protein activation in cells (36, 53). The underlying principle of this system is that Gαi subunits are constitutively attached to cellular membranes, whereas GIV is predominantly in the cytosol, such that only upon recruitment of GIV (or its GBA motif in isolation) to membranes can it activate G-proteins (36, 53). Experimentally, membrane recruitment is achieved via chemically-induced dimerization (CID) of FKBP and FRB domains with rapamycin (54) (Fig. 8A, left). FRB is fused to the first 11 aa of Lyn (Lyn11–FRB), which targets it to the plasma membrane (55, 56), whereas FKBP is fused to a GBA sequence (FKBP–GBA) and remains in the cytosol. Thus, rapamycin induces the translocation of FKBP–GBA to the plasma membrane, where Lyn11–FRB localizes. The effect of this translocation on G-protein activation is monitored in real time by bioluminescence resonance energy transfer (BRET) using a genetically-encoded biosensor system. In this system, dissociation of Gαβγ heterotrimers upon activation leads to the association of free Venus–Gβγ (BRET acceptor) with the C-terminal domain of its effector GRK3 fused to an enhanced luciferase (Nluc, BRET donor), which leads to an increase in BRET (Fig. 8A, right) (57, 58). Using this experimental system, the amplitude of G-protein activation upon GIV GBA translocation is only slightly lower than that obtained upon activation of a Gi-coupled GPCR like the muscarinic M4 receptor with saturating concentrations of ligand (36, 53). To assess the G-protein regulatory activity of PLCD4 and systematically compare it with other GBA proteins, we generated FKBP fusions of the GBA motifs of GIV, DAPLE, Calnuc, and NUCB2 as well as of PLCD4 (Fig. 8B), and we expressed them in HEK293T cells along with Lyn11–FRB and the components of the BRET biosensor. As in our previous report (36), addition of rapamycin induced a rapid increase in BRET in cells expressing FKBP–GBA GIV WT but not FKBP–GBA GIV FA (Fig. 8C), indicating GBA-dependent G-protein activation upon membrane translocation. Similarly, all the other GBA motifs, including PLCD4, led to an increase in BRET upon rapamycin stimulation that was not recapitulated by the corresponding FA mutants (Fig. 8C). However, the amplitude of the BRET responses was different depending on the specific GBA motif used (Fig. 8C). In fact, the order of potency of the GBA motifs in this readout matched very well with their affinity for Gαi3 determined by FP using the exact same 31-mer GBA sequences (Fig. 6A), with PLCD4 as the most potent activator and highest affinity binder. Also consistent with the relative affinity of different GBA motifs for Gαi3, we found that the corresponding GBA sequence of NOC2L only induced a marginal increase of BRET in this assay, which was almost negligible compared with PLCD4 (Fig. S4). One difference between the FKBP–GBA motif of PLCD4 and the other proteins is that the FA mutation failed to completely abolish the BRET response (Fig. 8C). This is not surprising given that this mutant still binds weakly to Gαi3 (Fig. 7C) and moderately activates G-protein signaling in yeast (Fig. 4, A and B). When PLCD4 mutants “FALA” and “LAFALA,” which completely disrupt Gαi3 binding and GEF activity in vitro (Fig. 7, C and D), were compared side-by-side with FKBP–PLCD4 GBA WT and the FA mutant, we observed no measurable G-protein activation upon rapamycin stimulation as determined by BRET (Fig. S5). These results confirm the specificity of the GBA-mediated activation of G-proteins by PLCD4.
Figure 8.
PLCD4 GBA motif activates G-protein signaling in cells. A, left, translocation of GBA sequences to membranes is controlled by CID using the FKBP–rapamycin–FRB system. FKBP-fused GBA is recruited to membranes upon rapamycin-induced binding of FKBP to the FRB domain that is fused to a membrane-targeting sequence (Lyn11). Right, G protein activation is determined by BRET. Dissociation of Gαi3/Gβγ trimers upon G-protein activation leads to the release of Venus-tagged Gβγ (V-Gβγ), which binds to the C-terminal domain of GRK3 fused to nanoluciferase (GRK3ct-Nluc) and causes an increase in BRET signal. B, immunoblot of FKBP-fused GBA motifs (WT or FA mutants) of human GIV (aa 1671–1701), DAPLE (aa 1661–1691), Calnuc (aa 303–333), NUCB2 (aa 304–334), or PLCD4 (aa 213–243) expressed in HEK293T cells used for the experiments shown in C. C, rapamycin-induced translocation of FKBP-fused WT GBA motifs (black) leads to G-protein activation as determined by BRET, and FA mutations (red) abolish or diminish this response. HEK293T cells expressing all the required assay components and transfected with equal amounts of plasmids encoding the FKBP–GBA fusions (0.1 μg per well) were treated with rapamycin (0.5 μm) at the indicated time point (arrow). Results are the average ± S.E. (shown only at 5-s intervals for clarity) of n = 4 expressed as increase in BRET after rapamycin stimulation (ΔBRET, baseline corrected by subtraction of the BRET values before stimulation). Lower right panel, scatter plot of ΔBRET 90 s after rapamycin addition for the indicated FKBP–GBA constructs. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with the respective WT using the Student's t test. The graph and immunoblot bands for FKBP–GBA PLCD4 have been reused in Fig. S4 for comparison with other results. D, left, translocation of GBA sequences to membranes is controlled by CID using the FKBP–rapamycin–FRB system as in A. Right, cAMP levels are measured by BRET using the Nluc–EPAC–VV sensor. Gαi3-GTP negatively regulates adenylyl cyclase (AC), thereby diminishing cAMP levels induced upon forskolin-mediated activation of AC. E, immunoblot of FKBP-fused GBA motifs of human GIV or PLCD4 expressed in HEK293T cells used for the experiments shown in F. F, rapamycin-induced translocation of FKBP–GBA GIV or FKBP–GBA PLCD4 inhibits forskolin-induced cAMP levels. HEK293T cells expressing the cAMP sensor Nluc–EPAC–VV, Lyn11–FRB, and the indicated FKBP-fused GBA constructs were treated at the indicated times (arrows) with forskolin alone (3 μm, black), forskolin followed by rapamycin (0.5 μm, red) or no compound (gray). One representative experiment is shown on the left graphs, and a scatter plot of rapamycin-induced inhibition of forskolin-stimulated cAMP levels from n = 4 is shown on the right. The scatter plot includes results of analogous experiments with cells expressing the α2A adrenergic receptor and stimulated with brimonidine (5 μm) instead of rapamycin for comparison. **, p < 0.01; ***, p < 0.001 compared with the control cells (−) using the Student's t test.
To further substantiate the conclusion that PLCD4 GBA activates G-protein signaling in cells, we investigated whether it regulates a signaling event downstream of G-protein activation (rather than measuring activation at the level of the G-protein as described above). The canonical effectors of G-proteins of the Gi family are adenylyl cyclases (AC), the enzymes responsible for the synthesis of cAMP. It is well-established that upon Gi activation, GTP-bound Gαi subunits bind AC to negatively modulate its activity (59). We used the same rapamycin-induced translocation system as above to trigger the action of FKBP-fused GBA motifs of PLCD4 and GIV (as a control) on G-proteins (Fig. 8D). The output detection system was a BRET-based biosensor (Nluc–EPAC–VV) that monitors intracellular cAMP levels in real time (Fig. 8D) (57, 60). Briefly, HEK293T cells were stimulated with forskolin to induce the direct activation of AC and were subsequently treated with rapamycin to translocate the GBA motifs to the plasma membrane. Although rapamycin had no effect on the forskolin-induced cAMP levels in control cells not expressing any FKBP–GBA fusion, it led to a substantial decrease of cAMP (∼25%) in cells expressing FKBP–GBA GIV or FKBP–GBA PLCD4 (Fig. 8, D–F). The response observed for FKBP–GBA PLCD4 was abolished when the same experiment was repeated using the Gαi-binding–deficient FALA mutant (Fig. S6). Although the amplitude of the GBA motif-mediated responses was not as marked as the one observed upon maximal activation of a Gi-coupled GPCR like the α2A-adrenergic receptor, which led to an ∼40–45% reduction in cAMP (Fig. 8F), these findings are consistent with the activation of Gi-dependent signaling by the GBA motifs of both GIV and PLCD4. Taken together, these results demonstrate that the GBA motif of PLCD4 can activate G-protein signaling in cells.
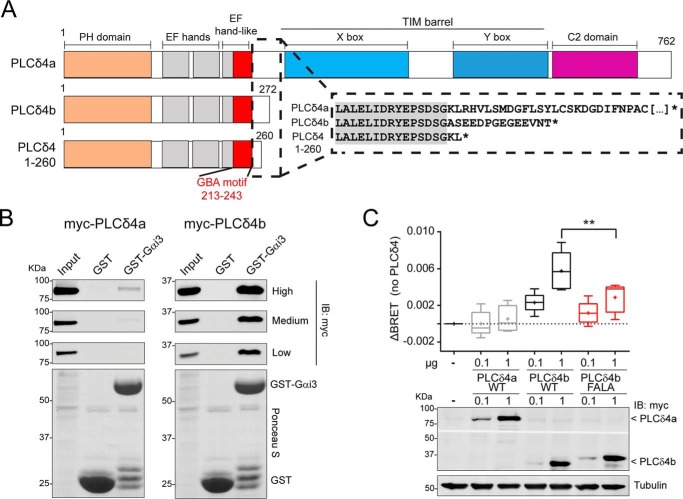
Gαi3 binds more strongly to PLCδ4b than to PLCδ4a
Although the results above strongly indicate that the GBA motif of PLCD4 leads to robust G-protein activation, we set out to characterize whether this was the case for the GBA motif in its native context of PLCD4 proteins. Based on the curated annotations for human PLCD4 in UniProt, this gene encodes for two different isoforms of PLCδ4, i.e. PLCδ4a (61) and the shorter isoform PLCδ4b (Fig. 9A) (62). PLCδ4a is considered the canonical isoform of PLCδ4 and contains all of the domains characteristic of this family (in order from the N to C terminus): the PH domain that can bind phosphoinositides, EF-hands, catalytic TIM barrel (with X- and Y-boxes) responsible for the phospholipase activity, and the C-terminal C2 domain (Fig. 9A). Interestingly, PLCδ4 contains two canonical EF-hands (although EF-hands in PLCδ or PLCβ isoforms do not bind Ca2+ (63–65)) followed by an EF-hand–like domain (203–237 amino acids) of uncharacterized function. The GBA motif of PLCD4 characterized in this work overlaps with this EF-hand–like domain, suggesting that G-protein regulation might be its actual function. It is also interesting that the PLCδ4b isoform contains this EF-hand–like/GBA sequence but is truncated before the catalytic TIM barrel, which makes it a protein without PLC activity but potentially with G-protein regulatory activity. This idea is supported by our results in yeast-based experiments (Fig. 4, A and B), which showed GBA-dependent G-protein activation by a PLCD4 construct that closely resembles PLCδ4b (i.e. corresponding to amino acids 1–260) (Fig. 9A).
Figure 9.
PLCδ4b binds Gαi3 more efficiently than PLCδ4a and activates G-proteins in cells. A, bar diagrams of human PLCδ4 isoforms and constructs. PLCδ4a contains (from N to C terminus) a PH domain, two EF-hands, an EF-hand–like domain that overlaps with the GBA motif, a catalytic TIM barrel (with X- and Y-boxes), and a C2 domain. PLCδ4b and PLCδ4(1–260) are truncated before the catalytic TIM barrel. Inset, partial sequence alignment of PLCδ4a, PLCδ4b, and PLCδ4(1–260). The C-terminal sequence of PLCδ4b differs from PLCδ4a in the 14 aa preceding its stop codon, and the PLCδ4(1–260) construct used in previous experiments (Figs. 2–5) is very similar to PLCδ4b. B, Gαi3 binding of two PLCδ4 isoforms. Approximately 30 μg of purified GST-Gαi3 or GST immobilized on GSH-agarose beads were incubated with lysates of HEK293T cells expressing either myc-PLCδ4a (left) or myc-PLCδ4b (right) in the presence of GDP. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. Three different capture intensities (low, medium, and high) are shown for the Myc immunoblotting. Input = 2.5% of the total amount of lysate added in each binding reaction. One representative experiment of at least three is shown. C, PLCδ4b, but not PLCδ4a, activates G-protein signaling in cells via its GBA motif. HEK293T cells were transfected with the indicated amounts of plasmids for the expression of myc-PLCδ4a (WT) or myc-PLCδ4b (WT or FALA mutants) and all the required components of the BRET-based G-protein activity assay described in Fig. 8A (Gαi3, V-Gβγ, and GRK3ct-Nluc). Results from n = 5 are presented as the increase in BRET (ΔBRET) compared with cells not expressing PLCδ4 in a box-and-whisker plot (median with boxes of 25–75%), and error bars indicate the range. Cross-marks indicate the average). An immunoblot of a representative experiment is shown below the graph. **, p < 0.01 compared with the respective WT using the Student's t test.
The finding of a GBA sequence that overlaps with an EF-hand fold is not novel, as Calnuc and NUCB2 also share this property (25). Moreover, results from our bioinformatics searches and peptide arrays (Fig. 1) showed an enrichment for GBA motifs located in EF-hands, i.e. 10 out of the 29 GBA hits in the peptide array correlated with EF-hands. These findings are explained well by the overlap of the consensus GBA motif sequence ψTψX(D/E)Fψ with the C-terminal segment of the consensus EF-hand sequence DXDXDGψ(D/S/T)XX(D/E)F (underlines indicate the overlap). However, our data also demonstrate that G-protein binding (and regulation) is not a widespread function of EF-hands because only one (i.e. PLCD4) of the 10 candidate GBA motifs turned out to have G-protein regulatory activity. Instead, G-protein regulatory activity seems to correlate with atypical EF-hand sequences, likely without Ca2+-binding function, as exemplified by the EF-hand–like of PLCD4. Interestingly, the second EF-hand of Calnuc, which is the one overlapping with its GBA motif, is also considered a noncanonical EF-hand based on sequence divergence (66, 67), and it only regulates G-proteins when it is not bound to Ca2+ (25).
To assess protein–protein binding of PLCδ4 isoforms to G-proteins, lysates of HEK293T cells expressing Myc-tagged PLCδ4a and PLCδ4b were used in pulldown assays with purified GST–Gαi3. We found that PLCδ4b binds Gαi3 much more robustly than PLCδ4a (Fig. 9B). A possible explanation for this difference is that the GBA motif is more accessible to the G-protein in the shorter isoform (in which it is located near the C-terminal end) than in the longer one (in which it is sandwiched between the EF-hands and the catalytic TIM barrel). Regardless of the explanation for the difference in Gαi3 binding among isoforms, these results suggest that PLCδ4b might be a better regulator of G-protein signaling in cells than PLCδ4a.
PLCδ4b but not PLCδ4a promote G-protein activation in cells
We used the BRET-based biosensor that directly monitors G-protein activity described above (i.e. V-Gβγ/GRK3ct-Nluc) to assess the function of PLCδ4a and PLCδ4b in cells. Myc-tagged constructs were expressed in HEK293T cells, and BRET was measured under steady-state conditions. Consistent with the protein–protein binding results, we found that expression of PLCδ4b but not PLCδ4a led to an increase in G-protein activity as determined by BRET (Fig. 9C). This BRET enhancement was not reproduced by the PLCδ4b FALA mutant (which is GEF-deficient, Fig. 7D), indicating that PLCδ4b activates G-proteins in mammalian cells via its GBA motif. Equivalent results were obtained using a construct consisting of PLCD4 aa 1–260 (Fig. S7), which is similar to PLCδ4b and identical to the construct used in yeast-based experiments (Figs. 2 and 4) and in in vitro protein-binding experiments (Figs. 3 and 5). These results further validate that PLCD4 exhibits consistent G-protein regulatory activity across different experimental systems.
To further substantiate that PLCδ4b activates G-protein signaling in cells, we investigated whether it regulates cAMP levels. For this, we fused full-length PLCδ4b (and PLCδ4a) to FKBP and tested the effect of recruiting the construct to the plasma membrane on forskolin-induced cAMP levels. We found that membrane recruitment of full-length PLCδ4b led to a decrease in cAMP levels (Fig. 10, A and B), much like the recruitment of its GBA motif (Fig. 8, E and F). PLCδ4b-mediated inhibition of cAMP was abolished upon introduction of the FALA mutation (Fig. S8), indicating that the observed response is GBA motif-dependent. The effect of full-length PLCδ4b was more modest than that of its GBA motif alone (∼12 versus ∼25% (Fig. 8), respectively), which might be explained by their relative levels of expression (Fig. S8). Based on our results from BRET-based G-protein activity assays in Fig. 9, we predicted that PLCδ4a would not inhibit forskolin-induced cAMP levels. Surprisingly, we found that, even though PLCδ4a did not inhibit cAMP, it caused an enhancement of forskolin-induced cAMP levels (Fig. 10, A and B). We reasoned that this enhancement of forskolin-induced cAMP levels was due to its catalytic PLC activity (which is absent in PLCδ4b). This is not far-fetched because several AC isoforms are activated by increased Ca2+ levels or Ca2+-dependent mechanisms (59), a direct downstream effect of PLC activity. We found that the effect of PLCδ4a on cAMP was indeed dependent on its PLC activity, as it was inhibited by the small molecule PLC inhibitor U73122 (Fig. 10D). However, U73122 had no effect on PLCδ4b-mediated inhibition of forskolin-induced cAMP levels (Fig. 10D), which is consistent with the lack of a catalytic PLC domain in this isoform and supports the specificity of the effect of the inhibitor on PLCδ4a. Taken together, these findings confirm that, instead of working as a phospholipase, PLCδ4b is a nonreceptor activator of heterotrimeric G-proteins that functions via a defined GBA motif.
Figure 10.
PLCδ4b inhibits forskolin-induced cAMP formation. A–C, BRET-based cAMP measurements upon rapamycin-mediated recruitment of FKBP-PLCδ4a or FKBP-PLCδ4b were performed exactly as described in Fig. 8D. HEK293T cells expressing the cAMP sensor Nluc–EPAC–VV, Lyn11-FRB, and the indicated FKBP-PLCδ4 constructs (0.25 μg DNA per well) were treated at the indicated times (arrows) with forskolin alone (3 μm, black), forskolin followed by rapamycin (0.5 μm, red), or no compound (gray). One representative experiment is shown in A, and a scatter plot of rapamycin-induced inhibition of forskolin-stimulated cAMP levels from n = 3 is shown in B. **, p < 0.01 compared with the control cells (−) using the Student's t test. An immunoblot of cell lysates from one representative experiment is shown in C. D, BRET-based cAMP measurements upon rapamycin-mediated recruitment of FKBP-PLCδ4a or FKBP-PLCδ4b were performed and quantified as described in A–C in the presence of the PLC inhibitor U73122 (+) (10 μm, 60 min preincubation) or DMSO (−). Results are the average ± S.E. of n = 3–4. **, p < 0.01; ns = not significant compared with control cells pretreated with DMSO (−) using the Student's t test.
Discussion
The main advances reported here are three: (i) the validation of a pipeline to identify and validate novel GBA motif–containing G-protein regulators; (ii) the discovery of PLCδ4b as a novel nonreceptor G-protein activator with a GBA motif; and (iii) a systematic characterization of the biochemical, structural, and signaling properties of all the GBA motifs described to date. Together, these findings advance our understanding of an understudied family of G-protein regulators with important biological functions and establish a framework for their discovery and characterization.
The existence of a specific sequence that defines a group of nonreceptor G-protein activators (i.e. the GBA motif) is useful but insufficient information to reliably identify novel members of this family of G-protein regulators based solely on sequence similarity. This contrasts with other families of G-protein regulators like RGS GAPs, as many RGS proteins were discovered based on sequence similarity. While the GBA motif is a short sequence found in only a few proteins, RGS proteins are defined by a longer sequence (i.e. the RGS box, of about 120 aa) present in a larger number of proteins, thereby providing a more robust training set for bioinformatics searches. The findings reported here help to overcome the shortcomings of sequence-based searches of GBA regulators in two ways. First is that increasing the number of GBA sequences that bind to G-proteins will be helpful to improve the training set for future bioinformatics searches. Second, and probably more useful, is the pipeline of high and medium throughput assays to filter out false positives from sequence searches. Dozens or hundreds of GBA motif candidates can be rapidly assessed for the essential biochemical property of binding to G-proteins by using peptide arrays, whereas yeast is used as a secondary system of facile genetic manipulation to test for G-protein signaling activity. It is tempting to speculate about the implementation of a similar pipeline to discover new members of another family of G-protein regulators such as GDIs that contain the so-called GoLoco motif. Much like the GBA motif family, these regulators are characterized by a short sequence (30–40 aa) present in a relatively small number of proteins, and the pipeline of assays described here is compatible with their characterization.
Related to the above, our comprehensive characterization of all the GBA motifs described to date in mammals (GIV, DAPLE, Calnuc, NUCB2, and now PLCD4) provides informative benchmarks and tools to assess the biochemical, structural, and signaling properties of this family. Consistent with prior data obtained through the characterization of GIV's GBA motif by mutagenesis (23, 29, 31), the potency of different GBA motifs in triggering G-protein activation in cells (Fig. 8) correlates with their affinity for G-proteins in vitro (Fig. 6). Likewise, all share a common binding site on the G-protein, although the newly identified GBA motif in PLCδ4 has slightly different requirements to establish the protein–protein interaction. Collectively, it appears that the C-terminal region of the GBA motif downstream of PLCδ4 Phe-227, which diverges in sequence from other GBA motifs (Fig. 6A), might establish different protein–protein contacts, as indicated by the requirement of simultaneous mutation of Phe-227 and Leu-228 to alanine to completely disrupt binding and by the different impact of the Gαi3 W258F mutation on PLCδ4 binding compared with other GBA motifs. Elucidating such nuanced differences in the mode of G-protein binding by GBA motifs will require atomic resolution structures.
The new GBA motif–containing protein we identified here, PLCδ4, presents a number of interesting properties. The most striking one is that we have been able to validate the G-protein regulatory activity for one of the two annotated isoforms, PLCδ4b. Although both PLCδ4a and PLCδ4b have been reported to be ubiquitously expressed in different tissues (61, 62), PLCδ4b appears to be a G-protein regulator rather than a phospholipase. This is because this isoform is composed only of the PH domain and the EF-hands, lacking the catalytic TIM barrel required for lipase activity. Interestingly, it has been previously reported that the PH domain of PLCδ4 in isolation, but not the full-length protein corresponding to PLCδ4a, localizes to the plasma membrane via phosphoinositide binding, and different manipulations that decrease phosphatidylinositol 4,5-bisphosphate levels lead to the release of PLCδ4 PH domain from the plasma membrane into the cytosol (68). Because PLCδ4b consists primarily of the PH domain with only a C-terminal extension containing the EF-hands, it is likely that its subcellular localization and regulation by phosphoinositides recapitulates the properties of the PH domain in isolation. This raises the interesting possibility that the G-protein regulatory activity of PLCδ4b might be controlled by alterations in phosphoinositide levels. We have recently shown that the main mechanism by which GBA proteins regulate G-protein signaling is by spatial proximity (36), i.e. G-protein activation occurs when the GBA motif is recruited to membranes in physical proximity to the target G-protein, Gαi, which is constitutively membrane-bound. Thus, if phosphoinositides regulate the localization of PLCδ4b at the plasma membrane, they might also impact its G-protein regulatory function. At a physiological level, it is not possible at this time to envision what function the GBA motif of PLCδ4 might have. PLCδ4 function has not been extensively characterized, but insights from mice in which the PLCδ4 gene has been knocked out indicate that it is required for male fertility (69, 70). However, this function is likely exerted by the PLCδ4a isoform, and we have not observed G-protein regulatory activity for this isoform. Although we cannot rule out that the GBA motif of PLCδ4a might become functional under specific circumstances that would make the GBA motif accessible to G-proteins (as PLC enzymes are believed to undergo significant conformational and/or spatial rearrangements (71, 72)), the only available evidence for G-protein regulatory activity is for the PLCδ4b isoform. Unfortunately, the function(s) of this isoform are still unknown. Further work will be required to elucidate the role of the GBA motif in the biological functions of PLCδ4.
Experimental procedures
Reagents and antibodies
Unless otherwise indicated, all reagents were of analytical grade and obtained from ThermoFisher Scientific or Sigma. Cell culture media and goat anti-rabbit and goat anti-mouse Alexa Fluor 680 antibodies were from Life Technologies, Inc. Fluorescein di-β-d-galactopyranoside was from Marker Gene Technologies, and the protein inhibitor mixture was from Sigma (catalog no. S8830). Leupeptin, pepstatin, and aprotinin were from Gold Biotechnology. All restriction endonucleases and E. coli strain BL21(DE3) were from ThermoFisher Scientific. E. coli strain DH5α was purchased from New England Biolabs. Pfu Ultra DNA polymerase was purchased from Agilent. Rabbit antibodies raised against Gαi3 (C-10) and Gβ (M-14) were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antibodies raised against α-tubulin (catalog no. T6074) and His tag (catalog no. H1029) were from Sigma and for HA tag (12CA5) were from Roche Applied Science. Rabbit antibodies for ppERK (catalog no. 4370) and mouse antibodies for Myc (catalog no. 9B11) were from Cell Signaling. Rabbit polyclonal antibody for RFP was from Abcam (catalog no. 62341). Goat anti-rabbit and goat anti-mouse IRDye 800 F(ab′)2 were from Li-Cor Biosciences (Lincoln, NE).
Bioinformatics searches and analyses
The search for proteins with a GBA motif in Homo sapiens was performed in ScanSite 3 using the QuickMatrix method (44). Briefly, we used the sequences of known GBA motifs of the proteins GIV, DAPLE, Calnuc, NUCB2, GBAS-1, and the synthetic peptides KB-752 and GSP (23, 25–27, 41, 43) to design the pattern as (VLIM)(T)(VLIM)X(DE)F(VLIM) and generate the corresponding position-specific scoring matrix (PSSM) to search in the H. sapiens proteome in the NCBI Protein GenPept/RefSeq database.
Synthesis of peptide libraries and Gαi3 overlay
Libraries of immobilized peptides were produced by automatic SPOT synthesis on continuous cellulose membrane supports (Whatman 50 cellulose membranes) using Fmoc (fluoren-9-ylmethoxycarbonyl) chemistry with the AutoSpot-Robot ASS 222 (Intavis Bioanalytical Instruments AG) (73). Candidate GBA motif peptides were synthesized as 24-mer peptides in which the invariable phenylalanine of the core of the GBA motif was in the 12th position. Individual peptide–cellulose complexes were solubilized and re-spotted on Celluspot® slides for subsequent probing. Slides were primed in binding buffer (4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 5 mm MgCl2, 1 mm DTT, 30 μm GDP, 1% (v/v) Triton X-100) and blocked for 1 h in the same buffer supplemented with 5% (w/v) BSA. Slides were incubated for 2 h at room temperature with rat His–Gαi3 at 20 μg/ml (∼0.5 μm) in the same buffer. After four washes, slides were sequentially incubated with primary (rabbit anti-Gαi3, 1: 250; 90 min) and secondary (goat anti-rabbit Alexa Fluor 680; 60 min) antibodies. As a control for nonspecific detection of the peptide sequence by the primary and/or secondary antibody, a replicate peptide array slide was processed in parallel under identical conditions except that no His–Gαi3 was added. Images were acquired, and spot intensity was quantified in an Odyssey IR scanner (Li-Cor). Specific binding was quantified by subtracting the signal intensity for each spot in the slide without Gαi3 incubation from the corresponding spot signals in the slide with Gαi3 incubation and normalized by dividing the resulting number by the intensity of the spot after Coomassie Blue staining. Images were processed using the ImageJ software (National Institutes of Health) and assembled for presentation using Photoshop and Illustrator (Adobe).
Plasmid constructs
Cloning of GBA candidates into plasmids for expression in yeast, bacteria, or, in some cases, mammalian cells was done using a ligation-independent cloning (LIC) system (74). Briefly, sequences corresponding to the GBA candidates indicated in Fig. S1 were amplified by PCR from plasmids obtained from OpenBiosystems, Addgene, GenScript, or individual investigators. The amplified fragments contained extensions at the 5′ and 3′ ends that made them compatible with any of the following LIC plasmids: pLIC-YES2 (yeast, described in Ref. 27), pLIC-GST (bacteria), and pLIC-myc (mammalian). pLIC-GST and pLIC-myc plasmids were kindly provided by J. Sondek (University of North Carolina at Chapel Hill) (75). pLIC-YES2-Ric-8A has been described before (27). Cloning of the pET28b-Gαi3 and pGEX-Gαi3 plasmids for the expression of rat His–Gαi3 or GST–Gαi3, respectively, in bacteria have been described previously (23). pET24a-Gαi3 for the expression of human Gαi3 has been described in Ref. 29. The GBA motif sequences of human GIV (aa 1671–1701), human DAPLE (aa 1661–1691), human Calnuc (aa 303–333), human NUCB2 (aa 304–334), or human PLCD4 (aa 213–243) inserted into the NruI/BamHI sites of the pmRFP-FKBP-pseudojanin plasmid (Addgene catalog no. 37999), which results in the replacement of the pseudojanin sequence by the GBA motifs to generate constructs that encode for RFP–FKBP–GBA fusions. Plasmids encoding the GBA motif of human PLCD4 (aa 213–243) or of human GIV (aa 1660–1701) fused to GST were generated by LIC using the pLIC-GST vector mentioned above. The pT7-5-Gαi1 plasmid (which bears an internal hexahistidine tag in the b/c loop of Gαi1) was kindly provided by H. Hamm (Vanderbilt University) (76), and the pbb131 plasmid encoding N-myristoyltransferase (NMT) (77) was a gift from Maurine Linder (Cornell University). The plasmid encoding Lyn11–FRB has been described previously (36). pcDNA3.1-A1R, pcDNA3.1-Venus(1–155)-Gγ2 (VN-Gγ2), and pcDNA3.1-Venus(155-239)-Gβ1 (VC-Gβ1) were kindly provided by N. Lambert (Augusta University, GA) (58). pcDNA3.1-masGRK3ct-Nluc and pcDNA3.1-Nluc–EPAC–VV (57) were gifts from K. Martemyanov (Scripps Research Institute, FL). The pcDNA3-Gαi3 plasmids for the expression of rat Gαi3 in mammalian cells and the pET28b-GIV(1660–1870) plasmid for the expression of His–GIV-CT in E. coli have been described previously (23, 42). Plasmids for the expression of N-terminally tagged myc-PLCδ4a, myc-PLCδ4b, and myc-PLCδ4(1–260) were generated by PCR amplification from full-length PLCD4 and insertion into pLIC-myc. The primers for myc-PLCδ4b included a 3′ extension to add the amino acids in its C terminus that diverge from myc-PLCδ4a and precede the stop codon. Plasmids encoding FKBP fusions of PLCδ4a and PLCδ4b were generated by PCR amplification and inserted into the NruI/BamHI sites of the pmRFP-FKBP-pseudojanin as described above for the isolated GBA motifs.
Yeast strains and manipulations
The previously described (48) Saccharomyces cerevisiae strain CY7967 [MATα GPA1(1–41)-Gαi3 far1Δ fus1p-HIS3 can1 ste14:trp1:LYS2 ste3Δ lys2 ura3 leu2 trp1 his3] (kindly provided by James Broach, Pennsylvania State University) was used for all yeast experiments. The main features of this strain are that the only pheromone-responsive GPCR (Ste3) is deleted, the endogenous Gα-subunit Gpa1 is replaced by a chimeric Gpa1(1–41)-human Gαi3 (36–354), and the cell cycle arrest-inducing protein far1 is deleted. In this strain, the pheromone-response pathway can be up-regulated by the ectopic expression of activators of human Gαi3 and does not results in the cell cycle arrest that occurs in the native pheromone response (47, 48). Plasmid transformations were carried out using the lithium acetate method. CY7967 was first transformed with a centromeric plasmid (CEN TRP) encoding the lacZ gene under the control of the FUS1 promoter, which is activated by the pheromone-response pathway. The PFUS1::lacZ-expressing strain was transformed with pLIC-YES2 plasmids (2 μm, URA) encoding each one of the GBA candidate constructs or Ric-8A as described under “Plasmid constructs.” Double transformants were selected in synthetic defined (SD)-TRP-URA media. Individual colonies were inoculated into 3 ml of SDGalactose-TRP-URA and incubated overnight at 30 °C to induce the expression of the proteins of interest under the control of the galactose-inducible promoter of pLIC-YES2. This starting culture was used to inoculate 20 ml of SDGalactose-TRP-URA at 0.3 OD600. Exponentially growing cells (∼0.7–0.8 OD600, 4–5 h) were pelleted to prepare samples for subsequent assays (see “β-Gal activity assay” and “Yeast protein immunoblotting” below).
β-Gal activity assay
This assay was performed as described previously (78) with minor modifications. Pellets corresponding to 0.5 OD600 (in duplicates) were washed once with PBS + 0.1% (w/v) BSA and resuspended in 200 μl of assay buffer (60 mm Na2PO4, 40 mm NaH2PO4, 10 mm KCl, 1 mm MgCl2, 0.25% (v/v) β-mercaptoethanol, 0.01% (w/v) SDS, 10% (v/v) chloroform) and vortexed. 100 μl were transferred to 96-well plates and reactions started by the addition of 50 μl of the fluorogenic β-gal substrate fluorescein di-β-d-galactopyranoside (100 μm final). Fluorescence (excitation, 485 ± 10 nm/emission, 528 ± 10 nm) was measured every 2 min for 90 min at 30 °C in a Biotek H1 synergy plate reader. Enzymatic activity was calculated from the slope of fluorescence (arbitrary units) versus time (minutes). At least three independent experiments were measured for each condition, and the results were normalized (fold activation) to the activity in controls (strains carrying an empty pLIC-YES2 plasmid).
Yeast protein immunoblotting
This assay was performed as described previously (78, 79) with minor modifications. Briefly, pellets corresponding to five OD600 were washed once with PBS + 0.1% BSA and resuspended in 150 μl of lysis buffer (10 mm Tris-HCl, pH 8.0, 10% (w/v) trichloroacetic acid (TCA), 25 mm NH4OAc, 1 mm EDTA). 100 μl of glass beads were added to each tube and vortexed at 4 °C for 5 min. Lysates were separated from glass beads by poking a hole in the bottom of the tubes followed by centrifugation onto a new set of tubes. The process was repeated after the addition of 50 μl of lysis buffer to wash the glass beads. Proteins were precipitated by centrifugation (10 min, 20,000 × g) and resuspended in 60 μl of solubilization buffer (0.1 m Tris-HCl, pH 11.0, 3% SDS). Samples were boiled for 5 min and centrifuged (1 min, 20,000 × g), and 50 μl of the supernatant were transferred to new tubes containing 12.5 μl of Laemmli sample buffer and boiled for 5 min. Proteins (∼15–20 μl per lane) were separated by SDS-PAGE, blocked in PBS supplemented with 5% BSA, and analyzed by sequential incubation with primary and secondary antibodies. Primary antibodies were diluted as follows: ppERK (which recognizes yeast ppFus3), 1:2500; Myc, 1:1000; and α-tubulin, 1:2500. Secondary antibodies (goat anti-mouse IRDye 800 F(ab′)2, Li-Cor Biosciences, and goat anti-rabbit Alexa Fluor 680, Life Technologies, Inc.) were used at 1:10,000. Images were acquired in an Odyssey IR scanner (Li-Cor), processed using ImageJ software (National Institutes of Health), and assembled for presentation using Photoshop and Illustrator (Adobe).
Protein expression and purification
All His-tagged and GST-tagged proteins were expressed in BL21(DE3) E. coli transformed with the corresponding plasmids by overnight induction at 23 °C with 1 mm isopropyl β-d-1-thio-galactopyranoside. Protein purification was carried out following previously described protocols (23, 42). Briefly, bacteria pelleted from 1 liter of culture were resuspended in 25 ml of buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 10 mm imidazole, 1% (v/v) Triton X-100 supplemented with protease inhibitor mixture (leupeptin 1 μm, pepstatin 2.5 μm, aprotinin 0.2 μm, phenylmethylsulfonyl fluoride 1 mm)). For rat or human Gαi3, this buffer was supplemented with 25 μm GDP and 5 mm MgCl2. After sonication (four cycles, with pulses lasting 20 s/cycle, and with 1-min interval between cycles to prevent heating), lysates were centrifuged at 12,000 ×g for 20 min at 4 °C. The soluble fraction (supernatant) of the lysate was used for affinity purification on HisPur cobalt or GSH-agarose resins (Pierce) and eluted with lysis buffer supplemented with 250 mm imidazole or with 50 mm Tris-HCl, pH 8, 100 mm NaCl, 30 mm reduced GSH, respectively. GST-tagged proteins were dialyzed overnight at 4 °C against PBS. For rat or human His–Gαi3, the buffer was exchanged for 20 mm Tris-HCl, pH 7.4, 20 mm NaCl, 1 mm MgCl2, 1 mm DTT, 10 μm GDP, 5% (v/v) glycerol using a HiTrap desalting column (GE Healthcare). Validation of the quality of the human Gαi3 mutants used in Fig. 7B by trypsin protection assays has been shown previously (29). All protein samples were aliquoted and stored at −80 °C. Myristoylated Gαi1 was purified from BL21(DE3) E. coli bacteria co-expressing NMT as described above for His–Gαi3, except that after the cobalt affinity purification step the eluate was subjected to ion-exchange chromatography in a HiTrapQ HP column connected to an AKTA FPLC. Bovine retinal Gβγ (i.e. Gβ1γ1) was purified as described previously. Briefly, holo-transducin was purified from rod outer segment membranes isolated from dark-adapted bovine retina as described previously (80). The Gβγ complex was separated from the α-subunit of transducin on a Hitrap Blue-Sepharose column (GE Healthcare) and further purified using anion-exchange chromatography using a Hitrap-Q column as described earlier (81). Gαi3–Gβγ heterotrimers were reconstituted by incubating rat His–Gαi3 (20–25 μm) with a 2-fold molar excess of Gβγ overnight at 4 °C in 20 mm Tris-HCl, pH 7.4, 20 mm NaCl, 1 mm MgCl2, 1 mm DTT, 10 μm GDP, 5% (v/v) glycerol. Reconstituted heterotrimers were aliquoted and frozen at −80 °C.
In vitro protein-binding assays
GST pulldown assays were carried out as described previously (25, 42) with minor modifications. For Fig. 3, 20–25 μg (∼1.5–2 μm final) of GST or GST-fused GBA candidates (described under “Plasmid constructs”) were immobilized on GSH-agarose beads for 90 min at room temperature in PBS. Beads were washed twice with PBS, resuspended in 250 μl of binding buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 0.4% (v/v) Nonidet P-40, 10 mm MgCl2, 5 mm EDTA, 2 mm DTT, 30 μm GDP), and incubated 4 h at 4 °C with constant tumbling in the presence of 5 μg (∼0.5 μm final) of rat His–Gαi3. Beads were washed four times with 1 ml of wash buffer (4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 0.1% (v/v) Tween 20, 10 mm MgCl2, 5 mm EDTA, 1 mm DTT, and 30 μm GDP), and resin-bound proteins were eluted with Laemmli sample buffer by incubation at 37 °C for 10 min. The conditions for Fig. 5 were the same except that His–Gαi3 was preincubated 3 h at 30 °C in binding buffer supplemented with GDP (30 μm), GDP + AlF4− (30 μm AlCl3, 10 mm NaF), or GTPγS (30 μm) for PLCD4 or 30 min at 4 °C with GDP or GDP + AlF4− for NOC2L. The wash buffer was also supplemented with the same nucleotides. For Fig. 7C and Fig. S3 10 μg of the indicated GST-fused protein and 1 μg of the G-protein were used. For Fig. 9B and Fig. S2, 30 μg of GST–Gαi3 or GST were used with the lysates of ∼75% of a 10-cm dish of HEK293T cells transfected with myc-PLCδ4a, myc-PLCδ4b, or other GBA proteins shown in Fig. S2. Proteins were separated by SDS-PAGE and transferred to PVDF membranes. After blocking with PBS supplemented with 5% nonfat milk, membranes were analyzed by Ponceau S staining (GST-fused proteins) or sequential incubation with primary and secondary antibodies. Primary anti-His or anti-Myc antibodies were used at 1:2500 or 1:1000 dilution, respectively, and secondary antibodies (goat anti-mouse IRDye 800 F(ab′)2, Li-Cor Biosciences) were used at 1:10,000. Immunoblot quantification was performed by IR imaging following the manufacturer's protocols using an Odyssey imaging system (Li-Cor Biosciences). All Odyssey images were processed using the ImageJ software (National Institutes of Health) and assembled for presentation using Photoshop and Illustrator (Adobe).
Peptide synthesis
Peptides corresponding to the GBA motif of human GIV (residues 1671–1701, KTGSPGSEVVTLQQFLEESNKLTSVQIKSSS), human DAPLE (residues 1662–1695, SASPSSEMVTLEEFLEESNRSSPTHDTPSCRDDL), human Calnuc (residues 303–333, NVDTNQDRLVTLEEFLASTQRKEFGDTGEGW), human NUCB2 (residues 304–334, EVDTNKDRLVTLEEFLKATEKKEFLEPDSWE), human PLCD4 (residues 213–243 ESFSADGQKLTLLEFLDFLQEEQKERDCTSE), or human NOC2L (residues 5–35) were synthesized using the in situ neutralization protocol for Boc-Solid Phase Peptide Synthesis on a p-methylbenzhydrylamine resin (Novabiochem, 0.67 mmol/g, 100–200 mesh). Following chain elongation, 5,6-carboxyfluorescein was activated with hexafluorophosphate azabenzotriazole tetramethyluronium (HATU) and N,N-diisopropylethylamine (DIEA) (4, 4, and 8 eq regarding the amount of peptidyl–resin, respectively) and coupled to the resin-bound peptides at 65 °C for 1 h to yield the carboxyfluorescein-labeled peptides. Peptides were cleaved from the resin using a solution of hydrofluoric acid (HF) containing 5% anisole for 1 h at 0 °C. Next, the HF solution containing the peptides was removed under vacuum, and the resulting residues were crushed out with Et2O and filtered. The collected solids were redissolved in a 50% CH3CN/H2O solution containing 0.1% of trifluoroacetic acid (TFA), frozen down, and lyophilized. Crude peptides were purified by reverse phase-HPLC using an XBridge BEH C18 OBD prep column (130 Å, 5 μm, 19 × 150 mm) at a flow of 20 ml/min using H2O (0.1% TFA) and CH3CN (0.1% TFA) as eluents. The identity and final purity (>97%) of the peptides were determined by analytical RP-HPLC and MS (ESI-TOF).
FP-based peptide-binding assays
Fluorescence polarization measurements were carried out in 384-well plates (Black OptiPlate-384F, PerkinElmer Life Sciences). His–Gαi3 proteins (0–32 μm) and fluorescently-labeled peptide (0.025 μm) were mixed at room temperature for 10 min in a final volume of 20 μl of binding buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 0.4% (v/v) Nonidet P-40, 10 mm MgCl2, 5 mm EDTA, 2 mm DTT) supplemented with 30 μm GDP. Fluorescence polarization (excitation, 485 ± 10 nm/emission 528 ± 10 nm) was measured every 2 min for 30 min at room temperature in a Biotek H1 synergy plate reader to ensure that the signals were stable in time. Fluorescence polarization at different times was averaged, normalized to maximal binding, and fitted to a one-site binding model to determine the equilibrium dissociation constant (Kd) using Prism (GraphPad).
Protein structure modeling and docking
A model of Gαi3 bound to a portion of PLCD4's GBA motif (aa 219–231) was generated by protein–protein docking using ICM version 3.8–3 (Molsoft LLC., San Diego). Independent human Gαi3 and PLCD4 GBA models were first generated by homology using the X-ray crystal structure of Gαi1 bound to the synthetic GBA peptide KB-752 (41) as a starting template (Protein Data Bank code 1Y3A; chains B and F for Gαi3 and PLCD4, respectively). The side chain positions of the separate homology models were energetically minimized using a Monte Carlo–based method after initial construction. Protein–protein docking of the PLCD4 peptide to Gαi3 was then conducted in silico using a two-stage fast Fourier transform method (ICM, Molsoft LLC.). The best scoring solution was chosen to represent the binding mode. Model images were generated with PyMOL Molecular Graphics System (Schrödinger, LLC.).
Steady-state GTPase assay
This assay was performed as described previously (23, 25, 42). Briefly, His–Gαi3 (100 nm) was preincubated with different concentrations of GST–PLCD4 GBA (aa 213–243) for 15 min at 30 °C in assay buffer (20 mm Na-HEPES, pH 8, 100 mm NaCl, 1 mm EDTA, 25 mm MgCl2, 1 mm DTT, 0.05% (w/v) C12E10). GTPase reactions were initiated at 30 °C by adding an equal volume of assay buffer containing 1 μm [γ-32P]GTP (∼50 cpm/fmol). Duplicate aliquots (25 μl) were removed at 15 min, and reactions were stopped with 975 μl of ice-cold 5% (w/v) activated charcoal in 20 mm H3PO4, pH 3. Samples were then centrifuged for 10 min at 10,000 × g, and 500 μl of the resultant supernatant were scintillation counted to quantify released [32P]Pi. Background [32P]Pi detected at 15 min in the absence of G-protein was subtracted from each reaction, and data were expressed as percentage of the Pi produced by His–Gαi3 in the absence of GST–PLCD4. Background counts were <5% of the counts detected in the presence of G-proteins. Experiments with G-protein heterotrimers were performed essentially the same way. Briefly, reconstituted Gαi3–Gβγ heterotrimers (50 nm/100 nm) were preincubated with His–GIV-CT (2 μm) or GST–PLCD4 GBA (6 μm) before starting the reactions by the addition of [γ-32P]GTP as described above.
BRET-based G-protein activation assays
BRET experiments were conducted as described previously (36, 82). HEK293T cells (ATCC®, CRL-3216) were seeded on 6-well plates (∼400,000 cells/well) coated with gelatin and after 1 day were transfected using the calcium phosphate method with plasmids encoding the following constructs (DNA amounts in parentheses): Venus(155–239)–Gβ1 (VC-Gβ1) (0.2 μg); Venus(1–155)–Gγ2 (VN-Gγ2) (0.2 μg); Gαi3 (1 μg) mass-GRK3ct-Nluc (0.2 μg); and Lyn11–FRB (3 μg) along with FKBP–GBA constructs (0.1 μg). Approximately 16–24 h after transfection, cells were washed and gently scraped in warm PBS, centrifuged (5 min at 550 × g), and resuspended in Tyrode's solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 0.37 mm NaH2PO4, 24 mm NaHCO3, 10 mm HEPES pH 7.4, 0.1% glucose) at a concentration of ∼106 cells/ml. 25,000 cells were added to a white opaque 96-well plate (Opti-Plate, PerkinElmer Life Sciences) and mixed with the nanoluciferase substrate Nano-Glo (Promega, final dilution 1:200) for 2 min before measuring luminescence. Polar STAR plate reader (BMG Labtech) was used to measure luminescence signals at 460 ± 20 and 528 ± 10 nm at 28 °C, and BRET was calculated as the ratio between the emission intensity at 528 ± 10 nm divided by the emission intensity at 460 ± 20 nm. For the kinetic measurements shown in Fig. 8C and Fig. S5, BRET was determined every 0.24 s, and results were presented as increase in BRET after subtraction of the basal signal measured for 30 s before any stimulation (ΔBRET (baseline)). For the steady-state BRET measurement shown in Fig. 9C and Fig. S7, results were presented as difference in BRET compared with the signal in cells not expressing PLCD4 (ΔBRET (no PLCD4)). Protein samples from BRET experiments were prepared by centrifugation of HEK293T cells and resuspension in lysis buffer (20 mm HEPES, pH 7.2, 5 mm Mg(CH3COO)2, 125 mm K(CH3COO), 0.4% Triton X-100, 1 mm DTT, and protease inhibitor mixture). After clearing by centrifugation at 14,000 × g at 4 °C for 10 min, protein concentration was determined by Bradford. Samples were supplemented with Laemmli sample buffer and boiled for 5 min. Proteins were separated by SDS-PAGE and transferred to PVDF membranes, which were sequentially incubated with primary and secondary antibodies (goat anti-rabbit or anti-mouse coupled to Alexa Fluor 680 or IRDye 800, 1:10,000). The primary antibodies were used at the following dilutions: α-tubulin 1:2500; Myc 1:1000; and RFP 1:1000. IR imaging of immunoblots was performed according to the manufacturer's protocols using an Odyssey IR Imaging System (Li-Cor Biosciences). All Odyssey images were processed using ImageJ software (National Institutes of Health) and assembled for presentation using Photoshop and Illustrator (Adobe).
Intracellular cAMP measurements
This assay was performed using the previously described BRET-based biosensor NLuc-EPAC-VV (51, 57). HEK293T cells were seeded, transfected, and harvested as described above (under “BRET-based G-protein activation assays”) except that the plasmids used were as follows (quantities in parentheses): Nluc–EPAC–VV (0.2 μg); Lyn11–FRB (3 μg); and FKBP–GBA constructs (0.1 μg). A Polar STAR plate reader (BMG Labtech) was used to measure luminescence signals at 460 ± 20 and 528 ± 10 nm at 28 °C every 4 s, and BRET was calculated as the ratio between the emission intensity at 528 ± 10 nm divided by the emission intensity at 460 ± 20 nm. Results were presented as the inverse of the BRET ratio after subtraction of the basal signal measured for 60 s before any stimulation (ΔBRET−1). Forskolin (3 μm) was added at 60 s and rapamycin (0.5 μm) at 360 s as indicated in the figures. Protein sample immunoblotting was performed as described under “BRET-based G-protein activation assays.”
Statistical analyses
Each experiment was performed at least three times. The data shown are presented as means with error bars representing the S.E. or as one representative result of each biological replicate (as indicated in the figure legends). Statistical significance between various conditions was assessed with Student's t test. p < 0.05 was considered significant.
Author contributions
M. M., S. B., V. D., J.-C. P., L. G.-N., and M. G.-M. formal analysis; M. M., S. B., V. D., J.-C. P., A. L., L. G.-N., and M. G.-M. investigation; M. M. and M. G.-M. writing-review and editing; J. B. B.-C. and G. S. B. resources; M. G.-M. conceptualization; M.G.-M. supervision; M. G.-M. funding acquisition; M. G.-M. writing-original draft; M. G.-M. project administration.
Supplementary Material
Acknowledgments
We thank Nevin Lambert (Augusta University) for providing numerous plasmids and for extensive discussions to set up and optimize the BRET assays and Ruth MacLeod for help making the peptide arrays. We also thank Richard A. Cerione and Sekar Ramachandran (Cornell University) for providing the purified Gβγ proteins.
This work was supported in part by National Institutes of Health Grants R01GM108733 and R01GM130120, American Cancer Society Grant RSG-13-362-01-TBE (to M. G.-M.), and by Medical Research Council Grant MR/M013944/1 (to G. S. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S8 and supporting Ref. 1.
- GPCR
- G-protein–coupled receptor
- GEF
- guanine nucleotide exchange factor
- GBA
- Gα-binding-and-activating
- GDI
- guanine nucleotide dissociation inhibitor
- GAP
- GTPase-activating protein
- GIV
- Gα interacting, vesicle-associated protein
- DAPLE
- Dvl-associating protein with a high frequency of leucines
- BRET
- bioluminescence resonance energy transfer
- AC
- adenylyl cyclase
- aa
- amino acid
- CID
- chemically-induced dimerization
- aa
- amino acid
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- FP
- fluorescence polarization
- PVDF
- polyvinylidene difluoride
- RGS
- regulators of G-protein signaling
- PSSM
- position-specific scoring matrix
- NMT
- N-myristoyltransferase
- HF
- hydrofluoric acid
- PH
- pleckstrin homology
- GST
- glutathione S-transferase
- RFP
- red fluorescent protein
- PLC
- phospholipase C
- LIC
- ligation-independent cloning
- FKBP
- FK506-binding protein.
References
- 1. Gilman A. G. (1987) G-proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- 2. de Mendoza A., Sebé-Pedrós A., and Ruiz-Trillo I. (2014) The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biol. Evol. 6, 606–619 10.1093/gbe/evu038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sato M., Blumer J. B., Simon V., and Lanier S. M. (2006) Accessory proteins for G-proteins: partners in signaling. Annu. Rev. Pharmacol. Toxicol. 46, 151–187 10.1146/annurev.pharmtox.46.120604.141115 [DOI] [PubMed] [Google Scholar]
- 4. Watson N., Linder M. E., Druey K. M., Kehrl J. H., and Blumer K. J. (1996) RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature 383, 172–175 10.1038/383172a0 [DOI] [PubMed] [Google Scholar]
- 5. Berman D. M., Wilkie T. M., and Gilman A. G. (1996) GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G-protein α subunits. Cell 86, 445–452 10.1016/S0092-8674(00)80117-8 [DOI] [PubMed] [Google Scholar]
- 6. De Vries L., Zheng B., Fischer T., Elenko E., and Farquhar M. G. (2000) The regulator of G-protein signaling family. Annu. Rev. Pharmacol. Toxicol. 40, 235–271 10.1146/annurev.pharmtox.40.1.235 [DOI] [PubMed] [Google Scholar]
- 7. Ross E. M., and Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G-proteins: regulators of G-protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 10.1146/annurev.biochem.69.1.795 [DOI] [PubMed] [Google Scholar]
- 8. Hepler J. R. (1999) Emerging roles for RGS proteins in cell signalling. Trends Pharmacol. Sci. 20, 376–382 10.1016/S0165-6147(99)01369-3 [DOI] [PubMed] [Google Scholar]
- 9. Druey K. M., and Kehrl J. H. (1997) Inhibition of regulator of G-protein signaling function by two mutant RGS4 proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 12851–12856 10.1073/pnas.94.24.12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kimple R. J., Kimple M. E., Betts L., Sondek J., and Siderovski D. P. (2002) Structural determinants for GoLoco-induced inhibition of nucleotide release by Gα subunits. Nature 416, 878–881 10.1038/416878a [DOI] [PubMed] [Google Scholar]
- 11. Peterson Y. K., Hazard S. 3rd, Graber S. G., and Lanier S. M. (2002) Identification of structural features in the G-protein regulatory motif required for regulation of heterotrimeric G-proteins. J. Biol. Chem. 277, 6767–6770 10.1074/jbc.C100699200 [DOI] [PubMed] [Google Scholar]
- 12. Blumer J. B., Oner S. S., and Lanier S. M. (2012) Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol. 204, 202–218 10.1111/j.1748-1716.2011.02327.x [DOI] [PubMed] [Google Scholar]
- 13. Bernard M. L., Peterson Y. K., Chung P., Jourdan J., and Lanier S. M. (2001) Selective interaction of AGS3 with G-proteins and the influence of AGS3 on the activation state of G-proteins. J. Biol. Chem. 276, 1585–1593 10.1074/jbc.M005291200 [DOI] [PubMed] [Google Scholar]
- 14. Peterson Y. K., Bernard M. L., Ma H., Hazard S. 3rd, Graber S. G., and Lanier S. M. (2000) Stabilization of the GDP-bound conformation of Giα by a peptide derived from the G-protein regulatory motif of AGS3. J. Biol. Chem. 275, 33193–33196 10.1074/jbc.C000509200 [DOI] [PubMed] [Google Scholar]
- 15. De Vries L., Fischer T., Tronchère H., Brothers G. M., Strockbine B., Siderovski D. P., and Farquhar M. G. (2000) Activator of G-protein signaling 3 is a guanine dissociation inhibitor for Gαi subunits. Proc. Natl. Acad. Sci. U.S.A. 97, 14364–14369 10.1073/pnas.97.26.14364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sjögren B., Blazer L. L., and Neubig R. R. (2010) Regulators of G-protein signaling proteins as targets for drug discovery. Prog. Mol. Biol. Transl. Sci. 91, 81–119 10.1016/S1877-1173(10)91004-1 [DOI] [PubMed] [Google Scholar]
- 17. Kimple A. J., Bosch D. E., Giguère P. M., and Siderovski D. P. (2011) Regulators of G-protein signaling and their Gα substrates: promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 63, 728–749 10.1124/pr.110.003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cismowski M. J., Ma C., Ribas C., Xie X., Spruyt M., Lizano J. S., Lanier S. M., and Duzic E. (2000) Activation of heterotrimeric G-protein signaling by a Ras-related protein. Implications for signal integration. J. Biol. Chem. 275, 23421–23424 10.1074/jbc.C000322200 [DOI] [PubMed] [Google Scholar]
- 19. Tall G. G., Krumins A. M., and Gilman A. G. (2003) Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J. Biol. Chem. 278, 8356–8362 10.1074/jbc.M211862200 [DOI] [PubMed] [Google Scholar]
- 20. Chan P., Gabay M., Wright F. A., and Tall G. G. (2011) Ric-8B is a GTP-dependent G-protein αs guanine nucleotide exchange factor. J. Biol. Chem. 286, 19932–19942 10.1074/jbc.M110.163675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee M. J., and Dohlman H. G. (2008) Coactivation of G-protein signaling by cell-surface receptors and an intracellular exchange factor. Curr. Biol. 18, 211–215 10.1016/j.cub.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Natochin M., Campbell T. N., Barren B., Miller L. C., Hameed S., Artemyev N. O., and Braun J. E. (2005) Characterization of the Gα(s) regulator cysteine string protein. J. Biol. Chem. 280, 30236–30241 10.1074/jbc.M500722200 [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Marcos M., Ghosh P., and Farquhar M. G. (2009) GIV is a nonreceptor GEF for Gαi with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 3178–3183 10.1073/pnas.0900294106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia-Marcos M., Ghosh P., and Farquhar M. G. (2015) GIV/Girdin transmits signals from multiple receptors by triggering trimeric G-protein activation. J. Biol. Chem. 290, 6697–6704 10.1074/jbc.R114.613414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garcia-Marcos M., Kietrsunthorn P. S., Wang H., Ghosh P., and Farquhar M. G. (2011) G protein binding sites on Calnuc (nucleobindin 1) and NUCB2 (nucleobindin 2) define a new class of G(α)i-regulatory motifs. J. Biol. Chem. 286, 28138–28149 10.1074/jbc.M110.204099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aznar N., Midde K. K., Dunkel Y., Lopez-Sanchez I., Pavlova Y., Marivin A., Barbazán J., Murray F., Nitsche U., Janssen K. P., Willert K., Goel A., Abal M., Garcia-Marcos M., and Ghosh P. (2015) Daple is a novel non-receptor GEF required for trimeric G-protein activation in Wnt signaling. eLife 4, e07091 10.7554/eLife.07091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coleman B. D., Marivin A., Parag-Sharma K., DiGiacomo V., Kim S., Pepper J. S., Casler J., Nguyen L. T., Koelle M. R., and Garcia-Marcos M. (2016) Evolutionary conservation of a GPCR-independent mechanism of trimeric G protein activation. Mol. Biol. Evol. 33, 820–837 10.1093/molbev/msv336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DiGiacomo V., Marivin A., and Garcia-Marcos M. (2018) When heterotrimeric G proteins are not activated by G protein-coupled receptors: structural insights and evolutionary conservation. Biochemistry 57, 255–257 10.1021/acs.biochem.7b00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Opakua A. I., Parag-Sharma K., DiGiacomo V., Merino N., Leyme A., Marivin A., Villate M., Nguyen L. T., de la Cruz-Morcillo M. A., Blanco-Canosa J. B., Ramachandran S., Baillie G. S., Cerione R. A., Blanco F. J., and Garcia-Marcos M. (2017) Molecular mechanism of Gαi activation by non-GPCR proteins with a Gα-binding and activating motif. Nat. Commun. 8, 15163 10.1038/ncomms15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DiGiacomo V., de Opakua A. I., Papakonstantinou M. P., Nguyen L. T., Merino N., Blanco-Canosa J. B., Blanco F. J., and Garcia-Marcos M. (2017) The Gαi-GIV binding interface is a druggable protein–protein interaction. Sci. Rep. 7, 8575 10.1038/s41598-017-08829-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Marcos M., Kietrsunthorn P. S., Pavlova Y., Adia M. A., Ghosh P., and Farquhar M. G. (2012) Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 1961–1966 10.1073/pnas.1120538109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin C., Ear J., Midde K., Lopez-Sanchez I., Aznar N., Garcia-Marcos M., Kufareva I., Abagyan R., and Ghosh P. (2014) Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol. Biol. Cell 25, 3654–3671 10.1091/mbc.e14-05-0978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Midde K. K., Aznar N., Laederich M. B., Ma G. S., Kunkel M. T., Newton A. C., and Ghosh P. (2015) Multimodular biosensors reveal a novel platform for activation of G-proteins by growth factor receptors. Proc. Natl. Acad. Sci. U.S.A. 112, E937–E946 10.1073/pnas.1420140112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez-Sanchez I., Dunkel Y., Roh Y. S., Mittal Y., De Minicis S., Muranyi A., Singh S., Shanmugam K., Aroonsakool N., Murray F., Ho S. B., Seki E., Brenner D. A., and Ghosh P. (2014) GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat. Commun. 5, 4451 10.1038/ncomms5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma G. S., Aznar N., Kalogriopoulos N., Midde K. K., Lopez-Sanchez I., Sato E., Dunkel Y., Gallo R. L., and Ghosh P. (2015) Therapeutic effects of cell-permeant peptides that activate G-proteins downstream of growth factors. Proc. Natl. Acad. Sci. U.S.A. 112, E2602–E2610 10.1073/pnas.1505543112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parag-Sharma K., Leyme A., DiGiacomo V., Marivin A., Broselid S., and Garcia-Marcos M. (2016) Membrane recruitment of the non-receptor protein GIV/Girdin (Gα-interacting, vesicle-associated protein/Girdin) is sufficient for activating heterotrimeric G protein signaling. J. Biol. Chem. 291, 27098–27111 10.1074/jbc.M116.764431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leyme A., Marivin A., Perez-Gutierrez L., Nguyen L. T., and Garcia-Marcos M. (2015) Integrins activate trimeric G-proteins via the nonreceptor protein GIV/Girdin. J. Cell Biol. 210, 1165–1184 10.1083/jcb.201506041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sasaki K., Kakuwa T., Akimoto K., Koga H., and Ohno S. (2015) Regulation of epithelial cell polarity by PAR-3 depends on Girdin transcription and Girdin-Gαi3 signaling. J. Cell Sci. 128, 2244–2258 10.1242/jcs.160879 [DOI] [PubMed] [Google Scholar]
- 39. Garcia-Marcos M., Ear J., Farquhar M. G., and Ghosh P. (2011) A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G-protein activity and growth factor signals. Mol. Biol. Cell 22, 673–686 10.1091/mbc.e10-08-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ghosh P. (2015) Heterotrimeric G-proteins as emerging targets for network based therapy in cancer: end of a long futile campaign striking heads of a hydra. Aging 7, 469–474 10.18632/aging.100781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnston C. A., Willard F. S., Jezyk M. R., Fredericks Z., Bodor E. T., Jones M. B., Blaesius R., Watts V. J., Harden T. K., Sondek J., Ramer J. K., and Siderovski D. P. (2005) Structure of Gα(i1) bound to a GDP-selective peptide provides insight into guanine nucleotide exchange. Structure 13, 1069–1080 10.1016/j.str.2005.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia-Marcos M., Ghosh P., Ear J., and Farquhar M. G. (2010) A structural determinant that renders Gα(i) sensitive to activation by GIV/girdin is required to promote cell migration. J. Biol. Chem. 285, 12765–12777 10.1074/jbc.M109.045161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Austin R. J., Ja W. W., and Roberts R. W. (2008) Evolution of class-specific peptides targeting a hot spot of the Gαs subunit. J. Mol. Biol. 377, 1406–1418 10.1016/j.jmb.2008.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ehrenberger T., Cantley L. C., and Yaffe M. B. (2015) Computational prediction of protein–protein interactions. Methods Mol. Biol. 1278, 57–75 10.1007/978-1-4939-2425-7_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. The UniProt Consortium. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weiss T. S., Chamberlain C. E., Takeda T., Lin P., Hahn K. M., and Farquhar M. G. (2001) Gαi3 binding to calnuc on Golgi membranes in living cells monitored by fluorescence resonance energy transfer of green fluorescent protein fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 14961–14966 10.1073/pnas.261572098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cismowski M. J., Takesono A., Ma C., Lanier S. M., and Duzic E. (2002) Identification of modulators of mammalian G-protein signaling by functional screens in the yeast Saccharomyces cerevisiae. Methods Enzymol. 344, 153–168 10.1016/S0076-6879(02)44712-X [DOI] [PubMed] [Google Scholar]
- 48. Cismowski M. J., Takesono A., Ma C., Lizano J. S., Xie X., Fuernkranz H., Lanier S. M., and Duzic E. (1999) Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat. Biotechnol. 17, 878–883 10.1038/12867 [DOI] [PubMed] [Google Scholar]
- 49. Kleuss C., Raw A. S., Lee E., Sprang S. R., and Gilman A. G. (1994) Mechanism of GTP hydrolysis by G-protein α subunits. Proc. Natl. Acad. Sci. U.S.A. 91, 9828–9831 10.1073/pnas.91.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., and Sprang S. R. (1994) Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science 265, 1405–1412 10.1126/science.8073283 [DOI] [PubMed] [Google Scholar]
- 51. Leyme A., Marivin A., Maziarz M., DiGiacomo V., Papakonstantinou M. P., Patel P. P., Blanco-Canosa J. B., Walawalkar I. A., Rodriguez-Davila G., Dominguez I., and Garcia-Marcos M. (2017) Specific inhibition of GPCR-independent G-protein signaling by a rationally engineered protein. Proc. Natl. Acad. Sci. U.S.A. 114, E10319–E10328 10.1073/pnas.1707992114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mukhopadhyay S., and Ross E. M. (2002) Quench-flow kinetic measurement of individual reactions of G-protein-catalyzed GTPase cycle. Methods Enzymol. 344, 350–369 10.1016/S0076-6879(02)44727-1 [DOI] [PubMed] [Google Scholar]
- 53. Maziarz M., and Garcia-Marcos M. (2017) Rapid kinetic BRET measurements to monitor G-protein activation by GPCR and non-GPCR proteins. Methods Cell Biol. 142, 145–157 10.1016/bs.mcb.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bayle J. H., Grimley J. S., Stankunas K., Gestwicki J. E., Wandless T. J., and Crabtree G. R. (2006) Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 13, 99–107 10.1016/j.chembiol.2005.10.017 [DOI] [PubMed] [Google Scholar]
- 55. Kovárová M., Tolar P., Arudchandran R., Dráberová L., Rivera J., and Dráber P. (2001) Structure-function analysis of Lyn kinase association with lipid rafts and initiation of early signaling events after Fcϵ receptor I aggregation. Mol. Cell. Biol. 21, 8318–8328 10.1128/MCB.21.24.8318-8328.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Inoue T., Heo W. D., Grimley J. S., Wandless T. J., and Meyer T. (2005) An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods 2, 415–418 10.1038/nmeth763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Masuho I., Ostrovskaya O., Kramer G. M., Jones C. D., Xie K., and Martemyanov K. A. (2015) Distinct profiles of functional discrimination among G-proteins determine the actions of G-protein-coupled receptors. Sci. Signal. 8, ra123 10.1126/scisignal.aab4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hollins B., Kuravi S., Digby G. J., and Lambert N. A. (2009) The C terminus of GRK3 indicates rapid dissociation of G-protein heterotrimers. Cell. Signal. 21, 1015–1021 10.1016/j.cellsig.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sadana R., and Dessauer C. W. (2009) Physiological roles for G-protein-regulated adenylyl cyclase isoforms: insights from knockout and overexpression studies. Neurosignals 17, 5–22 10.1159/000166277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Klarenbeek J. B., Goedhart J., Hink M. A., Gadella T. W., and Jalink K. (2011) A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PloS one 6, e19170 10.1371/journal.pone.0019170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee S. B., and Rhee S. G. (1996) Molecular cloning, splice variants, expression, and purification of phospholipase C-δ4. J. Biol. Chem. 271, 25–31 10.1074/jbc.271.1.25 [DOI] [PubMed] [Google Scholar]
- 62. Leung D. W., Tompkins C., Brewer J., Ball A., Coon M., Morris V., Waggoner D., and Singer J. W. (2004) Phospholipase Cδ-4 overexpression upregulates ErbB1/2 expression, Erk signaling pathway, and proliferation in MCF-7 cells. Mol. Cancer 3, 15 10.1186/1476-4598-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Harden T. K., Waldo G. L., Hicks S. N., and Sondek J. (2011) Mechanism of activation and inactivation of Gq/phospholipase C-β signaling nodes. Chem. Rev. 111, 6120–6129 10.1021/cr200209p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Essen L. O., Perisic O., Cheung R., Katan M., and Williams R. L. (1996) Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature 380, 595–602 10.1038/380595a0 [DOI] [PubMed] [Google Scholar]
- 65. Nakashima S., Banno Y., Watanabe T., Nakamura Y., Mizutani T., Sakai H., Zhao Y., Sugimoto Y., and Nozawa Y. (1995) Deletion and site-directed mutagenesis of EF-hand domain of phospholipase C-δ1: effects on its activity. Biochem. Biophys. Res. Commun. 211, 365–369 [DOI] [PubMed] [Google Scholar]
- 66. Lin P., Yao Y., Hofmeister R., Tsien R. Y., and Farquhar M. G. (1999) Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J. Cell Biol. 145, 279–289 10.1083/jcb.145.2.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. de Alba E., and Tjandra N. (2004) Structural studies on the Ca2+-binding domain of human nucleobindin (calnuc). Biochemistry 43, 10039–10049 10.1021/bi049310a [DOI] [PubMed] [Google Scholar]
- 68. Lee S. B., Várnai P., Balla A., Jalink K., Rhee S. G., and Balla T. (2004) The pleckstrin homology domain of phosphoinositide-specific phospholipase Cδ4 is not a critical determinant of the membrane localization of the enzyme. J. Biol. Chem. 279, 24362–24371 10.1074/jbc.M312772200 [DOI] [PubMed] [Google Scholar]
- 69. Fukami K., Yoshida M., Inoue T., Kurokawa M., Fissore R. A., Yoshida N., Mikoshiba K., and Takenawa T. (2003) Phospholipase Cδ4 is required for Ca2+ mobilization essential for acrosome reaction in sperm. J. Cell Biol. 161, 79–88 10.1083/jcb.200210057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fukami K., Nakao K., Inoue T., Kataoka Y., Kurokawa M., Fissore R. A., Nakamura K., Katsuki M., Mikoshiba K., Yoshida N., and Takenawa T. (2001) Requirement of phospholipase Cδ4 for the zona pellucida-induced acrosome reaction. Science 292, 920–923 10.1126/science.1059042 [DOI] [PubMed] [Google Scholar]
- 71. Lyon A. M., and Tesmer J. J. (2013) Structural insights into phospholipase C-β function. Mol. Pharmacol. 84, 488–500 10.1124/mol.113.087403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gresset A., Sondek J., and Harden T. K. (2012) The phospholipase C isozymes and their regulation. Subcell. Biochem. 58, 61–94 10.1007/978-94-007-3012-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Frank R. (2002) The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports–principles and applications. J. Immunol. Methods 267, 13–26 10.1016/S0022-1759(02)00137-0 [DOI] [PubMed] [Google Scholar]
- 74. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., and Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 10.1006/prep.2001.1603 [DOI] [PubMed] [Google Scholar]
- 75. Cabrita L. D., Dai W., and Bottomley S. P. (2006) A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol. 6, 12 10.1186/1472-6750-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Preininger A. M., Van Eps N., Yu N. J., Medkova M., Hubbell W. L., and Hamm H. E. (2003) The myristoylated amino terminus of Gα(i)(1) plays a critical role in the structure and function of Gα(i)(1) subunits in solution. Biochemistry 42, 7931–7941 10.1021/bi0345438 [DOI] [PubMed] [Google Scholar]
- 77. Mumby S. M., and Linder M. E. (1994) Myristoylation of G-protein α subunits. Methods Enzymol. 237, 254–268 10.1016/S0076-6879(94)37067-2 [DOI] [PubMed] [Google Scholar]
- 78. Hoffman G. A., Garrison T. R., and Dohlman H. G. (2002) Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 344, 617–631 10.1016/S0076-6879(02)44744-1 [DOI] [PubMed] [Google Scholar]
- 79. Cox J. S., Chapman R. E., and Walter P. (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 10.1091/mbc.8.9.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kroll S., Phillips W. J., and Cerione R. A. (1989) The regulation of the cyclic GMP phosphodiesterase by the GDP-bound form of the α subunit of transducin. J. Biol. Chem. 264, 4490–4497 [PubMed] [Google Scholar]
- 81. Ramachandran S., and Cerione R. A. (2011) A dominant-negative Gα mutant that traps a stable rhodopsin-Gα-GTP-βγ complex. J. Biol. Chem. 286, 12702–12711 10.1074/jbc.M110.166538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Marivin A., Leyme A., Parag-Sharma K., DiGiacomo V., Cheung A. Y., Nguyen L. T., Dominguez I., and Garcia-Marcos M. (2016) Dominant-negative Gα subunits are a mechanism of dysregulated heterotrimeric G-protein signaling in human disease. Sci. Signal. 9, ra37 10.1126/scisignal.aad2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.