Abstract
Long noncoding RNAs (lncRNAs) are vital players in cancers, including hepatocellular carcinoma (HCC). We previously identified an lncRNA, GAS8-AS1, that is located in intron 2 of GAS8. However, its involvement in HCC is still largely unknown. In this study, we report that both GAS8-AS1 and its host gene GAS8 act as HCC tumor suppressors. We found that expression of GAS8-AS1 or GAS8 is significantly decreased in HCC tissues and is associated with a poor prognosis among HCC patients. Interestingly, lncRNA GAS8-AS1 could promote GAS8 transcription. We detected a CpG island in the GAS8 promoter, but lncRNA GAS8-AS1 did not affect DNA methylation at this GAS8 promoter site. Moreover, we identified two GAS8-AS1–interacting proteins, mixed-lineage leukemia 1 (MLL1), a histone 3 Lys-4 (H3K4) methyltransferase, and its partner WD-40 repeat protein 5 (WDR5). RNA pulldown, ChIP, and RNA immunoprecipitation assays revealed that GAS8-AS1 is required for maintaining the GAS8 promoter in an open chromatin state by recruiting the MLL1/WDR5 complex and for enhancing RNA polymerase II activity and GAS8 transcription. Of note, GAS8-AS1–dependent GAS8 hyperactivation inhibited malignant transformation of hepatocytes. Our results provide important insights into how lncRNA GAS8-AS1 suppresses HCC development and suggest potential strategies for treating patients with liver cancer.
Keywords: long noncoding RNA (long ncRNA, lncRNA); hepatocellular carcinoma; histone modification; tumor suppressor gene; apoptosis; GAS8; GAS8-AS1; MLL1; WDR5; epigenetics
Introduction
Hepatocellular carcinoma (HCC)2 is a lethal malignancy with the highest morbidity in Asia and Sub-Saharan Africa (1, 2). Worldwide, China alone bears more than 50% of the whole burden (1). The evidently differential prevalence of HCC elucidates that multiple genetic and environmental factors might be involved in its development and progression. There are several main epidemiological risk factors, such as chronic hepatitis B or C virus infection, excessive drinking, and exposure to dietary aflatoxin B (2). Like other cancers, carcinogenesis and progression of HCC are associated with the accumulation of genetic and epigenetic alterations. However, the detailed molecular pathogenesis mechanisms of these alterations in HCC remain to be investigated.
Originally, long noncoding RNAs (lncRNAs), an emerging class of >200-nt-long transcripts mostly not translated into proteins, were considered as transcriptional noises of human genome (3). Accumulated evidence demonstrated that lncRNAs are vital players in diverse cellular processes from normal development to disease processes (4–6). It is now estimated that the human genome encodes about 28,000 distinct lncRNAs, many of which are still being discovered and are yet to be functionally annotated on the basis of the ENCODE project (7–9). In particular, it has become increasingly apparent that lncRNAs may play a critical role in regulating chromatin dynamics, gene expression, growth, differentiation, and development (10, 11). lncRNAs could act as scaffolds that bring together multiple proteins to form functional ribonucleoprotein (RNP) complexes and regulate the function, stability, or activity of these RNPs (12–14). For example, lncRNA HOTAIR acts as a scaffold to recruit Polycomb repressive complex 2 (PRC2) complex proteins required for chromatin remodeling and, thus, regulates H3 lysine 27 methylation and gene expression (15). As a result, increased expression of HOTAIR leads to a genome-wide reprogramming of PRC2 function and enhanced tumorigenesis as well as metastatic progression (15–19).
We identified a significantly mutated lncRNA gene, GAS8-AS1, as a tumor suppressor in our previous study (20). However, its role in HCC is still largely unknown. In this study, we show that lncRNA GAS8-AS1 directly interacts with mixed lineage leukemia 1 (MLL1), which can specifically methylate histone 3 Lys-4 (H3K4) via forming a complex with WD-40 repeat protein 5 (WDR5) and other components and have pivotal roles in the transcriptional regulation of multiple downstream genes (21–25). Interactions between lncRNA GAS8-AS1 and MLL1/WDR5 complex induce H3K4 methylation of tumor suppressor GAS8 promoter, facilitate RNA polymerase II (Pol II) recruitment, and consequently activate GAS8 expression. GAS8-AS1–dependent GAS8 hyperactivation inhibits malignant transformation of hepatocytes. Significantly suppressed expression of GAS8-AS1 is commonly observed in HCC and was associated with poor overall survival (OS) of HCC patients. Interestingly, restored expression of GAS8-AS1 leads to HCC cells to be more sensitive to sorafenib, a commonly used targeted therapy drug in advanced HCC patients. Our data reveal that lncRNA GAS8-AS1 is a novel HCC tumor suppressor in vivo and in vitro and provide a rationale for targeting GAS8-AS1, either alone or in combination with sorafenib, when treating HCC.
Results
Clinical significance of lncRNA GAS8-AS1 and GAS8 expression in HCC tissues
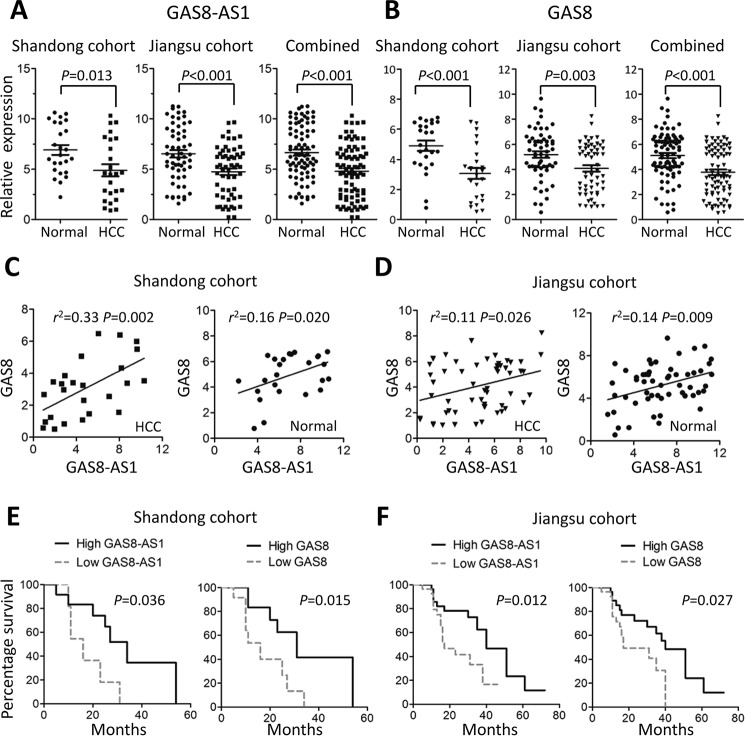
We detected GAS8-AS1 and GAS8 expression in 82 pairs of HCC tissues and adjacent normal tissue specimens. Significantly decreased GAS8-AS1 and GAS8 expression in HCC tissues was found compared with normal tissues in Shandong cohort (both p < 0.05) (Fig. 1, A and B), which was validated in Jiangsu cohort and combined data (all p < 0.01) (Fig. 1, A and B). Interestingly, we found significantly positive correlation between GAS8-AS1 and GAS8 expression in tissue specimens from both Shandong cohort (Spearman's correlation: HCC tissues, r2 = 0.33, p = 0.002; normal tissues, r2 = 0.16, p = 0.020) (Fig. 1C) and Jiangsu cohort (Spearman's correlation: HCC tissues, r2 = 0.11, p = 0.026; normal tissues, r2 = 0.14, p = 0.009) (Fig. 1D).
Figure 1.
lncRNA GAS8-AS1 and GAS8 expression in HCC and normal tissue specimens. A and B, GAS8-AS1 or GAS8 expression was quantified using qRT-PCR in 82 tumor-normal pairs (Shandong cohort, n = 25; Jiangsu cohort, n = 57). Significantly elevated expression of both GAS8-AS1 and GAS8 expression in HCC was found in HCC tissues. All data of GAS8-AS1 or GAS8 expression were normalized to β-actin mRNA expression levels. C and D, positive correlations between GAS8-AS1 and GAS8 expression in Shandong cohort or Jiangsu cohort. E and F, survival analyses of GAS8-AS1 or GAS8 expression of HCC patients from Shandong cohort or Jiangsu cohort. Survival curves were constructed using the Kaplan–Meier method and evaluated using the log-rank test.
We also examined whether differential GAS8-AS1 and GAS8 expression impact HCC prognosis. As shown in Fig. 1E, patients with high GAS8-AS1 expression in HCC tissues had significantly longer OS time than those with low GAS8-AS1 expression in Shandong cohort (median OS, 34.0 versus 16.0 months; p = 0.036). Similar results were also found when patients were grouped with GAS8 expression in HCC tissues (median OS, 31.0 versus 16.0 months; p = 0.015) (Fig. 1E). We also evaluated the prognostic value of GAS8-AS1 and GAS8 expression in Jiangsu cohort and observed consistent results (GAS8-AS1: median OS, 40.0 versus 17.0 months; p = 0.012; GAS8: 39.0 versus 18.0 months; p = 0.027) (Fig. 1F). These data elucidated that GAS8-AS1 and GAS8 may be tumor suppressors during HCC development.
lncRNA GAS8-AS1 and GAS8 suppress HCC proliferation
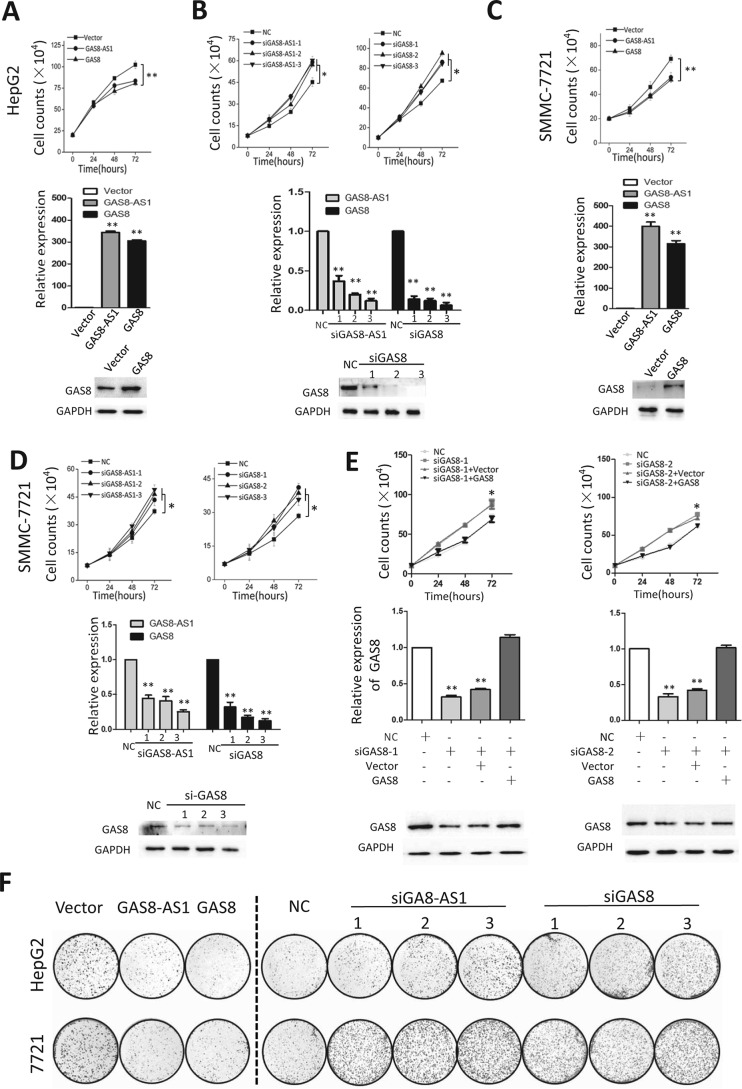
To investigate how GAS8-AS1 and GAS8 are involved in hepatocarcinogenesis, we next investigated their impacts on biological behaviors of HCC cells. We first examined whether ectopic GAS8-AS1 and GAS8 could modulate cellular proliferation (Fig. 2, A–D). We found that overexpression of GAS8-AS1 can significantly reduce viability of HepG2 and SMMC7721 cells (Fig. 2, A and C). For example, there was 18.4 or 22.3% reduced viability of HepG2 or SMMC7721 cells at 72 h after transfection (both p < 0.01). Similarly, overexpression of GAS8 can significantly inhibit growth of HCC cells (Fig. 2, A and C). On the contrary, lncRNA GAS8-AS1 down-regulation with different siRNAs (siGAS8-AS1-1, siGAS8-AS1-2, and siGAS8-AS1-3) did significantly accelerate proliferation of both HepG2 and SMMC7721 cells compared with cells transfected with NC RNA (all p < 0.05) (Fig. 2, A and C). Similar results were observed in HepG2 and SMMC7721 cells with silenced GAS8 expression (all p < 0.05) (Fig. 2, A and C). Interestingly, GAS8 overexpression could reverse the growth acceleration phenotype induced by GAS8-AS1 silencing (Fig. 2E). We also monitored how GAS8-AS1 and GAS8 influence colony formation and found that both genes could significantly suppress colony formation of both HepG2 and SMMC7721 cells (all p < 0.01) (Fig. 2F and Fig. S1). These are in line with cell viability assays and supporting the tumor suppressor nature of GAS8-AS1 and GAS8 in HCC (Fig. S1).
Figure 2.
lncRNA GAS8-AS1 or GAS8 inhibits cell proliferation of HepG2 or SMMC7721 cells. A and B, overexpression of GAS8-AS1 or GAS8 inhibits HepG2 cell growth. On the contrary, silencing expression of GAS8-AS1 or GAS8 with their siRNAs (siGAS8-AS1-1/-2/-3 or siGAS8-1/-2/-3) significantly accelerates proliferation of HepG2 cells. Cell number was counted at 24, 48, and 72 h after transfection. C and D, similar results were observed in SMMC7721 cells. E, endogenous GAS8-AS1 expression was first silenced using various siRNAs (siGAS8-AS1–1/-2), and GAS8 was then overexpressed using its expression construct in HepG2 cells. Interestingly, GAS8 overexpression could reverse the growth acceleration phenotype induced by GAS8-AS1 silencing. F, colony formation assays indicate that both genes significantly suppress colony formation of both HCC cell lines. At the 12th day after transfection, colony number in each well was counted. All results of the mean of triplicate assays with S.D. (error bars) are presented. *, p < 0.05; **, p < 0.01.
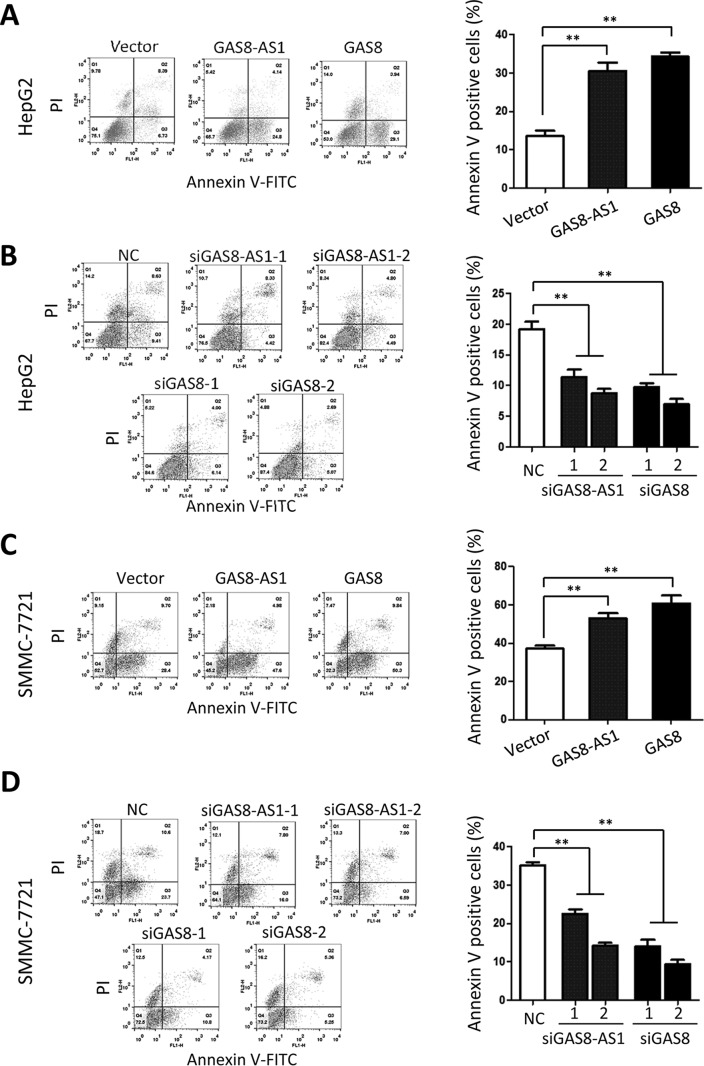
lncRNA GAS8-AS1 and GAS8 induce apoptosis of HCC cells via sensitizing cells to sorafenib
To gain insight into the functional relevance of lncRNA GAS8-AS1 and GAS8 on cell proliferation suppression, we detected how they influence apoptosis and cell cycle progression of HCC cells (Fig. 3 and Figs. S2–S4). As shown in Fig. S2, ectopic lncRNA GAS8-AS1 and GAS8 are able to induce significantly increased apoptosis of both HepG2 and SMMC7721 cell lines. Interestingly, lncRNA GAS8-AS1 or GAS8 significantly sensitizes HCC cells to sorafenib, a widely used targeted drug treating advanced HCC (Fig. 3, A and C). On the contrary, HCC cells with suppressed GAS8-AS1 or GAS8 expression showed much less apoptosis rates compared with the NC RNA controls (Fig. S2). Similar results were observed in HCC cells treated with sorafenib (Fig. 3, B and D). However, neither GAS8-AS1 nor GAS8 could induce obvious cell cycle arrest (Figs. S3 and S4).
Figure 3.
lncRNA GAS8-AS1 and GAS8 induce apoptosis of HepG2 or SMMC7721 cells via sensitizing them to sorafenib. HepG2 or SMMC7721 cells were transfected with expression constructs or siRNAs of GAS8-AS1 or GAS8 first. At the 24th hour after transfection, HCC cells were treated with 1 μmol/liter sorafenib, and apoptosis was determined with a FACSCalibur flow cytometer. All results of the mean of triplicate assays with S.D. (error bars) are presented. *, p < 0.05; **, p < 0.01.
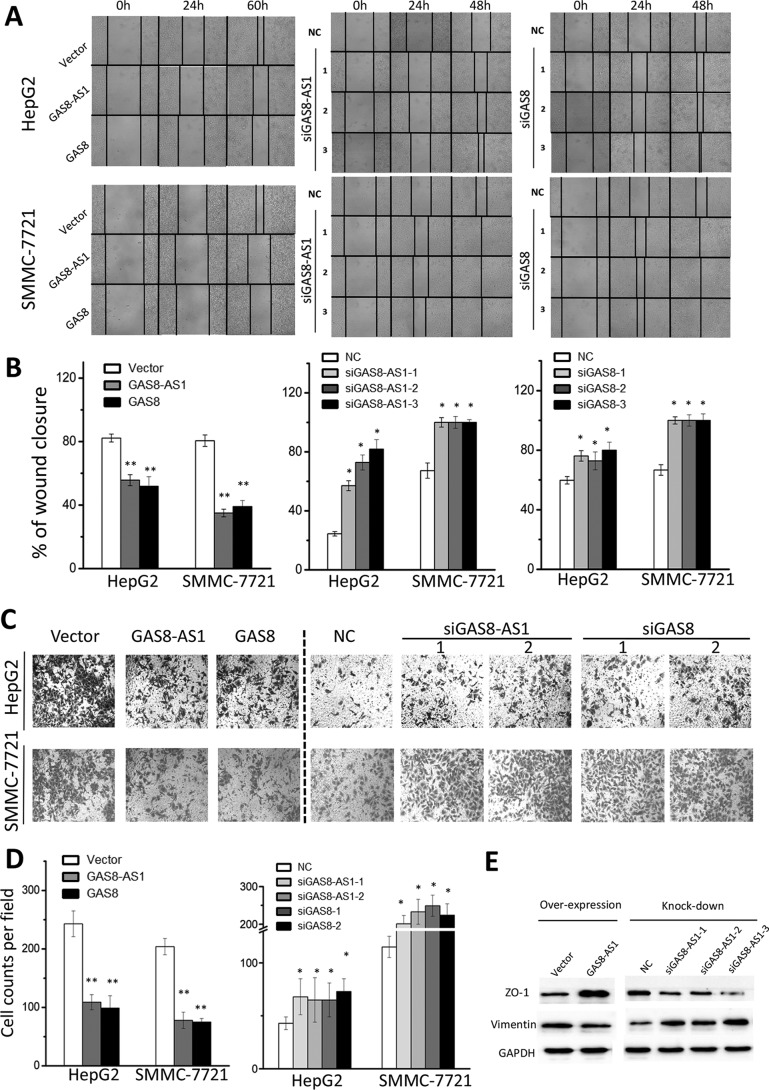
lncRNA GAS8-AS1 and GAS8 inhibit migration and invasion of HCC cells
We then investigated whether lncRNA GAS8-AS1 and GAS8 impact HCC invasion and metastasis. The wound-healing assays indicated that lncRNA GAS8-AS1 and GAS8 impaired the motility of the HepG2 and SMMC7721 cells compared with the control cells (Fig. 4, A and B). In contrast, siRNAs of GAS8-AS1 and GAS8 were capable of significantly accelerating migration of HCC cells (Fig. 4, A and B). Next, how lncRNA GAS8-AS1 and GAS8 influence invasiveness of HCC cells was examined using the Matrigel invasion assay system. lncRNA GAS8-AS1 and GAS8 could significantly reduce invasion ability of HCC cells (Fig. 4, C and D). Consistent with this, silencing lncRNA GAS8-AS1 or GAS8 also led to enhanced invasion ability of HCC cells (Fig. 4, C and D). To exclude the possibility that cell death contributes to the decrease in cell migration and invasion, we performed both migration and invasion assays within 24 h (Fig. S5), which is in line with results at 48 h. To evaluate the mechanistic rationale, we examined expression changes of two markers of epithelial-to-mesenchymal transition after overexpression or silencing of GAS8-AS1 in HCC cells. Interestingly, GAS8-AS1 could significantly increase ZO-1 expression and reduce vimentin expression. Additionally, silencing GAS8-AS1 induced ZO-1 expression and promote vimentin expression (Fig. 4E).
Figure 4.
lncRNA GAS8-AS1 and GAS8 suppress migration and invasion ability of HepG2 or SMMC7721 cells. A, GAS8-AS1 and GAS8 inhibit wound healing in HepG2 or SMMC7721 cells. Wound fields were observed directly after the removal of inserts (0 h), and cell migration was followed for 24, 48, or 60 h. B, wound-healing area in HepG2 or SMMC7721 cells was presented by histogram. C, GAS8-AS1 and GAS8 suppress invasion ability of HepG2 or SMMC7721 cells. Cells on the lower surface of the chamber were stained by crystal violet at 48 h after transfection. D, cell counts data of HepG2 or SMMC7721 cells are presented as a histogram. E, expression changes of two markers (ZO-1 and vimentin) of epithelial-to-mesenchymal transition were examined after overexpression or silencing of GAS8-AS1 in HepG2 cells. All results of the mean of triplicate assays with S.D. (error bars) are presented. *, p < 0.05; **, p < 0.01.
lncRNA GAS8-AS1 recruits MLL1/WDR5 complex to modulate H3K4 methylation of GAS8 promoter and activates its expression
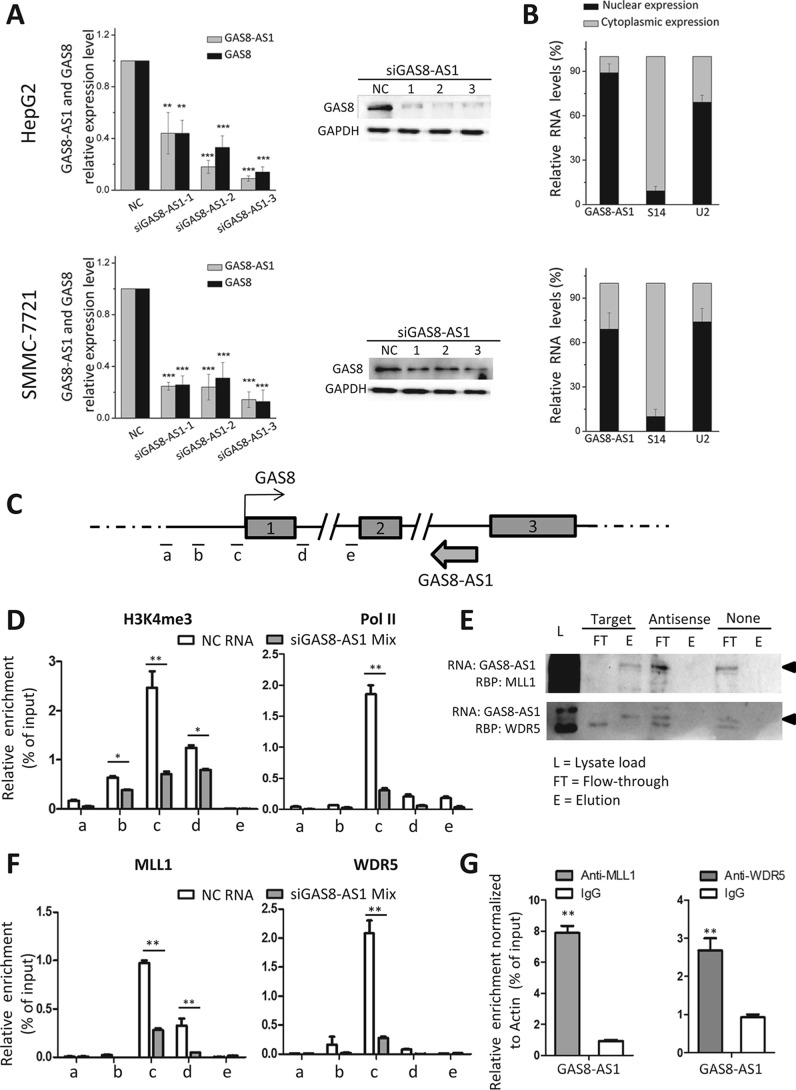
Positive correlation between lncRNA GAS8-AS1 and GAS8 mRNA in tissue specimens suggested that expression of GAS8 is functionally linked to transcription of GAS8-AS1. Therefore, we first examined whether silencing endogenous lncRNA GAS8-AS1 impacts expression of GAS8 in HCC cells (Fig. 5A). Indeed, knockdown of GAS8-AS1 in HepG2 or SMMC7721 cells led to decreased mRNA and protein expression of GAS8 (Fig. 5A), demonstrating that lncRNA GAS8-AS1 might play an important regulatory role in expression control of GAS8 in HCC cells. To exclude the possibility that those siGAS8-AS1 duplexes may target the GAS8 pre-mRNA, we designed and synthesized three siRNAs targeting intron 1 of GAS8 (Table S2). It was found that these siRNA duplexes could not significantly change GAS8 mRNA expression (Fig. S6).
Figure 5.
lncRNA GAS8-AS1 activates GAS8 expression via recruiting the MLL1/WDR5 complex to the GAS8 promoter and modulating promoter H3K4me3. A, knockdown of GAS8-AS1 with its siRNAs in HepG2 or SMMC7721 cells leads to significantly decreased mRNA and protein expression of GAS8. B, lncRNA GAS8-AS1 predominantly locates in the nuclear fraction in HCC cells. C, DNA fragments a, b, c, d, and e around the promoter region of GAS8 are detected using ChIP-qPCR. D, ChIP-qPCR results showed reduced H3K4me3 and RNA Pol II occupancy at the GAS8 promoter after silenced expression of endogenous lncRNA GAS8-AS1. E, RNA pulldown was performed by incubating in vitro–transcribed biotinylated transcripts of GAS8-AS1 or antisense GAS8-AS1 (negative control) with nucleus HepG2 cell extracts followed by detection of the presence of the GAS8-AS1 RNP complex by Western blotting. Both the GAS8-AS1 RNP complex components, MLL1 and WDR5, were detected in GAS8-AS1 pulldown, but not in GAS8-AS1 antisense pulldown. F, knockdown of GAS8-AS1 significantly inhibits MLL1/WDR5-mediated H3K4me3 of the GAS8 promoter in HepG2 cells. G, RNA-IP assays suggested that GAS8-AS1 may physically interact with the MLL1/WDR5 complex. Results of the mean of triplicate assays with S.D. (error bars) are presented. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We then evaluated cellular localization of lncRNA GAS8-AS1 in HCC cells to determine its detailed regulatory mechanisms on GAS8. After RNA was extracted separately from nuclear and cytoplasmic fractions of HCC cells, lncRNA GAS8-AS1 expression was examined alone with S14 and U2 RNA. lncRNA GAS8-AS1 was predominantly found in the nuclear fraction, as was U2 RNA in HCC cells (Fig. 5B). As a result, we speculated that lncRNA GAS8-AS1 may impact epigenetics modification of a promoter region of GAS8 (i.e. DNA methylation or histone modification) and, thus, regulate its expression. There is a predicted CpG island in the promoter of GAS8 and covering a part of the 5′-flanking region, whole exon 1, and a part of intron 1 (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/)3 (37). Considering that altered transcripted lncRNA may interfere with neighborhood DNA methylation, we detected whether suppressed GAS8-AS1 influences DNA methylation of the neighborhood GAS8 CpG island DNA methylation in HCC. As shown in Fig. S7, we found that silencing endogenous lncRNA GAS8-AS1 did not impact DNA methylation of the GAS8 CpG island in both HepG2 and HeLa cells.
There are multiple peaks of H3K4 methylation modifications (especially H3K4me3 modification) at the promoter of GAS8 in HepG2 cells according to the ENCODE ChIP-Seq data (Fig. S8). We then tested the hypothesis that lncRNA GAS8-AS1 may impact H3K4me3 at the GAS8 promoter. After knockdown expression of endogenous lncRNA GAS8-AS1 (Fig. S9), significantly reduced H3K4me3 at the GAS8 promoter was observed (Fig. 5, C and D), indicating that endogenous lncRNA GAS8-AS1 expression is essential in maintaining hyper-H3K4 trimethylation of the GAS8 promoter. In support of this, silencing GAS8-AS1 led to decreased occupancy of RNA Pol II at the GAS8 promoter (Fig. 5, C and D), which may be due to reduced H3K4me3 modification at the GAS8 promoter.
Accumulated evidence demonstrated that some lncRNAs alter epigenetic signatures through interactions with chromatin-remodeling enzymes. Accordingly, we predicted potential histone methyltransferase candidates that may interact with lncRNA GAS8-AS1 by the RPISeq software (http://pridb.gdcb.iastate.edu/RPISeq/)3 (38). With lncRNA HOTTIP as the positive control, we found that GAS8-AS1 may interact with MLL1, a key H3K4 methyltransferase, and its partner WDR5 (Fig. S10). We reasoned that lncRNA GAS8-AS1 might guide the MLL1/WDR5 complex to the GAS8 promoter. To verify this hypothesis, we performed RNA pulldown to determine whether GAS8-AS1 and MLL1 or WDR5 form a RNP complex. In vitro-transcribed biotinylated transcripts of GAS8-AS1 or antisense GAS8-AS1 (negative control) were incubated with nucleus HepG2 cell extracts. The presence of MLL1 and WDR5 was examined by Western blotting. Pulldown with GAS8-AS1, but not with an antisense GAS8-AS1, was able to detect both proteins (Fig. 5E), indicating that MLL1 and WDR5 associate with GAS8-AS1 in the nucleus. Consistent with this, ChIP assays indicate that both MLL1 and WDR5 can directly bind to the promoter of GAS8 (Fig. 5F). In support of the notion that lncRNA GAS8-AS1 recruits the MLL1/WDR5 complex to the GAS8 promoter, knockdown of GAS8-AS1 significantly inhibited MLL1-mediated H3K4me3 of the GAS8 promoter and prevented GAS8 expression in HCC cells (Fig. 5F). To examine whether GAS8-AS1 physically interacts with the MLL1/WDR5 complex, we detected GAS8-AS1 in RNA extracted from the precipitated MLL1 protein or WDR5 protein in HepG2 cells. We found that there was an 8.5- or 2.9-fold enrichment for MLL1 or WDR5 compared with IgG (Fig. 5G). On the contrary, ectopic lncRNA GAS8-AS1 increased H3K4me3 modification, Pol II occupancy, and binding of MLL1 or WDR5 to the GAS8 promoter in HCC cells (Fig. S11).
lncRNA GAS8-AS1 and GAS8 inhibit HCC growth in vivo
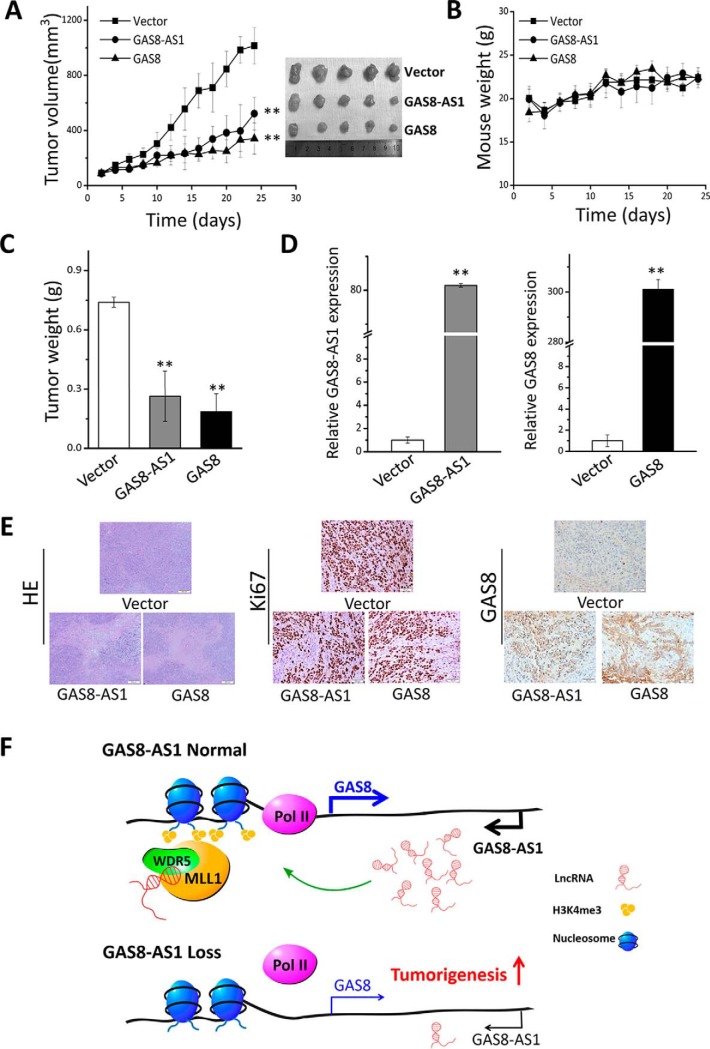
We evaluated the in vivo anti-cancer effects of GAS8-AS1 and GAS8 using HepG2 xenografts. It was found that tumor proliferation from xenografts with ectopic lncRNA GAS8-AS1 or GAS8 was significantly suppressed compared with the control HCC xenografts (both p < 0.01) (Fig. 6A). However, there were no significant differences in mouse weight between GAS8-AS1, GAS8, and the control groups (Fig. 6B). Significantly reduced tumor weights of the GAS8-AS1 or GAS8 stable transfected xenografts were observed compared with those of the control xenografts (both p < 0.01) (Fig. 6C). In support of that, elevated expression of GAS8-AS1 or GAS8, decreased Ki67 IHC staining, and increased GAS8 protein expression were found in the GAS8-AS1 or GAS8 xenografts (Fig. 6, D and E). A working model summarizing relationships between the lncRNA GAS8-AS1-GAS8 axis and tumorigenesis is shown in Fig. 6F.
Figure 6.
lncRNA GAS8-AS1 and GAS8 inhibit HCC growth in vivo. A, growth of tumors from GAS8-AS1 or GAS8-up-regulated HepG2 xenografts was significantly suppressed. B, there were no significant differences of mice weight between GAS8-AS1, GAS8, and control groups. C, significantly decreased tumor weights of the GAS8-AS1 or GAS8 xenografts were found. D, evidently increased expression of GAS8-AS1 or GAS8 was found in the GAS8-AS1 or GAS8 xenografts. E, representative pictures of HE staining, Ki67 IHC staining, and GAS8 IHC staining of xenografts. F, a working model summarizing relationships between the lncRNA GAS8-AS1-GAS8 axis and tumorigenesis **, p < 0.01. Error bars, S.D.
Discussion
We first identified lncRNA GAS8-AS1 as a significantly mutated gene in thyroid cancer (20). However, little is known about its involvement and downstream signaling in HCC development. In the current study, we find that lncRNA GAS8-AS1 recruits the MLL1/WDR5 complex to the GAS8 promoter, induces elevated H3K4me3 modification, facilitates RNA Pol II enrichment, and consequently activates GAS8 expression for the first time. Significantly positive correlations between expressions of these two genes in tissue specimens support the key regulatory role of lncRNA GAS8-AS1 on GAS8 expression. A series of in vitro and in vivo gene gain or loss assays prove the tumor suppressor nature of GAS8-AS1 and GAS8 during hepatocarcinogenesis. In support of this, we observed significantly reduced expression of both genes in human HCC tissues compared with normal tissues.
GAS8, previously known as “growth arrest–specific 11,” locates in chromosome 16, which is sometimes deleted in breast and prostate cancer (26). GAS8 is commonly up-regulated in growth-arrested cells and is a candidate tumor suppressor. Consistently, we found that GAS8 can inhibit malignant characteristics, including proliferation, invasion, and metastasis, in HCC and thyroid cancer (20). Interestingly, a genome-wide local ancestry approach identifies GAS8 and its genetic variants associated with chemotherapeutic susceptibility (27). In line with this, we also observed joint effects between GAS8 and sorafinib on inducing apoptosis of HCC cells. This could be due to the possibility that GAS8, an evolutionarily conserved microtubule-binding protein (28), may interfere with microtubule function because multiple anti-cancer drugs, including taxol, also target microtubule function. Additionally, GAS8 has also been proved to be involved in cytoskeleton function (29), male gametogenesis (30), motile cilia function (31), and primary ciliary dyskinesia (32, 33).
In all, we found that lncRNA GAS8-AS1 is required to maintain the GAS8 promoter in an open chromatin state. In accord with this notion, knockdown of GAS8-AS1 results in reduced MLL1/WDR5 binding, decreased H3K4me3 levels, diminished RNA Pol II functions, and gene silencing of GAS8. Thus, lncRNA GAS8-AS1 may function as part of a surveillance mechanism that maintains activation of GAS8 promoter and transcription, which thus prevents carcinogenesis. Our results elucidate the prevalent involvement of regulatory lncRNAs in cancers and provide pathogenic insights into HCC development and treatment.
Materials and methods
Patient cohorts
A total of 82 HCC patients were enrolled in this study. All patients received curative surgery for HCC in Shandong Cancer Hospital (Shandong cohort, n = 25, Jinan, Shandong Province, China) and Huaian No. 2 Hospital (Jiangsu cohort, n = 57, Huaian, Jiangsu Province, China) between April 2009 and October 2016. Patients did not receive any local or systemic anticancer treatments (i.e. chemotherapy or radiotherapy) before the surgery. All patients were postoperatively followed after surgery until January 2017, with a median follow-up of 13 months for Shandong cohort (range, 5–54 months) or 18 months for Jiangsu cohort (range, 6–70 months). The basic clinical-pathological characteristics of the patients are summarized in Table S1. This study was approved by the institutional review boards of Shandong Cancer Hospital and Huaian No. 2 Hospital for human or animal studies. At recruitment, written informed consent was obtained from each subject. The studies abide by the Declaration of Helsinki principles.
Quantitative RT-PCR (qRT-PCR) and Western blotting
Total RNA was isolated from surgically removed tissues or HCC cells with TRIzol reagent (Invitrogen). After treatment with RNase-free DNase to remove genomic DNA (Invitrogen), RNA samples were then reverse-transcribed into cDNAs using PrimeScript RT Master Mix (TaKaRa, Japan). GAS8-AS1, GAS8, and β-actin mRNAs were measured through qRT-PCR using SYBR Premix Ex Taq (TaKaRa). The GAS8-AS1 and GAS8 RNA expression was calculated relative to the β-actin expression (20, 34–36).
Total cellular proteins were separated with SDS-polyacrylamide gel and transferred to a polyvinylidene fluoride membrane (Millipore). The polyvinylidene fluoride membrane was then incubated with anti-GAS8 antibody (Abcam, ab209415) or anti-GAPDH antibody (0411, Santa Cruz Biotechnology, Inc., sc-47724). GAS8 and GAPDH proteins were visualized with enhanced chemiluminescence reagents (Millipore).
Cell proliferation, apoptosis, and cell cycle analyses
For gene overexpression, HepG2 and SMMC7721 cells were transfected with pcDNA3.1-GAS8-AS1 (GAS8-AS1), pcDNA3.1-GAS8 (GAS8), or pcDNA3.1 (Vector). To silence expression of endogenous GAS8-AS1 and GAS8, these HCC cells were transfected with GAS8-AS1 siRNA (siGAS8-AS1-1, siGAS8-AS1-2, and siGAS8-AS1-3), GAS8 siRNA (siGAS8-1, siGAS8-2, and siGAS8-3) (Table S2), or NC RNA (Genepharma), respectively. Cells were harvested and counted at 24, 48, and 72 h after transfection. HCC cells were treated with 1 μmol/liter sorafenib at 24 h after transfection and collected after another 24 h. Apoptosis was determined using the Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit (Invitrogen) with a FACSCalibur flow cytometer (BD Biosciences). For cell cycle analyses, transfected cells were dyed with PI and detected with the FACSCalibur flow cytometer.
Colony formation assays
A total of 2000 HepG2 or 3000 SMMC7721 cells were seeded into a 6-well cell culture plate and transfected with different gene expression constructs (GAS8-AS1, GAS8, or vector) or siRNAs (siGAS8-AS1–1/-2/-3, siGAS8–1/-2/-3, or NC RNA). After 12 days, cells were washed with cold PBS twice and fixed with 3.7% formaldehyde. After cells were dyed with crystal violet, colony number in each well was counted.
Wound-healing and transwell assays
The wound-healing and transwell assays were performed using HepG2 or SMMC7721 cells transfected with different gene expression constructs or siRNAs as described previously (35, 36). In wound-healing assays, the average extent of wound closure was quantified after the HepG2 or SMMC7721 cell layer was scratched at different time points. For transwell assays, HCC cells that migrated from BD Matrigel to the lower wells through pores were stained with 0.2% crystal violet solution and counted.
Subcellular fractionation
The cytosolic and nuclear fractions of SMMC7721 or HepG2 cells were separately isolated using the nuclear/cytoplasmic Isolation Kit (Biovision, San Francisco, CA) according to the manufacturer's instructions.
Quantitative DNA methylation analyses
Primers were designed to cover most CpG sites of the CpG island located near the GAS8 promoter region. The CpG island includes a part of the 5′-flanking region, whole exon 1, and a part of intron 1. Genomic DNA samples were modified by bisulfite reaction, and the mass spectra were collected using a MassARRAY Compact MALDI-TOF (Sequenom, San Diego, CA). The EpiTYPER software was used to generate the spectra's methylation ratios (Sequenom).
ChIP
All ChIP assays were performed using the EZ-ChIP chromatin immunoprecipitation kit (Millipore). HepG2 cells transfected with the GAS8-AS1 expression construct, vector, GAS8-AS1 siRNAs mixture (siGAS8-AS1-mix), or NC RNA were cross-linked in 1% formaldehyde. Genomic DNA was extracted from the fixed-chromatin cells and then subjected to IP with antibodies against H3K4me3 (Millipore, 17-614), Pol II (Millipore, 05-623B), MLL1 (Bethyl, A300-374), WDR5 (Abcam, ab56919), or normal mouse IgG (Millipore, 12-1371B). Purified DNA was analyzed by qPCR with five pairs of primers shown in Table S2.
RNA pulldown
GAS8-AS1 was amplified by PCR and cloned into a modified pMD19-T vector (TaKaRa) with inserted T7 promoter before and after the TA cloning site to prepare a plasmid construct as the template for RNA synthesis. The constructs were linearized and transcribed in vitro with biotin RNA labeling mix and T7 RNA polymerase (MEGAscript T7 transcript kit, Thermo Fisher Scientific) and purified with the RNeasy minikit (Qiagen). Sense or antisense biotinylated GAS8-AS1 RNAs were incubated with nucleus extracts at 4 °C for 1 h. Streptavidin magnetic beads were then washed with 100 μl of 1× Wash Buffer three times. Proteins bound were recovered by further incubating streptavidin magnetic beads with 50 μl of elution buffer at 37 °C for 30 min. The retrieved proteins were then detected by Western blotting with either antibodies of MLL1 (Bethyl, A300-374) or WDR5 (Abcam, ab56919).
RNA IP
RNA IP assays were performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) with the MLL1 (Bethyl, A300-374) and WDR5 (Abcam, ab56919) antibodies or nonspecific purified rabbit IgG (Millipore, pp64b). The MLL1 or WDR5 antibody was then recovered by magnetic beads protein A/G. lncRNA GAS8-AS1 and β-actin RNA levels in the precipitates were measured by qRT-PCR.
HCC xenografts
To evaluate the in vivo anti-tumor role of GAS8-AS1 and GAS8, we first transfected HepG2 cells with pcDNA3.1-GAS8-AS1 (GAS8-AS1), pcDNA3.1-GAS8 (GAS8), or pcDNA3.1 (Vector). After G418 (Geneticin) selection, we isolated stable cell clones with relative high expression of GAS8-AS1 or GAS8. A total of 3 × 108 HepG2 cells with stable transfection of GAS8-AS1 or GAS8 or pcDNA3.1 vector were inoculated subcutaneously into fossa axillaris of 5-week-old male nude BALB/c mice (Vital River Laboratory, Beijing, China) (n = 5/group). Tumor volumes were measured every 2 days after tumor volumes were ≥90 mm3. All procedures involving mice were approved by the institutional review board of Shandong Cancer Hospital.
Statistics
Student's t test was used to compare differences between two groups. The difference between three or more groups was calculated using one-way analysis of variance. The significance of association between GAS8-AS1 and GAS8 expression was calculated using Spearman's correlation. Kaplan–Meier plots and Cox proportional hazard regression analyses were performed to examine association between GAS8-AS1 or GAS8 expression and OS of HCC patients. A p value of <0.05 was used as the criterion of statistical significance. All analyses were performed with GraphPad Prism version 5 (GraphPad Software, Inc.) or the SPSS software package version 16.0 (SPSS Inc.).
Author contributions
M. Y. contributed to conception and design. W. P. and M. Y. contributed to acquisition of data or analysis and interpretation of data. M. Y., W. P., N. Z., W. L., J. L., L. Z., and Y. L. drafted the article.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grant 31671300 and Taishan Scholars Program of Shandong Province Grant tsqn20161060. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S11.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HCC
- hepatocellular carcinoma
- lncRNA
- long noncoding RNA
- MLL1
- mixed lineage leukemia 1
- H3K4
- histone 3 Lys-4
- H3K4me3
- H3K4 trimethylation
- WDR5
- WD-40 repeat protein 5
- RNP
- ribonucleoprotein
- PRC2
- Polycomb repressive complex 2
- Pol II
- polymerase II
- OS
- overall survival
- NC RNA
- negative control RNA
- qRT-PCR
- quantitative RT-PCR
- qPCR
- quantitative PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., and Parkin D. M. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2. El-Serag H. B., and Rudolph K. L. (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 10.1053/j.gastro.2007.04.061 [DOI] [PubMed] [Google Scholar]
- 3. Esteller M. (2011) Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 4. Fatica A., and Bozzoni I. (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 5. Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B. E., van Oudenaarden A., Regev A., Lander E. S., and Rinn J. L. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., Cabili M. N., Jaenisch R., Mikkelsen T. S., Jacks T., Hacohen N., et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mattick J. S., and Rinn J. L. (2015) Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7 10.1038/nsmb.2942 [DOI] [PubMed] [Google Scholar]
- 8. Sanfilippo P. G., and Hewitt A. W. (2014) Translating the ENCyclopedia Of DNA Elements Project findings to the clinic: ENCODE's implications for eye disease. Clin. Exp. Ophthalmol. 42, 78–83 10.1111/ceo.12150 [DOI] [PubMed] [Google Scholar]
- 9. Tragante V., Moore J. H., and Asselbergs F. W. (2014) The ENCODE project and perspectives on pathways. Genet. Epidemiol. 38, 275–280 10.1002/gepi.21802 [DOI] [PubMed] [Google Scholar]
- 10. Bhan A., Soleimani M., and Mandal S. S. (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res. 77, 3965–3981 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng W. X., Koirala P., and Mo Y. Y. (2017) LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta R. A., Shah N., Wang K. C., Kim J., Horlings H. M., Wong D. J., Tsai M. C., Hung T., Argani P., Rinn J. L., Wang Y., Brzoska P., Kong B., Li R., West R. B., van de Vijver M. J., Sukumar S., and Chang H. Y. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., Miyano S., and Mori M. (2011) Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- 14. Zhang X., Zhou L., Fu G., Sun F., Shi J., Wei J., Lu C., Zhou C., Yuan Q., and Yang M. (2014) The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis 35, 2062–2067 10.1093/carcin/bgu103 [DOI] [PubMed] [Google Scholar]
- 15. Pan W., Liu L., Wei J., Ge Y., Zhang J., Chen H., Zhou L., Yuan Q., Zhou C., and Yang M. (2016) A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol. Carcinog. 55, 90–96 10.1002/mc.22261 [DOI] [PubMed] [Google Scholar]
- 16. Zhu H., Lv Z., An C., Shi M., Pan W., Zhou L., Yang W., and Yang M. (2016) Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci. Rep. 6, 31969 10.1038/srep31969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mercer T. R., Dinger M. E., and Mattick J. S. (2009) Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 18. Wang K. C., and Chang H. Y. (2011) Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ulitsky I., and Bartel D. P. (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan W., Zhou L., Ge M., Zhang B., Yang X., Xiong X., Fu G., Zhang J., Nie X., Li H., Tang X., Wei J., Shao M., Zheng J., Yuan Q., et al. (2016) Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum. Mol. Genet. 25, 1875–1884 10.1093/hmg/ddw056 [DOI] [PubMed] [Google Scholar]
- 21. Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., and Allis C. D. (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872 10.1016/j.cell.2005.03.036 [DOI] [PubMed] [Google Scholar]
- 22. Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., and Roeder R. G. (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 10.1038/nsmb1128 [DOI] [PubMed] [Google Scholar]
- 23. Ruthenburg A. J., Wang W., Graybosch D. M., Li H., Allis C. D., Patel D. J., and Verdine G. L. (2006) Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13, 704–712 10.1038/nsmb1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao F., Townsend E. C., Karatas H., Xu J., Li L., Lee S., Liu L., Chen Y., Ouillette P., Zhu J., Hess J. L., Atadja P., Lei M., Qin Z. S., Malek S., Wang S., and Dou Y. (2014) Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 53, 247–261 10.1016/j.molcel.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alicea-Velázquez N. L., Shinsky S. A., Loh D. M., Lee J. H., Skalnik D. G., and Cosgrove M. S. (2016) Targeted disruption of the interaction between WD-40 repeat protein 5 (WDR5) and mixed lineage leukemia (MLL)/SET1 family proteins specifically inhibits MLL1 and SETd1A methyltransferase complexes. J. Biol. Chem. 291, 22357–22372 10.1074/jbc.M116.752626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitmore S. A., Settasatian C., Crawford J., Lower K. M., McCallum B., Seshadri R., Cornelisse C. J., Moerland E. W., Cleton-Jansen A. M., Tipping A. J., Mathew C. G., Savnio M., Savoia A., Verlander P., Auerbach A. D., et al. (1998) Characterization and screening for mutations of the growth arrest-specific 11 (GAS11) and C16orf3 genes at 16q24.3 in breast cancer. Genomics 52, 325–331 10.1006/geno.1998.5457 [DOI] [PubMed] [Google Scholar]
- 27. Wheeler H. E., Gorsic L. K., Welsh M., Stark A. L., Gamazon E. R., Cox N. J., and Dolan M. E. (2011) Genome-wide local ancestry approach identifies genes and variants associated with chemotherapeutic susceptibility in African Americans. PLoS One 6, e21920 10.1371/journal.pone.0021920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishimura N., Araki K., Shinahara W., Nakano Y., Nishimura K., Higashio H., and Sasaki T. (2008) Interaction of Rab3B with microtubule-binding protein Gas8 in NIH 3T3 cells. Arch. Biochem. Biophys. 474, 136–142 10.1016/j.abb.2008.03.032 [DOI] [PubMed] [Google Scholar]
- 29. Hill K. L., Hutchings N. R., Grandgenett P. M., and Donelson J. E. (2000) T lymphocyte-triggering factor of african trypanosomes is associated with the flagellar fraction of the cytoskeleton and represents a new family of proteins that are present in several divergent eukaryotes. J. Biol. Chem. 275, 39369–39378 10.1074/jbc.M006907200 [DOI] [PubMed] [Google Scholar]
- 30. Yeh S. D., Chen Y. J., Chang A. C., Ray R., She B. R., Lee W. S., Chiang H. S., Cohen S. N., and Lin-Chao S. (2002) Isolation and properties of Gas8, a growth arrest-specific gene regulated during male gametogenesis to produce a protein associated with the sperm motility apparatus. J. Biol. Chem. 277, 6311–6317 10.1074/jbc.M106941200 [DOI] [PubMed] [Google Scholar]
- 31. Lewis W. R., Malarkey E. B., Tritschler D., Bower R., Pasek R. C., Porath J. D., Birket S. E., Saunier S., Antignac C., Knowles M. R., Leigh M. W., Zariwala M. A., Challa A. K., Kesterson R. A., Rowe S. M., et al. (2016) Mutation of growth arrest specific 8 reveals a role in motile cilia function and human disease. PLoS Genet. 12, e1006220 10.1371/journal.pgen.1006220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Olbrich H., Cremers C., Loges N. T., Werner C., Nielsen K. G., Marthin J. K., Philipsen M., Wallmeier J., Pennekamp P., Menchen T., Edelbusch C., Dougherty G. W., Schwartz O., Thiele H., Altmüller J., et al. (2015) Loss-of-function GAS8 mutations cause primary ciliary dyskinesia and disrupt the nexin-dynein regulatory complex. Am. J. Hum. Genet. 97, 546–554 10.1016/j.ajhg.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeanson L., Thomas L., Copin B., Coste A., Sermet-Gaudelus I., Dastot-Le Moal F., Duquesnoy P., Montantin G., Collot N., Tissier S., Papon J. F., Clement A., Louis B., Escudier E., Amselem S., and Legendre M. (2016) Mutations in GAS8, a gene encoding a nexin-dynein regulatory complex subunit, cause primary ciliary dyskinesia with axonemal disorganization. Hum. Mutat. 37, 776–785 10.1002/humu.23005 [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Li M., Wang Z., Han S., Tang X., Ge Y., Zhou L., Zhou C., Yuan Q., and Yang M. (2015) Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J. Biol. Chem. 290, 3925–3935 10.1074/jbc.M114.596866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ren Y., Shang J., Li J., Liu W., Zhang Z., Yuan J., and Yang M. (2017) The long noncoding RNA PCAT-1 links the microRNA miR-215 to oncogene CRKL-mediated signaling in hepatocellular carcinoma. J. Biol. Chem. 292, 17939–17949 10.1074/jbc.M116.773978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang X., Yu D., Ren Y., Wei J., Pan W., Zhou C., Zhou L., Liu Y., and Yang M. (2016) Integrative functional genomics implicates EPB41 dysregulation in hepatocellular carcinoma risk. Am. J. Hum. Genet. 99, 275–286 10.1016/j.ajhg.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chojnacki S., Cowley A., Lee J., Foix A., and Lopez R. (2017) Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553 10.1093/nar/gkx273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muppirala U. K., Honavar V. G., and Dobbs D. (2011) Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics 12, 489 10.1186/1471-2105-12-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.