Homologs of the metazoan RNA repair enzymes RtcB and RtcA occur widely in eubacteria, suggesting a selective advantage. Although the enzymatic activities of the eubacterial RtcB and RtcA have been well characterized, the physiological roles remain largely unresolved. Here we report stress responses that activate expression of the σ54-dependent RNA repair operon (rsr-yrlBA-rtcBA) of S. Typhimurium and demonstrate that expression of the operon impacts cell survival under MMC-induced stress. Characterization of the requirements for activation of this tightly regulated operon provides clues to the possible functions of operon components in vivo, enhancing our understanding of how this human pathogen copes with environmental stressors.
KEYWORDS: NtrC, RNA repair, RtcR, SOS response, Salmonella Typhimurium, sigma54, stress response
ABSTRACT
The σ54 regulon in Salmonella enterica serovar Typhimurium includes a predicted RNA repair operon encoding homologs of the metazoan Ro60 protein (Rsr), Y RNAs (YrlBA), RNA ligase (RtcB), and RNA 3′-phosphate cyclase (RtcA). Transcription from σ54-dependent promoters requires that a cognate bacterial enhancer binding protein (bEBP) be activated by a specific environmental or cellular signal; the cognate bEBP for the σ54-dependent promoter of the rsr-yrlBA-rtcBA operon is RtcR. To identify conditions that generate the signal for RtcR activation in S. Typhimurium, transcription of the RNA repair operon was assayed under multiple stress conditions that result in nucleic acid damage. RtcR-dependent transcription was highly induced by the nucleic acid cross-linking agents mitomycin C (MMC) and cisplatin, and this activation was dependent on RecA. Deletion of rtcR or rtcB resulted in decreased cell viability relative to that of the wild type following treatment with MMC. Oxidative stress from peroxide exposure also induced RtcR-dependent transcription of the operon. Nitrogen limitation resulted in RtcR-independent increased expression of the operon; the effect of nitrogen limitation required NtrC. The adjacent toxin-antitoxin module, dinJ-yafQ, was cotranscribed with the RNA repair operon but was not required for RtcR activation, although YafQ endoribonuclease activated RtcR-dependent transcription. Stress conditions shown to induce expression the RNA repair operon of Escherichia coli (rtcBA) did not stimulate expression of the S. Typhimurium RNA repair operon. Similarly, MMC did not induce expression of the E. coli rtcBA operon, although when expressed in S. Typhimurium, E. coli RtcR responds effectively to the unknown signal(s) generated there by MMC exposure.
IMPORTANCE Homologs of the metazoan RNA repair enzymes RtcB and RtcA occur widely in eubacteria, suggesting a selective advantage. Although the enzymatic activities of the eubacterial RtcB and RtcA have been well characterized, the physiological roles remain largely unresolved. Here we report stress responses that activate expression of the σ54-dependent RNA repair operon (rsr-yrlBA-rtcBA) of S. Typhimurium and demonstrate that expression of the operon impacts cell survival under MMC-induced stress. Characterization of the requirements for activation of this tightly regulated operon provides clues to the possible functions of operon components in vivo, enhancing our understanding of how this human pathogen copes with environmental stressors.
INTRODUCTION
Salmonella enterica subsp. enterica serovar Typhimurium, the most common serotype of Salmonella associated with gastrointestinal disease in humans, has been extensively studied to reveal the adaptive strategies and virulence factors that lead to its success as a human pathogen (reviewed in reference 1). In the armory of transcriptional regulatory systems that are key to S. Typhimurium persistence in the host, the regulon of the alternative sigma factor σ54 (σN or RpoN) contributes numerous adaptation functions in response to conditions encountered during infection. These responses include nitrogen limitation, nitric oxide stress, loss of cell envelope integrity, toxic levels of zinc, and availability of alternative nutrient sources (reviewed in reference 2). While σ54 is like other alternative σ factors in that it interacts with RNA polymerase core (E) and directs the holoenzyme (Eσ) to a specific promoter sequence, σ54 is distinct in structure, conserved promoter elements, and its requirement for an activator that hydrolyzes ATP to initiate transcription (reviewed in reference 3). Each σ54-dependent promoter has an associated activator, called a bacterial enhancer binding protein (bEBP), that stimulates transcription from a nearby DNA enhancer sequence in response to a specific environmental stimulus. The bEBP-ATP-enhancer complex contacts the Eσ54 closed complex through DNA looping, whereupon ATP hydrolysis stimulates the conformational transition to an open complex for transcription initiation. Most bEBPs have an N-terminal regulatory domain that detects the environmental stimulus, a highly conserved AAA+ ATPase domain that interacts with Eσ54, and a C-terminal DNA binding domain that is specific for recognition of its associated enhancer sequence. In many bacteria, there are multiple bEBPs that control the expression of different σ54-dependent operons in response to a variety of cellular or environmental signals; the S. Typhimurium genome encodes 13 bEBPs (reviewed in reference 2).
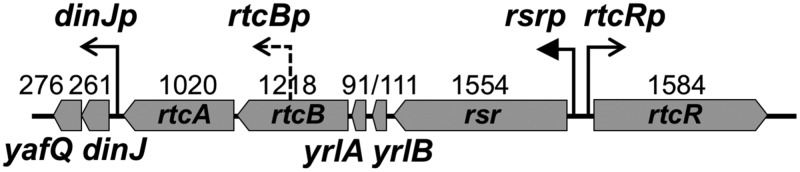
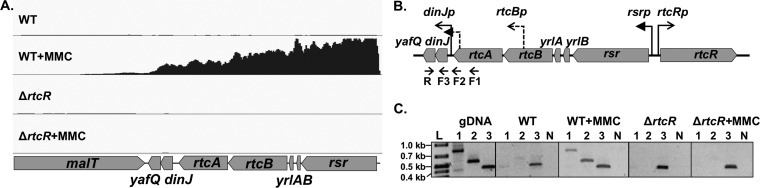
The σ54 regulon of S. Typhimurium includes a putative RNA repair operon (rsr-yrlBA-rtcBA) (4, 5) that encodes homologs of metazoan and archaeal RNA ligase (RtcB) and RNA 3′-phosphate cyclase (RtcA) (6) as well as metazoan Ro60 (Rsr) and Y RNAs (YrlA and YrlB) that form ribonucleoprotein complexes involved in noncoding-RNA quality control (7, 8) (Fig. 1). The environmental conditions that activate expression of the S. Typhimurium predicted RNA repair operon were heretofore uncharacterized, but previous work with constitutively active bEBP variants that contain deletions of the N-terminal regulatory region (the promiscuous DctD250 [4] and the enhancer-specific RtcR-ΔN [7]) showed that the promoter upstream of this operon, rsrp, is dependent on both σ54 and RtcR. The rtcR gene is divergently transcribed relative to the RNA repair operon from a σ70-dependent promoter, rtcRp (9, 10) (Fig. 1). Based on amino acid sequence similarity of the RtcR regulatory domain with the clustered regularly interspaced short palindromic repeat (CRISPR)-associated Rossmann fold (CARF) domains, it is predicted that RtcR is activated by binding of a nucleoside or modified nucleotide ligand (11).
FIG 1.
Organization of the RNA repair and dinJ-yafQ operons. The RNA repair operon carries rsr, yrlB, yrlA, rtcB, and rtcA, transcribed from a σ54-dependent promoter (rsrp). Transcription from rsrp is controlled by RtcR, whose gene is divergently transcribed from a σ70-type promoter (rtcRp). The toxin-antitoxin module, dinJ-yafQ, is carried immediately downstream of the RNA repair operon and is expressed from a σ70-type promoter (dinJp). A predicted σ70-type promoter within rtcB (rtcBp) generates a short transcript that does not extend into rtcA under the 22 infection-relevant growth conditions assessed by Kröger et al. (9). The length (in bp) is indicated above each gene.
Bacterial homologs of RtcB and RtcA were first identified in Escherichia coli, encoded within an RtcR-regulated σ54-dependent operon (12). Expression of the E. coli rtcBA operon is induced by conditions that disrupt translation, including treatment with ribosome binding antibiotics, such as minocycline (13). E. coli RtcB and RtcA exhibit the same biochemical activities as the metazoan homologs in vitro (6, 12, 14). Metazoan RtcB catalyzes the formation of a 3′-5′ phosphodiester linkage between RNA molecules bearing 2′,3′-cyclic phosphate or 3′-phosphate and 5′-hydroxyl termini (14, 15), and it functions in splicing of tRNAs (16) and XBP1 mRNA, which is required for the unfolded-protein response in animals (17). RtcA repairs 3′-phosphate or 2′-phosphate ends of cleaved RNA to 2′,3′-cyclic phosphates, which can serve as substrates for RtcB-mediated ligation (reviewed in reference 18), but the role for RtcA in RNA metabolic reactions in metazoans and bacteria is unknown. E. coli RtcB and RtcA have been shown to also utilize DNA substrates in vitro; RtcB adds a guanylyl “cap” to a 3′-phosphate end of nicked DNA (6, 19), and RtcA adenylylates DNA 5′-phosphate ends (20). In vivo analyses of RtcB activity in E. coli suggest that at least one substrate for RtcB RNA ligase activity is 16S rRNA (13, 21). The work of Temmel et al. (21) indicated that RtcB ligates cleaved 16S RNA within specialized ribosomes generated by endoribonuclease MazF of the MazEF toxin-antitoxin (TA) system in the stress response to amino acid starvation, suggesting a role for RtcB in recovery from the stress response; however, a recent study by Culviner and Laub (22) refutes the formation of specialized ribosomes in the MazF-mediated stress response, thus leaving the precise role of RtcB in E. coli unresolved.
RtcB, RtcA, and RtcR of S. Typhimurium share 88%, 68%, and 84% amino acid identity, respectively, with the E. coli proteins, but it is notable that the RNA repair operon of S. Typhimurium 14028s additionally encodes homologs of the Ro60 protein (Rsr) and Y RNAs (YrlA and YrlB), which are absent in the E. coli genome (6). In Deinococcus radiodurans, Rsr and YrlA form a ribonucleoprotein complex with polynucleotide phosphorylase (PNPase) to direct starvation-induced rRNA degradation (23), and Rsr functions with RNase PH and RNase II to process 23S rRNAs during growth at elevated temperature (24). In S. Typhimurium, Rsr and YrlA form a complex with PNPase, and Rsr associates with YrlB, but not within the Rsr-YrlA-PNPase complex (7). The functions of these ribonucleoprotein complexes in S. Typhimurium are unknown. Although the D. radiodurans genome encodes an RtcB homolog, rtcB is not in an operon with rsr; in addition, σ54 is not encoded in genomes of the Deinococcus genus (25). Therefore, analogies between S. Typhimurium and D. radiodurans cannot be made for the regulation of the RNA repair genes.
Defining the stress conditions and cellular pathways that regulate transcription of the RNA repair operon in S. Typhimurium will allow determination of the signal for RtcR activation and characterization of the physiological roles of the genes in the RNA repair operon. In this work, we show that exposure to direct nucleic acid-damaging agents, i.e., mitomycin C (MMC) or cisplatin (CDDP), results in high levels of RtcR-dependent transcription of the RNA repair operon, and this activation of RtcR requires RecA, most likely for its role in activating the SOS response. Peroxide stress, which also activates the SOS response, induces expression of the RNA repair operon that is mostly dependent on RtcR. However, nitrogen limitation results in expression of the RNA repair operon that is dependent on the bEBP NtrC instead of RtcR. Comparative transcriptome sequencing (RNA-seq) and Northern blot analyses indicate that the regulon of RtcR comprises primarily the RNA repair operon and the adjacent cotranscribed toxin-antitoxin (TA) module, dinJ-yafQ. Deletion of dinJ-yafQ does not prevent activation of RtcR, but overexpression of YafQ does result in a moderate level of RtcR-dependent expression of the RNA repair operon. An assay of cell viability following MMC treatment for wild-type (WT), ΔrtcR, and ΔrtcB strains indicates that RtcB functions in cell survival under these stress conditions. A direct comparison of conditions that activate RtcR and/or transcription of the rsr-yrlBA-rtcBA operon in S. Typhimurium versus the rtcBA operon in E. coli suggests that there are significant differences regarding cellular responses to different types of stress, resulting in differential expression of their respective RNA repair systems.
RESULTS AND DISCUSSION
Stress conditions and bEBPs that induce expression of the RNA repair operon.
The RNA repair operon, rsr-yrlBA-rtcBA, of S. Typhimurium has previously been shown to be expressed from a σ54-dependent promoter (rsrp) (4, 5) that can be activated by a constitutively active variant of the bEBP RtcR (7); nevertheless, conditions that activate the native RtcR to stimulate transcription of the operon have not been determined. Kröger and coworkers (9) performed RNA-seq-based analyses of the S. Typhimurium strain 4/74 transcriptome under a variety of infection-related stress conditions, including early- to late-stationary-phase growth and exposure to peroxide, nitric oxide, low/high pH/temperature, low Fe2+/Mg2+, or anaerobic/oxygen shock; no significant increase in transcription of the genes of the RNA repair operon (rsr, rtcB, or rtcA) was detected under any condition tested. However, in a microarray analysis of the ArcA regulon of S. Typhimurium strain 14028s by Morales et al. (26), transcript levels for rsr, rtcB, and rtcA increased 6.4-, 5.9-, and 2.1-fold, respectively, with peroxide treatment compared to those in the untreated control. The study did not assess the dependence of transcription on σ54 or RtcR, so it could be from an as-yet-unidentified σ70-type promoter or through activation of a bEBP other than RtcR. The difference in the expression levels of the RNA repair operon under peroxide stress between the studies by Kröger et al. (9) and Morales et al. (26) may reflect differences in the medium (phosphate-carbon-nitrogen [PCN] minimal medium and LB broth, respectively) and the timing of treatment with 1 mM H2O2 (12 min and 20 min, respectively).
Since the rsr-yrlAB-rtcBA operon encodes proteins predicted to be involved in RNA repair, processing, and/or degradation, we hypothesized that conditions causing RNA cleavage or damage are likely to generate the signal to activate RtcR and, in turn, transcription of the operon. Thus, transcription of rtcA or rtcB was assayed by quantitative reverse transcriptase PCR (qRT-PCR) under stress conditions that are known to cause nucleic acid cleavage/damage either directly or indirectly (Fig. 2): exposure to mitomycin C (MMC), peroxide stress, translation inhibition by chloramphenicol (CHL), cell wall synthesis inhibition by cefotaxime (CTX), nitrogen limitation, carbon starvation, amino acid starvation, and iron limitation. Each of these stress conditions has widespread effects on cellular processes; their impact on nucleic acids is highlighted here. MMC, a member of the mitomycin antibiotic family, is enzymatically reduced in the cell to a bifunctional alkylating agent that reacts with deoxyguanosine (dG) to form MMC-mono-dG adducts, intrastrand biadducts at -GpG-, and interstrand cross-links within the sequence 5′-CpG-3′ in DNA (reviewed in reference 27) and directly modifies RNA, including formation of RNA-MMC adducts (28). A primary result of MMC treatment of cells is the degradation of rRNA and ribosome decomposition (29, 30). H2O2, which is generated in the bacterial cell through aerobic metabolism and produced by host neutrophils and macrophages during the oxidative burst in response to S. Typhimurium infection (reviewed in references 31 and 32), causes direct damage to nucleic acids through oxidants produced in the Fenton reaction (33, 34) and leads to turnover of rRNA (35). Nitrogen, carbon, amino acid, and iron limitation also causes ribosome degradation and turnover of rRNA (36). Treatment with bactericidal antibiotics, including cefotaxime and chloramphenicol, can lead to substantial oxidative damage through a cellular death pathway (37, 38). In addition, treatment with chloramphenicol and MMC, as well as carbon and amino acid starvation, induces bacterial TA systems in which ribonucleolytic toxins cleave mRNA, tRNA, or rRNA (reviewed in reference 39). The S. Typhimurium genome (chromosome and pSLT virulence plasmid) encodes at least 19 type II and seven type I TA systems (40), some of which cleave RNA molecules, yielding 2′,3-cyclic phosphate ends and 5′-OH ends that are the substrates for RtcB ligase activity (15).
FIG 2.
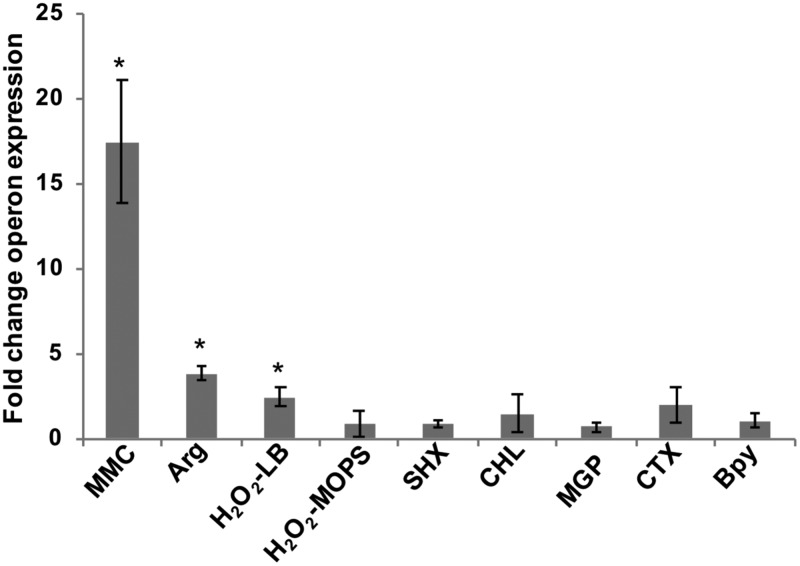
Expression of the RNA repair operon under conditions that directly or indirectly cause RNA damage. Relative transcript levels for the RNA repair operon were assayed by qRT-PCR for WT S. Typhimurium cultures at mid-log phase, treated versus untreated with the following stress conditions: 3 μM mitomycin C (MMC), 2.5 mM Arg as the sole nitrogen source, 1 mM H2O2 in LB or MOPS minimal medium, 0.4 mg/ml serine hydroxamate (SHX), 30 μg/ml chloramphenicol (CHL), 1% methyl-α-d-glucopyranoside (MGP), 2 μg/ml cefotaxime (CTX), or 250 μM 2,2′-bipyridyl (Bpy). The fold change operon expression is the ratio of normalized operon transcript levels for treated versus untreated samples (*, P < 0.02). For all stress conditions except peroxide stress, rtcA transcript levels were normalized to rpoD transcript levels; for peroxide stress conditions, rtcB transcript levels were normalized to kdgR transcript levels. Data shown are from three biological replicates for each growth condition; error bars represent ±1 standard deviation. The positive control in these qRT-PCR assays was the WT strain containing a multicopy expression vector encoding a constitutively active variant of RtcR (pDS183); the WT(pDS183) strain showed an 83-fold-higher level of rtcA expression than the untreated WT strain (P < 0.01) (these data are not shown in the graph).
MMC treatment produced the largest increase in expression of the RNA repair operon, 17-fold upregulation (P < 0.01); peroxide stress and nitrogen limitation resulted in more modest increases in RNA repair operon transcript levels of 2.5-fold (P < 0.02) and 3.5-fold (P < 0.02), respectively (Fig. 2). Consistent with previous S. Typhimurium transcriptome data, treatment for 20 min with 1 mM H2O2 in LB broth resulted in increased expression of the RNA repair operon, as was seen in the study by Morales et al. (26), but such treatment in morpholinepropanesulfonic acid (MOPS) minimal medium, which is similar to the PCN minimal medium used by Kröger and coworkers (9), did not (Fig. 2). As described in Materials and Methods, the qRT-PCR assays for expression of the RNA repair operon utilized primer pairs to measure transcript levels for rtcA relative to the stably expressed reference gene rpoD under all conditions except peroxide stress. Because expression of rpoD changed under peroxide stress, another reference gene whose expression was not affected by peroxide treatment, kdgR, was utilized; to measure the RNA repair operon transcript levels, a primer pair for amplification of rtcB was used.
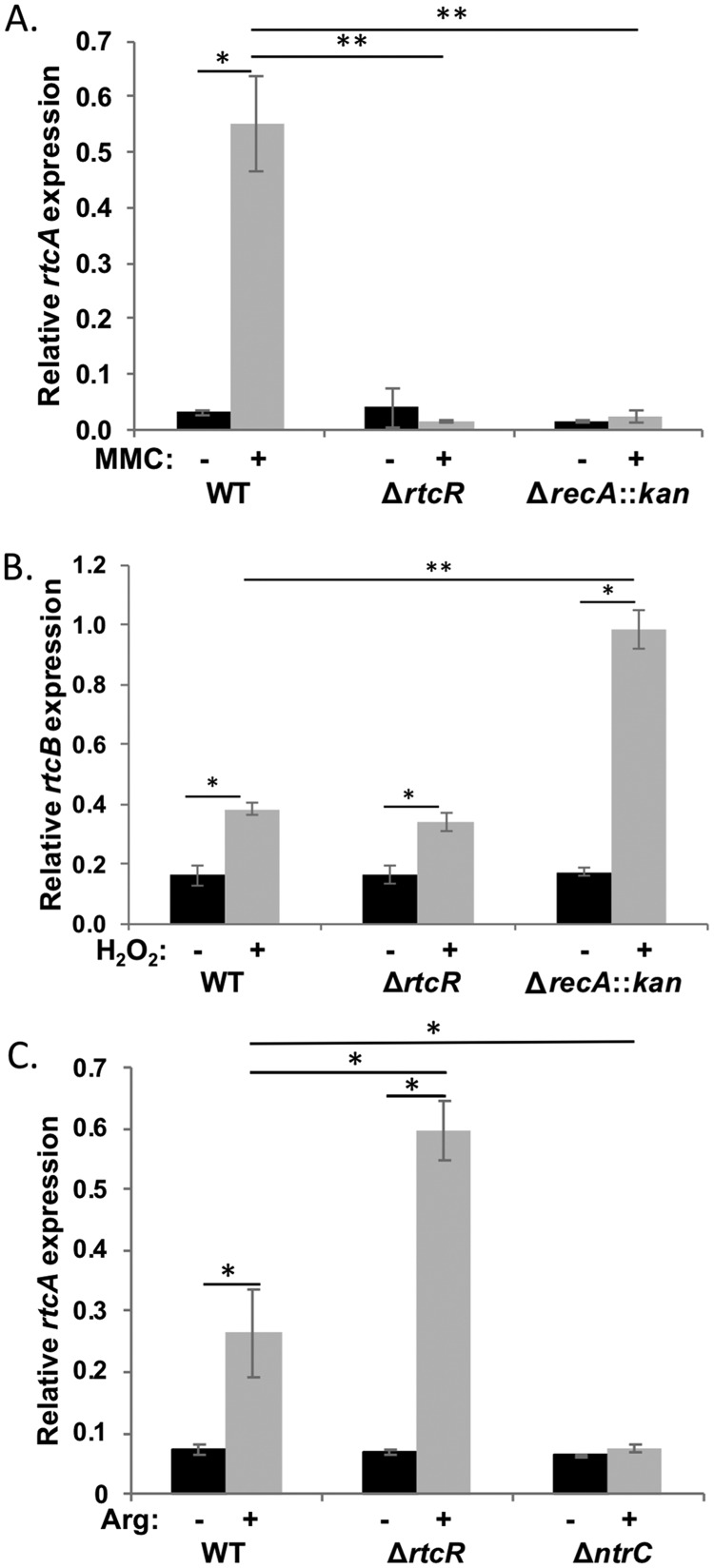
To determine whether the upregulation of RNA repair operon expression under these three stress conditions is dependent on RtcR, transcript levels for rtcA or rtcB were measured in a ΔrtcR mutant and a WT strain (Fig. 3). MMC-induced expression required RtcR. There was no increase in rtcA expression for the ΔrtcR strain with MMC treatment; the level of rtcA expression in the MMC-treated ΔrtcR strain was 36-fold lower than that in the MMC-treated WT strain (P < 0.001) (Fig. 3A). Treatment with 1 mM H2O2 upregulated expression of rtcB in the ΔrtcR strain by 2.1-fold (P < 0.01), which is not significantly different from the H2O2-induced expression in the WT strain (Fig. 3B), indicating that this level of expression of rtcB under peroxide stress is not RtcR dependent. Under nitrogen limitation, the RNA repair operon was induced 8.8-fold in the ΔrtcR strain (P < 0.01) and the transcript level for rtcA was 2.2-fold higher than that in the WT strain (P < 0.01) (Fig. 3C), revealing that upregulation of the RNA repair operon under nitrogen-limiting conditions is actually lower in the presence of RtcR.
FIG 3.
Assessment of RtcR and RecA dependence of RNA repair operon expression induced by MMC, peroxide stress, and nitrogen limitation. (A) Transcript levels of rtcA and the reference gene rpoD were assessed by qRT-PCR for mid-log-phase cultures of S. Typhimurium 14028s WT, ΔrtcR, and ΔrecA::kan strains in MOPS medium, untreated and treated for 90 min with 3 μM MMC; rtcA transcript levels were normalized to rpoD transcript levels (relative rtcA expression). (B) Transcript levels of rtcB and the reference gene kdgR were assessed by qRT-PCR for mid-log-phase cultures of S. Typhimurium 14028s WT, ΔrtcR, and ΔrecA::kan strains in LB, untreated and treated for 20 min with 1 mM H2O2; rtcB transcript levels were normalized to kdgR transcript levels (relative rtcB expression). (C) Cultures of S. Typhimurium 14028s WT, ΔrtcR, and ΔntrC strains were grown to mid-log phase in MOPS medium with ammonium chloride as the nitrogen source; cultures were then split, and cells were washed and resuspended in MOPS medium with either 14.3 mM ammonium chloride (−Arg) or 2.5 mM arginine (+Arg) as the nitrogen source. After 90 min of treatment, total RNA was harvested, and transcript levels of rtcA were assessed by qRT-PCR and normalized to rpoD transcript levels (relative rtcA expression). For all panels, significant differences in relative rtcA or rtcB expression between treated and untreated samples and/or between strains are indicated (*, P < 0.02; **, P < 0.003). Data shown are for three biological replicates for each strain; error bars represent ±1 standard deviation.
As shown in Fig. 1, there are two promoters associated with the RNA repair operon: the σ54-dependent promoter (rsrp), which is the only known promoter that expresses the full operon (4, 5, 9), and the σ70-type promoter within rtcB (rtcBp). The dependence on the bEBP RtcR for MMC-induced expression of rtcA at the end of the operon (Fig. 3A) indicates that transcription is from rsrp and is supported by experiments presented in the next two sections. Since increased expression of rtcB and rtcA under H2O2 treatment and nitrogen-limiting conditions, respectively, was not RtcR dependent, transcription of the operon was further characterized for these conditions to resolve which operon-associated promoter is activated. H2O2-induced expression is assessed further in the next section. Whether nitrogen limitation induces expression of rtcA from rtcBp or expression of the operon from rsrp is further addressed here. Since rsrp is a σ54-dependent promoter, it requires a bEBP to activate expression of the operon. NtrC is a bEBP that is activated by nitrogen limitation and controls expression of several σ54-dependent operons whose products allow growth during nitrogen starvation (41, 42). Because rtcA was upregulated by nitrogen limitation but independently of RtcR (Fig. 3C), we speculated that NtrC could be required for upregulation of the RNA repair operon during nitrogen limitation. To determine if that is the case, a ΔntrC strain was utilized in the qRT-PCR assays. Under nitrogen-limiting conditions, the expression of rtcA in the ΔntrC strain was nearly unchanged relative to that under nitrogen-excess conditions and was 3.5-fold lower than that in the WT strain (P < 0.02) (Fig. 3C). To ensure that NtrC was activated by the nitrogen-limiting conditions, expression of NtrC-regulated glnK (41) was measured by qRT-PCR (see Fig. S1 in the supplemental material). Expression of glnK was significantly increased by the treatment in WT cells (28-fold, P < 0.0001), and this induction of glnK expression was NtrC dependent, as indicated by the 1,336-fold-lower expression of glnK in the ΔntrC strain versus the WT under nitrogen-limiting conditions (P < 0.0001) (Fig. S1). Relative transcript levels for the first gene in the RNA repair operon, rsr, were assessed by qRT-PCR for the WT strain grown under nitrogen-limiting and nitrogen-rich conditions; rsr transcription was upregulated under nitrogen-limiting conditions by 3.1-fold (±1.1; P < 0.03). Taken together, these results indicate that expression of the RNA repair operon is from rsrp and regulated by NtrC, under nitrogen starvation. The regulation by NtrC may be direct or indirect, but the increased expression of rtcA in the absence of RtcR under nitrogen-limiting conditions (Fig. 3C) is consistent with overlapping enhancer binding sites for NtrC and RtcR. The enhancer sites for RtcR have not yet been defined in any bacteria, and only a few NtrC binding sites have been assessed in S. Typhimurium (43). To predict potential NtrC binding sites in the S. Typhimurium genome, including any that may be associated with the RNA repair operon, the motif-based algorithm Motif Locator (44) was utilized to search the S. Typhimurium genome sequence for motif sites based on a position-specific score matrix generated from DNA sequence motifs associated with NtrC binding in E. coli (see Materials and Methods). Of the 165 motif sequences in the S. Typhimurium genome identified by the algorithm (see Data Set S1 in the supplemental material), one is within the rsr gene in the RNA repair operon (GenBank accession number CP001363.1, genome position 3697006 to 3697025 [GTTGCGCTTTTTCAGTTCGA]), which could regulate the operon promoter since bEBP-enhancer complexes act on Eσ54 at the promoter from a distance by DNA looping (45; reviewed in reference 3). As the potential interactions between the NtrC and RtcR regulons in S. Typhimurium are further explored, the genomic binding sites for both of these bEBPs will be physically defined and characterized.
The activation of RNA repair operon transcription by NtrC is likely to be linked to the potential role of Rsr in the starvation stress response. In D. radiodurans, Rsr acts in complex with the exoribonuclease polynucleotide phosphorylase (PNPase) during stationary phase and interacts with ribosomes to play a role in starvation-induced rRNA degradation (23). Rsr in S. Typhimurium has also been shown to associate with PNPase in a ribonucleoprotein complex that includes YrlA (7); thus, the increased expression of rsr under nitrogen-limiting conditions may be part of a starvation stress response that directs degradation of ribosomes and rRNA (36).
A RecA-mediated process is involved in RtcR-mediated activation of the RNA repair operon.
MMC and H2O2, each of which causes direct damage to nucleic acids, both induced expression of the RNA repair operon in S. Typhimurium (Fig. 2 and 3A and B). As described previously, reduced MMC alkylates guanines and causes inter- and intrastrand cross-links in DNA (27), while H2O2 generates reactive species that cause base oxidation and single- and double-stranded breaks in nucleic acids (33); all of these DNA lesions can result in induction of the SOS response (46, 47). The SOS response (reviewed in reference 48) is a DNA repair network that is widespread in bacteria and includes many proteins involved in sensing and repairing DNA damage. Expression of genes in the SOS network is regulated by the LexA repressor; induction of the SOS response occurs when LexA undergoes RecA-dependent cleavage. RecA is activated to stimulate LexA autocleavage when it forms filaments on single-stranded DNA that is exposed during processing of nicks/breaks in double-stranded DNA or when DNA lesions block replication/transcription. The SOS response is proportionate to the extent of the DNA damage, and there is a temporal modulation of SOS response functions (e.g., nucleotide excision repair, homologous recombination, and DNA repair polymerase) in the bacterial cell (48–50).
To determine whether the SOS response is involved in signaling for activation of transcription of the RNA repair operon, qRT-PCR was performed to compare transcription in WT and ΔrecA::kan S. Typhimurium 14028s strains that were untreated or treated with MMC or H2O2 (Fig. 3A and B). Transcription of the RNA repair operon in the ΔrecA::kan strain was not upregulated after MMC treatment, and the level of rtcA expression in the treated ΔrecA::kan strain was 24-fold lower than that in the treated WT strain (P < 0.001) (Fig. 3A). However, expression of the RNA repair operon following treatment with H2O2 was induced 5.6-fold in the ΔrecA::kan strain (P < 0.003), and the level of rtcB transcription in treated ΔrecA::kan strain samples was 2.5-fold higher than that in the treated WT strain (P < 0.01) (Fig. 3B). These results indicate that a RecA-mediated process, most likely induction of the SOS response, is needed for the RtcR-dependent 17-fold increase in RNA repair operon expression following MMC treatment but not for the H2O2-induced 2.5-fold increase in expression of the RNA repair operon, which is independent of RtcR (Fig. 3). Pattern Locator (51) was utilized to search the rsr-rtcR intergenic region, containing the σ70-type rtcRp and the σ54-dependent rsrp, for LexA binding site motifs (52, 53). No predicted LexA binding site was identified in this region, and rtcR transcription is unchanged upon treatment with MMC (see the RNA-seq analysis described below), suggesting that the SOS response is indirectly involved in inducing the operon, perhaps generating the signal for activation of RtcR.
Since RtcR appears to be activated by the RecA-dependent SOS response following MMC treatment, it is surprising that peroxide stress, which also induces the SOS response, did not activate RtcR to stimulate expression of rtcB. Two plausible reasons for why the peroxide treatment does not activate RtcR are proposed here and examined below: (i) the type of DNA damage induced by MMC results in the signal for RtcR activation during the SOS response, such as the release of MMC-DNA adducts during nucleotide excision repair, or (ii) the level of DNA damage, which modulates the timing and intensity of expression of genes involved in the SOS response (48, 50, 54), is not sufficient from treatment with 1 mM H2O2 for 20 min to induce expression of the SOS response pathway that generates the signal for activation of RtcR. This level of peroxide treatment induces expression of the OxyR regulon, which results in scavenging of reactive oxygen species that cause DNA lesions (55); thus, it is likely that there is a limited SOS response during the peroxide treatment. The enhanced expression of the RNA repair operon in the ΔrecA::kan strain under peroxide stress is consistent with increased oxidative DNA damage since RecA-mediated recombination is central to the protection provided by the SOS response to this level of H2O2 exposure (47). RtcB DNA-capping activity may be the operon-encoded function that is needed in the absence of RecA-mediated repair. A 3′-PO4 end at a DNA break cannot be repaired by classical DNA ligases or extended by DNA polymerases, including the error-prone DNA polymerases of the SOS response; RtcB transfers GMP from a covalent RtcB-GMP intermediate to the 3′-PO4 terminus to generate a DNA 3′pp5′G cap, which blocks exonuclease activity and can be extended by DNA polymerases, such as polymerase II (Pol II) in SOS repair (19).
To test whether MMC-specific nucleic acid damage, such as MMC-DNA adducts, is responsible for the activation of RtcR, expression of the RNA repair operon in WT, ΔrtcR, and ΔrecA::kan S. Typhimurium xylE reporter strains was assessed following treatment with cisplatin [cis-diaminedichloroplatinum(II)] versus MMC. Cisplatin causes cross-links in nucleic acids similar to those seen with MMC and induces the SOS response (56, 57), but cisplatin is structurally and reactively dissimilar to MMC (58). In the cell, the chloride atoms of cisplatin are replaced by H2O, and the aquated cisplatin species react with N-7 atoms of purines in DNA and RNA, leading to intramolecular cross-links and, to a lesser extent, intermolecular cross-links (56, 57). The xylE reporter strains used in this assay have rsr replaced with xylE (Δrsr::xylE) so that XylE activity reflects the transcriptional/translational regulation of the first gene in the RNA repair operon on the chromosome of S. Typhimurium 14028s. As shown in Fig. 4, the MMC-induced changes in XylE expression measured for the xylE reporter strains were nearly identical to those seen for rtcA expression in the qRT-PCR assays (Fig. 3A), with an 18-fold increase (P < 0.004) in XylE expression in the WT strain and no significant change in XylE expression in either the ΔrtcR or ΔrecA::kan mutant following MMC treatment (Fig. 4); these results indicate that rsrp is responsible for increased transcription of the full operon under nucleotide-damaging conditions. Treatment of the WT xylE reporter strain with cisplatin upregulates expression of XylE by 6-fold (P < 0.004), while treatment of the ΔrtcR and ΔrecA::kan S. Typhimurium xylE reporter strains results in no significant increase in expression of XylE. These results indicate that cisplatin-mediated nucleic acid damage also activates expression of the RNA repair operon in an RtcR- and RecA-dependent manner. Thus, the signal for RtcR activation is not specifically MMC-DNA adducts but is likely to be dependent on a strong induction of the SOS response. The level of expression from rsrp is dose dependent for MMC and cisplatin, showing a 2.1-fold increase in expression from 1.5 μM to 3 μM MMC and a 2.2-fold increase in expression from 55 μM to 110 μM cisplatin (see Fig. S2 in the supplemental material), which is consistent with the level of DNA damage correlating with the induction of the SOS response and activation of RtcR.
FIG 4.
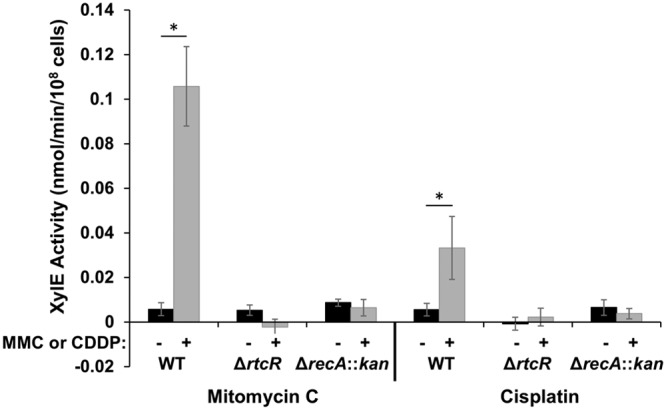
Comparison of RtcR and RecA dependence of RNA repair operon expression induced by MMC and cisplatin. Cultures of S. Typhimurium 14028s WT, ΔrtcR, and ΔrecA::kan reporter strains, which have xylE replacing the first gene of the RNA repair operon, were grown to mid-log phase in LB medium and split into portions that were untreated or treated with 3 μM MMC for 90 min or 110 μM cisplatin for 180 min. After treatment, the cells were harvested and assayed for XylE (catechol dioxygenase) activity over a period of time, as described in Materials and Methods. The activity is expressed in nanomoles of catechol oxidized per minute per 108 cells. Significant differences in XylE activity between treated and untreated samples are indicated (*, P < 0.004). Data shown are the average from at least three biological replicates for each strain; error bars represent ±1 standard deviation.
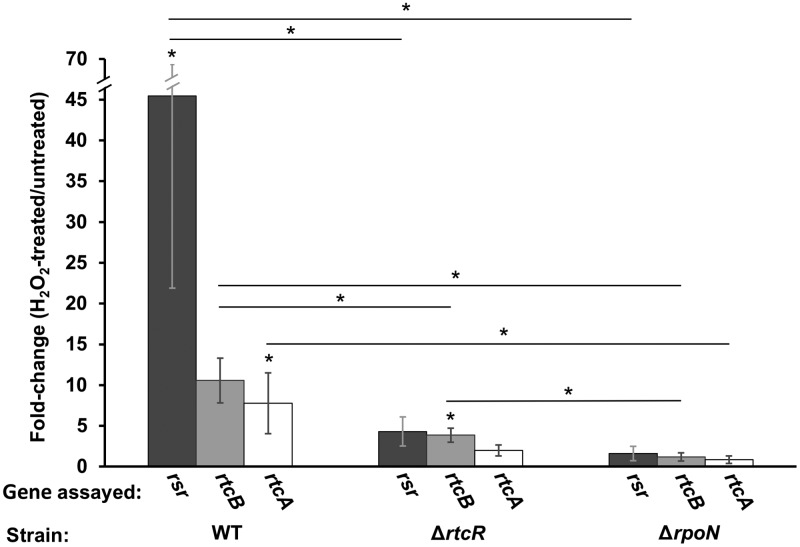
To examine the second proposed reason for why the 1 mM H2O2 treatment did not activate RtcR to express rtcB (Fig. 3B), i.e., the level of DNA damage from this peroxide treatment was not sufficient to generate the SOS-dependent signal for activation of RtcR, expression of rsr, rtcB, and rtcA was measured by qRT-PCR after a 20 min treatment with 7 mM H2O2 for WT, ΔrtcR, and ΔrpoN strains. This higher concentration of H2O2 has been shown to result in increased expression of SOS response genes (59). Following treatment with 7 mM H2O2, transcript levels for rsr, rtcB and rtcA increased in the WT strain by 41-, 11- and 7.2-fold, respectively (P values of 0.02, 0.06, and 0.03, respectively) (Fig. 5). This increase in expression of the full RNA repair operon was mostly dependent on RtcR; the fold change values for transcript levels of rsr, rtcB, and rtcA following H2O2 treatment were 11-, 2.7-, and 3.9-fold lower, respectively, in the ΔrtcR strain than in the WT strain (P values of 0.04, 0.02, and 0.06, respectively) (Fig. 5). Although expression of the operon was significantly reduced in the absence of RtcR, the transcript levels for rsr, rtcB, and rtcA increased following peroxide treatment (4.0-, 3.9-, and 1.9-fold, respectively; P values of 0.06, 0.04, and 0.1, respectively) (Fig. 5), which is consistent with the RtcR-independent expression seen for rtcB following 1 mM H2O2 treatment (Fig. 3B). However, all of the H2O2-induced expression of the RNA repair operon required σ54, since there was no significant change in transcript levels for rsr, rtcB and rtcA in the H2O2-treated versus untreated ΔrpoN strain (1.6-, 1.2-, and 0.86-fold changes; P values of 0.4, 0.7, and 0.6, respectively) (Fig. 5). Comparison of the fold change values for rsr, rtcB, and rtcA in the WT and ΔrpoN strains shows a significant decrease in H2O2-induced expression of all three genes in the absence of σ54 (28-, 8.9-, and 9.0-fold decreases, respectively; P values < 0.04). These results indicate that the full RNA repair operon is expressed from the σ54-dependent rsrp under peroxide stress and that the σ70-type promoter within rtcB is not responsible for the observed significant increase in transcript levels of rtcB or rtcA. RtcR- and σ54-dependent transcription from rsrp following treatment with 1 mM H2O2 was confirmed by qRT-PCR assays for rsr expression in WT, ΔrtcR, and ΔrpoN strains (see Fig. S3 in the supplemental material).
FIG 5.
Analysis of RtcR- and RpoN-dependent expression of the RNA repair operon genes during high-level peroxide exposure. Cultures of 14028s WT, ΔrtcR, or ΔrpoN strains were grown to mid-log phase in LB medium and split into two cultures; one culture was treated with 7 mM H2O2 for 20 min, while the other remained untreated. Transcript levels for rsr, rtcB, and rtcA were assessed by qRT-PCR, normalized to expression of the reference gene kdgR. The data shown denote the fold increase in gene expression for H2O2-treated versus untreated cells. Significant differences between treated and untreated cells and between different strains are indicated (*, P < 0.04); error bars represent ±1 standard deviation.
The RtcR-independent, σ54-dependent expression of RNA repair operon genes during peroxide stress suggests that an additional bEBP controls transcription from rsrp. In S. Typhimurium there are 13 identified bEBPs, including RtcR and NtrC, that control expression of the genes in the σ54 regulon (2, 4, 5). Analysis of the microarray data for S. Typhimurium 14028s transcriptome changes under peroxide stress after treatment with 1 mM H2O2 (26) revealed that the only significantly upregulated genes from the S. Typhimurium σ54 regulon were the genes of the RNA repair operon; thus, it appears that RtcR is the only known bEBP that is activated under this level of peroxide stress, since none of the genes that are controlled by the other bEBPs were significantly upregulated in expression.
RtcR-dependent transcriptome and RNA repair operon structure in S. Typhimurium.
The conditions that induce RtcR-dependent expression of the RNA repair operon are likely to produce the damaged nucleic acid substrate for the RNA repair proteins, as well as the signal for RtcR activation that may be generated from the damaged substrate. The impact of RtcR and the RNA repair operon on the transcriptome of S. Typhimurium was examined in the presence and absence of MMC-mediated stress using high-throughput transcriptome sequencing (RNA-seq). A strain deleted for rtcR, as well as strains deleted for structural components of the RNA repair system, rsr and rtcB, were utilized in the RNA-seq analysis. Total RNA was isolated from S. Typhimurium 14028s WT, ΔrtcR, Δrsr, and ΔrtcB strains cultured in the presence and absence of 3 μM MMC for 60 min and used to construct cDNA libraries for nonstranded RNA-seq after rRNA depletion. The libraries were sequenced on the Illumina Next-seq500 platform, and reads were mapped to the S. Typhimurium 14028s genome using Bowtie2 (60); the statistics for the RNA-seq are shown in Table S1 in the supplemental material. Cuffnorm (61) was used to obtain the number of fragments per kilobase of transcript per million fragments mapped (FPKM) for each biological replicate, and Pearson product-moment correlation coefficients (r) between the biological replicates for each strain and treatment condition were calculated (see Materials and Methods and Table S2 in the supplemental material). Pairwise comparisons of combined data for correlating biological replicates were analyzed with Cuffdiff (62); statistically significant differential gene expression met the criteria of a q value of <0.05 and a fold change of >3 (log2 fold change >1.585).
Activation of the SOS response by the MMC treatment used in these expression assays was confirmed by comparative analysis of values for the WT plus MMC (WT+MMC) versus WT, in which 453 gene transcripts were >3-fold upregulated and 95 gene transcripts were >3-fold downregulated (see Data Set S2 in the supplemental material). Of the 453 gene transcripts upregulated by MMC treatment, 22 genes overlap the 32 SOS response genes that have been shown to be directly regulated by LexA in E. coli (Table 1) (49, 63). In addition, four of the 26 TA systems encoded in the S. Typhimurium genome and pSLT virulence plasmid (40) were >3-fold upregulated: STM14_4236-STM14_4235 (yafQ-dinJ, type II), which is located immediately downstream of rtcA in the RNA repair operon; STM14_4402-STM14_4401 (tacT-tacA, type II); STM14_4845 (sehA of sehA-sehB, type II); and ysdB-ysdA (tisB-istR, type I) (Data Set S2). YafQ and SehA are both predicted RNase toxins that yield RNA cleavage products with 2′,3-cyclic phosphate ends and 5′-OH ends (64, 65), which are substrates for RtcB ligase activity (15). The comparative analysis of WT+MMC versus WT also confirmed that expression of rtcR from the σ70-type rtcRp was not significantly changed by MMC treatment (1.37-fold decrease; q value, 0.37), while expression of the RNA repair operon in the WT strain treated with MMC was highly induced (rsr, rtcB, and rtcA increased 191-, 158-, and 87-fold, respectively), suggesting activation of RtcR to stimulate transcription from σ54-dependent rsrp. A previous report by Benson et al. (66) that characterized the MMC-induced SOS response in S. Typhimurium using RNA fingerprinting by arbitrarily primed PCR did not identify the RNA repair operon as induced; however, only 20 genes were identified as upregulated, of which only two genes are in the LexA regulon. The difference in these results and our RNA-seq data reflects the disparity in the sensitivity of the assays for differential gene expression.
TABLE 1.
Known LexA-regulated genes induced by treatment with MMC in WT S. Typhimurium 14028s
| Locus tag | Genea | Log2 fold changeb |
|---|---|---|
| STM14_0117 | polB | 2.02 |
| STM14_0369 | dinP | 2.82 |
| STM14_0926 | uvrB | 1.78 |
| STM14_0953 | dinG | 1.80 |
| STM14_1215 | sulA | 4.48 |
| STM14_1331 | dinI | 4.62 |
| STM14_1589 | ydjQ | 2.97 |
| STM14_1605 | ydjM | 1.69 |
| STM14_2287 | yebG | 6.55 |
| STM14_2303 | ruvB | 3.36 |
| STM14_2304 | ruvA | 4.46 |
| STM14_2422 | umuC | 5.55 |
| STM14_2423 | umuD | 6.03 |
| STM14_2551 | sbmC | 4.66 |
| STM14_3289 | recN | 4.24 |
| STM14_3417 | recA | 4.11 |
| STM14_4583 | ysdA | 6.77 |
| STM14_4584 | ysdB | 6.70 |
| STM14_5094 | lexA | 3.17 |
| STM14_5095 | dinF | 3.62 |
| STM14_5112 | uvrA | 2.06 |
| STM14_5114 | ssbB | 3.14 |
The 22 genes listed here exhibited significant (>3-fold) differential gene expression and overlap 32 SOS response genes known to be controlled by LexA in E. coli (49, 63). Five of the E. coli SOS response genes (hokE, hlyE, molR, dinQ, and dinD) have no homolog in S. Typhimurium 14028s, and expression of five E. coli LexA-regulated genes (ybfE, ftsK, dinS, uvrD, and symE) was not increased by >3-fold.
Differential expression of S. Typhimurium 14020s WT+MMC versus untreated WT with P values of <0.05.
The gene transcripts that were significantly up- or downregulated in ΔrtcR+MMC compared to WT+MMC (Table 2) reflect transcriptional regulation of gene expression by RtcR or regulation of transcript levels by the products of RtcR-dependent operons. The differentially expressed genes in ΔrtcR+MMC versus WT+MMC comprise the following: the genes of the RNA repair operon, rsr, rtcB, and rtcA; the dinJ-yafQ TA operon and malT, which are located immediately adjacent to the RNA repair operon (Fig. 6A); rhoL (a pseudogene within leader transcript region of the Rho transcription termination factor gene); rpmE2 (encoding alternative 50S ribosomal protein L31); STM14_1523 (encoding a hypothetical protein); and the tRNA genes valV and gltUVW. The Y-like RNA genes, yrlA and yrlB, which are part of the RNA repair operon, are not annotated in the S. Typhimurium genome and so do not appear in the assigned reads. Since the relative transcript levels for rsr (313-fold down) and rtcB (227-fold down) were very low in ΔrtcR+MMC versus WT+MMC, gene transcripts that are significantly up- or downregulated in ΔrtcR+MMC versus WT+MMC and in Δrsr+MMC or ΔrtcB+MMC versus WT+MMC are likely to be regulated by Rsr or RtcB and not directly regulated by RtcR. rhoL was downregulated in both Δrsr+MMC and ΔrtcR+MMC, and rpmE2, STM14_1523, and gltUVW were downregulated in both ΔrtcB+MMC and ΔrtcR+MMC. This comparison of the RNA-seq results suggests that the RtcR regulon in S. Typhimurium comprises the RNA repair operon, the dinJ-yafQ operon, and possibly valV. Although malT appeared to be downregulated in the absence of RtcR under MMC-induced stress conditions, visualization of the mapped reads with Integrated Genome Viewer showed that this is most likely due to read-through transcription from rsrp into the 3′ end of the convergently transcribed malT gene (Fig. 6A). A limitation of the protocol used for the RNA-seq is that strand-specific analysis cannot be performed; as a consequence, the regulation of small RNAs (sRNAs) by RtcR, RtcB, or Rsr could not be assessed. Comparative analysis of gene expression for the mutant strains versus the WT when not treated with MMC is provided in Data Set S3 in the supplemental material; there are only a few gene transcripts that are significantly up- or downregulated in both the untreated mutant strains versus WT and the MMC-treated mutant strains versus WT+MMC: rhoL in the Δrsr mutant versus the WT and gltUW and argX in the ΔrtcB mutant versus the WT. Engl et al. (13) performed comparative RNA-seq analysis for E. coli ΔrtcB and ΔrtcA mutants versus the WT strain in the absence of cellular stress (13); the only overlap seen with the RtcR- or RtcB-dependent changes in gene expression in S. Typhimurium (untreated or treated with MMC) was the downregulation of serW (encoding Ser-tRNA), gltU (encoding Gln-tRNA), ffs (encoding 4.5S RNA of the signal recognition particle), and ugpA (encoding a subunit in the ABC transporter) in the ΔrtcB mutant compared to the WT. The differences between the RNA repair systems of E. coli and S. Typhimurium are addressed below.
TABLE 2.
Differentially expressed genes in RNA-seq comparative analysis of RNA repair mutants and WT S. Typhimurium 14028s under MMC-induced stressa
| Strains and locus tag | Gene | Protein function | Log2 fold change |
|---|---|---|---|
| ΔrtcR+MMC vs WT+MMC | |||
| STM14_0553 | rpmE2 | 50S ribosomal protein L31 type B | −2.31 |
| STM14_1523 | Hypothetical protein | −2.14 | |
| STM14_1721 | valV | tRNA-Val | −8.64b |
| STM14_3257 | gltW | tRNA-Glu | −3.27 |
| STM14_4234 | malT | Transcriptional regulator of maltose regulon | −2.51 |
| STM14_4235 | yafQ | Toxin of YafQ-DinJ TA system | −4.89 |
| STM14_4236 | din | Antitoxin to YafQ | −5.11 |
| STM14_4237 | rtcA | RNA repair, RNA 3′-terminal-phosphate cyclase | −6.79 |
| STM14_4238 | rtcB | RNA repair, RNA-splicing ligase | −7.83 |
| STM14_4239 | rsr | RNA repair, Ro 60-related ribonucleoprotein | −8.29 |
| STM14_4687 | gltU | tRNA-Glu | −2.70 |
| STM14_4714 | rhoL | Pseudogene in rho operon leader region | −1.85 |
| STM14_5022 | gltV | tRNA-Glu | −2.88 |
| Δrsr+MMC vs WT+MMC | |||
| STM14_4101 | Hypothetical protein | −1.61 | |
| STM14_4714 | rhoL | Pseudogene in rho operon leader region | −1.97 |
| STM14_4813 | Putative cytoplasmic protein | −1.70 | |
| STM14_2542 | pduS | Propanediol utilization polyhedral body protein | 1.94 |
| STM14_2686 | Putative sugar transporter | 1.59 | |
| ΔrtcB+MMC vs WT+MMC | |||
| STM14_0553 | rpmE2 | 50S ribosomal protein L31 type B | −2.38 |
| STM14_0554 | rpmJ_1 | 50S ribosomal protein L36 | −1.62 |
| STM14_1523 | Hypothetical protein | −2.39 | |
| STM14_2730 | Hypothetical protein | −6.81b | |
| STM14_3257 | gltW | tRNA-Glu | −2.34 |
| STM14_3536 | ygbE | Putative cytochrome oxidase | −1.92 |
| STM14_3551 | cysH | Phosphoadenosine phosphosulfate reductase | −1.64 |
| STM14_3588 | Hypothetical protein | −3.54b | |
| STM14_4424 | Hypothetical protein | −3.90b | |
| STM14_4687 | gltU | tRNA-Glu | −2.76 |
| SM14_5022 | gltV | tRNA-Glu | −2.01 |
| STM14_1645 | Hypothetical protein | 4.19b | |
| STM14_4729 | argX | tRNA-Arg | 1.80 |
Genes >3-fold differentially expressed with a q value of <0.05, arranged by locus tag with downregulated genes listed first. The log2 fold change for the deleted gene in each mutant strain was −8.1 to −12 (not included in the table).
The FPKM in the mutant strain was 0; a FPKM value of 1 was assigned to calculate a value for log2 fold change.
FIG 6.
Transcript analysis of RNA repair and dinJ-yafQ operons. RNA-seq data and endpoint RT-PCR analysis were used to assess the extent of the RtcR-dependent transcript initiated from rsrp. (A) RNA-seq data for the genome region carrying contiguous genes which showed RtcR-dependent upregulation are presented as reads mapped with Bowtie2 in the Integrated Genome Viewer (IGV). The y axis for each panel in the IGV reflects number of reads (range, 0 to 20,000 FPKM), and the x axis is the genome position (drawn to scale). One biological replicate of the two or three replicates used in the RNA-seq analysis is shown. (B) The RNA repair operon map is shown with the σ70-type promoter for the dinJ-yafQ operon, dinJp, and the potential promoters for RtcR-dependent expression of the dinJ-yafQ operon: the σ54-dependent promoter for the RNA repair operon (rsrp) and the Eσ54 binding site within rtcA. The reverse (R) and forward (F) primers for RT-PCR analysis of the dinJ-yafQ transcript(s) are indicated below the operon map. (C) For the RT-PCR analysis of the dinJ-yafQ transcript(s), RNA was isolated from MMC-treated and untreated cultures of WT and ΔrtcR strains and reverse transcribed with a gene-specific primer located in yafQ (R). cDNA was used in PCR with the R primer and forward primers F1, F2, F3, located in rtcA, overlapping the intergenic region downstream of rtcA, and in dinJ, respectively. PCR products from WT, WT+MMC, ΔrtcR, and ΔrtcR+MMC cDNA and from WT genomic DNA (gDNA) for the positive control were run on a 1.2% agarose gel. Lanes: L, 1-kb Plus ladder (Thermo Scientific); 1, F1/R primer pair; 2, F2/R primer pair; 3, F3/R primer pair; N, no-RT control with F3/R primer pair. Expected fragment sizes: F1/R, 812 bp; F2/R, 555 bp; F3/R, 424 bp. Results for one biological replicate are shown; two additional biological replicates gave identical results.
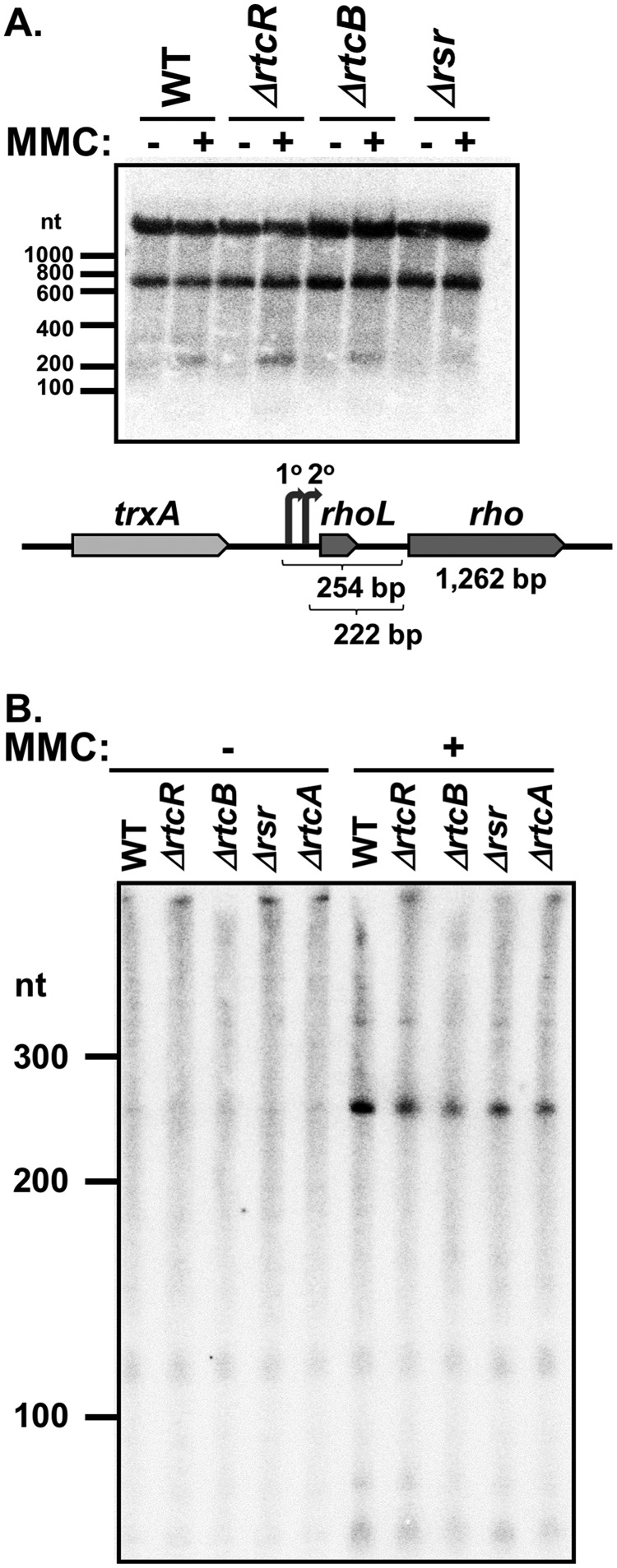
Northern blot analysis was performed for the potential RtcR regulon gene transcripts (genes in the RNA repair and dinJ-yafQ operons and tRNA gene valV) and for gene transcripts that were differentially expressed in both untreated and MMC-treated mutant strains versus the WT (rhoL and the tRNA genes gltUVW and argX) to further assess relative expression levels, possible changes in transcript processing or repair, and operon structure (Table 3 and Fig. 7 and 8; see Fig. S4 in the supplemental material). The mutant strain-dependent >3-fold changes in transcript levels for valV, gltUVW, and argX in the RNA-seq comparative analyses were not seen in the Northern analyses (Table 3). The discrepancy between the RNA-seq data and the Northern analyses may be due to inconsistency in reverse transcription of the highly structured tRNAs in the RNA-seq protocol or may reflect that transcripts from multiple loci were assayed in the Northern blots for valVW and gltUVW due to the extensive homology between gene copies. In addition, gltU, gltV, and gltW are located within three rRNA operons, and since rRNAs were removed during the library preparation, the downregulation observed may be due to unequal removal of unprocessed transcripts between samples. The Northern blot analyses did not reveal any cleavage products or processing intermediates for these tRNAs in the RNA repair mutant strains (Fig. S4). The transcripts that hybridized to the probe for rhoL (Fig. 7A) included the following: an ∼1,500-nucleotide (nt) transcript that is the appropriate length for the full rho mRNA, based on the S. Typhimurium transcriptome data from Kröger et al. (9); an ∼700-nt transcript, which may result from Rho-dependent intragenic termination (67); and an ∼260-nt transcript that corresponds to the leader region, carrying rhoL, which is involved in the Rho-dependent attenuation of rho transcription in E. coli (67) (Fig. 7A). The ∼260-nt transcript increased significantly after MMC treatment (Fig. 7B), and the fold change in ΔrtcR+MMC and Δrsr+MMC versus WT+MMC was close to that seen in the RNA-seq analysis (Table 3). Since the levels of the full rho transcript did not change significantly in either the RNA-seq or Northern analyses in the mutant strains or upon MMC treatment, the significance of the altered levels of the leader region transcript is not clear.
TABLE 3.
Northern blot analysis of transcript levels for selected differentially expressed genes in RNA-seq comparative analysis of RNA repair mutants and WT S. Typhimurium 14028s under MMC-induced stress
| Strains and gene | Fold change by: |
|
|---|---|---|
| RNA-seq | Northern blotting (mean ± SD)a | |
| ΔrtcR+MMC vs WT+MMC | ||
| valb | 0.003 | 0.6 ± 0.06 |
| gltUVWc | 0.1 | 0.7 ± 0.1 |
| yafQ | 0.03 | 0.09 ± 0.01 |
| dinJ | 0.03 | 0.05 ± 0.02 |
| rtcA | 0.009 | NQd |
| rtcB | 0.004 | NQ |
| rsr | 0.003 | NQ |
| rhoL | 0.3 | 0.6 ± 0.2 |
| Δrsr+MMC vs WT+MMC:rhoL | 0.3 | 0.4 ± 0.1 |
| ΔrtcB+MMC vs WT+MMC | ||
| gltUVWc | 0.2 | 0.7 ± 0.1 |
| argX | 4 | 1 ± 0.0 |
Transcript levels in each strain were normalized to the lpp transcript level, which was unchanged between strains, for calculation of the fold change between mutant and WT strains.
The Northern blot probe annealed to transcripts from valV and valW (tRNA-Val), but only valV showed a significant change in transcript levels in the comparative RNA-seq analysis of ΔrtcR+MMC versus WT+MMC.
The Northern blot probe annealed to transcripts from gltU, gltV, and gltW (tRNA-Glu); the indicated RNA-seq fold change is the average of the gltU, -V, and -W fold change values.
NQ, not quantifiable the transcript level in the mutant strain was too low to detect in quantitation, so the fold change could not be calculated.
FIG 7.
Northern blot analysis of MMC-induced differential expression of rhoL transcripts in RNA repair mutant strains. (A) The Northern blot was prepared with equal amounts of total steady-state RNA from S. Typhimurium 14028s WT, ΔrtcR, ΔrtcB, and Δrsr strains (untreated and treated with MMC) electrophoresed on a 1% agarose gel. The membrane was probed for rhoL transcripts. The genomic region carrying rhoL and rho is shown, with the primary and secondary promoters for the rho operon depicted by curved arrows (9) and the lengths of the leader regions and rho gene indicated. (B) The Northern blot was prepared with equal amounts of total steady-state RNA from S. Typhimurium 14028s WT, ΔrtcR, ΔrtcB, Δrsr, and ΔrtcA strains (untreated and treated with MMC) electrophoresed on an 8% acrylamide gel. The membrane was probed for rhoL transcripts. The Northern blots in panels A and B are representative of results from three biological replicates. The nucleotide (nt) lengths indicated for each Northern blot are from the RiboRuler low-range ladder (Thermo Fisher) included on the gels.
FIG 8.
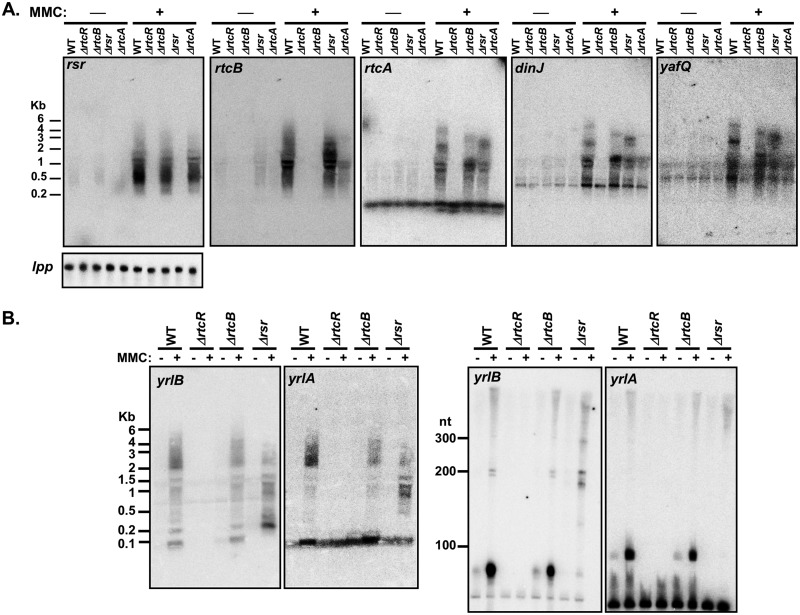
Northern blot analysis of gene transcripts from the RNA repair and dinJ-yafQ operons. (A) Northern blots were prepared with equal amounts of total steady-state RNA from S. Typhimurium 14028s WT, ΔrtcR, ΔrtcB, Δrsr, and ΔrtcA strains (untreated and treated with MMC) electrophoresed on a 1% agarose gel. The membranes were probed for rsr, rtcB, rtcA, dinJ, and yafQ transcripts. Every membrane with RNA from different biological replicates was probed for lpp, which was used for normalization in transcript quantitation; one representative Northern blot is shown. The probe for rtcA appears to cross-hybridize to another gene transcript that is expressed in the absence of MMC treatment. (B) Northern blots were prepared with equal amounts of total steady-state RNA from S. Typhimurium 14028s WT, ΔrtcR, ΔrtcB, and Δrsr strains (untreated and treated with MMC) electrophoresed on a 1% agarose gel (left two blot images) and an 8% acrylamide gel (right two blot images). The membrane was probed for yrlA and yrlB transcripts. The yrlA probe appears to cross-hybridize with a tRNA (most likely Asn-tRNA, based on significant homology with the probe), which obscures the yrlA band at 91 nt on the blot from the agarose gel, but the tRNA is resolved from the yrlA transcript in the acrylamide gel. The Northern blots in panels A and B are representative of results from two or three biological replicates. The nucleotide (nt) lengths indicated for the Northern blots from agarose gels and acrylamide gels are from the RiboRuler high-range ladder and low-range ladder, respectively, which were included on the gels.
The Northern blot analyses confirmed the very low levels of expression of rsr, rtcB, and rtcA in the absence of activated RtcR. Furthermore, the Northern blot and RNA-seq analyses demonstrated no significant change in rtcA transcript levels for the ΔrtcB strain versus the WT strain under untreated or MMC-treated conditions; this provides further evidence that the σ70-type promoter within rtcB (rtcBp), which is deleted in the ΔrtcB strain, does not impact expression of downstream genes under these experimental conditions (Table 2; Fig. 8). Northern blots also confirmed the >10-fold decrease in expression of dinJ and yafQ in ΔrtcR+MMC versus WT+MMC that was seen in the RNA-seq analysis (Table 3; Fig. 8A). In addition, the Northern blots for gene transcripts from the RNA repair and dinJ-yafQ operons (Fig. 8) indicate that a polycistronic transcript of ∼5,000 nt is produced, which is consistent with the length of a read-through transcript from rsrp through the dinJ-yafQ operon (Fig. 1 and 6A), and it is apparently processed to yield multiple RNA fragments, including the noncoding RNAs YrlA (91 nt) and YrlB (111 nt) (Fig. 8B). YrlA and YrlB were previously detected in S. Typhimurium expressing a constitutively active RtcR variant, and YrlA was shown to form a ribonucleoprotein complex with Rsr and PNPase (7). The decreased levels of YrlA and YrlB in Δrsr+MMC (Fig. 8B) are consistent with previous findings that Y RNAs are unstable in the absence of Ro60 in metazoans (8), but it is also possible that sequences within rsr are required for their processing.
Although RtcR-dependent expression of the dinJ-yafQ operon appears to be due to transcription read-through from the σ54-dependent promoter of the RNA repair operon (rsrp), RtcR-dependent transcription of the TA operon could additionally initiate from a recently identified σ54 binding site at the 3′ end of rtcA, which is oriented toward the 5′ end of dinJ (5) (Fig. 6B). There is also a σ70-type promoter, dinJp, located in the intergenic region between rtcA and dinJ (9). A potential LexA binding site overlapping the transcription start site (TSS) for dinJp (GenBank accession number CP001363.1, genome position 3694144 to 3694163 [TACTGTGTCTCTATACAGTT]) was identified using Pattern Locator (51), suggesting that expression of dinJ-yafQ from dinJp might be activated during the SOS response. The RtcR-dependent dinJ-yafQ transcript was mapped using endpoint RT-PCR with RNA harvested from S. Typhimurium WT and ΔrtcR strains grown in the presence and absence of MMC. Three different forward primers (F1, F2, and F3) were utilized in combination with the reverse primer in yafQ (R) in PCR to determine whether the transcript initiates upstream of the σ54 binding site in rtcA (F1/R; 812 bp), or downstream of the σ54 binding site and upstream of the σ70-type promoter for dinJ-yafQ (F2/R; 555 bp); the F3 primer is located within dinJ, so it should produce a product regardless of the position of the TSS (F3/R; 424 bp) (Fig. 6B). For the RNA isolated from WT and ΔrtcR untreated cultures and from the ΔrtcR+MMC culture, a PCR product was obtained only with the F3/R primer pair (Fig. 6B), indicating that in the absence of activated RtcR, the dinJ-yafQ operon is transcribed only from the dinJp σ70-type promoter. In the WT+MMC samples, PCR products were observed for all three primer pairs, indicating that dinJ-yafQ is transcribed as part of the RNA repair operon from the RtcR-, σ54-dependent rsrp. Visualization of the RNA-seq mapped reads for the WT and ΔrtcR libraries from MMC-treated and untreated cultures reflects the results of this RT-PCR transcript analysis (Fig. 6A). The RT-PCR analysis also shows that dinJ-yafQ is expressed from dinJp in the absence of MMC, indicating that LexA does not repress expression in the absence of the SOS response. This result is consistent with the regulation of the dinJ-yafQ operon in E. coli, which is not significantly activated by the SOS response (<2-fold) despite the presence of a LexA binding site overlapping one of the operator sites for the DinJ-YafQ complex that autoregulates transcription of the operon (68, 69). However, the dinJ-yafQ operon is not cotranscribed with the rtcBA RNA repair operon in E. coli, since it is located elsewhere on the chromosome.
YafQ can activate RtcR-dependent transcription but is not required for RtcR activation.
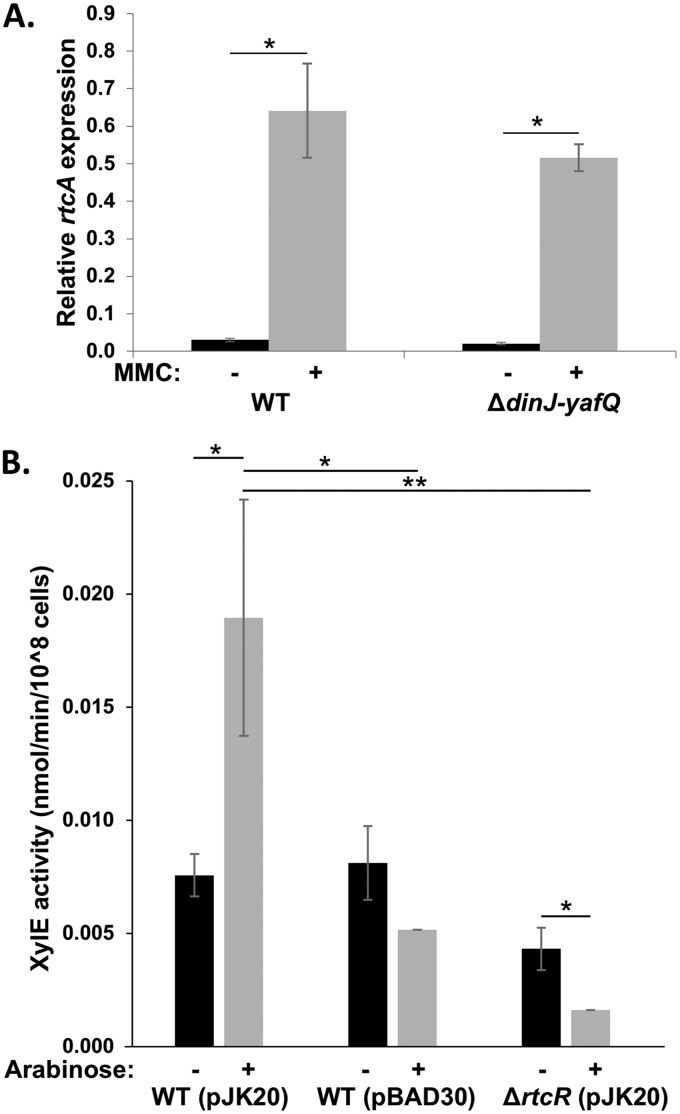
It has been proposed that the close proximity of the dinJ-yafQ operon to the RNA repair operon in S. Typhimurium may reflect the need for coexpression with the genes encoding a repair pathway for RNAs damaged by YafQ (6). YafQ and other TA systems have been shown to play an important role in bacterial persistence under stress conditions by inducing a state of physiological dormancy (70, 71), and RNA repair may be needed for rapid recovery from the stasis induced by RNase toxins. During the SOS response, the coexpressed antitoxin DinJ is degraded by Lon and ClpXP, resulting in increased levels of active YafQ (68). Although the substrate for YafQ has not been demonstrated in S. Typhimurium, S. Typhimurium YafQ (YafQST) is likely to show the same specificity as E. coli YafQ (YafQEc) for cleaving ribosome-associated mRNAs at AAA (Lys) codons (68), based on conservation of the basic residues that are proposed to directly interact with 16S rRNA on the ribosome and the catalytic residues for mRNA hydrolysis defined for YafQEc (64). YafQEc endoribonuclease activity yields RNA fragments with 2′,3-cyclic phosphate ends, which are potentially the ligand that binds the CARF-like domain of the regulatory region of RtcR (2). To determine whether RNA cleavage by YafQST generates a specific signal responsible for activating RtcR, qRT-PCR was performed with RNA harvested from WT and ΔdinJ-yafQ cells treated with MMC to assay for expression of the RNA repair operon. Expression of rtcA was upregulated in both strains after MMC treatment, and there was no significant difference in expression of the operon between the strains (Fig. 9A). However, induction of YafQST expression from a pBAD30-derived plasmid (pJK20) in the WT strain, which stops cell growth within an hour (see Fig. S5 in the supplemental material), activates transcription of the RNA repair operon 2.4-fold compared to that in the uninduced WT strain containing pJK20 (Fig. 9B), and this transcription activation requires RtcR (Fig. 9B). Thus, while a YafQST cleavage product is not the sole source for the signal that activates RtcR, YafQST endoribonuclease activity can induce RtcR-dependent transcription of the RNA repair operon and may generate substrates for the RNA repair system.
FIG 9.
dinJ-yafQ is not required for RtcR-dependent induction of the operon, but YafQ overexpression leads to induction of the operon in an RtcR-dependent manner. (A) Cultures of WT and ΔdinJ-yafQ strains at mid-log phase in MOPS minimal medium were treated with 3 μM MMC for 90 min, total RNA was harvested, and transcript levels of rtcA and rpoD were determined by qRT-PCR. Relative rtcA expression is the rtcA transcript levels normalized to rpoD transcripts. Significant differences in relative rtcA expression between treated and untreated samples are indicated (*, P < 0.02); there was no significant difference in rtcA expression between the WT and ΔdinJ-yafQ strains (P > 0.05). Data shown are from three biological replicates for each strain; error bars represent ±1 standard deviation. (B) YafQ expression from pJK20 was induced in reporter strains JEK17 (WT) and JEK41 (ΔrtcR::kan) by the addition of 0.2% arabinose to cultures at mid-log phase. pBAD30 was used as an empty-vector control. At 90 min after induction, uninduced (black bars) and induced (gray bars) cells were harvested and assayed for XylE activity. Each bar represents the average from three biological replicates; error bars represent ±1 standard deviation. Significant differences between induced and uninduced, pJK20 and pBAD30, and WT and ΔrtcR strains are denoted (*, P < 0.05; **, P < 0.01).
The RNA repair system confers a survival advantage following MMC treatment.
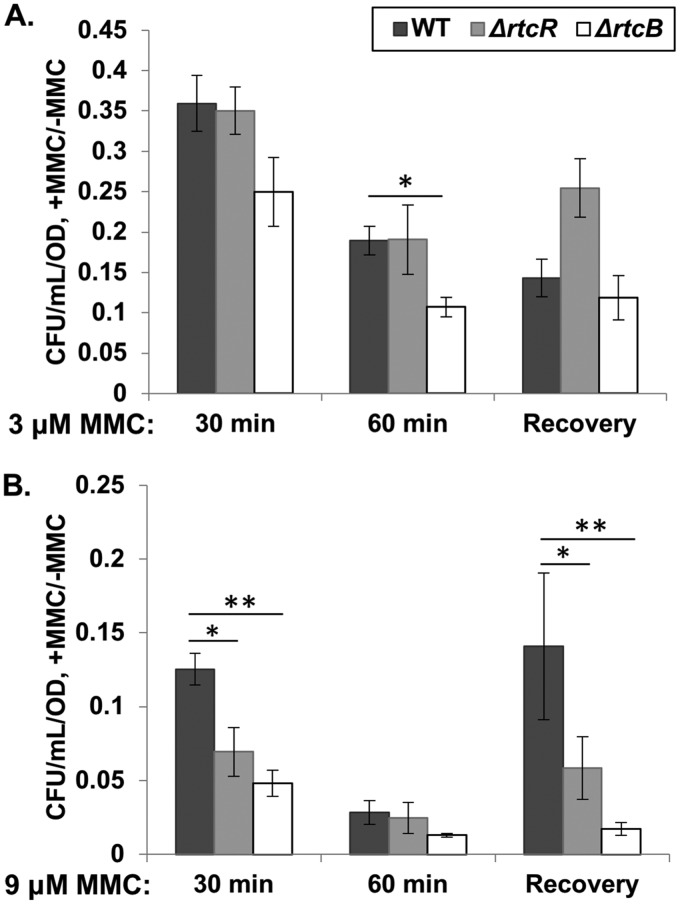
S. Typhimurium is exposed to various stresses during infection of its host and transmission that cause nucleic damage, such as reactive oxygen species and UV, respectively; under these stress conditions, the SOS response is induced and is important for survival (reviewed in references 72 to 74). To determine whether the presence of the RNA repair operon provides a survival advantage to cells under stress conditions that induce the SOS response and activate RtcR, a viability assay was conducted with S. Typhimurium 14028s WT, ΔrtcR, and ΔrtcB strains treated with MMC, which induces the SOS response (66) (Table 1) and optimally activates RtcR (Fig. 1 and 3). Mid-logarithmic-phase growth phase cultures were treated with or without MMC for 30 and 60 min and cell viability assayed (Fig. 10) (see Materials and Methods). After treatment with 3 μM MMC for 60 min, which induces significant increases in expression of RNA repair operon genes (Data Set S2; Fig. 6A), the ΔrtcB strain showed a 1.8-fold (P < 0.05) difference in cell viability compared to the WT, but the ΔrtcR strain did not show a significant difference in cell viability compared to the WT (Fig. 10A). However, after treatment with 9 μM MMC for 30 min, the ΔrtcR strain showed a 1.8-fold decrease in cell viability (P < 0.05) and the ΔrtcB strain exhibited a 2.6-fold decrease in cell viability (P < 0.01) compared to the WT strain (Fig. 10B). By 60 min, all strains treated with 9 μM MMC were reduced to approximately 2% viability without significant differences in cell viability between strains.
FIG 10.
Viability of WT and mutant cells after treatment with MMC. Cell viability was determined for WT, ΔrtcR, and ΔrtcB strains in aerobic culture following treatment with 3 μM (A) or 9 μM (B) MMC for 30 and 60 min and after a recovery phase, in which cells from the 30-min MMC treatment were washed, subcultured to an OD600 of 0.1 in fresh MOPS medium, and incubated for another 1 to 2 h. The CFU/ml, determined for dilutions plated on LB agar, were standardized to the OD600 for each sample, and CFU/ml/OD values for MMC-treated samples were normalized to those for untreated samples. Significant differences in rtcR or rtcB mutants relative to the WT strain are indicated (*, P < 0.05; **, P < 0.01). The data shown are from 6 biological replicates for each strain; error bars represent standard error of the mean.
Cell viability was also assessed for the WT and mutant strains after a 1-hour recovery period in the absence of MMC; this recovery period followed the 30-min treatment with 3 μM or 9 μM MMC (Fig. 10) (see Materials and Methods). There was no significant increase in cell viability for the WT strain following incubation in MOPS minimal medium without MMC compared to that measured immediately after the 30-min treatment with either level of MMC (Fig. 10A and B). These results suggest that restoration of cell division in the small fraction of cells that can recover from the extensive MMC-mediated nucleic acid damage, as reflected in colony growth on LB medium, may require more than 1 to 2 h of incubation in MOPS minimal medium.
Overall, the cell viability results suggest that activities of RtcB in both RNA and DNA repair (6, 19), and possibly other functions of the RNA repair operon, provide a survival advantage under the stress conditions induced by MMC-mediated nucleic acid damage.
Comparison of the rtc operons in S. Typhimurium and E. coli.
Induction conditions have been recently reported for the rtcBA operon in E. coli (13). As in S. Typhimurium, the operon is under the transcriptional control of a σ54-dependent promoter that is activated by RtcR. However, while nucleotide damage seems to be a predominant trigger for induction in S. Typhimurium, the rtcBA operon in E. coli has primarily shown increased expression in response to direct insults to the translation apparatus, such as ribosome-targeting antibiotics or toxin-mediated cleavage of tRNAs (13). S. Typhimurium and E. coli are closely related organisms, yet their respective RNA repair operons have several distinct differences. Notably, the E. coli genome does not contain homologs of rsr and the two yrl genes, while these genes are found in the RNA repair operons of nearly all sequenced strains of S. Typhimurium (6, 7). Additionally, the dinJ-yafQ module is located directly downstream of the rsr-yrlBA-rtcBA operon in most S. Typhimurium strains, but in E. coli this TA operon is distantly located on the chromosome. Given the divergence of the RNA repair operons in these two species, we asked whether the operons may be differentially regulated by RtcR as well.
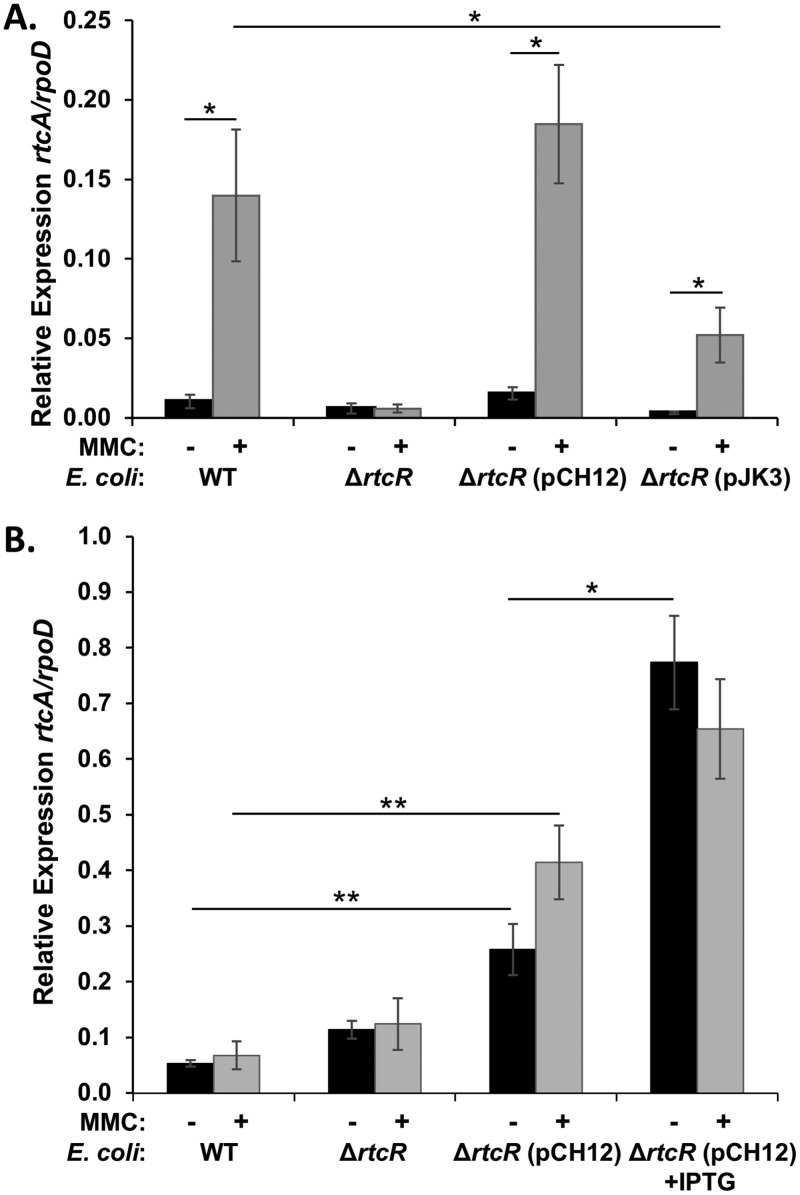
MMC strongly induces transcription of the RNA repair operon in S. Typhimurium. In previous studies of the SOS response to MMC-induced stress in E. coli, the rtcBA operon was not induced (63). Since there is variability in the induction of the SOS response in different E. coli strains (75), the E. coli strain used by Engl et al. (13) to characterize expression of the RNA repair operon, MG1655, and a ΔrtcR mutant of MG1655 (ACK206) were used in qRT-PCR assays for RNA repair operon transcription in the presence or absence of MMC-induced stress. The baseline levels of rtcA transcription were higher in E. coli than in S. Typhimurium in both the WT (6-fold) and ΔrtcR (36-fold) strains (Fig. 11), which is consistent with previous observations in E. coli that phenotypes related to RtcB expression can be seen even when no stress is present and that rtcB expression is not significantly reduced in a ΔrtcR strain (13). As seen in Fig. 11, there is no upregulation of the operon in response to MMC treatment in E. coli, which is in stark contrast to the case in S. Typhimurium.
FIG 11.
Complementation assays for RtcRST and RtcREc. Cultures of S. Typhimurium (A) or E. coli (B) were treated with 3 μM MMC (gray bars) or left untreated (black bars), and cells were harvested after 90 min; RNA was isolated, and qRT-PCR was performed to assess rtcA transcript levels relative to a reference gene, rpoD. (A) Expression of rtcRST and rtcREc from lacp on pCH12 and pJK3, respectively, in the S. Typhimurium ΔrtcR strains was induced with 50 μM IPTG. (B) Expression of rtcRST from lacp on pCH12 in the E. coli ΔrtcR strain was at a baseline low level in the absence of glucose or highly induced with 1 mM IPTG. Each bar represents the average from three biological replicates; error bars are ±1 standard deviation. Significant differences are denoted (*, P < 0.05; **, P < 0.01).
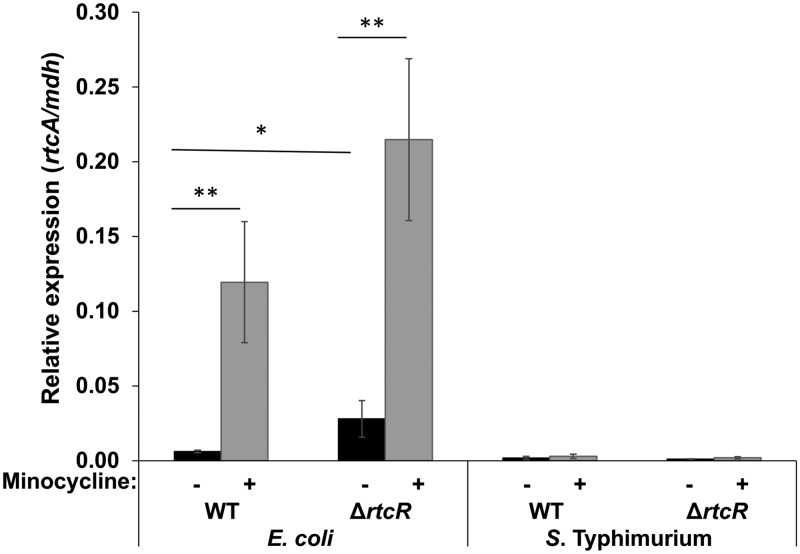
To further compare stress responses that may impact expression of the RNA repair operons of E. coli and S. Typhimurium, transcription of rtcA was assessed following treatment with minocycline, a tetracycline-class antibiotic that was reported to significantly induce expression of the E. coli operon (13). qRT-PCR assays confirmed that rtcA transcription was upregulated in E. coli (19.3-fold) after exposure to minocycline (Fig. 12); however, this response was independent of RtcR, as operon expression was also significantly upregulated in the ΔrtcR strain following minocycline treatment. Whether RtcR was required for minocycline-induced expression of the E. coli rtcBA operon was not assessed in the study by Engl et al. (13), and, based on global TSS mapping in E. coli by Thomason et al. (10), there are multiple σ70-type promoters that potentially generate transcripts through rtcB and/or rtcA. Minocycline treatment of S. Typhimurium did not induce expression of the RNA repair operon (Fig. 12). The dissimilar expression of the RNA repair operons of E. coli and S. Typhimurium in response to the same stress conditions suggests that these closely related species have distinct responses to some stressors, generating alternative signals to control transcriptional regulators of the RNA repair operon. Alternatively, the transcriptional regulators of the RNA repair operon in these species may differ with regard to the stress signals to which they respond.
FIG 12.
RNA repair operon expression in response to minocycline treatment. Cultures at mid-log phase were treated with 40 μg/ml minocycline (gray bars) or left untreated (black bars) and harvested after 2 h. Total RNA was extracted, and qRT-PCR was performed to assess rtcA transcript levels relative to the reference gene mdh. Each bar represents the average from 3 to 5 biological replicates; error bars represent ±1 standard deviation. Significant differences between treated and untreated or WT and ΔrtcR strains are denoted (*, P < 0.05; **, P < 0.1).
Although RtcR from E. coli (RtcREc) and RtcR from S. Typhimurium (RtcRST) share a high degree of overall amino acid sequence identity (84%), there are 30 substitutions (15 nonconservative substitutions) in the 185-amino-acid (aa) sequences of the regulatory domains (see Fig. S6 in the supplemental material) that could potentially affect signal ligand specificity and activation of RtcR. To test whether the lack of RtcR-dependent induction of the E. coli RNA repair operon in response to MMC treatment is due to differences in RtcR structure, functional complementation assays were performed with the expression vector pSRK-Tc (76) containing rtcREc (pJK3) or rtcRST (pCH12) in S. Typhimurium and E. coli ΔrtcR strains (Fig. 11). In the S. Typhimurium ΔrtcR strain expressing RtcREc, MMC treatment induced transcription of the RNA repair operon 16-fold relative to that in the untreated strain, which is comparable to the relative expression (MMC treated versus untreated) seen for S. Typhimurium ΔrtcR expressing RtcRST (12-fold) and the WT strain (14-fold) (Fig. 11A). The expression level of rtcA normalized to rpoD in the RtcREc-complemented ΔrtcR strain is ∼3-fold lower than in the WT or RtcRST-complemented ΔrtcR strain, which might indicate a difference in the affinity of the DNA binding domain of RtcREc for the S. Typhimurium enhancer sequences associated with the RNA repair operon (Fig. 11A). In contrast to the complementation results in S. Typhimurium, expression of RtcRST in the E. coli ΔrtcR strain does not result in MMC-dependent induction of the RNA repair operon (Fig. 11B). However, expressing RtcRST in the E. coli ΔrtcR strain does increase expression of the operon regardless of MMC treatment and in a concentration-dependent manner [Fig. 11B, compare ΔrtcR(pCH12) with ΔrtcR(pCH12)+IPTG].
These complementation results indicate that RtcREc and RtcRST can respond to the same cellular signal in S. Typhimurium (Fig. 11A), thus indicating that the altered regulation of the RNA repair operon in E. coli versus S. Typhimurium is not due to divergences in RtcR structure or signal recognition. The variances in RtcR-dependent expression of the RNA repair operon between these two Enterobacteriaceae species may reflect distinct cellular responses to particular stress conditions and/or different levels of a signaling molecule under the same growth conditions. There are many potential reasons for the different responses to the stress conditions that appear to activate RtcR for expression of the RNA repair operons in E. coli and Salmonella, but one of particular importance may be the absence of a MazEF-related TA system in S. Typhimurium based on an extensive characterization of all TA modules in S. Typhimurium (40) and a search for orthologs of MazF in 23 S. enterica serovars using OrthoDB (77). The mazEF TA module is proposed to play important roles in the stress response for E. coli. Under stress conditions in E. coli, such as amino acid starvation, the endoribonuclease activity of MazF on 16S rRNA may generate a specialized stress ribosome that translates leaderless MazF-cleaved mRNAs; there is evidence that during recovery from stress, RtcB repairs cleaved 16S RNA in the stress ribosome, thereby restoring recognition and translation of canonical mRNAs (21). Although recent work suggests that MazF does not generate a significant population of specialized ribosomes or leaderless mRNAs in response to stress (22), it is still evident that RtcB plays a role in 16S RNA stability in E. coli under stress conditions (13, 21). In addition, a MazEF-mediated cell death pathway that is active in some E. coli strains interferes with the SOS response (75). Simply considering this one feature that is not shared by E. coli and S. Typhimurium, it is apparent that substrates for an RNA repair system and pathways for the generation of the signal to activate RtcR are likely to differ between even closely related species.
Concluding remarks.
Here we report stress conditions and cellular processes that activate the cognate bEBP for the S. Typhimurium RNA repair operon, RtcR, giving some insight into possible physiologically relevant roles for components of the RNA repair operon. The highest levels of transcription of the operon were observed under conditions that induce the SOS response, i.e., treatment with the nucleic acid cross-linking agents MMC and cisplatin and with hydrogen peroxide. RtcR activation was found to be dependent on RecA. The level of expression of the RNA repair operon increased with higher concentrations of MMC, cisplatin, or hydrogen peroxide, suggesting that the intensity of the SOS response to greater levels of DNA damage produced more signal for RtcR activation. Although MMC, as well as cisplatin and hydrogen peroxide, damages both RNA and DNA, the role of the RecA-mediated SOS response in activating RtcR suggests that the DNA-modifying activities of RtcB may contribute to the survival advantage conferred by RtcB following MMC treatment. A 3′-PO4 end at a DNA break cannot be repaired by classic DNA ligases or extended by DNA polymerases, including the error-prone DNA polymerases of the SOS response (19). RtcB has a DNA-capping activity that allows repair of DNA breaks containing a 3′-PO4 end. RtcB transfers GMP to the 3′-PO4 terminus (DNA 3′pp5′G); this modified end blocks exonuclease activity and can be extended by DNA polymerases, such as Pol II in SOS repair (19).
Cotranscription of the dinJ-yafQ TA module with the RNA repair operon, as well as the activation of RtcR by overexpression of the endoribonuclease YafQ, suggests that YafQ may generate a substrate for the RNA repair system. As the role of toxins of TA modules in generating persister cells in bacterial populations becomes clearer (reviewed in reference 71), it is an appealing hypothesis that RNA repair may be involved in recovery from a persister state associated with activation of YafQ and other endoribonuclease toxins, such as SehA, during the SOS response.
The stimulation of RNA repair operon transcription by NtrC under nitrogen-limiting conditions is a rare example of a single σ54-dependent promoter apparently being responsive to two different bEBPs and suggests that components of the operon are involved in responding to very different types of stresses. In addition, comparison of stress conditions that activate expression of the S. Typhimurium and E. coli RNA repair operons suggests that differences in the stress responses of bacteria, even highly related bacteria, impact the regulation of their RNA repair operons and probably generate alternative nucleic acid substrates for the repair enzymes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids are listed in Table S3 in the supplemental material with description/construction and source/reference. The wild-type (WT) S. Typhimurium stain used in this study is ATCC 14028s; the WT E. coli strain is MG1655 (courtesy of S. Kushner). All other strains used in this study were derived from these two strains, unless otherwise indicated. All oligonucleotides utilized in strain and plasmid constructions, reverse transcription, and PCR assays are listed by number in Table S4 in the supplemental material with name and sequence.
λ-Red recombineering (78) was used to construct functional deletion mutants of S. Typhimurium 14028s: DJS101 (ΔrtcR, leaving 17 codons at the start and 14 codons at the end), ANE004 (Δrsr, with a deletion from the start to the stop codon), DJS102 (ΔrtcB, leaving 21 codons at the start and 13 codons at the end), CH06 (ΔdinJ-yafQ, with a deletion from the start codon of dinJ to the stop codon of yafQ), and CH01 (ΔntrC, with a deletion that begins 2 bp upstream of the start codon and leaves 8 codons at the end). Oligonucleotides 1 and 2 (rtcR), 3 and 4 (rsr), 5 and 6 (rtcB), 7 and 8 (dinJ-yafQ), and 9 and 10 (ntrC) were used to amplify the kanamycin resistance cassette (kan) from pKD4 (78). Substitutions were confirmed by PCR using gene-flanking primers (oligonucleotides 15 and 16, 17 and 18, 19 and 20, 21 and 22, and 25 and 26, respectively). The substitution mutations were transduced by P22 HT int into a clean S. Typhimurium 14028s background, and the aph (kan) gene was removed by introducing the FLP expression plasmid pCP20 (79). Excision of aph in each mutant was confirmed by kanamycin sensitivity, colony PCR with the same primers used to verify insertion of the cassette, and sequencing of the PCR product (Genewiz, South Plainfield, NJ). Plasmid pCP20 was removed from the deletion strains with growth at 37°C in the absence of selective pressure, utilizing the temperature-sensitive replication origin (growth maintaining the plasmid was at 30°C). The reporter strain JEK17 (14028s Δrsr::xylE) was constructed similarly, except that overlap extension (80) was used to generate a xylE-kan fusion PCR product with flanking regions of homology to the regions that flank rsr on the chromosome. kan was PCR amplified from template pKD4 (oligonucleotides 13 and 14), and the xylE gene of Pseudomonas putida was PCR amplified from template pSB306 (81) (oligonucleotides 11 and 12); overlap PCR with these products generated the xylE-kan fragment used to transform S. Typhimurium 14028s. Construction of JEK21 (ΔrtcR Δrsr::xylE-kan) was similar to that of JEK17, except λ-Red recombination was carried out in DJS101; aph could not be excised due to the active FRT site still present within the ΔrtcR locus. The S. Typhimurium 14028s ΔrecA::kan mutant, DS103, was created by transduction of the marked deletion from JE10649 (S. Typhimurium LT2 metE205 ara-9 ΔrecA::kan) using P22 HT int, and the deletion was verified by UV sensitivity (82) and colony PCR with oligonucleotides 23 and 24. Construction of JEK26 (JEK17 ΔrecA::kan) was similar, but the donor strain for the transduction was the ΔrecA::kan mutant strain (SGD_recA_Kan) from the S. Typhimurium 14028s single-gene deletion mutant library (83) (BEI Resources, American Type Culture Collection), and the recipient strain was JEK17. JEK41 (JEK17 ΔrtcR::kan) was constructed similarly to S. Typhimurium 14028s ΔrtcR (DJS101), except λ-Red recombination was carried out in strain JEK17 instead of the WT. The E. coli ΔrtcR strain (ACK206) was generated by transducing MG1655 rph-1 with P1 lysate grown in JW3385 (BW25113 ΔrtcR, with a deletion from the second codon to the seventh codon from the C terminus) (84) and confirmed by PCR (oligonucleotides 27 and 28). aph was removed by introducing pCP20.
All plasmids were generated using enzymes purchased from New England BioLabs (NEB), Ipswich, MA. For pDS171 and pDS185, PCR products containing 197 bp of rpoD (oligonucleotides 29 and 30) and 180 bp of rtcA (oligonucleotides 31 and 32), respectively, were generated with OneTaq from S. Typhimurium 14028s genomic DNA and TOPO cloned into pCR2.1 and pCR4 (Invitrogen, Carlsbad, CA), respectively. For pDS183, a PCR product encoding a truncated copy of S. Typhimurium RtcR beginning at Leu179 was generated (oligonucleotides 51 and 52) and TOPO cloned into pCR4 (pDS181); the BamHI-HindIII fragment from pDS181 was introduced into the expression vector pKH66 (85). pCH11 was created by PCR amplifying the full-length rtcRST gene from S. Typhimurium 14028s genomic DNA (oligonucleotides 53 and 54) and TOPO cloning the PCR product into pCR2.1. To create pCH12, the NdeI-XhoI fragment from pCH11 was introduced into the low-copy-number expression vector pSRK-Tc (76); pJK3 was constructed similarly to pCH12, except rtcREc was PCR amplified from MG1655 genomic DNA (oligonucleotides 55 and 56), and the PCR ends were digested with NdeI and XhoI prior to ligation with pSRK-Tc. To construct pJK20, yafQ was PCR amplified from S. Typhimurium 14028s genomic DNA (oligonucleotides 57 and 58); the ends were digested with EcoRI and HindIII, and the product was introduced into pBAD30. All plasmid constructs were confirmed by DNA sequencing (Genewiz, South Plainfield, NJ). All plasmids were first replicated in E. coli DH5α and then moved through MS1868, a restriction-negative/modification-positive mutant of S. Typhimurium LT2 (leuA414 hsdL Fels2), before introduction into S. Typhimurium 14028s.
Growth media and conditions.
Unless otherwise noted, bacteria were grown at 37°C with aeration. Growth was in either Luria-Bertani (LB) broth (Fisher Scientific, Fair Lawn, NJ) or MOPS minimal medium (86). Solid medium contained 1.5% agar (Fisher Scientific). Unless otherwise indicated, all supplements and antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). H2O2 was purchased from Fisher Scientific. Antibiotics were added to media for maintenance of plasmids or assay for chromosomal markers in S. Typhimurium and E. coli at the following concentrations (LB/MOPS; in μg/ml): ampicillin, 80/50; kanamycin, 50/50; tetracycline, 10/10.
Cultures of WT or mutant S. Typhimurium 14028s or E. coli MG1655 strains for assays of transcripts by quantitative reverse transcriptase PCR (qRT-PCR) or Northern blotting under various stress conditions were grown in MOPS medium or LB broth (as indicated) overnight from a single colony and then subcultured to an optical density at 600 nm (OD600) of 0.05 in fresh medium. Treatment was done at the mid-log growth phase (OD600 of 0.4) at 37°C with the following conditions for the indicated time periods (all cultures were split before treatment for untreated and treated samples): mitomycin C (MMC) (3 μM) for 90 min, carbon starvation (1% methyl-α-d-glucopyranoside [MGP] as sole C source) for 90 min, nitrogen starvation (2.5 mM arginine as sole nitrogen source instead of 14.3 mM ammonium chloride) for 90 min, peroxide stress (1 mM H2O2) for 20 min, amino acid starvation (0.4 mg/ml serine hydroxamate [SHX]) for 90 min, translation inhibition (30 μg/ml chloramphenicol [CHL]) for 90 min, cell wall stress (2 μg/ml cefotaxime [CTX]) for 90 min, iron limitation (250 μM 2,2′-bipyridyl [Bpy]) for 90 min, cisplatin (CDDP) (110 μM) for 180 min, and minocycline (40 μg/ml) for 120 min. For α-d-glucopyranoside and arginine treatments, mid-exponential-phase cells were pelleted via centrifugation and resuspended in MOPS medium containing α-d-glucopyranoside as the C source or arginine as the sole N source for the 90-min treatment.
RNA extraction, reverse transcription, and quantitative PCR.
For qRT-PCR and Northern blotting, cultures were treated as described above and RNA was harvested by the RNAsnap method for Gram-negative bacteria (87). Briefly, cell pellets were resuspended in 1/10 volume of RNA extraction solution (18 mM EDTA, 0.025% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol [β-ME], 95% formamide) and incubated for 7 min at 95°C. Cellular debris was pelleted at 16,000 × g for 5 min at room temperature. The supernatant was removed to a new tube and extracted with acid phenol-chloroform, followed by a chloroform extraction, and then ethanol precipitated as described in reference 87. Pellets were rehydrated in RNase-free H2O. RNA was quantified in a NanoDrop 2000 or Invitrogen Qubit RNA BR assay (Thermo Fisher Scientific), and quality was assessed via visualization on 1% agarose gels. RNA samples were treated with Ambion Turbo DNase (Thermo Fisher Scientific) or DNase I (NEB) according to the manufacturer's instructions. For each experiment, an equal amount of RNA from each sample (2 to 8 μg) was treated in the DNase I reactions. DNase I was inactivated, and the RNA samples were either purified with Zymo RNA clean-and-concentrate columns (Zymo, Irvine, CA) or ethanol precipitated. Samples were tested for DNA contamination by PCR with oligonucleotides 29 and 30 (for Salmonella) or 43 and 44 (for E. coli) in the absence of reverse transcriptase and were retreated if an amplification product was seen on a 1.5% agarose gel. RNA was quantified by NanoDrop or Qubit RNA BR assay.
Reverse transcription was carried out either separately with an iScript cDNA synthesis kit using random hexamers (Bio-Rad; Hercules, CA) or as part of the Bio-Rad iTaq Universal SYBR green one-step kit for qRT-PCR using gene-specific primers. For iScript cDNA synthesis, an equal amount of RNA, ∼1 μg, was used, and cDNA was quantified using the Qubit single-stranded DNA (ssDNA) assay kit.
Quantitative PCR was performed on an iCycler or CFX Connect instrument (Bio-Rad) using iQ SYBR green supermix (Bio-Rad) (if reverse transcription was performed separately) or the Bio-Rad iTaq Universal SYBR green one-step kit in 20-μl reaction mixtures, according to the manufacturer's instructions. An equal amount of cDNA or RNA was used as the template in each reaction. Negative controls were prepared by omitting DNA/RNA from the reaction mixture. Standard curves were generated on each plate using serial 10-fold dilutions (to 1:10,000) of pDS171 (rpoD), pDS185 (rtcA), or 14028s/MG1655 genomic DNA (GenElute bacterial genomic DNA kit; Sigma-Aldrich) digested with EcoRI-HF (NEB); the starting quantity (SQ) of transcript was calculated by the Bio-Rad CFX software relative to the defined quantity of the standard curve dilutions. Data for each biological replicate indicate the average from three technical replicates. To determine relative gene expression under most growth conditions, rtcA transcript levels (qRT-PCR with oligonucleotides 31 and 32 or 33 and 34 for S. Typhimurium and oligonucleotides 45 and 46 for E. coli) were divided by rpoD transcript levels (qRT-PCR with oligonucleotides 29 and 30 for S. Typhimurium and oligonucleotides 43 and 44 for E. coli) for each sample. The relative rtcA expression level was then compared between strains and treatments. Peroxide-treated and nitrogen-limited samples were also assessed for rsr transcript levels (qRT-PCR with oligonucleotides 80 and 81). For peroxide- and minocycline-treated samples, rpoD levels were not consistent between treated and untreated cells (data not shown); therefore, other reference genes that exhibited constant expression in treated and untreated cells were utilized: for peroxide treatment, kdgR (qRT-PCR with oligonucleotides 37 and 38) was used for normalizing rtcB expression (qRT-PCR with oligonucleotides 35 and 36), and for minocycline treatment, mdh (qRT-PCR with oligonucleotides 49 and 50) was used for normalizing rtcA expression. For NtrC activation, glnA levels were measured with oligonucleotides 39 and 40. Data presented indicate the average ± 1 standard deviation from three biological replicates. Results were analyzed using Student t test 2-tailed, paired analysis.
Assays for cisplatin, MMC, and YafQ induction of the RNA repair operon in XylE reporter strains.
Cultures of JEK17 (14028s Δrsr::xylE), JEK26 (JEK17 ΔrecA::kan), and JEK21 (14028s ΔrtcR Δrsr::xylE-kan) were prepared and treated with MMC or cisplatin as described above. After MMC and cisplatin treatment for 90 and 180 min, respectively, whole-cell XylE assays were carried out as previously described (81). Treated cells were pelleted and washed once in 50 mM phosphate buffer (pH 7.4). Cells were then diluted in phosphate buffer to an OD600 of 1.0. The number of cells per milliliter was determined by performing serial dilutions and enumerating colonies on LB plates. The XylE reaction was initiated by adding 100 μl of the cell suspension to 900 μl of catechol solution (10 mM catechol, 50 mM phosphate buffer [pH 7.4]), and the conversion of catechol to 2-hydroxymuconic semialdehyde was monitored continuously at room temperature (22°C), with readings taken every 10 s for 2 min using a Thermo Scientific Genesys 20 spectrophotometer at λ = 375 nm. Each individual culture was assayed for 2 or 3 technical replicates. The XylE activity was calculated as the nanomoles of catechol oxidized per minute per 108 cells during linear activity, based on the Beer-Lambert law and using an extinction coefficient of 44,000 liters mol−1 cm−1 for 2-hydroxymuconic semialdehyde. Simplified, the equation is as follows: XylE activity = [1,000 × (ΔOD375/min)]/(44 × number of cells × 108).
To test the effects of YafQ on expression of the RNA repair operon, pJK20 or pBAD30 was transformed into JEK17 and JEK41 (JEK17 ΔrtcR). YafQ activity was confirmed by cessation of growth during growth curve assays. Cells were grown overnight in LB plus 100 μg/ml carbenicillin plus 0.2% glucose, subcultured 1:100, and grown to mid-log phase (OD600 = 0.4). At this point, cultures were split and one of each pair was induced with 0.2% arabinose. OD600 readings were taken from the induced/uninduced cultures every hour for 5 h following induction. For XylE assays, YafQ was expressed as described above, and cells were harvested 90 min after induction, at which point XylE activity was assayed as described above.
RNA-sequencing library preparation and data analysis.
Overnight cultures of S. Typhimurium WT, ΔrtcR, Δrsr, and ΔrtcB strains grown in MOPS medium (three biological replicates each) were diluted 1:40 in fresh MOPS medium and grown to an OD600 of 0.4. Cultures were split, and half of each were treated with MMC for 60 min. RNA was prepared by RNAsnap, as described above. After DNase I treatment, rRNA was removed from the samples with a Ribo-Zero rRNA removal kit (Gram-negative bacteria) (Epicentre), and ethanol precipitation was used to concentrate the rRNA-depleted samples. Before preparing the libraries, a portion of each sample was reverse transcribed and assayed via qPCR for rtcA expression in MMC-treated samples compared to untreated samples, as described above (rtcA expression was confirmed as increased in the MMC-treated wild-type, Δrsr, and ΔrtcB strains and unchanged in the ΔrtcR strain [data not shown]). Reverse transcription and library preparation were conducted as described in the Illumina TruSeq v2 manual, using SuperScript III reverse transcriptase to generate cDNA; this method did not provide a stranded library. The libraries were sequenced on the Illumina Next-seq500 platform at the Georgia Genomics Facility, producing 75-bp single-end reads. For each library (24 libraries comprise three biological replicates for each strain and condition), the total number of reads sequenced exceeded 14.9 million and the number of reads that mapped uniquely to the S. Typhimurium chromosome exceeded 8.0 million, with the exception of biological replicate 3 for the untreated ΔrtcR strain, for which there were fewer than 3.8 million total reads and 1.6 million reads that mapped uniquely (see Table S1 in the supplemental material). Haas et al. (88) demonstrated for bacterial RNA-seq data that 5 to 10 million non-rRNA fragments are sufficient to detect all but a few of the most poorly expressed genes in diverse bacteria under a range of growth conditions; thus, the RNA-seq data for all the biological replicates provide excellent coverage of the transcriptome, with the exception of ΔrtcR replicate 3, which was removed from the data set before further analysis. For the remaining libraries, an average of 89% of the sequenced reads mapped to the S. Typhimurium chromosome.
Illumina reads were mapped to the S. Typhimurium 14028s genome (assembly, GCA_000022165.1) using Bowtie2 (version 2.2.3) (60) with default parameters. Cuffnorm (61) was used to obtain the number of fragments per kilobase of transcript per million fragments mapped (FPKM) for each biological replicate, and Pearson correlations were calculated between all replicates. The FPKM data for WT+MMC 1 (biological replicate 1 of the WT treated with MMC), ΔrtcR+MMC 1, and Δrsr+MMC 2 gave Pearson r values of ≈0.9 or lower compared to the other two biological replicates (see Table S2 in the supplemental material), and thus each of these replicates was removed from further analysis; all remaining biological replicates correlated well (r > 0.94). Pairwise comparisons of aligned reads were made using Cuffdiff (version 2.2.1) (62) with default parameters. The significance of log2 fold change values is reflected in a q value of <0.05, which is a false-discovery rate (FDR)-adjusted P value in the Cuffdiff statistical analysis program. Additionally, a cutoff 3-fold differential expression (log2 fold change >1.585) between WT and mutant samples treated with MMC or between MMC-treated and untreated samples was applied. For some genes identified as significantly differentially expressed, the FPKM value in either the WT or mutant strain was reported as zero. Log2 fold change values cannot be calculated for genes with an FPKM of zero. Because these genes can be expressed, as evidenced by FPKM values greater than zero in some strains, we assigned a value of one to these genes for the purpose of calculating log2 fold change values.
In silico analyses for LexA and NtrC DNA binding sites.
The program Pattern Locator (51) was used for in silico analysis of the rsr-yrlBA-rtcBA-dinJ-yafQ operon regulatory regions for a LexA binding site. The pattern searched was based on the consensus LexA binding site (52, 53).
E. coli genomic sequences associated with the confirmed peaks from NtrC chromatin immunoprecipitation assays with sequencing (ChIP-seq) (42) were used in the motif discovery tool MEME (89), and the 29 NtrC DNA binding motif sites (see Data Set S1 in the supplemental material) were then utilized to generate the position-specific score matrix in Motif Locator for the search of the S. Typhimurium 14028s genome sequence using the algorithm described in (44) to identify similar motif sites (Data Set S1).
Northern analysis.
The levels of specific transcripts in different genetic backgrounds under various conditions were determined using either agarose or polyacrylamide Northern analyses. Total steady-state RNA for Northern analysis was isolated as described above (for cultures treated with 3 μM MMC for 90 min or untreated). Northern analyses using polyacrylamide and agarose gels were carried out as described previously (90, 91). Briefly, total RNA (10 μg for acrylamide gels and 20 μg for agarose gels) were initially dried in a Speed Vac. The dried RNA sample was resuspended in 5 μl of 2× RNA loading dye (Fermentas; Thermo Fisher) for acrylamide gels and separated in either 6% or 8% polyacrylamide gels containing 8 M urea in Tris-borate-EDTA (TBE) buffer as described previously (90). For agarose gels, the dried RNA sample was resuspended in 5 μl RNase-free water and 5 μl of glyoxal loading dye (Ambion) and separated in 1% agarose gel in BPTE buffer (91). RiboRuler high-range or low-range ladders (Thermo Fisher) were included as size standards for each gel. RNAs from gels were transferred to nylon transfer membranes (Nytran SPC; GE Healthcare) by electroblotting.
Membranes were probed with transcript-specific oligonucleotide probes (gene transcript, oligonucleotide probe: valV, 69; argX, 70, gltUVW, 71; rhoL, 72 lpp, 73; rsr, 74, rtcB, 75, rtcA, 76; dinJ, 77; yafQ, 65; yrlB, 78; yrlA, 79) using PerfectHyb Plus hybridization buffer (Sigma) at appropriate temperature. The probes were labeled with 32P as described previously (90). A single membrane was successively hybridized with several different probes after stripping the previous probe, as described previously (90). The lpp transcript was used to normalize all the quantification data since its expression was not changed after MMC treatment or between mutant and WT strains. All Northern blots were scanned using a PhosphorImager (Typhoon; GE Healthcare) and analyzed using ImageQuant TL 8.1 software (GE Healthcare). All quantification data (relative fold increase or decrease) presented represent an average of two or three independent biological replicates.
Transcript analysis for dinJ-yafQ operon structure.
RNA was prepared by RNAsnap, as described above, from S. Typhimurium 14028s WT and ΔrtcR cells grown in MOPS with and without 60 min of MMC treatment. Reverse transcription was carried out using the SuperScript III first-strand synthesis system (Invitrogen), according to the manufacturer's instructions, using a gene-specific primer (oligonucleotide 65); a no-RT control was included for each RNA sample. Endpoint PCR was performed with Taq polymerase in standard Taq buffer (NEB) using oligonucleotide 65 with each of the forward primers (oligonucleotides 66 to 68) to assess the extent of the transcript containing dinJ-yafQ. Genomic DNA was used as the template for a positive control with each primer pair. PCR products were visualized on 1.2% agarose gels.
Cell viability assays following MMC treatment.
Cultures of S. Typhimurium 14028s WT, ΔrtcR, and ΔrtcB strains were grown in MOPS medium to an OD600 of 0.3 and split into treated and untreated samples, and MMC was added to treated samples to a final concentration of 3 μM or 9 μM. Cells were pelleted and resuspended in MOPS medium without MMC and then plated on LB agar at 30 and 60 min after MMC addition to assay for viable cells. The OD600 was measured at each time point. The cultures were grown overnight at 37°C, and CFU were enumerated. CFU/ml was calculated and normalized to OD600 to represent cell viability. The data are presented as the ratio of the cell viability of treated samples to the cell viability of untreated samples at each time point and are from six biological replicates.
RtcR functional complementation.
S. Typhimurium and E. coli WT strains and ΔrtcR strains with or without RtcREc or RtcRST expression plasmids (pJK3 or pCH12, respectively) were grown overnight and then subcultured to an OD600 of 0.05 in fresh LB; 10 μg/ml tetracycline was included when necessary to maintain the plasmids. Cultures were grown at 37°C with shaking to OD600 of 0.1, at which point they were induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (Gold Biotechnology, USA) at the concentrations specified in the figure legends. Incubation was resumed and continued until cultures reached mid-log phase (OD600 = 0.4). At this time, the cultures were split in two; one of each pair remained untreated as a control, and the other was treated with MMC at a concentration of 3 μM. All cultures were grown for an additional 90 min. At the end of treatment, 1 ml cells was harvested for transcriptional analysis. Cells were treated with RNAprotect (Qiagen, Hilden, Germany) according to the manufacturer's instructions prior to RNA extraction as described above. qRT-PCR to assess rtcA expression was performed as described above.
Accession number(s).
The RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (92) and are accessible through GEO Series accession number GSE82167 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82167).
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the perceptive suggestions and methodological advice provided by Timothy Hoover, Zachary Lewis, Russell Karls, and Diana Downs. We thank the following students for their assistance in strain and plasmid construction and in performing assays for expression of the RNA repair operon: David Thoms and Judy Chen, who were participants in the NSF Research Experience for Undergraduates site for Research in Prokaryotic Biology at the University of Georgia (UGA) (DBI-1460671); Alexandria Obi Okafor and Isabella Tondi-Resta, who received UGA Center for Undergraduate Research Opportunities Assistantships; and Amber Enriquez and Zahilys Soto-Arocho, who contributed to this work during their UGA Microbiology and ILS graduate program rotations, respectively.
This work was supported by National Institutes of Health grant AI117102 to A.C.K.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00476-18.
REFERENCES
- 1.Fàbrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartman CE, Samuels DJ, Karls AC. 2016. Modulating Salmonella Typhimurium's response to a changing environment through bacterial enhancer-binding proteins and the RpoN regulon. Front Mol Biosci 3:41. doi: 10.3389/fmolb.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev 76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels DJ, Frye JG, Porwollik S, McClelland M, Mrázek J, Hoover TR, Karls AC. 2013. Use of a promiscuous, constitutively-active bacterial enhancer-binding protein to define the σ54 (RpoN) regulon of Salmonella TyphimuriumLT2. BMC Genomics 14:602. doi: 10.1186/1471-2164-14-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bono AC, Hartman CE, Solaimanpour S, Tong H, Porwollik S, McClelland M, Frye JG, Mrázek J, Karls AC. 2017. Novel DNA binding and regulatory activities for σ54 (RpoN) in Salmonella enterica serovar Typhimurium 14028s. J Bacteriol 199:e00816-. doi: 10.1128/JB.00816-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das U, Shuman S. 2013. 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19:1355–1362. doi: 10.1261/rna.039917.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Taylor DW, Fowler CC, Galán JE, Wang HW, Wolin SL. 2013. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell 153:166–177. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolin SL, Belair C, Boccitto M, Chen X, Sim S, Taylor DW, Wang HW. 2013. Non-coding Y RNAs as tethers and gates: insights from bacteria. RNA Biol 10:1602–1608. doi: 10.4161/rna.26166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Thomason MK, Bischler T, Eisenbart SK, Förstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. 2015. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L. 2014. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet 5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genschik P, Drabikowski K, Filipowicz W. 1998. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J Biol Chem 273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- 13.Engl C, Schaefer J, Kotta-Loizou I, Buck M. 2016. Cellular and molecular phenotypes depending upon the RNA repair system RtcAB of Escherichia coli. Nucleic Acids Res 44:9933–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka N, Chakravarty AK, Maughan B, Shuman S. 2011. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lührmann R, Söll D, Martinez J. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 17.Filipowicz W. 2014. Making ends meet: a role of RNA ligase RTCB in unfolded protein response. EMBO J 33:2887–2889. doi: 10.15252/embj.201490425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipowicz W. 2016. RNA 3′-terminal phosphate cyclases and cyclase-like proteins. Postepy Biochem 62:327–334. [PubMed] [Google Scholar]
- 19.Das U, Chauleau M, Ordonez H, Shuman S. 2014. Impact of DNA3′pp5′G capping on repair reactions at DNA 3′ ends. Proc Natl Acad Sci U S A 111:11317–11322. doi: 10.1073/pnas.1409203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarty AK, Shuman S. 2011. RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J Biol Chem 286:4117–4122. doi: 10.1074/jbc.M110.196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Temmel H, Müller C, Sauert M, Vesper O, Reiss A, Popow J, Martinez J, Moll I. 2017. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res 45:4708–4721. doi: 10.1093/nar/gkw1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Culviner PH, Laub MT. 2018. Global analysis of the E. coli toxin MazF reveals widespread cleavage of mRNA and the inhibition of rRNA maturation and ribosome biogenesis. Mol Cell 70:868–880. doi: 10.1016/j.molcel.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurtmann EJ, Wolin SL. 2010. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci U S A 107:4022–4027. doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL. 2007. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev 21:1328–1339. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales EH, Collao B, Desai PT, Calderon IL, Gil F, Luraschi R, Porwollik S, McClelland M, Saavedra CP. 2013. Probing the ArcA regulon under aerobic/ROS conditions in Salmonella enterica serovar Typhimurium. BMC Genomics 14:626. doi: 10.1186/1471-2164-14-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasz M. 1995. Mitomycin C: small, fast and deadly (but very selective). Chem Biol 2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 28.Snodgrass RG, Collier AC, Coon AE, Pritsos CA. 2010. Mitomycin C inhibits ribosomal RNA: a novel cytotoxic mechanism for bioreductive drugs. J Biol Chem 285:19068–19075. doi: 10.1074/jbc.M109.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki H, Kilgore WW. 1967. Decomposition of ribosomal particles in Escherichia coli treated with mitomycin C. J Bacteriol 94:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki H, Kilgore WW. 1967. Effects of mitomycin C on macromolecular synthesis in Escherichia coli. J Bacteriol 93:675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storz G, Imlay JA. 1999. Oxidative stress. Curr Opin Microbiol 2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 32.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med 192:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henle ES, Linn S. 1997. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J Biol Chem 272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 34.Liu M, Gong X, Alluri RK, Wu J, Sablo T, Li Z. 2012. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol Chem 393:123–132. doi: 10.1515/hsz-2011-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wurtmann EJ, Wolin SL. 2009. RNA under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol 44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basturea GN, Zundel MA, Deutscher MP. 2011. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA 17:338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Zhao X. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother 53:1395–1402. doi: 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda H, Inouye M. 2017. Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) 9:E140. doi: 10.3390/toxins9040140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobato-Márquez D, Moreno-Córdoba I, Figueroa V, Díaz-Orejas R, García-del Portillo F. 2015. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci Rep 5:9374. doi: 10.1038/srep09374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci U S A 97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. 2014. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun 5:4115. doi: 10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferro-Luzzi Ames G, Nikaido K. 1985. Nitrogen regulation in Salmonella Typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J 4:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mrázek J, Xie S, Guo X, Srivastava A. 2008. AIMIE: a web-based environment for detection and interpretation of significant sequence motifs in prokaryotic genomes. Bioinformatics 24:1041–1048. doi: 10.1093/bioinformatics/btn077. [DOI] [PubMed] [Google Scholar]
- 45.Belitsky BR, Sonenshein AL. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci U S A 96:10290–10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenyon CJ, Walker GC. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A 77:2819–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imlay JA, Linn S. 1987. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol 169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel B. 2005. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3:e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman N, Vardi S, Ronen M, Alon U, Stavans J. 2005. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol 3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mrázek J, Xie S. 2006. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 22:3099–3100. doi: 10.1093/bioinformatics/btl551. [DOI] [PubMed] [Google Scholar]
- 52.Wade JT, Reppas NB, Church GM, Struhl K. 2005. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev 19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seitzer P, Wilbanks EG, Larsen DJ, Facciotti MT. 2012. A Monte Carlo-based framework enhances the discovery and interpretation of regulatory sequence motifs. BMC Bioinformatics 13:317. doi: 10.1186/1471-2105-13-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishna S, Maslov S, Sneppen K. 2007. UV-induced mutagenesis in Escherichia coli SOS response: a quantitative model. PLoS Comput Biol 3:e41. doi: 10.1371/journal.pcbi.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goerlich O, Quillardet P, Hofnung M. 1989. Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol 171:6141–6147. doi: 10.1128/jb.171.11.6141-6147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hostetter AA, Chapman EG, DeRose VJ. 2009. Rapid cross-linking of an RNA internal loop by the anticancer drug cisplatin. J Am Chem Soc 131:9250–9257. doi: 10.1021/ja809637e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharya R, Beck DJ. 2002. Survival and SOS induction in cisplatin-treated Escherichia coli deficient in Pol II, RecBCD and RecFOR functions. DNA Repair (Amst) 1:955–966. doi: 10.1016/S1568-7864(02)00147-7. [DOI] [PubMed] [Google Scholar]
- 58.Sheng J, Gan J, Huang Z. 2013. Structure-based DNA-targeting strategies with small molecule ligands for drug discovery. Med Res Rev 33:1119–1173. doi: 10.1002/med.21278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asad NR, Asad LM, Silva AB, Felzenszwalb I, Leitao AC. 1998. Hydrogen peroxide effects in Escherichia coli cells. Acta Biochim Pol 45:677–690. [PubMed] [Google Scholar]
- 60.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol 35:1560–1572. [DOI] [PubMed] [Google Scholar]
- 64.Maehigashi T, Ruangprasert A, Miles SJ, Dunham CM. 2015. Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res 43:8002–8012. doi: 10.1093/nar/gkv791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, Gorvel JP, Meresse S. 2013. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog 9:e1003827. doi: 10.1371/journal.ppat.1003827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Benson NR, Wong RM, McClelland M. 2000. Analysis of the SOS response in Salmonella enterica serovar Typhimurium using RNA fingerprinting by arbitrarily primed PCR. J Bacteriol 182:3490–3497. doi: 10.1128/JB.182.12.3490-3497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto Y, Shigesada K, Hirano M, Imai M. 1986. Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: localization and function of its attenuators. J Bacteriol 166:945–958. doi: 10.1128/jb.166.3.945-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, Woychik NA. 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol 71:1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- 69.Ruangprasert A, Maehigashi T, Miles SJ, Giridharan N, Liu JX, Dunham CM. 2014. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J Biol Chem 289:20559–20569. doi: 10.1074/jbc.M114.573006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu Y, Kwan BW, Osbourne DO, Benedik MJ, Wood TK. 2015. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ Microbiol 17:1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- 71.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 72.Fang FC, Frawley ER, Tapscott T, Vazquez-Torres A. 2016. Bacterial stress responses during host infection. Cell Host Microbe 20:133–143. doi: 10.1016/j.chom.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 74.Fatica MK, Schneider KR. 2011. Salmonella and produce: survival in the plant environment and implications in food safety. Virulence 2:573–579. doi: 10.4161/viru.2.6.17880. [DOI] [PubMed] [Google Scholar]
- 75.Kalderon Z, Kumar S, Engelberg-Kulka H. 2014. The SOS response is permitted in Escherichia coli strains deficient in the expression of the MazEF pathway. PLoS One 9:e114380. doi: 10.1371/journal.pone.0114380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, Seppey M, Loetscher A, Kriventseva EV. 2017. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res 45:D744–D749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 80.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 81.Benoit SL, Maier RJ. 2011. Mua (HP0868) is a nickel-binding protein that modulates urease activity in Helicobacter pylori. mBio 2:e00039-. doi: 10.1128/mBio.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clark AJ, Margulies AD. 1965. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci U S A 53:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang HJ, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hughes KT, Youderian P, Simon MI. 1988. Phase variation in Salmonella: analysis of Hin recombinase and hix recombination site interaction in vivo. Genes Dev 2:937–948. doi: 10.1101/gad.2.8.937. [DOI] [PubMed] [Google Scholar]
- 86.Maloy S, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 87.Stead MB, Agrawal A, Bowden KE, Nasir R, Mohanty BK, Meagher RB, Kushner SR. 2012. RNAsnap: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res 40:e156. doi: 10.1093/nar/gks680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J. 2012. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13:734. doi: 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mohanty BK, Giladi H, Maples VF, Kushner SR. 2008. Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol 447:3–29. doi: 10.1016/S0076-6879(08)02201-5. [DOI] [PubMed] [Google Scholar]
- 91.Burnett WV. 1997. Northern blotting of RNA denatured in glyoxal without buffer recirculation. Biotechniques 22:668–671. doi: 10.2144/97224st01. [DOI] [PubMed] [Google Scholar]
- 92.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.