Abstract
Brain and muscle ARNT-like protein-1 (BMAL-1) is an important component of the cellular circadian clock. Proteins such as epidermal (EGF) or nerve growth factor (NGF) affect the cellular clock via extracellular signal–regulated kinases-1/2 (ERK-1/2) in NIH3T3 or neuronal stem cells, but no such data are available for the insulin-like growth factor-1 (IGF-1). The hypothalamus expresses receptors for all three growth factors, acts as a central circadian pacemaker, and releases hormones in a circadian fashion. However, little is known about growth factor–induced modulation of clock gene activity in hypothalamic cells. Here, we investigated effects of IGF-1, EGF, or NGF on the Bmal-1 promoter in two hypothalamic cell lines. We found that only IGF-1 but not EGF or NGF enhanced activity of the Bmal-1 promoter. Inhibition of ERK-1/2 activity did not affect IGF-1–induced Bmal-1 promoter activation and all three growth factors similarly phosphorylated ERK-1/2, questioning a role for ERK-1/2 in controlling BMAL-1 promoter activity. Of note, only IGF-1 induced sustained phosphorylation of glycogen synthase kinase-3β (GSK-3β). Moreover, the GSK-3β inhibitor lithium or siRNA-mediated GSK-3β knockdown diminished the effects of IGF-1 on the Bmal-1 promoter. When IGF-1 was used in the context of temperature cycles entraining hypothalamic clock gene expression to a 24-h rhythm, it shifted the phase of Bmal-1 promoter activity, indicating that IGF-1 functions as a zeitgeber for cellular hypothalamic circadian clocks. Our results reveal that IGF-1 regulates clock gene expression and that GSK-3β but not ERK-1/2 is required for the IGF-1–mediated regulation of the Bmal-1 promoter in hypothalamic cells.
Keywords: circadian clock, insulin-like growth factor (IGF), growth factor, glycogen synthase kinase 3 (GSK-3), aging
Introduction
The cellular circadian clock is present in each one of our cells, with genes, proteins, and metabolites oscillating every 24 h. It acts as a biological timekeeper and manifold higher order physiological processes are subject to regulation by the circadian clock. Sleep/wake cycles, core body temperature, metabolism, and neuroendocrine secretion are controlled by diverse, cell- or organ-specific daily clocks (1–4).
Much is known about the molecular components of the cellular clock in mammals, which features a network of feedback loops that span transcriptional, translational, and post-translational levels (5, 6). Briefly, BMAL-13 and its partner CLOCK bind E-boxes within the promoter of many regulatory clock genes (Periods, Cryptochromes, Rev-Erbs, and RORs), thereby enhancing their expression. Several of the clock gene products exert a negative feedback on their own expression by inhibiting the transcriptional activation by BMAL-1 and CLOCK. The Bmal-1 promoter is controlled by two orphan nuclear receptor proteins: REV-ERBα and the retinoid orphan receptor A (RORA), with RORA activating and REV-ERBα repressing Bmal-1 expression. Because BMAL-1 is an activator of Rev-Erbα, this represents an additional feedback pathway. Through a combination of repression and activation, BMAL-1 expression rises and falls as part of the circadian cycle (7).
As known for other transcription factors, post-translational modifications of clock components are critical for their function. Clock proteins are heavily phosphorylated, thus, second messenger-dependent kinases regulate the clock on the molecular level (8). cAMP-dependent kinase A-mediated activation of the cAMP-response element–binding protein (CREB) or ERK-1/2 has been shown to regulate PER expression (9–12). GSK-3β has been described to modulate expression or cellular location of CLOCK, REV-ERBα, or PER proteins (13–18).
Although the key role of growth factors for cell proliferation and differentiation has been established, their role in the regulation of the cellular clock is not fully understood. Serum (a mixture of growth factors and cytokines) is well known to induce expression of clock genes (12, 19, 20). Induction of the c-FOS protein and activation of CREB, both acting on Per-1 or −2, has been proposed to be the molecular link between serum-induced signaling and clock gene expression (12). EGF or NGF enhanced expression of Per-2 in fibroblasts or neuronal stem cells via ERK-1/2 (9–11, 21–25). Of note, their relative IGF-1 has not yet been reported to regulate the cellular clock although it also activates ERK-1/2 in numerous cell lines (26). Likewise, despite the high impact of GSK-3β on clock gene expression described above, no effects on clock gene expression by growth factors via this kinase have been reported, although functional interactions between GSK-3β and growth factors are well established (26–28).
Rhythms of circadian clock genes, as described above, occur ex vivo in cell culture, showing the cell-based nature of the circadian clock. Similar to organismal circadian clocks, cellular clocks in constant conditions show a free-running approximately 24-h rhythm. Cellular and organismal clocks can also be entrained to an external clue. In such a case, the clock adopts to the rhythm provided by this clue, which is then called zeitgeber (German for time emitter). Daily 24-h oscillations in core body temperature have been proposed to be such a zeitgeber for cellular and organismal clocks (29–32). Clock gene expression regulating hormones are, thus, able to act as a zeitgeber for the cellular clock as shown for serum (12). EGF, did not entrain the clock in fibroblasts but in neuronal stem cells (9, 21). NGF has been shown to entrain the clock in the suprachiasmatic nucleus of Syrian hamster (22, 25).
Here we aimed at analyzing effects of IGF-1 on clock gene expression and compared its effects to other growth factors such as EGF and NGF. The hypothalamus is crucial for the generation of circadian rhythms, undergoes daily rhythmicity and expresses receptors for all three growth factors (22, 33–37). Thus, we used murine hypothalamic cells to investigate effects of growth factors on clock gene expression. We found that IGF-1 regulates Bmal-1 expression, in contrast to NGF and EGF, in these cells. It does so not via CREB, c-FOS, or ERK-1/2 but rather via GSK-3β. When IGF-1 was used in the context of an entraining temperature cycle, it shifted the phase of Bmal-1 promoter activity, indicating that it functions as a zeitgeber for cellular circadian clocks via GSK-3β.
Results
IGF-1 but not EGF or NGF regulates clock gene expression in hypothalamic cells
Hypothalamic mHypoA-2/10 cells were used to investigate the effects of growth factors on Bmal-1 promoter activity (38–40). These cells have recently been shown to express receptors for melanocortins and neuropeptide Y as well as the precursor of thyroliberin in a T3-dependent manner (39). Thus, mHypoA-2/10 cells resemble specialized neurons of the paraventricular nucleus. To measure promoter activity under constant conditions, we stably expressed a plasmid encoding the Bmal-1 promoter (containing binding sites for REV-ERBα and RORA) fused to a luciferase gene (Bmal-1-luc) in mHypoA-2/10 cells (mHypoA-2/10-Bmal-1-luc cells). Light emission in the presence of luciferin or luciferin ethyl ester was monitored in living cells (29). Although luminescence in mHypoA-2/10-Bmal-1-luc cells was detectable in lysates, it was too low to be measured in living cells (Fig. S1). We thus used protein lysates for measurements of Bmal-1 promoter activity in these cells.
We first determined if hypothalamic cell cultures are capable of rhythmic clock gene expression. mHypoA-2/10-Bmal-1-luc cells were entrained by a protocol including serum starvation (1 h) or serum stimulation (40%, 30 min), followed by a 48-h incubation in 0.1% serum. Samples were harvested at 4-h intervals over these 2 days revealing two cycles of expression from the Bmal-1 promoter (Fig. 1A). These data suggest that the cellular clock in mHypoA-2/10-Bmal-1-luc cells was synchronized by a serum shock, showing that this cell line is a suitable model for the analysis of growth factor–induced effects on clock-regulated gene expression using the Bmal-1 promoter as a proxy. Because growth factor–induced signaling might depend on the cellular context, we also investigated the effects of growth factors on Bmal-1 promoter activity in hypothalamic cells producing the gonadotropin-releasing hormone, GT1–7 cells (41). Expression of the Bmal-1 reporter transiently in GT1–7 cells revealed that in these cells the Bmal-1 promoter was also synchronized by a serum shock (Fig. 1B). Thus, mHypoA-2/10-Bmal-1-luc and GT1–7 cells transiently expressing the Bmal-1-luc reporter are suitable for the analysis of growth factor–induced effects on the Bmal-1 promoter.
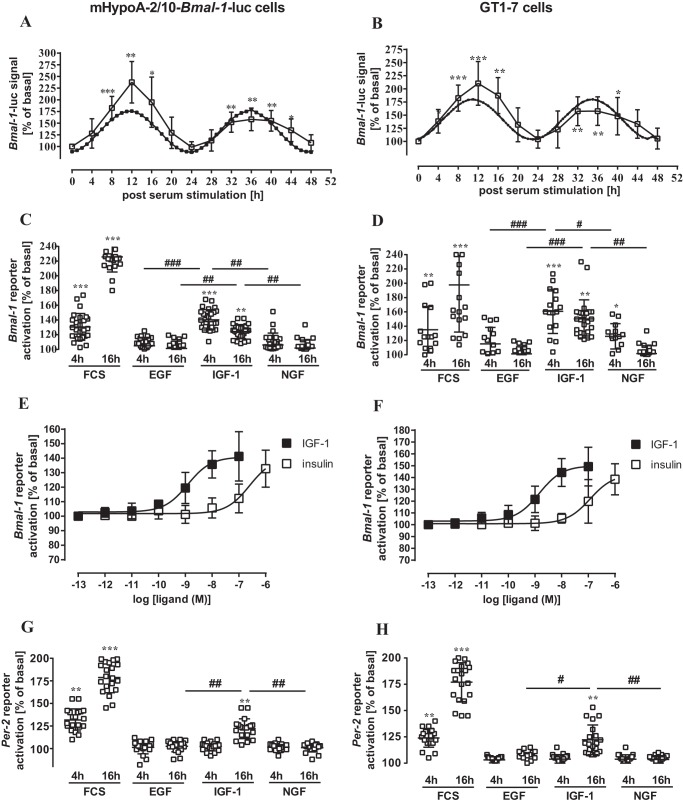
Figure 1.
IGF-1, but not EGF of NGF, induced Bmal-1 or Per-2 promoter activity in hypothalamic cells. In A, mHypoA-2/10-Bmal-1-luc cells and in B, GT1–7 cells transiently expressing the Bmal-1 reporter were serum-starved (1 h), stimulated or not for 30 min with 40% of serum, and maintained for 48 h in 0.1% serum. Lysates were analyzed at 4-h intervals. Data of treated cells were normalized to untreated control cells (basal). Data of 5 (A) or 3 (B) experiments were compiled and presented as the mean ± S.D. Asterisks indicate significant differences to time point 0 (one-way ANOVA). A significant Cosinor fit with a p value of 0.03 (r2 = 0.61) was determined in A and with a p value of 0.02 (r2 = 0.73) in B (Circwave; dotted line). In C and E, mHypoA-2/10-Bmal-1-luc cells and in D and F, GT1–7 cells transiently expressing the Bmal-1 reporter, and in E, mHypoA-2/10 or F, GT1–7 cells transiently expressing the Per-2 reporter are shown. In C, D, G, and H, cells were serum-starved (1 h), stimulated or not for 30 min with 40% of serum or 100 nm EGF, IGF-1, or NGF, and lysates were analyzed after 4 and 16 h. In E and F, cells were stimulated with increasing concentrations of IGF-1 or insulin and analyzed after 4 h. Data from 5 independent experiments, each performed in quadruplicates, were compiled by setting values of untreated cells to 100% and are shown as the mean ± S.D. Asterisks indicate significant differences relative to 100% (one-sample Student's t test) and hash marks between growth factors (one-way ANOVA).
We next used the same protocol but replaced serum with IGF-1, EGF, or NGF. We first focused on acute Bmal-1 reporter activation and analyzed the effects of growth factors on both cell types 4 and 16 h post-stimulation. Strikingly, only IGF-1 activated the Bmal-1 promoter at both time points in either cell line (Fig. 1, C and D), highlighting a hitherto unappreciated functional interaction between IGF-1 and the Bmal-1 promoter. EGF failed to induce Bmal-1 reporter activation at any time point in both cell lines. NGF promoted significant Bmal-1 promoter activity only after 4 h in GT1–7 cells.
The IGF-1 concentration used (100 nm) activates both IGF and insulin receptors (42). A recent study revealed that insulin inactivates BMAL-1 by phosphorylation and subsequent nuclear extraction via insulin receptors in hepatic cells (43). Thus, to discriminate between insulin and IGF-1 receptors, we performed concentration-response curves with both hormones (Fig. 1 E and F). EC50 values of IGF-1 for activation of the BMAL-1 promoter were about 2 log orders lower compared with insulin in both cell lines (mHypoA-2/10-BMAL-luc: 1.2 ± 0.4 nm for IGF-1 and 245 ± 37 nm for insulin; GT1–7 cells: 1.5 ± 0.5 nm for IGF-1 and 117 ± 49 nm for insulin), clearly suggesting that IGF and not insulin receptors are involved.
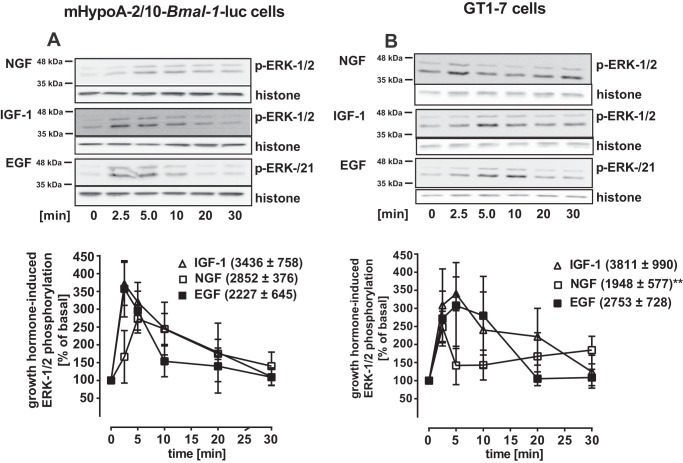
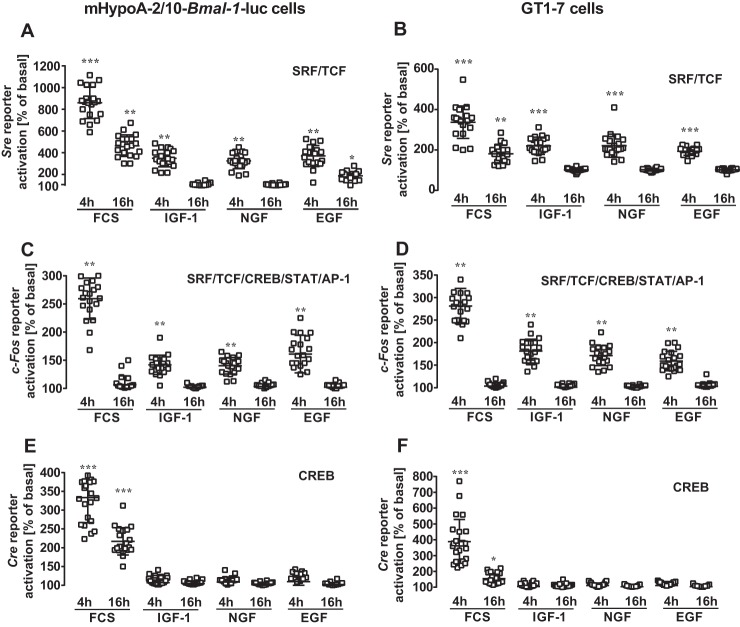
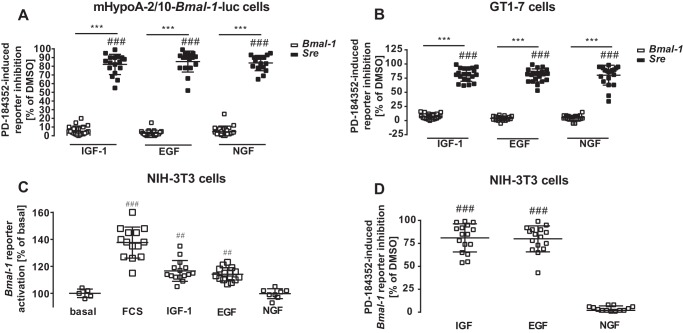
Lack of Bmal-1 promoter activation by EGF is surprising in light of its effects on clock gene expression via the Per-2 promoter in other cell lines (9, 21, 22, 25). Note that PER-2 hinders the Bmal-1 suppressor REV-ERBα from its inhibitory actions on this promoter. Thus activation of the Per-2 promoter should subsequently enhance Bmal-1 promoter activity. Similar, because BMAL-1 activates E-box elements of the Per-2 promoter, activation of the Bmal-1 promoter should subsequently enhance Per-2 promoter activity (Fig. 9C). Hence, we next tested the effect of all three growth factors on a Per-2 luciferase reporter in hypothalamic cells (Fig. 1, G and H). As expected from the functional interactions between BMAL-1 and the Per-2 promoter, IGF-1 induced significant Per-2 promoter activity 16 h but not at 4 h post-stimulation, indicating that it affects first the Bmal-1 and subsequently the Per-2 promoter. EGF and NGF failed again to activate this clock gene reporter at any time and in either cell line, suggesting that both growth factors do not affect clock gene expression in hypothalamic cells at all. These results could be because these cells do not express NGF or EGF receptors. Thus, we next analyzed growth factor–induced phosphorylation of ERK-1/2 and used a luciferase reporter gene that incorporated serum-response elements (Sre) and thus would respond to activated serum-response factor (SRF) and ternary complex factors (TCF). There were no significant differences between EGF- and IGF-1–induced ERK-1/2 phosphorylation in both cell types (Fig. 2, A and B), suggesting that a lack of EGF receptor expression is not the cause for absent EGF-induced Bmal-1 or Per-2 promoter activation. NGF-induced phosphorylation of ERK-1/2 was significantly increased relative to unstimulated cells, but significantly weaker compared with IGF-1 in GT1–7 cells. All three growth factors similarly induced luciferase expression in cells containing the Sre reporter (Fig. 3, A and B, Fig. S2). These results indicate that appropriate receptors for all of these growth factors were expressed on these cells and were functional. To identify where in the cognate signal transduction pathways of IGF-1 acts differently from NGF and EGF, two additional reporter genes were used. A reporter containing the entire c-Fos promoter (harboring SRF/TCF-binding elements as well as binding sites for signal transducers and activators of transcription and for the activating protein-1) fused to luciferase was transfected into hypothalamic cells. As for the Sre reporter, treatments with serum or all three growth factors showed similarly increased gene expression (Fig. 3, C and D). When a reporter consisting of cAMP-response elements (Cre) (containing binding sites for CREB) fused to the luciferase gene was expressed, serum activated gene expression, but all three growth factors failed to induce luciferase activity in both cell types (Fig. 3, E and F). So far, our data suggest that IGF-1–promoted circadian clock gene expression in hypothalamic cells is unique among the three tested growth factors and distinct from what is known in other cell models, because it does not correlate with the Cre promoter activation, c-FOS induction, or ERK-1/2 phosphorylation. To exclude a role for ERK-1/2, we used PD-184352, an inhibitor of the ERK-1/2 kinases mitogen-activated protein kinase-1/2. PD-184352 did not inhibit the effects of IGF-1 on the Bmal-1 promoter, indicating that ERK-1/2 signaling is indeed not involved in this process (Fig. 4, A and B). In contrast the inhibitor strongly decreased Sre reporter activation by all growth factors (Fig. 4, A and B). We next asked whether the distinct effects of IGF-1 on the Bmal-1 reporter are also found in cell models, where ERK-1/2-mediated activation of clock gene expression by EGF has previously been shown. Thus, we tested the Bmal-1 reporter in NIH-3T3 fibroblasts for its sensitivity toward growth factors (9). Indeed, IGF-1 and EGF induced reporter activation in these cells (Fig. 4C) in a PD-184352-dependent manner (Fig. 4D). Hence, it appears that IGF-1–induced circadian gene expression in hypothalamic cells is unique among growth factors and cell types.
Figure 9.
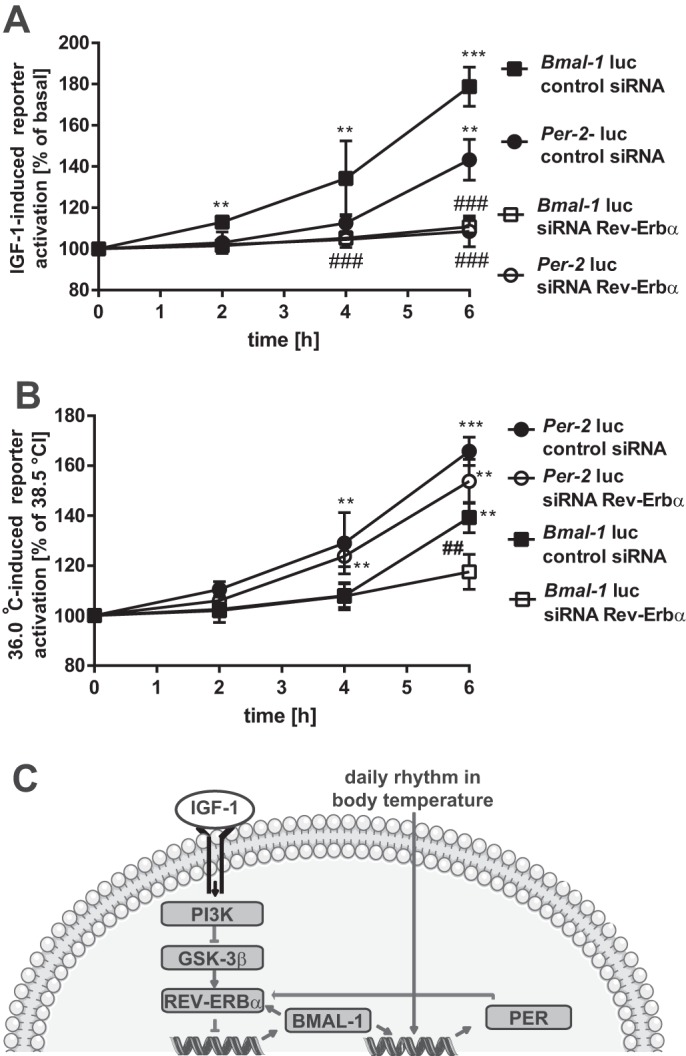
IGF-1 and temperature induce Bmal-1 and Per-2 promoter activity with distinct kinetics. A and B, Bmal-1-Luc or Per-2-Luc reporter expressing mHypoA-2/10 cells co-expressing a control/scrambled siRNA or a target-specific siRNA against REV-ERBα were serum-starved for 1 h followed by stimulation with IGF-1 (100 nm) for 30 min (A) or in (B) temperature-stimulated (36.0 and 38.5 °C). Luciferase activity was determined 2, 4, and 6 h post-stimulation. Asterisks indicate a significant difference relative to basal/control samples; hash marks between cells expressing the random or the selective siRNA (two-way ANOVA). C, a cartoon depicting a model illustrating distinct IGF-1- and temperature-induced signaling pathway leading to Bmal-1 or Per-2 promoter activation, respectively, is shown.
Figure 2.
IGF-1, EGF, or NGF similarly induced ERK-1/2 phosphorylation in hypothalamic cells. ERK-1/2 phosphorylation was determined by Western blotting in mHypoA-2/10 (A) and GT1–7 cells (B) after stimulation with NGF, IGF-1, or EGF (100 nm) for the indicated periods of time after 1 h of serum starvation. One of 5 representative blots is shown; data were quantified by ImageJ, compiled by setting values of untreated cells to 100% and shown in the graphs below. The area under the curve (restricted to 100%) is given in parentheses. Asterisks indicate significant differences between EGF or NGF and IGF-1 (two-sample Student's t test).
Figure 3.
IGF-1, EGF, or NGF similarly induced Sre, c-Fos, or Cre reporter activation in hypothalamic cells. Growth factor–induced reporter activation was measured in mHypoA-2/10 (A, C, and E) or GT1–7 (B, D, and F) cells carrying the Sre (A and B), c-Fos (C and D), or Cre (E and F) luciferase reporter. After 1 h of serum starvation, cells were stimulated using 100 nm of the indicated peptide or 40% of serum for 30 min followed by a washing step. Lysates were analyzed 4 and 16 h post-stimulation. Data from 5 independent experiments, each performed in quadruplicates, were compiled by setting values of untreated cells to 100% and are shown as the mean ± S.D. Asterisks indicate significant differences relative to 100% (one-sample Student's t test).
Figure 4.
ERK-1/2 activity is required for IGF-1–induced Bmal-1 promoter activation in NIH3T3 but not in hypothalamic cells. In A, mHypoA-2/10 and B, GT1–7 cells expressing either the Bmal-1 or the Sre reporter were serum-starved for 1 h in the presence of 10 μm PD-184352 or 0.1% DMSO (control) followed by stimulation with IGF-1, EGF, or NGF (100 nm) for 30 min. Luciferase activity was determined 4 h post-stimulation. Data from 3 to 5 independent experiments performed in quadruplicates were compiled and are shown as the mean ± S.D. Hash marks indicate significant differences to DMSO (one-sample Student's t test) and asterisks between cells expressing different reporters (two-way ANOVA). In C and D, Bmal-1 reporter activation in NIH-3T3 cells was measured. In C, cells were serum-starved for 1 h, stimulated with serum (40%) or IGF-1, EGF, or NGF for 30 min and reporter activity was measured after 4 h. D, cells were serum-starved for 1 h in the presence of 10 μm Pd-184352 or as a control with 0.1% DMSO followed by stimulation with IGF-1, EGF, or NGF (100 nm) for 4 h. Hash marks indicate significant differences to 100% (one-sample Student's t test).
PI3K/GSK-3β signaling is required for IGF-1–induced Bmal-1 promoter activity in hypothalamic cells
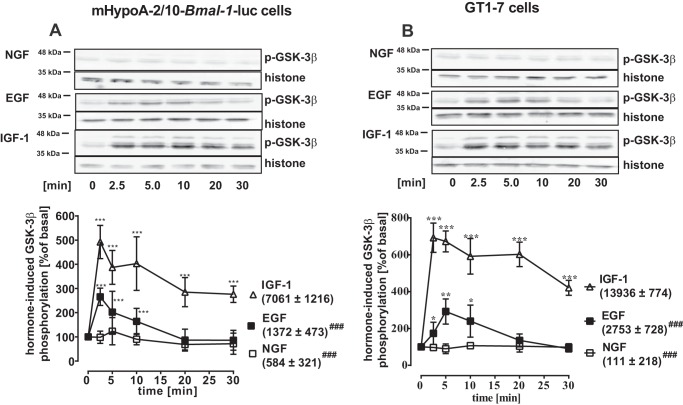
GSK-3β acts on the molecular circadian clock and is an established component of growth factor signaling (13, 15, 26–28, 44). Thus, we next investigated functional interactions between this kinase and growth factors. Phosphorylation of Ser-9 has been reported to inhibit GSK-3β activity. Analysis of hypothalamic cells revealed that NGF failed to phosphorylate GSK-3β in both cell lines (Fig. 5, A and B). EGF weakly phosphorylated this enzyme transiently. In contrast, IGF-1 induced high levels of GSK-3β phosphorylation persisting for at least 30 min after stimulation, with the area under the curve ∼5 times larger in the case of IGF-1 compared with EGF stimulation in both cell types (Fig. 5, A and B). Thus, sustained phosphorylation of GSK-3β may represent a specific signaling pathway engaged by IGF-1 in hypothalamic cells consistent with its function on clock gene expression.
Figure 5.
IGF-1, EGF, or NGF differently induced GSK-3β phosphorylation in hypothalamic cells. GSK-3β phosphorylation was determined in mHypoA-2/10 (A) and GT1–7 (B) cells by Western blotting after stimulation of cells with EGF, IGF-1, or NGF (100 nm) for the indicated periods of time after 1 h of serum starvation. One of 5 representative blots is shown; data were quantified by ImageJ, compiled by setting values of untreated cells to 100% and shown as the mean ± S.D. in the graphs below. Asterisks indicate significant differences relative to time point 0 (one-way ANOVA). The area under the curve (restricted to 100%) is given in parentheses. Hash marks indicate significant differences between of EGF of NGF and IGF-1 (two-sample Student's t test).
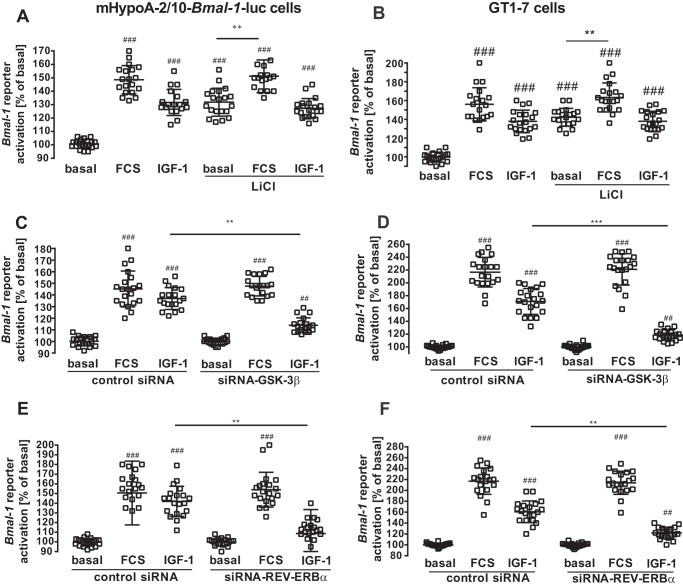
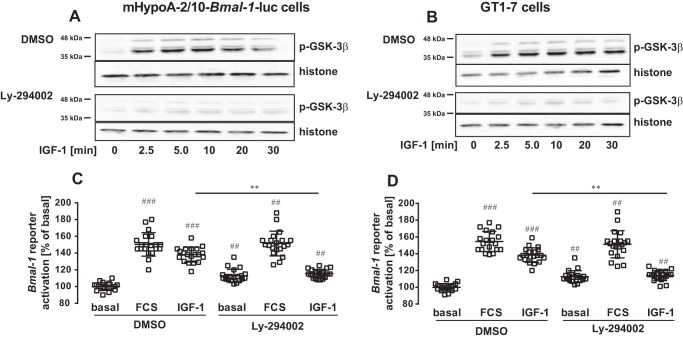
Lithium has been shown to inhibit GSK-3β and is proposed to lengthen the free running period of circadian rhythms (13–18). LiCl enhanced basal Bmal-1 reporter gene activity but prevented an additional increase by IGF-1, suggesting that IGF-1 engages a GSK-3β-mediated pathway to modulate Bmal-1 promoter activity (Fig. 6A and B). Because lithium also affects other cellular targets besides GSK-3β, we tested this hypothesis by silencing GSK-3β expression with siRNA (45). Down-regulation of GSK-3β expression significantly inhibited IGF-1–induced Bmal-1 promoter activation (Fig. 6, C and D, Fig. S3A). REV-ERBα represses Bmal-1 expression and GSK-3β stabilizes the REV-ERBα protein (17). To test whether REV-ERBα is involved in IGF-1–induced Bmal-1 reporter activation, we also employed a siRNA approach to knock down the expression of the Bmal-1 repressor (Fig. 6, E and F, Fig. 3B). This maneuver blocked the effects of IGF-1 on the Bmal-1 promoter, supporting a model in which IGF-1–induced inhibition of GSK-3β destabilizes REV-ERBα and thereby increases Bmal-1 promoter activity. We next analyzed the effects of the established GSK-3β kinase, phosphoinositide 3-kinase (PI3K), on IGF-1–induced Bmal-1 promoter activation. The specific PI3K inhibitor Ly-294002 blocked IGF-1–induced GSK-3β phosphorylation and Bmal-1 promoter expression (Fig. 7, A–D), indicating that IGF-1–induced PI3K activation leads to sustained GSK-3β inactivation and increased activity of the Bmal-1 promoter.
Figure 6.
GSK-3β expression is required for IGF-1–induced Bmal-1 promoter activation in hypothalamic cells. In A and B, mHypoA-2/10-Bmal-1-luc or GT1–7 cells transfected with the Bmal-1 reporter were serum-starved in the presence or absence 10 mm of LiCl followed by stimulation with serum (40%) or IGF-1 (100 nm) for 30 min. Luciferase activity was determined 4 h post-stimulation. Data from 5 independent experiments performed in quadruplicates were compiled by setting basal values to 100% and are shown as the mean ± S.D. Hash marks indicate significant differences to basal (one-sample Student's t test) and asterisks between inhibitor alone and with FCS (two-way ANOVA). C–F, mHypoA-2/10-Bmal-1-luc (C and E) or GT1–7 (D and F) cells expressing a control (scrambled) or a target-specific siRNA against GSK-3β (C and D) or REV-ERBα (E and F) were serum-starved for 1 h and then stimulated with serum (40%) or IGF-1 (100 nm) for 30 min. GT1–7 cells were also co-transfected with the Bmal-1 reporter plasmid. Luciferase activity was determined 4 h post-stimulation. Data from 5 independent experiments were compiled and are shown as the mean ± S.D. Hash marks indicate significant differences to basal with control siRNA (one-sample Student's t test) and asterisks between IGF-1–treated cells (one-way ANOVA).
Figure 7.
PI3K activity is required for IGF-1–induced GSK-3β phosphorylation and Bmal-1 promoter activation in hypothalamic cells. GSK-3β phosphorylation was determined in mHypoA-2/10-Bmal-1-luc (A) or GT1–7 cells (B) by Western blotting in DMSO (0.1%) or Ly-294002 (20 μm) pretreated cells that were stimulated with IGF-1 (100 nm) for the indicated period of time after 1 h of serum starvation. One of 5 representative blots is shown. C, mHypoA-2/10-Bmal-1-luc or D, Bmal-1 reporter expressing GT1–7 cells were serum-starved for 1 h with DMSO (0.1%) with or without the PI3K inhibitor Ly-294002 (20 μm) and then stimulated with serum (40%) or IGF-1 (100 nm) for 30 min. Luciferase activity was determined 4 h post-stimulation. Data of 5 independent experiments performed in quadruplicates were compiled and are shown as the mean ± S.D. Hash signs indicate significant differences to DMSO (one-sample Student's t test) and asterisks between IGF-1–treated cells (one-way ANOVA).
IGF-1 is a zeitgeber in hypothalamic cells
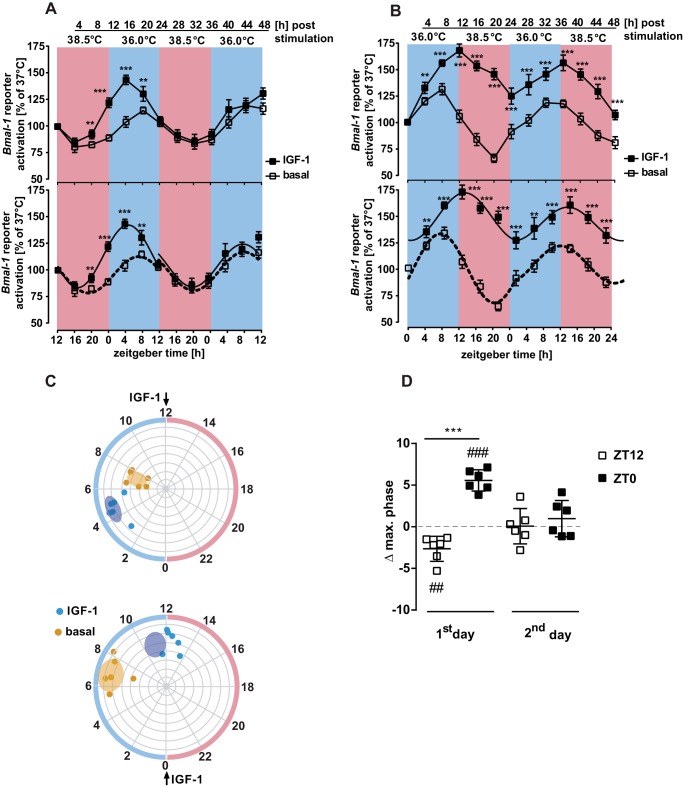
After having established the enhancing effects of IGF-1 on clock genes in hypothalamic cells, we next asked whether IGF-1 would modify the entrained phase of clock gene expression. Temperature is a near universal zeitgeber for circadian clocks, with 24-h temperature cycles within a physiological range synchronizing the expression of circadian clocks (30, 31, 46). Thus, mHypoA-2/10-Bmal-1-luc cells were either exposed to 24-h temperature cycles (12 h at 38.5 and 12 h at 36.0 °C or 12 h at 36.0 and 12 h at 38.5 °C) or, as a control, kept at constant temperature of 37.0 °C. Luciferase activity was measured every 4 h for 48 h. As shown in Fig. 8, A and B, temperature cycles synchronize oscillations of Bmal-1 promoter activity with peaks in luciferase expression in the low temperature phase. Thus, we used temperature-entrained cultures to investigate if IGF-1 is capable of advancing or delaying the phase of Bmal-1 luciferase expression by applying IGF-1 to cells at ZT12 and ZT0. Note that for mice, ZT12 corresponds to high core body temperature which occurs in the dark period in a 12-h light/12-h dark cycle. Addition of IGF-1 at the low to high temperature transition (ZT12) led to a 2.7 ± 0.6-h phase advance (Fig. 8, A, C, and D). Adding the growth factor at the high to low temperature transition (ZT0) resulted in a 5.5 ± 0.5-h phase delay (Fig. 8, B–D). Hence, IGF-1 acts as a zeitgeber for hypothalamic cells, as demonstrated by the change in the phase of temperature-entrained Bmal-1 promoter activity.
Figure 8.
IGF-1 is a zeitgeber in temperature entraining cells. A and B, after 1 h of serum starvation mHypoA-2/10-Bmal-1-luc cells were treated with IGF-1 (100 nm) for 30 min at 37.0 °C, after which they were washed and placed into temperature-controlled incubators set either to 12 h 38.5/12 h 36.0 °C (A) or 12 h 36.0/12 h 38.5 °C in B for 48 h. Cells were sampled for luciferase activity in 4-h intervals. Data were normalized relative to time point-specific control cells kept at 37.0 °C. Asterisks indicate significant differences relative to basal at the given time point (two-way ANOVA). In the lower panels, the same data were analyzed by Circwave 1.4 using a 1-harmonic fit for each 24-h interval. All fits were significant (ZT12: 1st day basal r2 = 0.65, IGF-1 r2 = 0.79, 2nd day basal r2 = 0.51, IGF-1 r2 = 0.46; ZT0: 1st day basal r2 = 0.76, IGF-1 r2 = 0.56, 2nd day basal r2 = 0.51, IGF-1 r2 = 0.3, p < 0.001 for all). C, polar plots indicate the phase of maximum Bmal-1 expression on the circular axis and the amplitude of expression on the radial axis. On the outer circle pink indicates warm phase and blue shows the cold phase. The upper plot indicates the application of IGF-1 at ZT12 and the lower plot at ZT0. D, the phase shift of maximal Bmal-1 activity by IGF-1 application on day 1 and day 2 are shown. Asterisks indicate significant differences relative to basal (two-sample Student's t test) and hash marks refer to a difference relative to zero (one sample Student's t test). Data from six experiments were compiled and shown here as mean ± S.D.
One might assume that IGF-1 acts as zeitgeber in temperature-entrained cells by modifying distinct transcriptional regulators within the clock. As shown in Fig. 1, IGF-1 first activated the Bmal-1 (4 h) and subsequently the Per-2 (16 h) reporter, suggesting that Bmal-1 is the primary target of IGF-1 within the clock. To further test this hypothesis, we performed a checkerboard experiment whereby kinetics (2, 4, and 6 h) of IGF-1 or temperature-induced Bmal-1 and Per-2 promoter activation was compared with and without REV-ERBα (modulated by siRNA application). In the presence of REV-ERBα, IGF-1 induced significant activity of the Bmal-1 promoter after 2 h and of the Per-2 promoter after 6 h, whereas temperature first activated the Per-2 and subsequently the Bmal-1 promoter (Fig. 9, A and B). When REV-ERBα expression was suppressed by siRNA, IGF-1 failed to activate Bmal-1 and the Per-2 promoter. Thus, the sequence of transcription factors affected by IGF-1 is REV-ERBα, BMAL-1, and PER-2 (Fig. 9C). Temperature still activated the Per-2 promoter in the absence of REV-ERBα, supporting the concept that temperature primarily affects the Per-2 promoter independently of the Bmal-1 suppressor (Fig. 9, A and B). However, temperature-induced Bmal-1 promoter activation was significantly inhibited, indicating the following sequence of temperature-controlled transcription factor activation: PER-2, REV-ERBα, and BMAL-1 (Fig. 9C).
Discussion
The IGF-1 system, consisting of the hormone itself, its receptors and binding proteins (IGFBP) is crucial for the growth, development, and plasticity of the central nervous system (37). IGF-1 is produced by many cell types in the central nervous system and IGF receptors are widely distributed (37). Here, we provide the first evidence that IGF-1 directly affects the expression of the circadian gene Bmal-1 and influences circadian clock gene expression in hypothalamic cells. Thus, regulation of hypothalamic circadian rhythms can be considered as a new biological role for IGF-1.
GSK-3β has previously been reported to affect circadian rhythms via multiple mechanisms (13–18). This role for GSK-3β in clock gene expression has so far either been demonstrated by overexpression or pharmacologically (lithium). Thus, it remained elusive whether GSK-3β is also involved in the regulation of clock gene expression, when regulated by a physiological stimulus. Here, we show for the first time that hormone-controlled regulation of GSK-3β activity via PI3K affects circadian clock gene expression. First, this finding is supported by the strong correlation between the propensity of a given growth factor to phosphorylate GSK-3β and its ability to activate the Bmal-1 promoter. Second, down-regulation of GSK-3β via siRNA or inhibition by LiCl inhibited IGF-1–induced Bmal-1 activation. Because both effects, GSK-3β phosphorylation and Bmal-1 reporter activation, were also blunted by a specific PI3K inhibitor, we suggest a new mechanistic model in which IGF-1 influences circadian rhythms via PI3K-promoted inhibition of GSK-3β (Fig. 9C).
Interestingly, in both hypothalamic cell types analyzed this pathway is neither affected by EGF nor NGF, even though receptors for both growth factors are functionally expressed. EGF and NGF have been reported to regulate clock gene expression in various cell lines via ERK-1/2-mediated expression of PER-2 (9–11, 21–24). Thus, either the Bmal-1 promoter is differentially regulated depending on the cell type or growth factors per se affect the Bmal-1 promoter differentially independent of the cellular context. We show that IGF-1 and EGF induce Bmal-1 reporter expression via ERK-1/2 in NIH-3T3 fibroblasts. Thus, ERK-1/2 appears to be key for growth factor-induced clock gene expression in fibroblasts. Of note, lithium affects clock gene expression in NIH3T3 cells and EGF or IGF-1 phosphorylate GSK-3β in these cells (47–49). However, for yet unknown reasons, growth factor–mediated GSK-3β activity is apparently less important for clock gene expression in NIH3T3 cells. On the contrary, our data indicate that in hypothalamic cells ERK-1/2 appears to be less important for growth factor-induced clock gene expression, whereas GSK-3β is pivotal. Because EGF and NGF only weakly phosphorylated GSK-3β in hypothalamic cell models, this situation allows hypothalamic cells to exclusively link IGF-1 to the Bmal-1 promoter. At this point we can only speculate why in both hypothalamic cells EGF and NGF only poorly phosphorylate GSK-3β. Although EGF and NGF receptors mainly interact with the GRB2 adapter protein, IGF receptors prefer insulin receptor substrates. Thus, different functions and/or expression levels of both adapter proteins in distinct cells will affect growth factor signaling differentially. Furthermore, recent data propose a role for Gαi proteins in EGF- but not IGF-1–induced PI3K activation, suggesting that distinct expression of Gαi subtypes could also underlie different interactions between receptor tyrosine kinases and PI3K (50).
Our work not only reveals that IGF-1 regulates Bmal-1 promoter activity via PI3K/GSK-3β but also suggests that IGF-1 changes the phase of temperature-entrained Bmal-1 promoter activity and thus acts as cellular circadian zeitgeber. When IGF-1 was applied prior to the cold-warm transition (ZT12), it induced an ∼2 h advance in the Bmal-1 promoter activity phase. In clear contrast, it promoted a ∼5 h delay when given before the warm-cold transition (ZT0). Thus, IGF-1 shows phase-specific modulation of clock gene expression similar to other zeitgebers. Recently serum IGF-1 levels in WT mice and mice deficient for the clock genes cryptochrome-1 and −2 were determined. A peak in IGF-1 serum levels was found at ZT0, where effects of IGF-1 on the Bmal-1 promoter in hypothalamic cells were most effective and advanced the phase of clock gene expression (51). Of note, circadian IGF-1 serum peaks were dramatically reduced in animals deficient for both cryptochrome genes. These results together with the data obtained here, suggest that the cellular clock regulates circadian IGF-1 levels and that the hormone feeds back on the clock by regulating its phase. Recent work by Shibata and colleagues (52) clearly shows that application of IGF-1 to liver modifies the expression of the clock gene Per-2. In vitro, the free running period is shortened. In vivo the phase of rhythmic gene expression is advanced following administration of the hormone at ZT5. Thus, serum IGF-1 levels could be a zeitgeber for both circadian entrainment of the liver based on feeding time and temperature-mediated entrainment in the hypothalamus. It will be interesting to discover what cell types integrate rhythmic IGF-1 expression for tuning circadian clock-regulated gene expression.
In addition to IGF-1, its receptors and binding proteins also show daily oscillations (53–55). Thus, the net effect of the IGF system on the cellular clock in vivo may depend on the circadian rhythms of IGF, IGF receptors, and IGFBP's in combination. Interestingly, EGF serum levels and EGF receptor expression and its signaling also show diurnal patterns with peaks in the resting phase, suggesting that feedback loops between the circadian clock and growth factors are not unique for IGF-1 (56, 57), at least in the liver.
We provide data on the cellular level indicating that IGF-1 affects clock gene expression and thus potentially modulates clock-dependent processes in cells. Assuming that our data obtained on the cellular level can be extrapolated to the living human organism or to behavior, the impact of IGF-1 on the clock would be time-of-life specific. IGF-1 serum levels increase throughout childhood, reaching a plateau after puberty and subsequently declining in adults and the elderly (58–66). Changes in the phase of circadian rhythm has been observed in adolescent and elderly individuals (25, 26, 67–71). Re-synchronization of the “aged clock” may even slow down the aging process (72). Thus, the new link between IGF-1 and circadian rhythm, suggested by our data, provides an intriguing molecular mechanistic explanation for age-related changes in circadian rhythms and may offer a new strategy for therapeutic intervention.
Experimental procedures
Materials
IGF-1, EGF, NGF, Ly-2940002, and PD-184352 were from Sigma. Antibody against p-ERK-1/2 (sc-514302) or p-GSK-3β (sc-11757) and the siRNA's against GSK-3β (sc-35525), REV-ERBα (sc-61459), or the random siRNA (sc-37003) were from Santa Cruz. The antibody against histone-3 (Ab-1791) was from Abcam. The Bmal-1 reporter plasmid (29) was from Addgene (number 46824), the Sre reporter was (73) from Dr. Susanne Mühlich, Walter-Straub-Institut, LMU, München, Germany, and the Cre reporter was (74) from Dr. Himmler, Bender GmbH, Vienna, Austria.
Cell culture and transfection
Hypothalamic or NIH-3T3 cells were routinely cultured in DMEM with 10% FBS, 2 mm l-glutamine and penicillin/streptomycin at 37 °C and 5% CO2. mHypoA-2/10 cells (Clu-176) were obtained from Cedarlane, GT1–7 cells were from Dr. Weiner, University of California, San Francisco, CA, and NIH-3T3 cells were from Sigma (38, 41). To obtain mHypoA-2/10 cells stably expressing the Bmal-1-Luc reporter (mHypoA-2/10-Bmal-1-luc cells), cells were co-transfected with the pABpuro-BluF9 plasmid and an empty pcDNA4 vector containing the resistance gene for zeocin. Transfected cells were then selected with 600 μg/ml of zeocin for 4 weeks. For transient expression of reporter plasmids, as for Figs. 1–3 and 4B, ∼20,000 GT1–7 cells per well were seeded in 12-well plates 24 h prior to transfection. 200 to 400-ng plasmids were transfected using TurbofectTM according to the manufacturer's protocol. After 24 h, cells were stimulated as described in the corresponding figure legends. To down-regulate expression of GSK-3β (sc-35525) or REV-ERBα (sc-61459), gene-specific siRNAs from Santa Cruz (Heidelberg, Germany) were used and tested against a control siRNA construct (sc-37003). To obtain sufficient transfection efficacy, siRNAs were used on mHypoA-2/10-Bmal-1-luc cells alone (Fig. 4) or together with the corresponding reporter plasmid into mHypoA-2/10 (Fig. 6) or GT1–7 cells (Fig. 4D) in conjunction with electroporation using the neon® transfection system from Invitrogen according to the manufacturer's protocol. Briefly, 500,000 cells together with 50 nm of the corresponding siRNA or with this siRNA and 2 μg of the reporter plasmid were pulsed with 1450 V for 30 ms and then placed in 24-well plates. To culture cells under changing temperature conditions, two FRIOCELL®-22 incubators were used. Cells were kept in DMEM/F-12 medium (50:50) containing 25 mm HEPES, 0.1% FCS, and penicillin/streptomycin. For experiments shown in Fig. 1, one incubator was set to a 12-h 38.5/36.0 °C and the second to 12-h 36.0/38.5 °C rhythm. A control incubator was kept at a constant 37 °C (Hereaus, T6030). Before cells were cultured in the anti-phase temperature cycles or at constant temperature, they were first serum-starved for 1 h, stimulated or not with IGF-1 for 30 min at 37 °C in DMEM/F-12 without FCS, and then washed and kept in DMEM/F-12 with 0.1% FCS for the duration of the experiment. For each time point, one 12-well plate (6 control and 6 IGF-1–stimulated cells) was used for each time point in temperature cycles and one 12-well plate with 6 unstimulated control cell pools at constant temperature. Cells were removed at the indicated time points. For analysis, cells from temperature cycles (unstimulated or IGF-1 treated) were normalized to those from unstimulated cells at a constant temperature by setting luciferase measured to 100%. For the experiments shown in Fig. 6, A and B, cells were kept in DMEM/F-12 without FCS at 37 °C for 18 h. One incubator was cooled down to 36 °C, whereas another was heated to 38.5 °C. For each time point, one 12-well plate was removed from each incubator.
Reporter assay
Cells were lysed in 100–200 μl of lysis buffer (25 mm Tris-HCl, pH 7.4, 4 mm EGTA, 8 mm MgCl2, 1 mm DTT, and 1% Triton X-100) and a volume of 90–180 μl was transferred to white-bottomed, 96-well plates. Luciferase activity was measured after automatically injecting the luciferase substrate (20–40 μl) using a FLUOstar® Omega plate reader. Total light emission was detected every second for 10 s post-injection and average emission between 2 and 10 s was determined.
Western blotting
Cells were seeded on 6-well plates (∼100,000/well), cultured for 1 day, stimulated as indicated in the figure legends, and then lysed in Laemmli buffer. Lysates were subjected to SDS-PAGE (10%), proteins were transferred to nitrocellulose by Western blotting and probed for total histone-3 proteins as a loading control and for GSK-3β or ERK-1/2 phosphorylation. Immune reactivity was quantified by densitometry, with the ratios between p-GSK-3β or p-ERK-1/2 and histone signals were calculated using ImageJ. Ligand-induced protein phosphorylation was normalized to unstimulated cells.
Quantification and statistical analysis
Values represent the mean ± S.D. of three to six independent experiments. Western blotting data are representative of a typical experiment repeated 5 times or more and also quantified by ImageJ. Statistical analysis was performed either by one-sample or two-sample Student's t test, or one-way or two-way ANOVA followed by Dunnett's post-test using the GraphPad Prism software. Circular wave fit was performed using the CircWave software version 1.4 (75). Data of fitted curves were then transferred to GraphPad Prism and further analyzed. One symbol indicates a p value of <0.05, two of <0.01, and three of <0.001.
Author contributions
A. B. and J. S. conceptualization; A. B. and L. M. data curation; A. B., J. S., M. G., and M. M. formal analysis; A. B. supervision; A. B., M. G., and M. M. validation; A. B. investigation; A. B. methodology; A. B., M. M., and T. G. writing-original draft; A. B. project administration; T. G. funding acquisition.
Supplementary Material
Acknowledgments
We are thankful to Dr. Stuart Peirson, Prof. Jean-Christophe Billeter, and Prof. Steve Brown for their critical reading of the manuscript and supplying useful comments.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- BMAL-1
- brain and muscle ARNT-like protein-1
- RORA
- retinoid orphan receptor A
- CREB
- cAMP-response element–binding protein
- EGF
- epidermal growth factor
- ERK-1/2
- extracellular-regulated kinases-1/2
- NGF
- nerve growth factor
- GSK-3β
- glycogen synthase kinase-3β
- IGF-1
- insulin-like growth factor-1–binding
- IGFBP
- insulin-like growth factor-1–binding protein
- PER
- period
- PI3K
- phosphoinositide 3-kinase
- SRF
- serum-response factor
- TCF
- ternary complex factor
- ZT
- zeitgeber time
- Sre
- serum-response element
- DMEM
- Dulbecco's modified Eagle's medium
- ANOVA
- analysis of variance.
References
- 1. Winfree A. T. (1982) Circadian timing of sleepiness in man and woman. Am. J. Physiol. 243, R193–R204 [DOI] [PubMed] [Google Scholar]
- 2. Lazar M. A. (2016) Rev-erbs: integrating metabolism around the clock (Sassone-Corsi P., and Christen Y., eds) A Time for Metabolism and Hormones Cham (CH), Springer, New York: [PubMed] [Google Scholar]
- 3. Morris C. J., Aeschbach D., and Scheer F. A. (2012) Circadian system, sleep and endocrinology. Mol. Cell. Endocrinol. 349, 91–104 10.1016/j.mce.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kalsbeek A., and Fliers E. (2013) Daily regulation of hormone profiles. Handb. Exp. Pharmacol. 2013, 185–226 [DOI] [PubMed] [Google Scholar]
- 5. Zheng X., and Sehgal A. (2012) Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 35, 574–585 10.1016/j.tins.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buhr E. D., and Takahashi J. S. (2013) Molecular components of the mammalian circadian clock. Handb. Exp. Pharmacol. 2013, 3–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park N., Kim H. D., Cheon S., Row H., Lee J., Han D. H., Cho S., and Kim K. (2015) A novel Bmal1 mutant mouse reveals essential roles of the C-terminal domain on circadian rhythms. PloS One 10, e0138661 10.1371/journal.pone.0138661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanselow K., Vanselow J. T., Westermark P. O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., and Kramer A. (2006) Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 20, 2660–2672 10.1101/gad.397006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akashi M., and Nishida E. (2000) Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14, 645–649 [PMC free article] [PubMed] [Google Scholar]
- 10. Vadigepalli R., Hao H., Miller G. M., Liu H., and Schwaber J. S. (2006) Epidermal growth factor receptor-induced circadian-time-dependent gene regulation in suprachiasmatic nucleus. Neuroreport 17, 1437–1441 10.1097/01.wnr.0000227989.15422.71 [DOI] [PubMed] [Google Scholar]
- 11. Balsalobre A., Marcacci L., and Schibler U. (2000) Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 10, 1291–1294 10.1016/S0960-9822(00)00758-2 [DOI] [PubMed] [Google Scholar]
- 12. Balsalobre A., Damiola F., and Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 10.1016/S0092-8674(00)81199-X [DOI] [PubMed] [Google Scholar]
- 13. Iitaka C., Miyazaki K., Akaike T., and Ishida N. (2005) A role for glycogen synthase kinase-3β in the mammalian circadian clock. J. Biol. Chem. 280, 29397–29402 10.1074/jbc.M503526200 [DOI] [PubMed] [Google Scholar]
- 14. Kinoshita C., Miyazaki K., and Ishida N. (2012) Chronic stress affects PERIOD2 expression through glycogen synthase kinase-3β phosphorylation in the central clock. Neuroreport 23, 98–102 10.1097/WNR.0b013e32834e7ec2 [DOI] [PubMed] [Google Scholar]
- 15. Martinek S., Inonog S., Manoukian A. S., and Young M. W. (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769–779 10.1016/S0092-8674(01)00383-X [DOI] [PubMed] [Google Scholar]
- 16. Iwahana E., Akiyama M., Miyakawa K., Uchida A., Kasahara J., Fukunaga K., Hamada T., and Shibata S. (2004) Effect of lithium on the circadian rhythms of locomotor activity and glycogen synthase kinase-3 protein expression in the mouse suprachiasmatic nuclei. Eur. J. Neurosci. 19, 2281–2287 10.1111/j.0953-816X.2004.03322.x [DOI] [PubMed] [Google Scholar]
- 17. Yin L., Wang J., Klein P. S., and Lazar M. A. (2006) Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311, 1002–1005 10.1126/science.1121613 [DOI] [PubMed] [Google Scholar]
- 18. Spengler M. L., Kuropatwinski K. K., Schumer M., and Antoch M. P. (2009) A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8, 4138–4146 10.4161/cc.8.24.10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fahrenkrug J., Georg B., Hannibal J., and Jørgensen H. L. (2012) Altered rhythm of adrenal clock genes, StAR and serum corticosterone in VIP receptor 2-deficient mice. J. Mol. Neurosci. 48, 584–596 [DOI] [PubMed] [Google Scholar]
- 20. Xiang S., Mao L., Duplessis T., Yuan L., Dauchy R., Dauchy E., Blask D. E., Frasch T., and Hill S. M. (2012) Oscillation of clock and clock controlled genes induced by serum shock in human breast epithelial and breast cancer cells: regulation by melatonin. Breast Cancer (Auckl) 6, 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mogi A., Yomoda R., Kimura S., Tsushima C., Takouda J., Sawauchi M., Maekawa T., Ohta H., Nishino S., Kurita M., Mano N., Osumi N., and Moriya T. (2018) Entrainment of the circadian clock in neural stem cells by epidermal growth factor is closely associated with ERK1/2-mediated induction of multiple clock-related genes. Neuroscience 379, 45–66 10.1016/j.neuroscience.2018.02.045 [DOI] [PubMed] [Google Scholar]
- 22. Pizzio G. A., Hainich E. C., Plano S. A., Ralph M. R., and Golombek D. A. (2005) Nerve growth factor-induced circadian phase shifts and MAP kinase activation in the hamster suprachiasmatic nuclei. Eur. J. Neurosci. 22, 665–671 10.1111/j.1460-9568.2005.04247.x [DOI] [PubMed] [Google Scholar]
- 23. Kramer A., Yang F. C., Snodgrass P., Li X., Scammell T. E., Davis F. C., and Weitz C. J. (2003) Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signalling. Novartis Found. Symp. 253, 250–262; discussion 102–109, 263–266, 281–284 [PubMed] [Google Scholar]
- 24. Baeza-Raja B., Eckel-Mahan K., Zhang L., Vagena E., Tsigelny I. F., Sassone-Corsi P., Ptacek L. J., and Akassoglou K. (2013) p75 neurotrophin receptor is a clock gene that regulates oscillatory components of circadian and metabolic networks. J. Neurosci. 33, 10221–10234 10.1523/JNEUROSCI.2757-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bina K. G., and Rusak B. (1996) Nerve growth factor phase shifts circadian activity rhythms in Syrian hamsters. Neurosci. Lett. 206, 97–100 10.1016/S0304-3940(96)12432-0 [DOI] [PubMed] [Google Scholar]
- 26. Denduluri S. K., Idowu O., Wang Z., Liao Z., Yan Z., Mohammed M. K., Ye J., Wei Q., Wang J., Zhao L., and Luu H. H. (2015) Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wee P., and Wang Z. (2017) Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 9, E52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crowder R. J., and Freeman R. S. (2000) Glycogen synthase kinase-3 beta activity is critical for neuronal death caused by inhibiting phosphatidylinositol 3-kinase or Akt but not for death caused by nerve growth factor withdrawal. J. Biol. Chem. 275, 34266–34271 10.1074/jbc.M006160200 [DOI] [PubMed] [Google Scholar]
- 29. Brown S. A., Fleury-Olela F., Nagoshi E., Hauser C., Juge C., Meier C. A., Chicheportiche R., Dayer J. M., Albrecht U., and Schibler U. (2005) The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 3, e338 10.1371/journal.pbio.0030338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saini C., Morf J., Stratmann M., Gos P., and Schibler U. (2012) Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 26, 567–580 10.1101/gad.183251.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buhr E. D., Yoo S. H., and Takahashi J. S. (2010) Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330, 379–385 10.1126/science.1195262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown S. A., Zumbrunn G., Fleury-Olela F., Preitner N., and Schibler U. (2002) Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 12, 1574–1583 10.1016/S0960-9822(02)01145-4 [DOI] [PubMed] [Google Scholar]
- 33. Cavadas C., Aveleira C. A., Souza G. F., and Velloso L. A. (2016) The pathophysiology of defective proteostasis in the hypothalamus: from obesity to ageing. Nat. Rev. Endocrinol. 12, 723–733 [DOI] [PubMed] [Google Scholar]
- 34. Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., and Hogenesch J. B. (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saper C. B., Scammell T. E., and Lu J. (2005) Hypothalamic regulation of sleep and circadian rhythms. Nature 437, 1257–1263 10.1038/nature04284 [DOI] [PubMed] [Google Scholar]
- 36. Kramer A., Yang F. C., Snodgrass P., Li X., Scammell T. E., Davis F. C., and Weitz C. J. (2001) Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294, 2511–2515 10.1126/science.1067716 [DOI] [PubMed] [Google Scholar]
- 37. Dyer A. H., Vahdatpour C., Sanfeliu A., and Tropea D. (2016) The role of insulin-like growth factor 1 (IGF-1) in brain development, maturation and neuroplasticity. Neuroscience 325, 89–99 10.1016/j.neuroscience.2016.03.056 [DOI] [PubMed] [Google Scholar]
- 38. Belsham D. D., Fick L. J., Dalvi P. S., Centeno M. L., Chalmers J. A., Lee P. K., Wang Y., Drucker D. J., and Koletar M. M. (2009) Ciliary neurotrophic factor recruitment of glucagon-like peptide-1 mediates neurogenesis, allowing immortalization of adult murine hypothalamic neurons. FASEB J. 23, 4256–4265 10.1096/fj.09-133454 [DOI] [PubMed] [Google Scholar]
- 39. Breit A., Besik V., Solinski H. J., Muehlich S., Glas E., Yarwood S. J., and Gudermann T. (2015) Serine-727 phosphorylation activates hypothalamic STAT-3 independently from tyrosine-705 phosphorylation. Mol. Endocrinol. 29, 445–459 10.1210/me.2014-1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Glas E., Mückter H., Gudermann T., and Breit A. (2016) Exchange factors directly activated by cAMP mediate melanocortin 4 receptor-induced gene expression. Sci. Rep. 6, 32776 10.1038/srep32776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mellon P. L., Windle J. J., Goldsmith P. C., Padula C. A., Roberts J. L., and Weiner R. I. (1990) Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron 5, 1–10 10.1016/0896-6273(90)90028-E [DOI] [PubMed] [Google Scholar]
- 42. Pollak M. (2012) The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer 12, 159–169 [DOI] [PubMed] [Google Scholar]
- 43. Dang F., Sun X., Ma X., Wu R., Zhang D., Chen Y., Xu Q., Wu Y., and Liu Y. (2016) Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat. Commun. 7, 12696 10.1038/ncomms12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Noguchi T., Lo K., Diemer T., and Welsh D. K. (2016) Lithium effects on circadian rhythms in fibroblasts and suprachiasmatic nucleus slices from Cry knockout mice. Neurosci. Lett. 619, 49–53 10.1016/j.neulet.2016.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Phiel C. J., and Klein P. S. (2001) Molecular targets of lithium action. Annu. Rev. Pharmacol. Toxicol. 41, 789–813 10.1146/annurev.pharmtox.41.1.789 [DOI] [PubMed] [Google Scholar]
- 46. Liu Y., Merrow M., Loros J. J., and Dunlap J. C. (1998) How temperature changes reset a circadian oscillator. Science 281, 825–829 10.1126/science.281.5378.825 [DOI] [PubMed] [Google Scholar]
- 47. Osland T. M., Fernøo J., Håvik B., Heuch I., Ruoff P., Lærum O. D., and Steen V. M. (2011) Lithium differentially affects clock gene expression in serum-shocked NIH-3T3 cells. J. Psychopharmacol. 25, 924–933 10.1177/0269881110379508 [DOI] [PubMed] [Google Scholar]
- 48. Eldar-Finkelman H., Seger R., Vandenheede J. R., and Krebs E. G. (1995) Inactivation of glycogen synthase kinase-3 by epidermal growth factor is mediated by mitogen-activated protein kinase/p90 ribosomal protein S6 kinase signaling pathway in NIH/3T3 cells. J. Biol. Chem. 270, 987–990 10.1074/jbc.270.3.987 [DOI] [PubMed] [Google Scholar]
- 49. Fang X., Yu S. X., Lu Y., Bast R. C. Jr, Woodgett J. R., and Mills G. B. (2000) Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl. Acad. Sci. U.S.A. 97, 11960–11965 10.1073/pnas.220413597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cao C., Huang X., Han Y., Wan Y., Birnbaumer L., Feng G. S., Marshall J., Jiang M., and Chu W. M. (2009) Gα(i1) and Gα(i3) are required for epidermal growth factor-mediated activation of the Akt-mTORC1 pathway. Sci. Signal. 2, ra17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chaudhari A., Gupta R., Patel S., Velingkaar N., and Kondratov R. (2017) Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol. Biol. Cell 28, 834–842 10.1091/mbc.e16-08-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ikeda Y., Kamagata M., Hirao M., Yasuda S., Iwami S., Sasaki H., Tsubosaka M., Hattori Y., Todoh A., Tamura K., Shiga K., Ohtsu T., and Shibata S. (2018) Glucagon and/or IGF-1 production regulates resetting of the liver circadian clock in response to a protein or amino acid-only diet. EBioMedicine 28, 210–224 10.1016/j.ebiom.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oscarsson J., Johannsson G., Johansson J. O., Lundberg P. A., Lindstedt G., and Bengtsson B. A. (1997) Diurnal variation in serum insulin-like growth factor (IGF)-I and IGF binding protein-3 concentrations during daily subcutaneous injections of recombinant human growth hormone in GH-deficient adults. Clin. Endocrinol. 46, 63–68 10.1046/j.1365-2265.1997.d01-1740.x [DOI] [PubMed] [Google Scholar]
- 54. Skjaerbaek C., Frystyk J., Kaal A., Laursen T., Møller J., Weeke J., Jørgensen J. O., Sandahl Christiansen J., and Orskov H. (2000) Circadian variation in serum free and total insulin-like growth factor (IGF)-I and IGF-II in untreated and treated acromegaly and growth hormone deficiency. Clin. Endocrinol. 52, 25–33 10.1046/j.1365-2265.2000.00876.x [DOI] [PubMed] [Google Scholar]
- 55. Heuck C., Skjaerbaek C., Orskov H., and Wolthers O. D. (1999) Circadian variation in serum free ultrafiltrable insulin-like growth factor I concentrations in healthy children. Pediatr. Res. 45, 733–736 10.1203/00006450-199905010-00021 [DOI] [PubMed] [Google Scholar]
- 56. Lauriola M., Enuka Y., Zeisel A., D'Uva G., Roth L., Sharon-Sevilla M., Lindzen M., Sharma K., Nevo N., Feldman M., Carvalho S., Cohen-Dvashi H., Kedmi M., Ben-Chetrit N., Chen A., et al. (2014) Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 5, 5073 10.1038/ncomms6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haus E., Dumitriu L., Nicolau G. Y., Bologa S., and Sackett-Lundeen L. (2001) Circadian rhythms of basic fibroblast growth factor (bFGF), epidermal growth factor (EGF), insulin-like growth factor-1 (IGF-1), insulin-like growth factor binding protein-3 (IGFBP-3), cortisol, and melatonin in women with breast cancer. Chronobiol. Int. 18, 709–727 10.1081/CBI-100106083 [DOI] [PubMed] [Google Scholar]
- 58. Juul A., Holm K., Kastrup K. W., Pedersen S. A., Michaelsen K. F., Scheike T., Rasmussen S., Müller J., and Skakkebaek N. E. (1997) Free insulin-like growth factor I serum levels in 1430 healthy children and adults, and its diagnostic value in patients suspected of growth hormone deficiency. J. Clin. Endocrinol. Metab. 82, 2497–2502 10.1210/jc.82.8.2497,10.1210/jcem.82.8.4137 [DOI] [PubMed] [Google Scholar]
- 59. Xu S., Gu X., Pan H., Zhu H., Gong F., Li Y., and Xing Y. (2010) Reference ranges for serum IGF-1 and IGFBP-3 levels in Chinese children during childhood and adolescence. Endocr. J. 57, 221–228 10.1507/endocrj.K09E-200 [DOI] [PubMed] [Google Scholar]
- 60. Bereket A., Turan S., Omar A., Berber M., Ozen A., Akbenlioglu C., and Haklar G. (2006) Serum IGF-I and IGFBP-3 levels of Turkish children during childhood and adolescence: establishment of reference ranges with emphasis on puberty. Hormone Res. 65, 96–105 10.1159/000091301 [DOI] [PubMed] [Google Scholar]
- 61. Juul A., Dalgaard P., Blum W. F., Bang P., Hall K., Michaelsen K. F., Müller J., and Skakkebaek N. E. (1995) Serum levels of insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) in healthy infants, children, and adolescents: the relation to IGF-I, IGF-II, IGFBP-1, IGFBP-2, age, sex, body mass index, and pubertal maturation. J. Clin. Endocrinol. Metab. 80, 2534–2542 [DOI] [PubMed] [Google Scholar]
- 62. Brabant G., von zur Mühlen A., Wüster C., Ranke M. B., Kratzsch J., Kiess W., Ketelslegers J. M., Wilhelmsen L., Hulthén L., Saller B., Mattsson A., Wilde J., Schemer R., Kann P., and German, KIMS Board (2003) Serum insulin-like growth factor I reference values for an automated chemiluminescence immunoassay system: results from a multicenter study. Horm. Res. 60, 53–60 10.1159/000071871 [DOI] [PubMed] [Google Scholar]
- 63. Elmlinger M. W., Kühnel W., Weber M. M., and Ranke M. B. (2004) Reference ranges for two automated chemiluminescent assays for serum insulin-like growth factor I (IGF-I) and IGF-binding protein 3 (IGFBP-3). Clin. Chem. Lab. Med. 42, 654–664 [DOI] [PubMed] [Google Scholar]
- 64. Palta M., LeCaire T. J., Sadek-Badawi M., Herrera V. M., and Danielson K. K. (2014) The trajectory of IGF-1 across age and duration of type 1 diabetes. Diabetes Metab. Res. Rev. 30, 777–783 10.1002/dmrr.2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mattila A. L., Perheentupa J., Pesonen K., and Viinikka L. (1985) Epidermal growth factor in human urine from birth to puberty. J. Clin. Endocrinol. Metab. 61, 997–1000 10.1210/jcem-61-5-997 [DOI] [PubMed] [Google Scholar]
- 66. Kamat A., Ghosh P. M., Glover R. L., Zhu B., Yeh C. K., Choudhury G. G., and Katz M. S. (2008) Reduced expression of epidermal growth factor receptors in rat liver during aging. J. Gerontol. A Biol. Sci. Med. Sci. 63, 683–692 10.1093/gerona/63.7.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Duffy J. F., and Czeisler C. A. (2002) Age-related change in the relationship between circadian period, circadian phase, and diurnal preference in humans. Neurosci. Lett. 318, 117–120 10.1016/S0304-3940(01)02427-2 [DOI] [PubMed] [Google Scholar]
- 68. Duffy J. F., Zitting K. M., and Chinoy E. D. (2015) Aging and circadian rhythms. Sleep Med. Clinics 10, 423–434 10.1016/j.jsmc.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sonntag W. E., Steger R. W., Forman L. J., and Meites J. (1980) Decreased pulsatile release of growth hormone in old male rats. Endocrinology 107, 1875–1879 10.1210/endo-107-6-1875 [DOI] [PubMed] [Google Scholar]
- 70. Khan A. S., Sane D. C., Wannenburg T., and Sonntag W. E. (2002) Growth hormone, insulin-like growth factor-1 and the aging cardiovascular system. Cardiovasc. Res. 54, 25–35 10.1016/S0008-6363(01)00533-8 [DOI] [PubMed] [Google Scholar]
- 71. Roenneberg T., Kuehnle T., Pramstaller P. P., Ricken J., Havel M., Guth A., and Merrow M. (2004) A marker for the end of adolescence. Curr. Biol. 14, R1038–1039 10.1016/j.cub.2004.11.039 [DOI] [PubMed] [Google Scholar]
- 72. Fonseca Costa S. S., and Ripperger J. A. (2015) Impact of the circadian clock on the aging process. Front. Neurol. 6, 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Muehlich S., Wang R., Lee S. M., Lewis T. C., Dai C., and Prywes R. (2008) Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol. Cell. Biol. 28, 6302–6313 10.1128/MCB.00427-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Himmler A., Stratowa C., and Czernilofsky A. P. (1993) Functional testing of human dopamine D1 and D5 receptors expressed in stable cAMP-responsive luciferase reporter cell lines. J. Recept. Res. 13, 79–94 10.3109/10799899309073647 [DOI] [PubMed] [Google Scholar]
- 75. Oster H., Damerow S., Hut R. A., and Eichele G. (2006) Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J. Biol. Rhythms 21, 350–361 10.1177/0748730406293053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.