Abstract
The repair of DNA double-strand breaks (DSBs) by homologous recombination (HR) is initiated by nucleolytic resection of the DNA break ends. The current model, being based primarily on genetic analyses in Saccharomyces cerevisiae and companion biochemical reconstitution studies, posits that end resection proceeds in two distinct stages. Specifically, the initiation of resection is mediated by the nuclease activity of the Mre11–Rad50–Xrs2 (MRX) complex in conjunction with its cofactor Sae2, and long-range resection is carried out by exonuclease 1 (Exo1) or the Sgs1–Top3–Rmi1–Dna2 ensemble. Using fully reconstituted systems, we show here that DNA with ends occluded by the DNA end-joining factor Ku70–Ku80 becomes a suitable substrate for long-range 5′–3′ resection when a nick is introduced at a locale proximal to one of the Ku-bound DNA ends. We also show that Sgs1 can unwind duplex DNA harboring a nick, in a manner dependent on a species-specific interaction with the ssDNA-binding factor replication protein A (RPA). These biochemical systems and results will be valuable for guiding future endeavors directed at delineating the mechanistic intricacy of DNA end resection in eukaryotes.
Keywords: DNA damage, DNA repair, homologous recombination, DNA helicase, DNA binding protein, DNA gap, DNA nick, Exo1, nuclease, Sgs1-Dna2, DNA end resection
Introduction
DNA double-strand breaks (DSBs)2 are induced by reactive metabolites and upon exposure to ionizing radiation or perturbations during DNA replication (1). If left unrepaired or repaired inappropriately, these lesions can cause chromosome aberrations and rearrangements, leading to senescence or apoptosis, cell transformation, and cancer (2–4). The majority of DSBs are eliminated via two mechanistically distinct pathways, namely nonhomologous end joining (NHEJ) and homologous recombination (HR) (5, 6). In NHEJ, DSB ends are often subject to limited nucleolytic attrition prior to rejoining, which leads to a loss of genetic information in the product of repair (7). By utilizing the undamaged sister chromatid to template repair, HR is geared toward restoring the original genetic content and is therefore considered error-free (8). NHEJ operates throughout the cell cycle (7, 9), whereas HR becomes an important DSB repair tool only when cells are in the S or G2 phase (5, 10–13).
As first revealed in studies done in the budding yeast Saccharomyces cerevisiae, a large number of DNA damage checkpoint and repair factors are temporally recruited to DSBs (14–16). The Ku70–Ku80 heterodimer (hereafter referred to as Ku) and the Mre11–Rad50–Xrs2 (MRX; MRE11–RAD50–NBS1 (MRN) in mammals) complex are among the first of such factors to associate with DSBs. The presence of Ku marks the DSB ends for rejoining by a protein complex that harbors DNA ligase IV (7). In addition to the cell cycle phase, the choice between NHEJ and HR also depends on the nature of the DSB ends. Specifically, the initiation of 5′ strand resection at DNA ends effectively prevents end rejoining by NHEJ while facilitating HR-mediated repair (17, 18). The activation of DNA end resection in the S and G2 phases is achieved via cyclin-dependent kinase–catalyzed phosphorylation of Sae2 (CtIP in mammals) (19, 20), which is an essential cofactor that activates the endonucleolytic scission of the 5′-terminated DNA strand by MRX/MRN proximal to a Ku-occluded DNA end (21, 22).
Following the initial strand nicking by MRX/MRN, long-range 5′–3′ resection occurs via one of two pathways being mediated by Exo1 (a 5′–3′ exonuclease) or the protein ensemble harboring the helicase Sgs1, the helicase/nuclease Dna2, and the Top3–Rmi1 complex (23, 24). The 5′ strand polarity and overall efficiency of long-range resection are influenced by the single-strand DNA-binding protein RPA (25, 26). Importantly, RPA also protects the resulting 3′-terminated single-stranded DNA (ssDNA) tail from nucleolytic attack, and the RPA–ssDNA complex serves to recruit the Mec1–Ddc2 (ATR–ATRIP in mammals) kinase complex for DNA damage checkpoint activation. For HR to proceed, RPA must first be replaced by the recombinase enzyme Rad51. RPA-Rad51 exchange on ssDNA is facilitated by HR mediators, such as Rad52 in yeast and the tumor suppressor BRCA2 in complex with the DSS1 protein in humans (27–31).
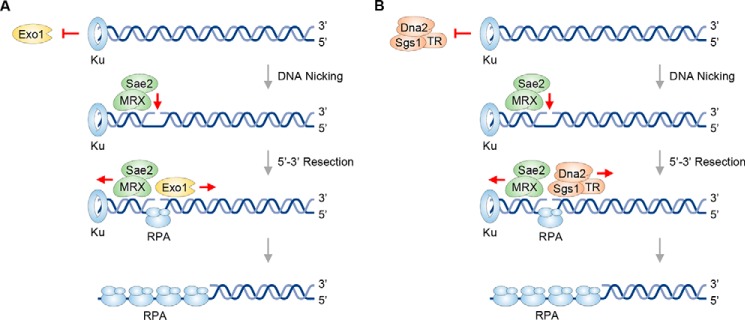
Previous studies from our laboratory and by others have provided direct biochemical evidence that Ku shields DNA ends from exonucleolytic digestion but triggers endonucleolytic scission by MRX–Sae2 (21, 22, 32). However, whether the DNA nick introduced by MRX–Sae2 serves directly as a suitable entry site for Exo1 or Sgs1–Dna2 to initiate long-range resection remains to be determined. Here, using reconstituted systems with highly purified end resection factors and plasmid-length DNA substrates, we show that 1) Ku binding to DNA ends poses an obstacle to Exo1 or Sgs1–Dna2, but this inhibitory effect is relieved by the introduction of a DNA nick proximal to one of the Ku-bound DNA ends; 2) both Exo1 and Sgs1–Dna2 can carry out long-range DNA end resection from a DNA nick with a 5′ to 3′ polarity; and 3) Sgs1 helicase is able to unwind double-stranded DNA (dsDNA) harboring a nick or a gap, which depends on its species-specific interaction with RPA. These findings support a model wherein a DNA nick created by MRX–Sae2 at Ku-blocked DSB ends can serve as the entry site for Exo1 or Sgs1–Dna2. These findings are likely relevant for understanding the mechanism of resection of other types of protein-blocked DSB ends, such as Spo11-conjugated ends at meiotic DSBs or arrested topoisomerase–DNA conjugates (33, 34).
Results
Relief of Ku-mediated end resection restriction by a DNA nick
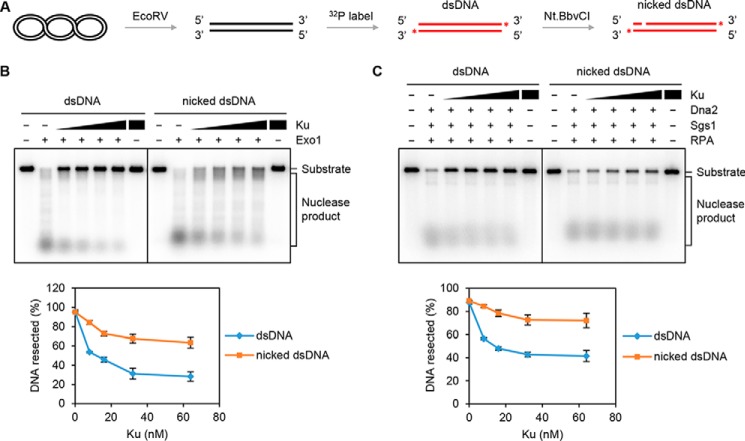
MRX–Sae2 incises the 5′-terminated DNA strand endonucleolytically within the vicinity of Ku-occluded DNA ends (21, 22). To mimic the endonucleolytically cleaved DNA generated by MRX–Sae2, we inserted an Nt.BbvCI nicking endonuclease recognition sequence proximal to the DNA end. An incision site 59 nucleotides (nt) away from the 5′-terminated DNA end was introduced by Nt.BbvCI digestion (Fig. 1A). We first tested the nuclease activity of Exo1 on the 3′-32P-labeled nicked dsDNA. As shown in Fig. S1A, Exo1 was able to resect dsDNA or nicked dsDNA with a comparable efficiency when there was no Ku at the DNA ends. Likewise, in the absence of Ku, Sgs1–Dna2 was also able to resect dsDNA with or without a nick equally well (Fig. S1B).
Figure 1.
The inhibitory effect of Ku on Exo1- or Dna2-mediated long-range resection is relieved by a nick. A, schematic of DNA substrate preparation (see “Experimental procedures” for details). The asterisk denotes the 32P label. B, activity of Exo1 (8 nm) on linear dsDNA without or with a nick (1 nm) prebound by Ku (8, 16, 32, and 64 nm). The results from three independent experiments were graphed with the error bars representing S.D. C, the activity of Dna2 (32 nm) on dsDNA or nick-containing dsDNA substrate prebound by Ku as in B was tested with Sgs1 (16 nm) and RPA (800 nm). The results were graphed as in B. See also Fig. S1.
We next tested the effect of Ku on Exo1- or Sgs1–Dna2-mediated long-range DNA end resection. On dsDNA without a nick, digestion by Exo1 or Sgs1–Dna2 was attenuated when we preincubated the DNA substrate with Ku, indicating that Ku restricts the access of Exo1 or Sgs1–Dna2 to DNA ends (Fig. 1, B and C). Importantly, the inhibitory effect of Ku on Exo1- or Sgs1–Dna2-mediated resection was significantly lessened upon introducing the site-specific nick into the DNA substrate (Fig. 1, B and C). Altogether, these results indicate that Ku protects DNA ends from nucleolytic digestion by Exo1 or Sgs1–Dna2 and also provide evidence that Exo1 and Sgs1–Dna2 can initiate DNA resection from a DNA nick. The latter premise is further validated in studies described below. Our previous findings indicate that MRX–Sae2 creates nicks ∼40 or ∼60 nt away from Ku-blocked DNA ends (21). To evaluate the effect of the distance between nick and DNA end on resection activity, DNA substrates with nicks in different positions (41, 59, or 82 nt away from DNA ends) were utilized. As shown in Fig. S1C, Exo1 processed these nicks equally well when DNA ends were blocked by Ku. Likewise, no obvious difference was seen with Sgs1–Dna2 on these DNA substrates (Fig. S1D).
DNA nick processing activity of Exo1 and Sgs1–Dna2
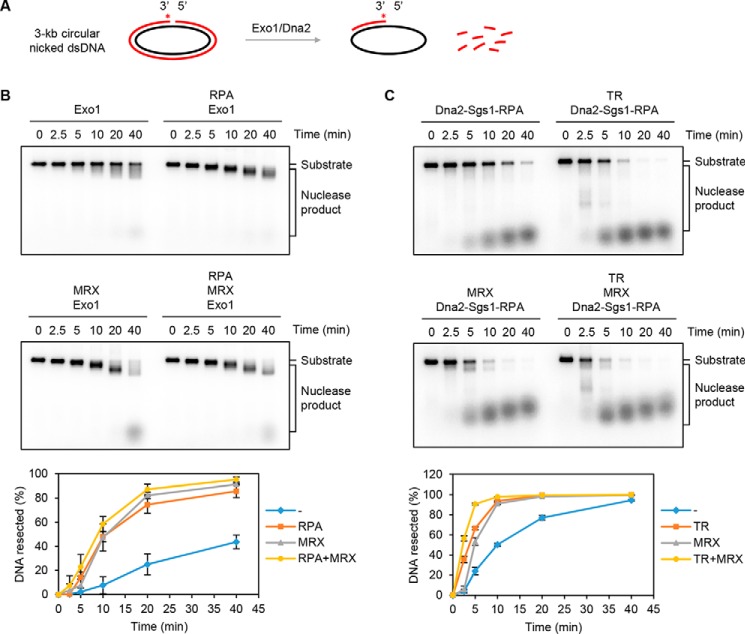
Based on the above observations (Fig. 1), we surmised that a DNA nick can serve as an entry site for Exo1- or Sgs1–Dna2-mediated resection. To eliminate the possibility of strand resection by Exo1 or Sgs1–Dna2 initiating from a free end, we constructed a circular dsDNA substrate that harbors a unique nick at which the 3′ terminus was labeled with 32P (Fig. 2A). As shown in Fig. S2A, Exo1 alone was able to digest the circular nicked dsDNA substrate. The cleavage pattern of Exo1 on this DNA substrate was similar to that seen with bacteriophage T7 exonuclease, which has a strict 5′–3′ polarity (Fig. S2B) and is known to catalyze 5′ nucleotide removal at a DNA nick (35, 36). We further tested Exo1 alone or in combination with RPA and/or MRX on the nick-containing DNA substrate. As shown in Fig. 2B, the digestion of the circular nicked dsDNA by Exo1 was enhanced by RPA or MRX. The stimulatory effect of RPA stems from its ability to sequester ssDNA to prevent the formation of nonproductive Exo1–ssDNA complexes (37), and MRX likely acts by recruiting Exo1 to the DNA substrate (38).
Figure 2.
Nuclease activity of Exo1 and Dna2–Sgs1–RPA on circular dsDNA with a nick. A, reaction schematic. The asterisk denotes the 32P label in the substrate. B, the circular nick-containing substrate (1 nm) was incubated with Exo1 (0.25 nm) in the presence of RPA (800 nm) and/or MRX (16 nm) for the indicated times. The results from three independent experiments were graphed with the error bars representing S.D. C, the activity of Dna2 (8 nm) was tested with combinations of Sgs1 (8 nm), RPA (800 nm), TR (8 nm), and MRX (16 nm) as indicated. The results were graphed as in B. See also Fig. S2.
Dna2, Sgs1, and RPA constitute the minimal set of proteins capable of long-range DNA end resection (25, 26), and the combination of these proteins was sufficient to digest the nicked substrate (Fig. 2C). In agreement with our previous observation (25), resection of DNA from the nick by Sgs1–Dna2–RPA was stimulated by Top3–Rmi1 (TR) or MRX, with an additive effect being seen upon combining the latter two components (Fig. 2C). Collectively, these results provide evidence that Exo1 and Sgs1–Dna2 can initiate long-range resection from a DNA nick and that this activity is up-regulated by known cofactors of these enzymatic entities.
Characteristics of Exo1- and Sgs1–Dna2-mediated resection from a DNA nick
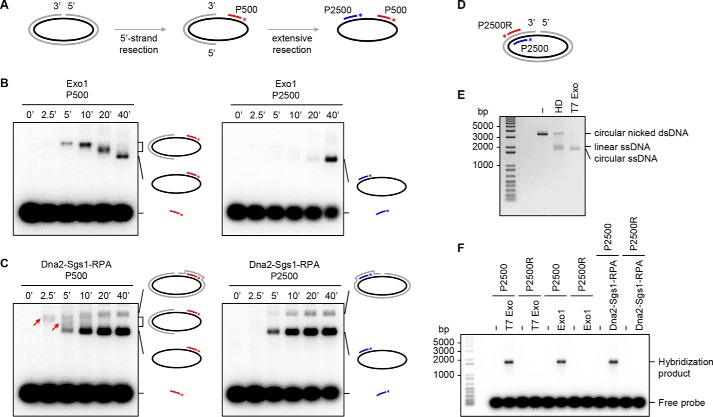
We used Southern blotting to determine the directionality of Exo1- or Sgs1–Dna2-mediated resection that is initiated from a DNA nick in plasmid DNA. Two radiolabeled probes were used: P500 and P2500 that are complementary to the circular DNA strand near and further away from the 5′ terminus of the DNA nick (Fig. 3A). A time-course analysis showed that, at the early time points, nuclease products generated by Exo1 were detectable by P500 but not by P2500 (Fig. 3B), indicating that it carries out 5′ strand resection from the nick. A 5′–3′ polarity of resection was also seen with Sgs1–Dna2 (Fig. 3C). The final reaction product, detectable by hybridization with either P500 or P2500 (Fig. 3B and 3C), has the same size as that generated by T7 exonuclease (Fig. 3, E and F). As expected, the final reaction product was not recognized by the radiolabeled probe P2500R that is complementary to the nicked DNA strand in the substrate (Fig. 3, D and F).
Figure 3.
Resection by Exo1 or Dna2–Sgs1–RPA initiated at a DNA nick occurs in the 5′ to 3′ direction. A, schematic of reaction and Southern blot analysis. P500 and P2500 are 20-nt probes that correspond to DNA segments in the nicked DNA strand 500 and 2500 nt from the 5′ terminus of the nick, respectively. The asterisk denotes the 32P label in the probes. B, the DNA nick-containing substrate (3 nm) was incubated with Exo1 (0.75 nm) for the indicated times and analyzed using the P500 or P2500 probe. C, resection mediated by Dna2 (24 nm) in conjunction with Sgs1 (24 nm) and RPA (2400 nm) was analyzed as in B. The red arrows denote ssDNA region exposed by 5′ strand resection at the early time points. D, probes used in the following experiments. P2500R is a 20-nt probe that is complementary to the indicated locale of the nicked DNA strand in the substrate. The asterisk denotes the 32P label in the probes. E, analysis of the nicked circular DNA substrate with or without heat denaturation (HD) or digestion with T7 exonuclease (Exo) by agarose gel electrophoresis and ethidium bromide staining. F, the nicked circular DNA substrate was incubated with T7 exonuclease, Exo1, or Sgs1–Dna2–RPA followed by hybridization with radiolabeled probe P2500 or P2500R.
Although Dna2 alone was able to digest circular ssDNA (39), this nuclease activity was undetectable when RPA was also present (Fig. 4, A and B). Taken together, these results support a model wherein Exo1 or Sgs1–Dna2–RPA specifically resects DNA in the 5′ to 3′ direction from the nick, generating a ssDNA circle that is protected by RPA from digestion.
Figure 4.
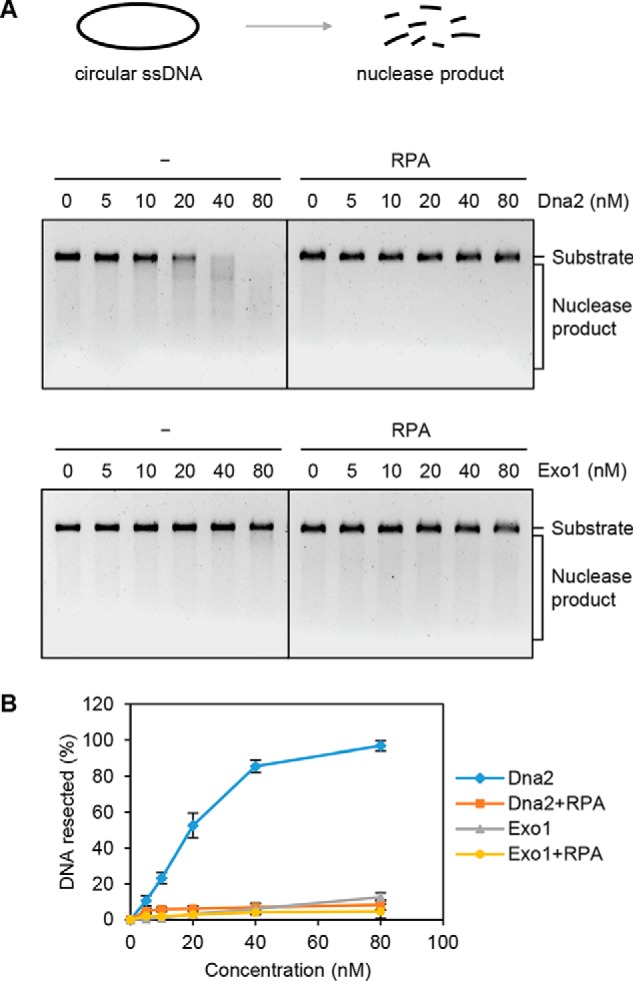
Activity of Dna2 and Exo1 on circular ssDNA. A, Dna2 or Exo1 was incubated with circular ssDNA φX174 (10 nm) in the absence or presence of RPA (8 μm). B, the results from three independent experiments (A) were graphed with the error bars representing S.D.
Sgs1 unwinds circular dsDNA with a nick or gap
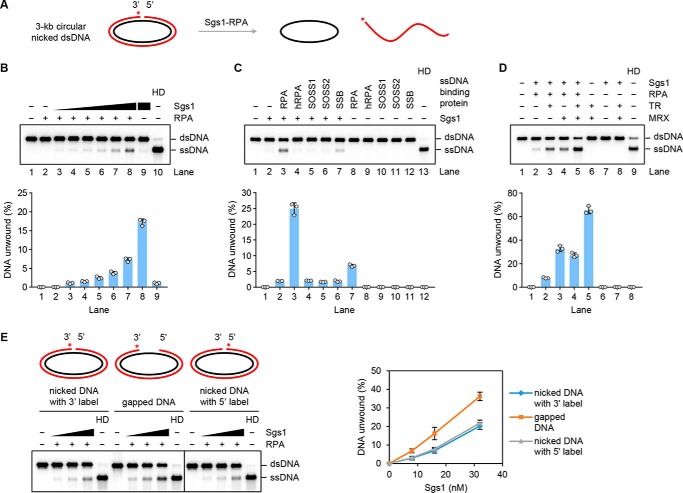
Sgs1 provides the helicase activity to unwind long stretches of duplex DNA during long-range end resection (25, 26). We therefore asked whether Sgs1 could release the strand that harbors the nick from the circular DNA substrate (Fig. 5A). Sgs1 could completely unwind a significant fraction of the nicked circular substrate when RPA was present (Fig. 5B). In contrast, the Mph1 helicase showed no activity on this substrate (Fig. S3A).
Figure 5.
Sgs1-mediated unwinding of circular dsDNA substrates with a nick or gap. A, reaction schematic involving the use of a nick-containing circular DNA substrate. The asterisk denotes the 32P label in the substrate. B, Sgs1 (1, 2, 4, 8, 16, and 32 nm) was tested on the nick-containing DNA substrate (1 nm) in the presence of RPA (800 nm). The results from three independent experiments were graphed with the error bars representing S.D. HD, heat denaturation. C, yeast RPA, hRPA, human SOSS1 and SOSS2, and E. coli SSB (800 nm each) were tested for their effect on unwinding of the nick-containing substrate by Sgs1 (32 nm) as in B. The results were graphed as in B. D, TR (4 nm) and MRX (8 nm) were tested, alone or in combination, for their effect on unwinding of the nick-containing substrate by Sgs1 (32 nm) and RPA (800 nm) as in B. The results were graphed as in B. E, the nick-containing or gapped circular dsDNA (0.5 nm each) was incubated with Sgs1 (8, 16, and 32 nm) and RPA (400 nm). The asterisk denotes the 32P label in the substrate. See also Fig. S3.
Consistent with published results (40, 41), affinity pulldown showed a robust physical interaction between Sgs1 and yeast RPA (Fig. S3C). In contrast, only a weak interaction of Sgs1 and human RPA (hRPA) was detected under the same reaction conditions (Fig. S3C). Importantly, the unwinding of the nicked DNA substrate by Sgs1 relies on its species-specific interaction with RPA as a much reduced level of fully unwound product occurred upon the substitution of yeast RPA with human RPA or with either of the human single-strand DNA-binding protein complexes SOSS1 and SOSS2 (Fig. 5C and Fig. S3B). The failure to stimulate Sgs1 is not due to protein inactivity as hRPA, SOSS1, and SOSS2 are fully capable of binding ssDNA (Fig. S3D). Although Escherichia coli SSB also supported DNA unwinding by Sgs1, its stimulatory effect was less than that of yeast RPA (Fig. 5C). Sgs1-mediated unwinding of the nicked substrate was also stimulated by TR and MRX individually (Fig. S3, E and F), and an additive effect was seen upon combining the latter two factors (Fig. 5D). We also verified that the stimulatory effect of TR and MRX on Sgs1-mediated unwinding is reliant on RPA (Fig. 5D).
A recent study from our laboratory has provided direct evidence that MRX–Sae2 utilizes its 3′ to 5′ exonuclease activity to create a DNA gap from an incision site (21). We therefore tested Sgs1 for the ability to unwind a 3-kb circular dsDNA containing a 46-nt DNA gap. As shown in Fig. 5E and Fig. S3G, Sgs1 was able to unwind the gapped substrate in the presence of RPA. Compared with the nicked DNA, the gapped substrate was unwound more efficiently by Sgs1 (Fig. 5E and Fig. S3G).
Discussion
By virtue of its abundance and high affinity for DNA ends, Ku is among the first protein factors that arrive at DSBs within the cellular setting where it not only promotes break repair by NHEJ (15, 16) but also restricts access of the end resection machinery in the G1 phase (42, 43). Recent studies have provided direct biochemical evidence that Ku shields DNA ends from exonucleolytic digestion but facilitates endonucleolytic scission by MRX–Sae2 (21, 22). The current model posits that the DNA nick introduced by MRX–Sae2 can be enlarged into a DNA gap via the 3′–5′ exonucleolytic action of this nuclease ensemble (21). However, whether DNA gap creation represents an obligatory step in long-range resection by Exo1 or the Sgs1–Dna2 complex or whether the DNA nick introduced by Mre11 is utilized as an entry site for either of the long-range resection nuclease entities is not known. This question has been addressed in our current study. Using reconstituted reactions with either Ku-bound linear dsDNA with a unique nick or nicked circular dsDNA, we have shown that resection by either Exo1 or Sgs1–Dna2 occurs efficiently and with the 5′ to 3′ polarity observed with linear dsDNA harboring free ends (25, 26). We have also shown that DNA unwinding by Sgs1 initiated from either a DNA nick or a DNA gap is enhanced by the TR subcomplex and by MRX. In the case of the circular substrate, the covalently closed ssDNA is refractory to the endonucleolytic action of Dna2 when RPA is present, which is consistent with the observation that internal sites of RPA-bound 3′-tailed ssDNA are resistant to Sgs1–Dna2. Fig. 6 presents models depicting how a DNA nick introduced by MRX–Sae2 helps overcome the inhibitory effect of Ku on the initiation of long-range 5′ strand resection by Exo1 or Sgs1–Dna2. Herein, MRX generates the substrate for the recruitment of Exo1 or Sgs1–Dna2. This resembles the situation in mammalian DNA replication fork repair wherein the endonuclease EEPD1 nicks the stalled fork and loads Exo1 onto that structure (44).
Figure 6.
Exo1- or Dna2-mediated 5′ strand resection from an entry site created by MRX–Sae2. Ku, while protecting the DNA end from exonucleolytic digestion by Exo1, promotes 5′ strand endonucleolytic cleavage by MRX–Sae2. The resulting nick can serve as an entry site for Exo1 (A) or Sgs1–TR–Dna2–RPA (B) to carry out long-range DNA end resection in the 5′–3′ direction.
The experimental systems described herein should be valuable for delineating mechanistic attributes of the eukaryotic resection machineries. For instance, it would be feasible to determine how DNA gap size may affect the recruitment and retention of Exo1 and the Sgs1–TR–Dna2 ensemble.
Experimental procedures
DNA substrates
The pBS-Nt.BbvCI plasmid was constructed by inserting the recognition sequence (CCTCAGC) for the DNA-nicking enzyme Nt.BbvCI into pBluescript II SK (−). To prepare the 3-kb linear dsDNA substrate, pBS-Nt.BbvCI plasmid DNA was digested with the restriction enzyme EcoRV (New England Biolabs). The resulting DNA was precipitated by adding isopropanol, washed with 70% (v/v) ethanol, and then dissolved in TE buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA). Then the DNA was 3′-labeled with [α-32P]cordycepin 5′-triphosphate (PerkinElmer Life Sciences) and terminal transferase (Roche Applied Science). The unincorporated radioactive nucleotide was removed using the QIAquick gel extraction kit (Qiagen). The radiolabeled DNA was treated with Nt.BbvCI (New England Biolabs) to introduce a DNA nick 59 nt away from the DNA end and then purified using the QIAquick gel extraction column. To prepare the linear DNA substrate with a DNA nick 41 or 82 nt away from the DNA end, pBS-Nt.BbvCI plasmid DNA was digested with SmaI or SalI (New England Biolabs) followed by the procedures as described above.
The circular dsDNA substrate with a nick was prepared by digesting pBS-Nt.BbvCI plasmid DNA with Nt.BbvCI followed by DNA precipitation with isopropanol and purified as above. Circular nicked dsDNA was either 3′-labeled with 32P as described above or 5′-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England Biolabs). Before 5′-end labeling, circular nicked dsDNA was dephosphorylated with shrimp alkaline phosphatase (New England Biolabs).
The pG46 plasmid was constructed by inserting five tandem repeats of Nt.BbvCI recognition sequences into pBluescript II SK (−). To prepare the circular gapped dsDNA substrate containing a 46-nt gap, pG46 plasmid DNA was digested by Nt.BbvCI followed by purification with a QIAquick gel extraction column at 80–85 °C. Circular gapped dsDNA was 3′-labeled with 32P as described above.
Expression and purification of SOSS1 and SOSS2 complexes
The coding sequences for hSSB1 with an N-terminal His6 tag, INTS3 with a C-terminal FLAG tag, and hSSBIP1 with a C-terminal Strep tag were amplified by PCR. The PCR products were inserted into the MacroBac 11A plasmid using ligation-independent cloning (45). To generate 11A-SOSS1 plasmid, the biobrick subcloning strategy was used to combine expression cassettes from 11A-hSSB1, 11A-INTS3, and 11A-hSSBIP1. Bacmid and baculovirus were prepared in E. coli DH10Bac and Spodoptera frugiperda Sf9 cells, respectively, following the manufacturer's protocol (Bac-to-Bac baculovirus expression system, Invitrogen). For SOSS1 expression, Trichoplusia ni High Five cells (1 × 106 cells/ml) were infected with high-titer P3 baculovirus. Cells were harvested after 46 h and stored at −80 °C. All the subsequent steps were carried out at 0–4 °C. The cell pellet (∼16 g from 1 liter of culture) was suspended in 50 ml of T buffer (25 mm Tris-HCl, 10% glycerol, 0.5 mm EDTA) with 300 mm KCl and protease inhibitors. After sonication for 1 min, the cell lysate was clarified by ultracentrifugation at 100,000 × g for 1 h. The supernatant was incubated with 4 ml of nickel-nitrilotriacetic acid-agarose (Qiagen) for 1.5 h with constant mixing followed by washing the resin with 100 ml of T buffer containing 300 mm KCl and 20 mm imidazole. Bound proteins were eluted with 8 ml of T buffer containing 300 mm KCl and 200 mm imidazole for 10 min with gentle agitation. The protein eluate was mixed gently with 2 ml of anti-FLAG M2 affinity gel (Sigma) for 2 h. After washing the resin with 100 ml of T buffer containing 150 mm KCl, SOSS1 was eluted with 6 ml of T buffer containing 100 mm KCl and 250 ng/μl FLAG peptide for 20 min. The protein eluate was fractionated in a 6-ml SP Sepharose column (GE Healthcare) with a 72-ml gradient of 50–350 mm KCl in T buffer. The SOSS1 peak fractions were pooled, concentrated in an Amicon Ultra 30K microconcentrator, and subjected to gel filtration in a 24-ml Superdex 200 column (GE Healthcare) in T buffer containing 300 mm KCl. The SOSS1 peak fractions were concentrated to ∼7 mg/ml and stored in small aliquots at −80 °C. The yield of highly purified SOSS1 was ∼1.5 mg.
The 11A-SOSS2 plasmid was generated by combining the expression cassettes of 11A-hSSB2, 11A-INTS3, and 11A-hSSBIP1. We followed the procedure developed for SOSS1 to express and purify the SOSS2 complex. The yield of highly purified SOSS2 was ∼1.5 mg.
Other proteins
Yeast Top3 and Rmi1 proteins were expressed in E. coli, purified to near homogeneity, and used for the assembly of the Top3–Rmi1 complex as described previously (25). Exo1 and Ku were expressed in insect and yeast cells, respectively, and purified to near homogeneity according to our published procedures (46, 47). Dna2 was purified from yeast cells overexpressing the protein as described previously (48). Sgs1 and Mph1 were expressed and purified from insect cells as reported before (25, 49). The yeast and human RPA complexes were purified as described previously (50, 51). E. coli SSB was purchased from Promega, and T7 exonuclease was purchased from New England Biolabs. Mre11, Rad50, and Xrs2 were expressed and purified from yeast, and the MRX complex was assembled according to our published procedures (52–54).
Nuclease reactions and Southern blotting hybridization
Nuclease reactions were performed with 1 nm DNA substrate and the indicated concentration of Exo1 or Dna2–Sgs1–RPA in 12 μl of R buffer (25 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm DTT, 100 μg/ml BSA, 2 mm ATP, 20 mm creatine phosphate, and 30 μg/ml creatine kinase) containing 100 mm KCl (final concentration). Where indicated, Ku was added prior to Exo1 or Dna2–Sgs1–RPA to occupy the DNA ends. After a 30-min incubation at 30 °C, the reactions were terminated by treatment with SDS (0.5%, w/v) and proteinase K (1 mg/ml) at 37 °C for 20 min. After adding 4 μl of 4× loading buffer (20 mm Tris-HCl, pH 7.5, 40% (v/v) glycerol, 2 mm EDTA, and 0.2% (w/v) orange G), the reaction mixtures were resolved in a 1% (w/v) agarose gel in TAE buffer (40 mm Tris acetate and 1 mm EDTA, pH 8.2). Gels were dried onto positively charged nylon membrane (GE Healthcare) and subjected to phosphorimaging analysis. Gel images were quantified using Quantity One software (Bio-Rad) based on the loss of the DNA substrate radioactivity. Nuclease reactions with circular ssDNA φX174 were analyzed by agarose gel electrophoresis and ethidium bromide staining. Gel images were captured using the G:BOX gel imaging system (Syngene) and quantified using TotalLab software based on the disappearance of the substrate band.
Southern blotting hybridization with radiolabeled oligonucleotide probes was performed to analyze the resection polarity of Exo1 or Dna2 on circular nicked dsDNA. First, a standard nuclease reaction was carried out as described above. Upon deproteinization with SDS and proteinase K, 3′-radiolabeled probe P500, P2500, or P2500R was added. The reaction mixtures were incubated in a thermocycler (being held at 70 °C for 5 min and then cooled to 20 °C over 125 min) and further analyzed as described above.
Helicase assay
Helicase assays were carried out in 12 μl of R buffer containing 100 mm KCl. The radiolabeled DNA substrate (1 nm) was incubated with the indicated concentrations of Sgs1, TR, and/or MRX either with or without RPA (800 nm). After a 30-min incubation at 30 °C, reaction mixtures were deproteinized and analyzed as described above.
Electrophoretic mobility shift assay
The indicated concentrations of hRPA, SOSS1, and SOSS2 were incubated with 20 nm 90-nt ssDNA in 12 μl of buffer (25 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm DTT, and 100 μg/ml BSA) containing 100 mm KCl (final concentration). After a 10-min incubation at 37 °C, the reaction mixtures were resolved in a 4% (w/v) native polyacrylamide gel in TAE buffer on ice. Gels were dried onto filter paper and subjected to phosphorimaging analysis.
Affinity pulldown assay
To test for interaction between Sgs1 and yeast or human RPA, the protein pairs (350 ng of each) were incubated on ice in 30 μl of T buffer containing 50 mm KCl and 20 mm imidazole for 1 h. Then 15 μl of nickel-nitrilotriacetic acid-agarose (Qiagen) was added to the reaction mixture to capture the protein complex. After periodic gentle mixing over 1 h, the supernatant was removed, and the resin was washed three times with 200 μl of the same buffer. Bound proteins were eluted from the resin in 40 μl of 1× SDS-PAGE loading buffer (30 mm Tris-HCl, pH 6.8, 10% (v/v) glycerol, 1% (w/v) SDS, and 0.01% (w/v) bromphenol blue). The supernatant, wash, and eluate fractions were subjected to Western blot analysis with antibodies as indicated.
Author contributions
W. W. investigation; W. W. writing-original draft; J. M. D., Y. K., X. X., D. S. K., A. S. M., K. A. N., E. A. W., E. Y. S., S. E. L., and R. H. resources; P. S. supervision; P. S. writing-review and editing.
Supplementary Material
This work was supported by National Institutes of Health Grants ES007061, CA220123, CA205224, GM071011, and GM109645. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
- DSB
- DNA double-strand break
- NHEJ
- nonhomologous end joining
- HR
- homologous recombination
- MRX
- Mre11–Rad50–Xrs2
- Ku
- Ku70–Ku80
- TR
- Top3–Rmi1
- ssDNA
- single-stranded DNA
- Exo1
- exonuclease 1
- RPA
- replication protein A
- MRN
- MRE11–RAD50–NBS1
- nt
- nucleotide(s)
- hRPA
- human RPA
- SSB
- single-strand DNA-binding protein
- hSSB
- human SSB
- CtIP
- CtBP-interacting protein
- ATR
- ataxia telangiectasia–mutated and Rad3-related
- ATRIP
- ATR-interacting protein.
References
- 1. Cannan W. J., and Pederson D. S. (2016) Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell. Physiol. 231, 3–14 10.1002/jcp.25048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pierce A. J., Stark J. M., Araujo F. D., Moynahan M. E., Berwick M., and Jasin M. (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol. 11, S52–S59 10.1016/S0962-8924(01)02149-3 [DOI] [PubMed] [Google Scholar]
- 3. White R. R., and Vijg J. (2016) Do DNA double-strand breaks drive aging? Mol. Cell 63, 729–738 10.1016/j.molcel.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konishi A., Shimizu S., Hirota J., Takao T., Fan Y., Matsuoka Y., Zhang L., Yoneda Y., Fujii Y., Skoultchi A. I., and Tsujimoto Y. (2003) Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell 114, 673–688 10.1016/S0092-8674(03)00719-0 [DOI] [PubMed] [Google Scholar]
- 5. Her J., and Bunting S. F. (2018) How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 293, 10502–10511 10.1074/jbc.TM118.000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Symington L. S., and Gautier J. (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- 7. Pannunzio N. R., Watanabe G., and Lieber M. R. (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 293, 10512–10523 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sung P., and Klein H. (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 10.1038/nrm2008 [DOI] [PubMed] [Google Scholar]
- 9. Sallmyr A., and Tomkinson A. E. (2018) Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 293, 10536–10546 10.1074/jbc.TM117.000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daley J. M., and Sung P. (2014) 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 34, 1380–1388 10.1128/MCB.01639-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karanam K., Kafri R., Loewer A., and Lahav G. (2012) Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 47, 320–329 10.1016/j.molcel.2012.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arnoult N., Correia A., Ma J., Merlo A., Garcia-Gomez S., Maric M., Tognetti M., Benner C. W., Boulton S. J., Saghatelian A., and Karlseder J. (2017) Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 10.1038/nature24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wright W. D., Shah S. S., and Heyer W. D. (2018) Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 293, 10524–10535 10.1074/jbc.TM118.000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lisby M., Barlow J. H., Burgess R. C., and Rothstein R. (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- 15. Yang G., Liu C., Chen S. H., Kassab M. A., Hoff J. D., Walter N. G., and Yu X. (2018) Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 46, 3446–3457 10.1093/nar/gky088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu D., Topper L. M., and Wilson T. E. (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178, 1237–1249 10.1534/genetics.107.083535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fouquin A., Guirouilh-Barbat J., Lopez B., Hall J., Amor-Guéret M., and Pennaneach V. (2017) PARP2 controls double-strand break repair pathway choice by limiting 53BP1 accumulation at DNA damage sites and promoting end-resection. Nucleic Acids Res. 45, 12325–12339 10.1093/nar/gkx881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daley J. M., Laan R. L., Suresh A., and Wilson T. E. (2005) DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 280, 29030–29037 10.1074/jbc.M505277200 [DOI] [PubMed] [Google Scholar]
- 19. Huertas P., and Jackson S. P. (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., and Jackson S. P. (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang W., Daley J. M., Kwon Y., Krasner D. S., and Sung P. (2017) Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 31, 2331–2336 10.1101/gad.307900.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reginato G., Cannavo E., and Cejka P. (2017) Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 31, 2325–2330 10.1101/gad.308254.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niu H., Raynard S., and Sung P. (2009) Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 23, 1481–1486 10.1101/gad.1824209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cejka P. (2015) DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 290, 22931–22938 10.1074/jbc.R115.675942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., Ira G., and Sung P. (2010) Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., and Kowalczykowski S. C. (2010) DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 10.1038/nature09355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jensen R. B., Carreira A., and Kowalczykowski S. C. (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 10.1038/nature09399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 10.1074/jbc.272.45.28194 [DOI] [PubMed] [Google Scholar]
- 29. Thorslund T., McIlwraith M. J., Compton S. A., Lekomtsev S., Petronczki M., Griffith J. D., and West S. C. (2010) The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 10.1038/nsmb.1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J., Doty T., Gibson B., and Heyer W. D. (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 10.1038/nsmb.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao W., Vaithiyalingam S., San Filippo J., Maranon D. G., Jimenez-Sainz J., Fontenay G. V., Kwon Y., Leung S. G., Lu L., Jensen R. B., Chazin W. J., Wiese C., and Sung P. (2015) Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA mimicry. Mol. Cell 59, 176–187 10.1016/j.molcel.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gnügge R., and Symington L. S. (2017) Keeping it real: MRX-Sae2 clipping of natural substrates. Genes Dev. 31, 2311–2312 10.1101/gad.310771.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tammaro M., Liao S., Beeharry N., and Yan H. (2016) DNA double-strand breaks with 5′ adducts are efficiently channeled to the DNA2-mediated resection pathway. Nucleic Acids Res. 44, 221–231 10.1093/nar/gkv969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garcia V., Phelps S. E., Gray S., and Neale M. J. (2011) Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244 10.1038/nature10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kerr C., and Sadowski P. D. (1972) Gene 6 exonuclease of bacteriophage T7. II. mechanism of the reaction. J. Biol. Chem. 247, 311–318 [PubMed] [Google Scholar]
- 36. Kerr C., and Sadowski P. D. (1972) Gene 6 exonuclease of bacteriophage T7. I. purification and properties of the enzyme. J. Biol. Chem. 247, 305–310 [PubMed] [Google Scholar]
- 37. Cannavo E., Cejka P., and Kowalczykowski S. C. (2013) Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 110, E1661–E1668 10.1073/pnas.1305166110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicolette M. L., Lee K., Guo Z., Rani M., Chow J. M., Lee S. E., and Paull T. T. (2010) Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17, 1478–1485 10.1038/nsmb.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fortini B. K., Pokharel S., Polaczek P., Balakrishnan L., Bambara R. A., and Campbell J. L. (2011) Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J. Biol. Chem. 286, 23763–23770 10.1074/jbc.M111.243071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cobb J. A., Bjergbaek L., Shimada K., Frei C., and Gasser S. M. (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22, 4325–4336 10.1093/emboj/cdg391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hegnauer A. M., Hustedt N., Shimada K., Pike B. L., Vogel M., Amsler P., Rubin S. M., van Leeuwen F., Guénolé A., van Attikum H., Thomä N. H., and Gasser S. M. (2012) An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 31, 3768–3783 10.1038/emboj.2012.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., Zhu Z., Paull T. T., Ira G., and Lee S. E. (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 10.1038/emboj.2010.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mimitou E. P., and Symington L. S. (2010) Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369 10.1038/emboj.2010.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim H. S., Nickoloff J. A., Wu Y., Williamson E. A., Sidhu G. S., Reinert B. L., Jaiswal A. S., Srinivasan G., Patel B., Kong K., Burma S., Lee S. H., and Hromas R. A. (2017) Endonuclease EEPD1 is a gatekeeper for repair of stressed replication forks. J. Biol. Chem. 292, 2795–2804 10.1074/jbc.M116.758235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gradia S. D., Ishida J. P., Tsai M. S., Jeans C., Tainer J. A., and Fuss J. O. (2017) MacroBac: new technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 592, 1–26 10.1016/bs.mie.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krasner D. S., Daley J. M., Sung P., and Niu H. (2015) Interplay between Ku and replication protein A in the restriction of Exo1-mediated DNA break end resection. J. Biol. Chem. 290, 18806–18816 10.1074/jbc.M115.660191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adkins N. L., Niu H., Sung P., and Peterson C. L. (2013) Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 20, 836–842 10.1038/nsmb.2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller A. S., Daley J. M., Pham N. T., Niu H., Xue X., Ira G., and Sung P. (2017) A novel role of the Dna2 translocase function in DNA break resection. Genes Dev. 31, 503–510 10.1101/gad.295659.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xue X., Papusha A., Choi K., Bonner J. N., Kumar S., Niu H., Kaur H., Zheng X. F., Donnianni R. A., Lu L., Lichten M., Zhao X., Ira G., and Sung P. (2016) Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1. Genes Dev. 30, 687–699 10.1101/gad.276139.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Komen S., Macris M., Sehorn M. G., and Sung P. (2006) Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 408, 445–463 10.1016/S0076-6879(06)08028-1 [DOI] [PubMed] [Google Scholar]
- 51. Sigurdsson S., Trujillo K., Song B., Stratton S., and Sung P. (2001) Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 276, 8798–8806 10.1074/jbc.M010011200 [DOI] [PubMed] [Google Scholar]
- 52. Chen L., Trujillo K., Ramos W., Sung P., and Tomkinson A. E. (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115 10.1016/S1097-2765(01)00388-4 [DOI] [PubMed] [Google Scholar]
- 53. Trujillo K. M., and Sung P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 276, 35458–35464 10.1074/jbc.M105482200 [DOI] [PubMed] [Google Scholar]
- 54. Trujillo K. M., Roh D. H., Chen L., Van Komen S., Tomkinson A., and Sung P. (2003) Yeast Xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J. Biol. Chem. 278, 48957–48964 10.1074/jbc.M309877200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.