Abstract
FK228 is an FDA-approved anticancer drug naturally produced by Chromobacterium violaceum No. 968 up to 19 mg/L in a pilot industry-scale batch fermentation. Here we report a genomics-guided discovery of Burkholderia thailandensis MSMB43 as a new and significantly better source of FK228. The genome of B. thailandensis MSMB43 was found to contain a functional biosynthetic gene cluster highly homologous to that of FK228 in C. violaceum No. 968, and the bacterium indeed produces authentic FK228. By simple fermentation in shaking flasks in a preferred M8 medium, B. thailandensis MSMB43 produced FK228 up to 67.7 mg/L; by fed-batch fermentation in a 20-L fermentor in M8 medium, B. thailandensis MSMB43 produced FK228 up to 115.9 mg/L, which is 95 fold higher than that of C. violaceum No. 968 under the same laboratory fermentation conditions. RT-PCR analysis indicated that the high FK228 yield of B. thailandensis MSMB43 was due to high expression of biosynthetic genes, represented by Bth_depA, during the fermentation process. Further genetic manipulation resulted in a recombinant strain, B. thailandensis MSMB43/pBMTL3-tdpR, which harbors a broad host-range vector expressing the thailandepsin biosynthetic pathway regulatory gene tdpR. This engineered strain produced up to 168.5 mg/L of FK228 in fed-batch fermentation in a 20-L fermentor in M8 medium. Therefore, the wild-type B. thailandensis MSMB43 or its engineered derivative could potentially be a good starting point for an industrial process to improve FK228 production for its expanding use in therapy.
Keywords: Burkholderia thailandensis MSMB43, Fermentation optimization, FK228, Genome mining, Natural product, Productivity
1. Introduction
FK228 (Fig. 1B), also know as depsipeptide (FR901228), romidepsin (NSC 630176), and marketed as Istodax® by Celgene Corporation, is an anticancer drug approved by the US Food and Drug Administration (FDA) first in 2009 for the treatment of cutaneous T-cell lymphoma [1], and then in 2011 for the treatment of peripheral T-cell lymphoma [2]. It is being aggressively pursued in numerous combination therapies against various types of cancer and other disease conditions [[3], [4], [5], [6], [7]]. The bicyclic structure of FK228 is composed of four amino acids (d-valine, d-cysteine, Z-dehydrobutyrine, l-valine) and a complex moiety (3S,4E)-3-hydroxy-7-mercapto-4-heptenoic acid (HMHA), which is likely built from one l-cysteine unit and two C2 units derived from malonyl coenzyme A. The signature disulfide linkage in FK228 is critical for structural stability when FK228 normally exists as a prodrug, and for bioactivity as a potent histone deacetylase (HDAC) inhibitor when FK228 is reduced in vivo [8]. The biosynthesis of FK228 in Chromobacterium violaceum No. 968 was extensively studied, revealing a thiotemplated, hybrid nonribosomal peptide synthetase (NRPS)-polyketide synthase (PKS) assembly line with multiple accessory enzymes and a stress-inducible transcriptional regulator [[9], [10], [11], [12], [13]].
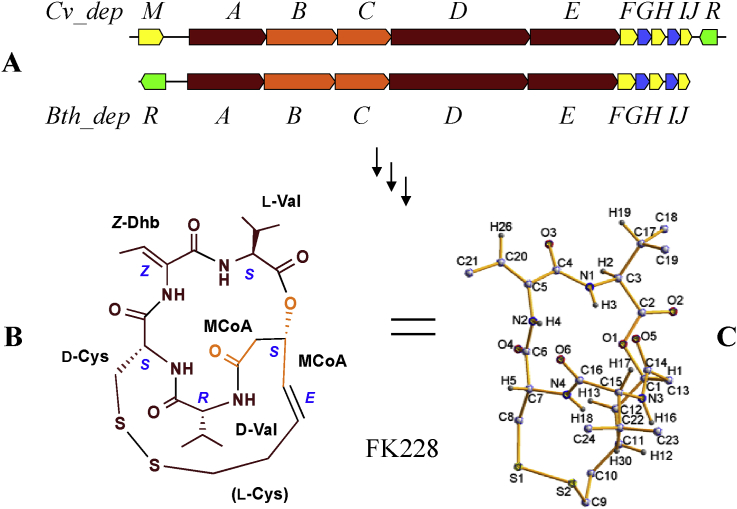
Fig. 1.
FK228 biosynthesis cluster and structure. (A) A graphic comparison between two highly homologous FK228 biosynthetic gene clusters from C. violaceum No. 968 (Cv_dep) and B. thailandensis MSMB43 (Bth_dep), respectively. Genes are color-coded to indicate functional categories of their encoded proteins. Dark red: nonribosomal peptide synthetases; orange: polyketide synthases; yellow: tailoring proteins; blue: resistance proteins; green: regulatory proteins. (B) A 2-dimensional structure of FK228, with molecular building blocks labeled. (C) A 3-dimensional molecular drawing of FK228.
Chemical synthesis of FK228 has been difficult due to challenges of asymmetric construction of the HMHA moiety, the formation of a 16-membered cyclic depsipeptide, and intramolecular oxidative coupling of two thiol moieties to form a 15-membered disulfide-containing ring [14,15]. Current commercial supply of FK228 is made possible through microbial fermentation of C. violaceum No. 968, although the exact market size of Istodax® and the yield of industrial production and conditions of FK228 are proprietary [16]. An original report of FK228 production up to 19 mg/L from a pilot industry-scale of bacterial fermentation was achieved [17].
Burkholderia species have been shown to be a productive source of diverse natural products [18], and multiple laboratories have isolated unique depsipeptides related to FK228 including burkholdacs [19], FR901375 [20,21], spiruchostatins [22,23] and thailandepsins [24,25]. In this paper, we report the discovery of a biosynthetic gene cluster in the genome of B. thailandensis MSMB43 which is highly homologous to that of FK228 in C. violaceum No. 968, and show that B. thailandensis MSMB43 produces a high titer of authentic FK228 in a defined growth medium through laboratory-scale fermentation. We suggest that B. thailandensis MSMB43 wild-type strain, or an engineered derivative of which, could potentially be a good starting point for an industrial process to improve FK228 production for its expanding use in therapy.
2. Materials and methods
2.1. Bacterial strains and genomic analysis
The bacterial strains and plasmids used in this study are listed in Table S1 (Supplemental Material). The original draft genome sequence of B. thailandensis MSMB43 was reported by T. D. Read and associates in 2010 [26], but the quality was found to be suboptimal for our natural product discovery purpose due to insufficient coverage. We thus resequenced the bacterial genome and obtained a significantly improved draft genome sequence, which contains at least 13 secondary metabolite biosynthetic gene clusters [27]. Sequence gaps within a targeted gene cluster were filled by nested PCR and Sanger sequencing [28]. Search for homologous gene and protein sequences was performed with Blast algorithms available at the NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi/) [29]. Analysis of the domain and module organization of PKSs and NRPS was carried out with popular web servers [30,31].
2.2. Genetic manipulation of B. thailandensis MSMB43
To create a series of B. thailandensis MSMB43 derivative strains (Table S1), vector pBMTL-3 and three gene-expression constructs [pBMTL3-depR, pBMTL3-spiR and pBMTL3-tdpR] were introduced into B. thailandensis MSMB43 cells via interspecies conjugation as follows. Donor strain E. coli S17-1/vector or construct was grown in Luria-Bertani (LB) media supplemented with 10 μg/mL tetracycline (Tc10) and 34 μg/mL chloramphenicol (Cm34) at 37 °C with shaking until the late mid-log phase (6–8 h); B. thailandensis MSMB43 was grown similarly with 50 μg/mL apramycin (Am50) at 30 °C until the late mid-log phase (6–8 h). Cells from 1 mL of each culture were collected by centrifugation at 4,000×g for 15 min at 4 °C, and the cell pellets were washed once with 1 mL LB medium. Cells were collected again by centrifugation and resuspended in 100 μL LB medium. Cell suspensions of two bacterial strains were pooled and spread evenly on a wet 0.45-μm nitrocellulose membrane (GE Healthcare, Pittsburgh, PA) on LB agar. After the plate had been incubated at 30 °C for 12–16 h, the membrane seeded with bacteria was used to print several LB agar plates containing Am50 and Cm50 to select for exconjugants.
2.3. Real-time quantitative PCR
B. thailandensis MSMB43 and C. violaceum No. 968 were grown for 48 or 24 h in LB broth as seed cultures, respectively. Then 1.6 mL of each seed culture was inoculated into 40 mL M8 medium for 72 h fermentation. Aliquots of culture were collected for RNA extraction, and the remaining cultures were used for quantification of the FK228 production levels of the two strains. Extraction of total RNA was performed using AxyPrep Multisource Total RNA Miniprep kit (AxyGen, CA, USA). RNA samples were digested with DNase I (Takara, Dalian, China) to remove residual DNA and then recovered using RNA Clean kit (Tiangen, Beijing, China). Absence of DNA contamination in RNA samples was confirmed with conventional PCR reactions. DNA-free RNA samples were then reverse transcribed to cDNA using reverse transcriptase M-MLV kit (Takara). Genes depA, depJ and depR were chosen for real-time PCR; their primer sequences were listed in Table S2. Real-time PCR was performed using SYBR Premix Ex Taq kit (Tiangen). A typical reaction of 20 μL contains 10 μL 2X Premix Ex Taq DNA polymerase, 4 pmol of each primer and 100-fold dilution (for 16s rDNA) or 10-fold dilution (for other genes) cDNA solution. PCR program was as followed: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 62 °C for 31 s. The expression levels of target genes were first normalized to reference 16s rDNA gene. Then, fold differences between the expression levels of genes in B. thailandensis MSMB43 relative to their counterparts in C. violaceum No. 968 were calculated using 2−ΔΔCt method [32].
2.4. Bacterial fermentation
For small volume fermentation in shaking flasks, a seed culture of 50 mL of B. thailandensis MSMB43 in LB medium containing Am50, or C. violaceum No. 968 in 1% Difco nutrient broth (NB) containing 200 μg/mL of ampicillin (Ap200), was cultivated at 30 °C for 24 h with agitation (200 rpm). Subsequently, 10 mL seed culture of each strain was inoculated into 300 mL NB/G medium (NB with 1% glucose; pH 7.0; systematically named as M1 medium) or M8 medium (0.5% glucose, 0.5% peptone, 0.3% NaCl, 0.12% Na2HPO4, and 0.05% KH2PO4; pH 7.0) in each of four 1-L flasks, and cultured at 30 °C for 120 h with agitation (150 rpm). Similarly for large volume batch fermentation, 250 mL seed culture was inoculated into 12 L M8 medium in each of two 20-L fermentors (BioFlo IV, New Brunswick Scientific Co., NJ) and fermented at 30 °C for 120 h under 20 L/min aeration and 200 rpm agitation; pH was maintained at 7.0 with 1 N NaCl or 1 M HCl. For large volume fed-batch fermentation, 3 L of 10X concentrated medium was fed between 24 h and 48 h on top of the batch fermentation. Fermentation of genetically altered B. thailandensis derivative strains was conducted similarly but with the addition of Cm50 to the medium to maintain the pBMTL-3 based plasmids. Furthermore, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium at 0 h and again at 48 h to a final concentration of 0.5 mM to induce gene expression.
2.5. Quantification of FK228 production by liquid chromatography-mass spectrometry (LC-MS)
All LC-MS analyses were performed on an Agilent 1100 series LC/MSD Trap SL mass spectrometer equipped with an Agilent Zorbax ODS C18 column (3.0 × 10 mm) and a UV detector. The solvent system consists of buffer A (water with 0.1% formic acid) and buffer B (acetonitrile with 0.1% formic acid). The LC program included a linear gradient from 40% buffer B to 100% buffer B in 10 min, a constant elution in 100% buffer B for 2 min, followed by a linear return to 40% of buffer B in 1 min. Flow rate was 0.5 mL/min and UV spectrum was monitored at 210 nm. The inert nitrogen gas flow rate, nebulizer pressure and drying temperature for the MS system were 10 L/min, 30 psi and 350 °C, respectively. MS was scanned in positive mode in a range of 100 to 1000 m/z; the target compound FK228 (540.1 m/z) was eluted at 3.6–4.5 min under those conditions. A standard curve of FK228 concentrations vs. ion signal peak areas was first generated by running a series of authentic FK228 solutions in acetonitrile (0.5, 1, 2, 4, 6, 8 and 10 μg/mL) on LC-MS in triplicates, and the mean value of peak areas of extracted FK228 ion signal were plotted against the concentrations (Fig. S1). The standard curve showed a good linear correlation between varying concentrations and peak areas of the extracted ion signal of FK228. Fermentation cultures (10 mL from each of the four flasks and then pooled together, or 40 mL from a fermentor) were sampled at 24, 48, 72, 96 and 120 h, respectively, and extracted three times with equal volume of ethyl acetate. Three extracts were pooled and concentrated to dryness with a rotary evaporator, and the resulting residue was dissolved in 1 mL of acetonitrile. This suspension was centrifuged at 8000 rpm for 3 min, and 10 μL of the supernatant or proper dilutions was subjected to LC-MS analysis as described above.
2.6. Isolation of FK228 from larger scale fermentation
Because FK228 was previously found to present mostly in the aqueous phase, fermentation broth was first centrifuged at 3800 rpm for 20 min to remove cell debris. The resulting supernatant was passed three times through a large glass column (i.d. 10 × 100 cm) packed 1/3 full with a mixture of Amberlite XAD16 resin (20–60 mesh, Sigma-Aldrich, USA) and Diaion HP-20 resin (Sigma-Aldrich) in a 1:1 ratio. Resin mixture was further washed with water, drained and eluted repeatedly with three volumes of ethyl acetate. Elution fractions were pooled and concentrated with a rotary evaporator, and the resulting oily residue was subjected to three steps of chromatographic separation to obtain FK228 with greater than 98% purity (Fig. S2).
2.7. Nucleotide sequence accession number
The nucleotide sequence of the newly identified FK228 biosynthetic gene cluster from B. thailandensis MSMB43 (Bth_dep) reported in this paper has been deposited in the GenBank database under accession number JX268530.
3. Results
3.1. B. thailandensis MSMB43 possesses a putative FK228 biosynthetic gene cluster (Bth_dep)
Bioinformatic analysis of the 7.2-Mb improved draft genome sequence of B. thailandensis MSMB43 revealed at least 13 putative gene clusters coding for the biosynthesis of a wide range of natural products [27]. Among them, one gene cluster (No. 13; annotated as Bth_dep) is highly homologous to that of FK228 from C. violaceum No. 968 (labeled in this paper as Cv_dep for distinguishing from Bth_dep; GenBank accession no. EF210776) [[9], [10], [11], [12]] (Fig. 1; Table 1). There are only two points of difference between these two gene clusters. First, depM is present in the Cv_dep gene cluster but not in the Bth_dep gene cluster; it is a relevant but nonessential gene encoding a putative aminotransferase for FK228 biosynthesis in C. violaceum No. 968 [12]. Second, Cv_depR encodes an OxyR-type transcriptional regulator and is located at the far right of the Cv_dep gene cluster, while Bth_depR encodes an AraC-type transcriptional regulator and is located at the far left of the Bth_dep gene cluster; these two regulatory genes or their deduced proteins do not have significant sequence homology. The remaining genes (depABCDEFGHIJ) in the two gene clusters are overwhelmingly homologous in terms of gene order, gene size, sequence at either nucleotide level or deduced amino acid level, and the predicted substrate specificity of NRPS or PKS enzymes. Therefore, it was conveniently assumed that, if this new Bth_dep gene cluster is functional and expressed, its translated hybrid NRPS-PKS assembly line with multiple accessory enzymes shall direct the biosynthesis of FK228 in B. thailandensis MSMB43.
Table 1.
A comparison of genes and deduced proteins between homologous FK228 biosynthetic (dep) gene clusters in two bacterial species.
|
dep Gene Cluster in C. violaceum No. 968 |
dep Gene Cluster in B. thailandensis MSMB43 |
Gene sequence identity/gap | Protein sequence similarity/identity | Confirmed or deduced protein function | ||
|---|---|---|---|---|---|---|
| Gene | Deduced protein (# of amino acids) | Gene | Deduced protein (# of amino acids) | |||
|
Cv_depM – |
Cv_DepM (389) – |
– Bth_depR |
– Bth_DepR (442) |
– – |
– – |
Aminotransferase AraC-type transcriptional regulator |
| Cv_depA | Cv_DepA (1,697) | Bth_depA | Bth_DepA (1,694) | 79%/2% | 77%/85% | NRPSa |
| Cv_depB | Cv_DepB (1,553) | Bth_depB | Bth_DepB (1,563) | 84%/1% | 84%/89% | PKSb |
| Cv_depC | Cv_DepC (1,183) | Bth_depC | Bth_DepC (1,187) | 83%/1% | 82%/88% | PKSb |
| Cv_depD | Cv_DepD (3,057) | Bth_depD | Bth_DepD (3,081) | 81%/2% | 82%/87% | NRPSa |
| Cv_depE | Cv_DepE (1,892) | Bth_depE | Bth_DepE (1,923) | 82%/2% | 80%/86% | NRPSa |
| Cv_depF | Cv_DepF (390) | Bth_depF | Bth_DepF (390) | 88%/<1% | 93%/96% | FadE2-like acyl-CoA dehydrogenase |
| Cv_depG | Cv_DepG (321) | Bth_depG | Bth_DepG (321) | 82%/<1% | 86%/92% | Phosphotransferase |
| Cv_depH | Cv_DepH (319) | Bth_depH | Bth_DepH (301) | 86%/1% | 86%/91% | FAD-dependent disulfide oxidoreductase |
| Cv_depI | Cv_DepI (304) | Bth_depI | Bth_DepI (304) | 81%/2% | 81%/88% | Esterase/Lipase |
| Cv_depJ | Cv_DepJ (254) | Bth_depJ | Bth_DepJ (266) | 72%/1% | 67%/79% | Type II thioesterase |
| Cv_depR | Cv_DepR (312) | – | – | – | – | OxyR-type transcriptional regulator |
NRPS.
–: not available.
: nonribosomal peptide synthetase; PKS.
Polyketide synthase.
3.2. Identification of cultivation conditions for Bth_dep gene cluster expression and FK228 production
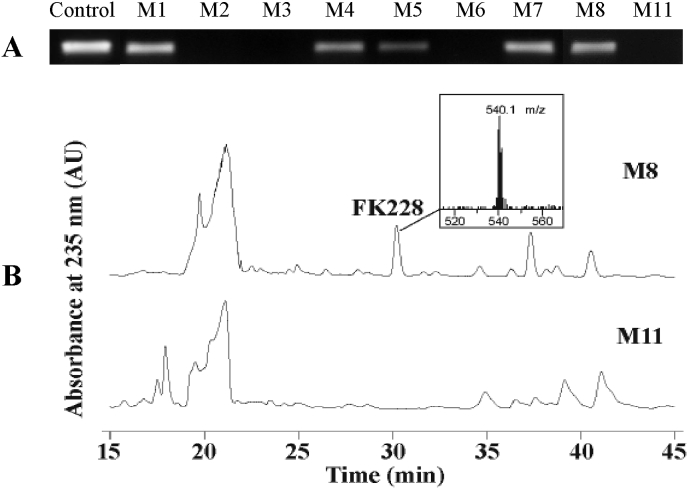
B. thailandensis MSMB43 was grown in a set of nine media previously described [24]. RT-PCR analysis showed that Bth_depJ, which is the last gene of the Bth_dep gene cluster and represents the gene cluster, is abundantly expressed in M1, M7 and M8 media, but not at all in M2, M3, M6 or M11 medium at 48 h of growth (Fig. 2A). Subsequently, comparative analysis of the ethyl acetate extracts of B. thailandensis MSMB43 cultures grown in the nine media by HPLC revealed a well-separated peak present in the extract of M8 culture but absent in that of M11 culture (Fig. 2B; M8 and M11 profiles chosen as examples for their clean background around the target peak). This peak was collected, vacuum-dried, and the material of which was subjected to LC-MS analysis, revealing a dominant compound with an ion signal of m/z 540.1 for [M + H]+ (Fig. 2B insert), consistent with our previous detection of FK228 with the same instrument under the same experimental conditions [9]. Further large-scale purification from the M8 culture afforded a batch of FK228 with greater than 98% purity. The chemical identity and structure of FK228 (Fig. 1B) was confirmed by 1H, 13C, 1H-1H COSY, 1H-13C HSQC, and 1H-13C HMBC NMR analyses (see respective 1H and 13C NMR spectra in Fig. S3 and Fig. S4). The extracted NMR data (Table S3) match to the originally reported NMR data of FK228 [33]. The absolute configuration of purified FK228 was previously shown by X-ray crystallography (Fig. 1C) [34] to be identical to that of the originally reported FK228 [33]. The Bth_dep gene cluster was thus concluded to be functional and B. thailandensis MSMB43 was found to be a new source of FK228.
Fig. 2.
Differential gene expression and FK228 production. (A) RT-PCR profiling of Bth_depJ (representing Bth_dep gene cluster) expression in nine different media. Amplification of a 283-bp DNA from genomic template was included as a positive control. (B) HPLC profiling of FK228 productivity in M8 and M11 media with UV absorbance monitored at 235 nm. Insert shows an LC-MS chromatography of FK228 with [M + H]+ = 540.1 m/z.
3.3. Identification of M8 as an optimal medium for FK228 production
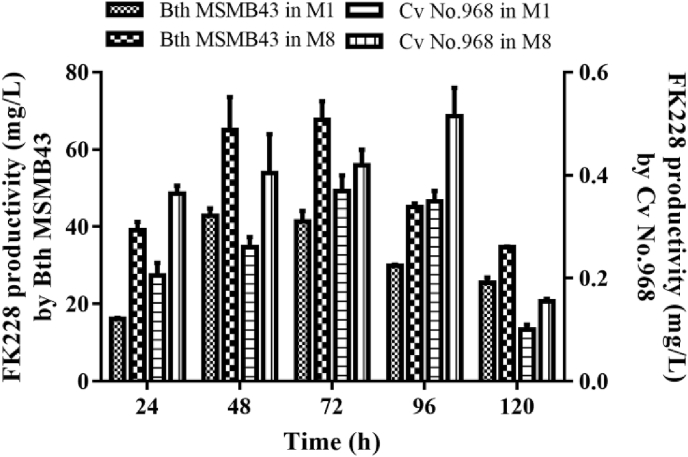
The original FK228-producer, C. violaceum No. 968, and the newly identified FK228-producer, B. thailandensis MSMB43, were cultivated side by side in M1 medium [the original NB/G medium [17]] and M8 medium in flasks for 120 h. The time-course of FK228 production by two strains in two media was assessed by LC-MS. It was found that both strains produced higher titers of FK228 in M8 medium than in M1 medium and the maximal FK228 productivity of both strains in M8 medium was achieved between 72 h and 96 h. The measured highest titer of FK228 produced by B. thailandensis MSMB43 in M8 medium at 72 h was 67.7 mg/L, which is strikingly 135-fold higher than the highest titer by C. violaceum No. 968 at 96 h (0.5 mg/L) (Fig. 3). M8 was thus selected as an optimal medium for further FK228 productivity investigations.
Fig. 3.
Time-course of FK228 production by B. thailandensis MSMB43 and C. violaceum No. 968 in M1 or M8 medium in small volumne fermentation in shaking flasks.
3.4. Improved FK228 production in a fed-batch fermentation
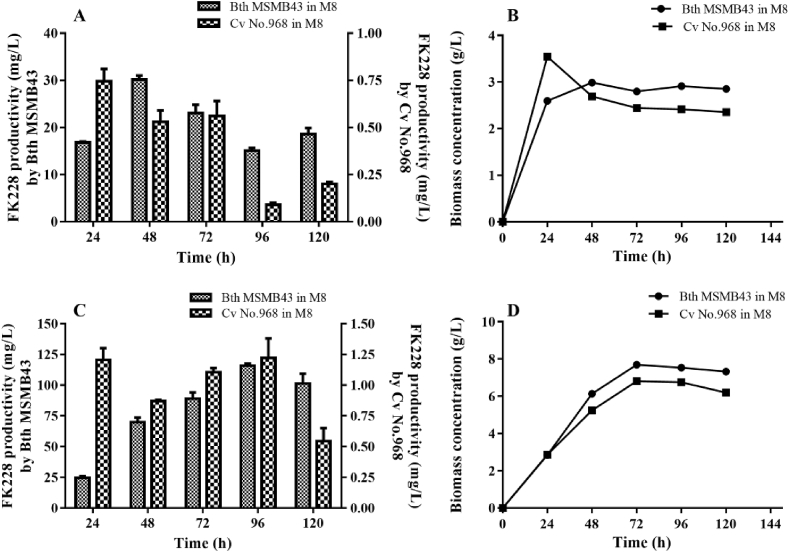
Both C. violaceum No. 968 strain and B. thailandensis MSMB43 strain were first batch-fermented in M8 medium side by side in two 20-L fermentors. The FK228 productivity of B. thailandensis MSMB43 reached the highest titer of 30.2 mg/L at 48 h, and that of C. violaceum No. 968 reached the highest titer of 0.8 mg/L at 24 h (Fig. 4A). Biomass quantification showed that C. violaceum No. 968 culture had already reached its maximal growth at 24 h, while B. thailandensis MSMB43 at 48 h (Fig. 4B), suggesting a rather quick depletion of limited nutrients from the medium. For this reason, fed-batch fermentation was subsequently performed for these two strains. As a result, the FK228 productivity of B. thailandensis MSMB43 reached the highest titer of 115.9 mg/L at 96 h, and that of C. violaceum No. 968 reached the highest titer of 1.2 mg/L at 96 h as well (Fig. 4C). There is a 96-fold yield difference of FK228 between the two strains. Biomass quantification showed that both cultures reached their maximal growth at 72 h (Fig. 4D), indicating that nutrient replenishment is critical for achieving higher FK228 titers. B. thailandensis MSMB43 was thus proven to be a significantly better source of FK228.
Fig. 4.
Time-course of FK228 production and biomass concentrations by B. thailandensis MSMB43 and C. violaceum No. 968 in M8 medium by fermentation in laboratory-scale fermentors. (A) FK228 production in batch fermentation; (B) biomass concentrations in batch fermentation; (C) FK228 production in fed-batch fermentation; (D) biomass concentrations in fed-batch fermentation.
3.5. High expression of depA in B. thailandensis MSMB43
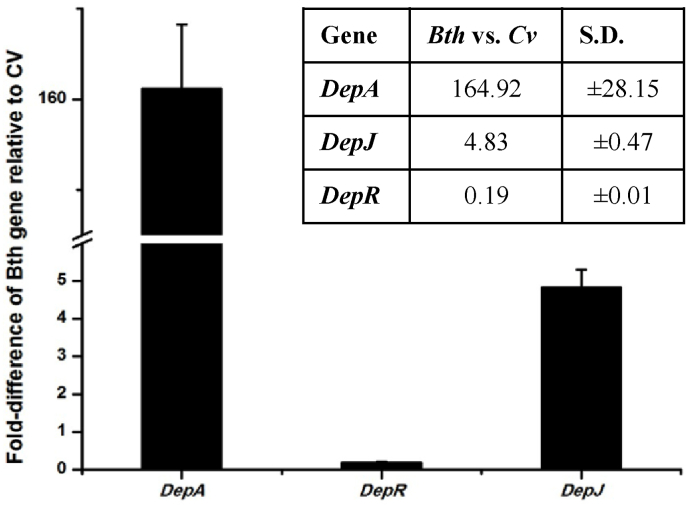
To probe the underlining mechanism for a high yield of FK228 production in B. thailandensis MSMB43, real-time PCR was performed to exam the expression of depA (representing the first committed biosynthetic gene of the gene cluster), depJ (representing the last committed tailoring gene of the gene cluster) and depR in both strains cultivated in M8 medium (Fig. 5). It revealed that the relative level of depA expression in B. thailandensis MSMB43 is about 164 fold higher than that in C. violaceum No. 968, which is consistent with the greatest difference of FK228 yield between these two strains grown in flasks in M8 (Fig. 3). For depJ, the relative expression level is only about 4.8 fold difference, suggesting that tailoring genes are not so critical for the high production of FK228 in B. thailandensis MSMB43. Surprisingly, the relative expression level of depR in B. thailandensis MSMB43 is only a fraction (0.19 fold) of that in C. violaceum No. 968, suggesting that either Bth_depR in B. thailandensis MSMB43 is much more efficient than Cv_depR in C. violaceum No. 968, or more likely these two regulatory genes have a totally different mode of regulating downstream biosynthetic genes.
Fig. 5.
Quantification by real-time PCR of relative gene expression levels (B. thailandensis MSMB43 vs. C. violaceum No. 968). Data are mean values of results from triplicate experiments, with error bars indicating standard deviation.
3.6. Further FK228 yield improvement through metabolic engineering
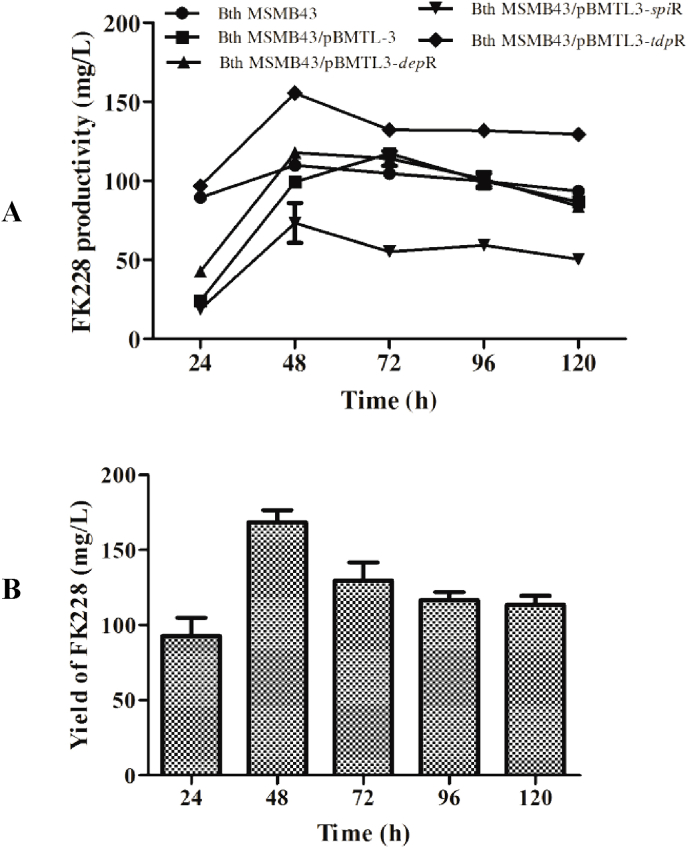
Because pathway regulatory genes often play a critical role in natural product biosynthesis, yet they are small and amenable to genetic manipulation, we tested whether overexpression of a heterologous regulatory gene in B. thailandensis MSMB43 have an impact on FK228 production. Three regulatory genes–depR from the FK228 biosynthetic gene cluster in C. vioaceum No. 968 [12], spiR from the spiruchostatin biosynthetic gene cluster in Pseudomonas sp. Q71576 [35] and tdpR from the thailandepsin biosynthetic gene cluster in B. thailandensis E264 [24]–were cloned into pBMTL-3, a broad host-range expression vector [36], the resultant constructs–pBMTL3-depR, pBMTL3-spiR and pBMTL3-tdpR, along with vector control–were independently introduced into B. thailandensis MSMB43 via conjugation to create recombinant strains Bth MSMB43/pBMTL3-depR, -spiR or -tdpR, and a control strain Bth MSMB43/pBMTL-3 (Table S1). Small volume fermentation in flasks in M8 medium showed that strain Bth MSMB43/pBMTL3-tdpR produced 155.4 mg/L FK228, the highest amount at 48 h (Fig. 6A). Time-course production of FK228 of this recombinant strain in fed-batch fermentation in M8 showed that FK228 reached the highest value of 168.5 mg/L at 48 h (Fig. 6B), indicating that genetic manipulation of metabolic pathway expression is effective in further improving the yield of FK228.
Fig. 6.
Yield improvement of FK228 through genetic manipulation. (A) Time-course of FK228 production by B. thailandensis MSMB43 wild type and derivtive strains in M8 medium in small volumne fermentation in shaking flasks. (B) Time-course of FK228 production by B. thailandensis MSMB43/pBMTL3-tdpR strain in M8 medium by fermentation in laboratory-scale fermentor.
4. Discussion
4.1. A new and significantly better source of FK228
Potent microbial natural products are usually produced in minute amount, which often creates a bottleneck issue in drug development process. FK228 was discovered in early 1990s from the culture broth of C. violaceum No. 968, and the reported best yield was 19 mg/L by a pilot-scale industrial process [17]. As FK228 had forged its way into clinics as a new class of drug for the treatment of multiple forms of T-cell lymphoma [1,2] and potentially other types of neoplasm through combination therapies, material supply for drug manufacturing may become a limiting factor. Previously we had tapped into the FK228 biosynthetic potential, achieved moderate 10%–34% yield increase of FK228 in C. violaceum No. 968 through overexpressing either the Cv_depR gene encoding a transcriptional regulator or the Cv_depH gene encoding a rate-limiting disulfide oxidoreductase [10,12]. Studies reported herein showed that B. thailandensis MSMB43 is not only a new source of FK228 but also a significantly better source of FK228. This bacterial strain produced 67.7 mg/L of FK228 by simple fermentation in shaking flasks; by fed-batch laboratory-scale fermentation B. thailandensis MSMB43 achieved the highest titer of 115.9 mg/L, which is 95-fold higher than that of C. violaceum No. 968 under the identical fermentation conditions. Moreover, the titer of FK228 production by B. thailandensis MSMB43 can be further improved up to 168.5 mg/L in fed-batch fermentation through overexpression of a heterologous regulatory gene depR. Therefore, the wild-type B. thailandensis MSMB43 or its engineered derivative could potentially be a good starting point for an industrial process to improve FK228 production for its expanding use in therapy.
4.2. Evolution of the FK228 biosynthetic gene cluster
Natural product biosynthetic gene clusters are among life's most diverse and rapidly evolving genetic elements [37]. One of the underlining mechanisms for the evolution of many natural product biosynthetic genes and gene clusters is horizontal gene transfer (HGT), often facilitated by mobile elements such as transposons [38,39]. A classic example of HGT is the transmission of NRPS genes responsible for the production of β-lactam antibiotics (e.g., penicillins and cephalosporins), evidently from bacteria to bacteria and from bacteria to fungi [40,41]. A more recent example is the andrimid gene clusters from Pantoea agglomerans CU2194 and Vibrionales bacterium AWAT-3; the only syntenic difference is the insertion of a single gene at the 3′ end of the latter gene cluster [37,42]. The striking similarity between the dep gene clusters from C. violeceum No. 968 and B. thailandensis MSMB43 revealed by this study suggests that the gene cluster most likely have been horizontally transferred from one species to another. Phylogenetically C. violeceum No. 968 belongs to Neisseriales, while B. thailandensis MSMB43 belongs to Burkholderiales; both, however, are within the order of betaproteobacteria. The flanking regions of the Cv_dep gene cluster in C. violeceum No. 968 does not contain apparent mobile elements but rather house-keeping genes [9], while the flanking regions of Bth_dep gene cluster in B. thailandensis MSMB43 contain several putative transposons at each side (data not shown). DNA statistics showed that both dep gene clusters have a G+C content of 69–70%, while the G+C content of the flanking regions is about 63% in C. violeceum No. 968, or 65% in B. thailandensis MSMB43, suggesting that the dep gene clusters might have originated from a third organism with a higher G+C genome. Along with the intriguing but unanswered question of evolution, it is equally intriguing yet unanswered about why various bacterial species produce a small molecule FK228 which targets histone deacetylases in eukaryotic epigenome [8].
Declarations of interest
The authors declare no financial or commercial conflict of interest.
Acknowledgements
We dedicate this paper to and in memory of our beloved friend and colleague, Prof. Keqiang Yang, who devoted his life to the research on Streptomyces physiology, antibiotic biosynthesis, and molecular regulation. This work was supported in parts by a Catalyst Award from the University of Wisconsin-Milwaukee Research Foundation and a Public Health Service grant (CA152212) from the National Cancer Institute to YQC, the National Science Foundation of China (31430002, 31770055, 31570031), the Fundamental Research Funds for the Central Universities (22221818014), the Major Basic Program of the Natural Science Foundation of Shandong Province (ZR2017ZB0206), and the Shandong Taishan Scholar Program of China to LZ.
We thank J. Gee (US Centers for Diseases Control, CDC) for facilitating the distribution of B. thailandensis MSMB43 strain from CDC to YQC; this strain was originally isolated by Dr. Bart Currie and associates at Darwin University, Australia [43]. We thank Holger Foersterling (University of Wisconsin-Milwaukee NMR Facility) for assistance with NMR analysis, and Ilia Guzei (University of Wisconsin-Madison Crystallography Facility) for collecting crystallographic data.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2018.10.011.
Contributor Information
Xueting Liu, Email: Liuxt2010@126.com.
Yi-Qiang Cheng, Email: YiQiang.Cheng@unthsc.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.StatBite F.D.A. FDA oncology drug product approvals in 2009. J Natl Cancer Inst. 2010;102(4):219. doi: 10.1093/jnci/djq030. [DOI] [PubMed] [Google Scholar]
- 2.Robey R.W., Chakraborty A.R., Basseville A., Luchenko V., Bahr J., Zhan Z., Bates S.E. Histone deacetylase inhibitors: emerging mechanisms of resistance. Mol Pharm. 2011;8(6):2021–2031. doi: 10.1021/mp200329f. PubMed PMID: 21899343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Habibi H., Atashi A., Abroun S., Noruzinia M. Synergistic effect of simvastatin and romidepsin on gamma-globin gene induction. Cell J. 2019;20(4):576–583. doi: 10.22074/cellj.2019.5589. PubMed PMID: 30124006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valdez B.C., Li Y., Murray D., Liu Y., Nieto Y., Champlin R.E., Andersson B.S. Combination of a hypomethylating agent and inhibitors of PARP and HDAC traps PARP1 and DNMT1 to chromatin, acetylates DNA repair proteins, down-regulates NuRD and induces apoptosis in human leukemia and lymphoma cells. Oncotarget. 2018;9(3):3908–3921. doi: 10.18632/oncotarget.23386. PubMed PMID: 29423093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y., Anderson J.L., Lewin S.R. Getting the “Kill” into “Shock and Kill”: strategies to eliminate latent HIV. Cell Host Microbe. 2018;23(1):14–26. doi: 10.1016/j.chom.2017.12.004. PubMed PMID: 29324227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrich A., Nabhan C. Use of class I histone deacetylase inhibitor romidepsin in combination regimens. Leuk Lymphoma. 2016;57(8):1755–1765. doi: 10.3109/10428194.2016.1160082. PubMed PMID: 27118119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losson H., Schnekenburger M., Dicato M., Diederich M. Natural compound histone deacetylase inhibitors (HDACi): synergy with inflammatory signaling pathway modulators and clinical applications in cancer. Molecules. 2016;21(11) doi: 10.3390/molecules21111608. PubMed PMID: 27886118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furumai R., Matsuyama A., Kobashi N., Lee K.H., Nishiyama M., Nakajima H., Tanaka A., Komatsu Y., Nishino N., Yoshida M., Horinouchi S. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62(17):4916–4921. PubMed PMID: 12208741. [PubMed] [Google Scholar]
- 9.Cheng Y.-Q., Yang M., Matter A.M. Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum No. 968. Appl Environ Microbiol. 2007;73(11):3460–3469. doi: 10.1128/AEM.01751-06. PubMed PMID: 17400765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C., Wesener S.R., Zhang H., Cheng Y.-Q. An FAD-dependent pyridine nucleotide-disulfide oxidoreductase is involved in disulfide bond formation in FK228 anticancer depsipeptide. Chem Biol. 2009;16(6):585–593. doi: 10.1016/j.chembiol.2009.05.005. PubMed PMID: 19549597. [DOI] [PubMed] [Google Scholar]
- 11.Wesener S.R., Potharla V.Y., Cheng Y.-Q. Reconstitution of the FK228 biosynthetic pathway reveals cross talk between modular polyketide synthases and fatty acid synthase. Appl Environ Microbiol. 2011;77(4):1501–1507. doi: 10.1128/AEM.01513-10. PubMed PMID: 21183648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potharla V.Y., Wesener S.R., Cheng Y.-Q. New insights into the genetic organization of the FK228 biosynthetic gene cluster in Chromobacterium violaceum no. 968. Appl Environ Microbiol. 2011;77(4):1508–1511. doi: 10.1128/AEM.01512-10. PubMed PMID: 21183645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao Y., Tong T., Xue J., Lin W., Deng Z., Cheng Y.-Q., Zhu D. Mechanistic studies of DepR in regulating FK228 biosynthesis in Chromobacterium violaceum no. 968. PloS One. 2018;13(4) doi: 10.1371/journal.pone.0196173. PubMed PMID: 29672625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greshock T.J., Johns D.M., Noguchi Y., Williams R.M. Improved total synthesis of the potent HDAC inhibitor FK228 (FR-901228) Org Lett. 2008;10(4):613–616. doi: 10.1021/ol702957z. PubMed PMID: 18205373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K.W., Wu J., Xing W., Simon J.A. Total synthesis of the antitumor depsipeptide FR-901,228. J Am Chem Soc. 1996;118(30):7237–7238. [Google Scholar]
- 16.VanderMolen K.M., McCulloch W., Pearce C.J., Oberlies N.H. Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J Antibiot (Tokyo) 2011;64(8):525–531. doi: 10.1038/ja.2011.35. PubMed PMID: 21587264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda H., Nakajima H., Hori Y., Fujita T., Nishimura M., Goto T., Okuhara M. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J Antibiot (Tokyo) 1994;47(3):301–310. doi: 10.7164/antibiotics.47.301. PubMed PMID: 7513682. [DOI] [PubMed] [Google Scholar]
- 18.Liu X., Cheng Y.-Q. Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biotechnol. 2014;41(2):275–284. doi: 10.1007/s10295-013-1376-1. PubMed PMID: 24212473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biggins J.B., Gleber C.D., Brady S.F. Acyldepsipeptide HDAC inhibitor production induced in Burkholderia thailandensis. Org Lett. 2011;13(6):1536–1539. doi: 10.1021/ol200225v. PubMed PMID: 21348454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masakuni O., Toshio G., Takashi F., Yasuhiro H., Hirotsugu U. 1991. inventorsFR901375 substance and production thereof patent Japan Patent JP 3141296 (A) [Google Scholar]
- 21.Chen Y., Gambs C., Abe Y., Wentworth P., Jr., Janda K.D. Total synthesis of the depsipeptide FR-901375. J Org Chem. 2003;68(23):8902–8905. doi: 10.1021/jo034765b. PubMed PMID: 14604360. [DOI] [PubMed] [Google Scholar]
- 22.Masuoka Y., Nagai A., Shin-ya K., Furihata K., Nagai K., Suzuki K., Hayakawa Y., Seto H. Spiruchostatins A and B, novel gene expression-enhancing substances produced by Pseudomonas sp. Tetrahedron Lett. 2001;42:41–44. [Google Scholar]
- 23.Klausmeyer P., Shipley S.M., Zuck K.M., McCloud T.G. Histone deacetylase inhibitors from Burkholderia thailandensis. J Nat Prod. 2011;74(10):2039–2044. doi: 10.1021/np200532d. PubMed PMID: 21967146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C., Henkes L.M., Doughty L.B., He M., Wang D., Meyer-Almes F.J., Cheng Y.-Q. Thailandepsins: bacterial products with potent histone deacetylase inhibitory activities and broad-spectrum antiproliferative activities. J Nat Prod. 2011;74(10):2031–2038. doi: 10.1021/np200324x. PubMed PMID: 21793558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., Flemming C.J., Cheng Y.-Q. Discovery and activity profiling of thailandepsins A through F, potent histone deacetylase inhibitors, from Burkholderia thailandensis E264. Med Chem Commun. 2012;3:976–981. doi: 10.1039/C2MD20024D. PubMed PMID: 21793558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S., Thomason M.K., Lentz S., Nolan N., Willner K., Gee J.E., Glass M.B., Inglis T.J., Merritt A., Levy A., Sozhamannan S., Mateczun A., Read T.D. High-redundancy draft sequencing of 15 clinical and environmental Burkholderia strains. J Bacteriol. 2010;192(23):6313–6314. doi: 10.1128/JB.00991-10. PubMed PMID: 20870763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuo Y., Liu L., Wang Q., Liu X., Ren B., Liu M., Ni P., Cheng Y.-Q., Zhang L. Revised genome sequence of Burkholderia thailandensis MSMB43 with improved annotation. J Bacteriol. 2012;194(17):4749–4750. doi: 10.1128/JB.00931-12. PubMed PMID: 22887659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Hijum S.A., Zomer A.L., Kuipers O.P., Kok J. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki356. (Web Server issue):W560-W566 PubMed PMID: 15980536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. PubMed PMID: 2231712. [DOI] [PubMed] [Google Scholar]
- 30.Ansari M.Z., Yadav G., Gokhale R.S., Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh359. (Web Server issue):W405-13. PubMed PMID: 15215420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blin K., Medema M.H., Kottmann R., Lee S.Y., Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017;45(D1):D555–D559. doi: 10.1093/nar/gkw960. PubMed PMID: 27924032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Shigematsu N., Ueda H., Takase S., Tanaka H., Yamamoto K., Tada T. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. II. Structure determination. J Antibiot (Tokyo) 1994;47(3):311–314. doi: 10.7164/antibiotics.47.311. PubMed PMID: 8175483. [DOI] [PubMed] [Google Scholar]
- 34.Liu X.Y., Wang C., Cheng Y.-Q. FK228 from Burkholderia thailandensis MSMB43. Acta Crystallogr Sect E Struct Rep Online. 2012;68(Pt 9):o2757–o2758. doi: 10.1107/S160053681203601X. [pii]. PubMed PMID: 22969639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potharla V.Y., Wang C., Cheng Y.-Q. Identification and characterization of the spiruchostatin biosynthetic gene cluster enable yield improvement by overexpressing a transcriptional activator. J Ind Microbiol Biotechnol. 2014;41(9):1457–1465. doi: 10.1007/s10295-014-1474-8. PubMed PMID: 24973954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch M.D., Gill R.T. Broad host range vectors for stable genomic library construction. Biotechnol Bioeng. 2006;94(1):151–158. doi: 10.1002/bit.20836. PubMed PMID: 16496398. [DOI] [PubMed] [Google Scholar]
- 37.Fischbach M.A., Walsh C.T., Clardy J. The evolution of gene collectives: how natural selection drives chemical innovation. Proc Natl Acad Sci U S A. 2008;105(12):4601–4608. doi: 10.1073/pnas.0709132105. PubMed PMID: 18216259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence J. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev. 1999;9(6):642–648. doi: 10.1016/s0959-437x(99)00025-8. PubMed PMID: 10607610. [DOI] [PubMed] [Google Scholar]
- 39.Toussaint A., Chandler M. Prokaryote genome fluidity: toward a system approach of the mobilome. Methods Mol Biol. 2012;804:57–80. doi: 10.1007/978-1-61779-361-5_4. PubMed PMID: 22144148. [DOI] [PubMed] [Google Scholar]
- 40.Liras P., Rodriguez-Garcia A., Martin J.F. Evolution of the clusters of genes for beta-lactam antibiotics: a model for evolutive combinatorial assembly of new beta-lactams. Int Microbiol. 1998;1(4):271–278. PubMed PMID: 10943374. [PubMed] [Google Scholar]
- 41.Brakhage A.A., Al-Abdallah Q., Tuncher A., Sprote P. Evolution of beta-lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry. 2005;66(11):1200–1210. doi: 10.1016/j.phytochem.2005.02.030. PubMed PMID: 15950251. [DOI] [PubMed] [Google Scholar]
- 42.Jin M., Fischbach M.A., Clardy J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J Am Chem Soc. 2006;128(33):10660–10661. doi: 10.1021/ja063194c. PubMed PMID: 16910643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gee J.E., Glass M.B., Novak R.T., Gal D., Mayo M.J., Steigerwalt A.G., Wilkins P.P., Currie B.J. Recovery of a Burkholderia thailandensis-like isolate from an Australian water source. BMC Microbiol. 2008;8:54. doi: 10.1186/1471-2180-8-54. PubMed PMID: 18384685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.