Flagellins are abundant bacterial proteins comprising the flagellar filaments that propel bacterial movement. Several members of the alphaproteobacterial group express multiple flagellins, in contrast to model systems, such as with Escherichia coli, which has one type of flagellin. The plant pathogen Agrobacterium tumefaciens has four flagellins, the abundant and readily detected FlaA and FlaB, and lower levels of FlaC and FlaD. Mutational analysis reveals that FlaA requires at least one of the other flagellins to function, as flaA mutants produce nonhelical flagella and cannot swim efficiently. Suppressor mutations can rescue this swimming defect through mutations in the remaining flagellins, including structural changes imparting helical shape to the flagella, and putative regulators. Our findings shed light on how multiple flagellins contribute to motility.
KEYWORDS: flagella, motility, flagellin, Agrobacterium tumefaciens
ABSTRACT
Rotary flagella propel bacteria through liquid and across semisolid environments. Flagella are composed of the basal body that constitutes the motor for rotation, the curved hook that connects to the basal body, and the flagellar filament that propels the cell. Flagellar filaments can be composed of a single flagellin protein, such as in Escherichia coli, or made up of multiple flagellins, such as in Agrobacterium tumefaciens. The four distinct flagellins FlaA, FlaB, FlaC, and FlaD produced by wild-type A. tumefaciens are not redundant in function but have specific properties. FlaA and FlaB are much more abundant than FlaC and FlaD and are readily observable in mature flagellar filaments, when either FlaA or FlaB is fluorescently labeled. Cells producing FlaA with any one of the other three flagellins can generate functional filaments and thus are motile, but FlaA alone cannot constitute a functional filament. In flaA mutants that manifest swimming deficiencies, there are multiple ways by which these mutations can be phenotypically suppressed. These suppressor mutations primarily occur within or upstream of the flaB flagellin gene or in the transcription factor sciP regulating flagellin expression. The helical conformation of the flagellar filament appears to require a key asparagine residue present in FlaA and absent in other flagellins. However, FlaB can be spontaneously mutated to render helical flagella in the absence of FlaA, reflecting their overall similarity and perhaps the subtle differences in the specific functions they have evolved to fulfill.
IMPORTANCE Flagellins are abundant bacterial proteins comprising the flagellar filaments that propel bacterial movement. Several members of the alphaproteobacterial group express multiple flagellins, in contrast to model systems, such as with Escherichia coli, which has one type of flagellin. The plant pathogen Agrobacterium tumefaciens has four flagellins, the abundant and readily detected FlaA and FlaB, and lower levels of FlaC and FlaD. Mutational analysis reveals that FlaA requires at least one of the other flagellins to function, as flaA mutants produce nonhelical flagella and cannot swim efficiently. Suppressor mutations can rescue this swimming defect through mutations in the remaining flagellins, including structural changes imparting helical shape to the flagella, and putative regulators. Our findings shed light on how multiple flagellins contribute to motility.
INTRODUCTION
Flagella typically function as a means of bacterial locomotion in liquid environments, using a discontinuous pattern of swimming composed of straight runs and directional changes known as tumbles. Other forms of motility include swarming, which also requires flagella, and gliding, twitching, and sliding, which do not. Flagella in most bacteria are extracellular helical filaments with a curved hook serving as a universal joint between the filament and the rotary basal body embedded in the cellular membrane (1). The basal body generates power and rotates the flagellum, and the hook is important for rendering the optimum angle for propulsion. Flagellar filaments are composed of up to 30,000 subunits of the protein flagellin (2). For bacteria with multiple flagella, these form a bundle to propel forward movement and unbundle to promote cellular reorientation (3). Flagellar rotation in enteric bacteria, such as Escherichia coli and Salmonella enterica serovar Typhimurium, is bidirectional (counterclockwise promotes bundling and straight swimming, and reversals to clockwise rotation disrupt the flagellar bundle, causing tumbles). However, in members of the family Rhizobiaceae, including Sinorhizobium meliloti and Agrobacterium tumefaciens, the rotation is unidirectional and clockwise, with disruption of bundles occurring due to discordance in flagellar rotation rates (4). The helical shape of the flagellar filament is of utmost importance for these dynamic aspects of propulsion (5).
Bacterial filaments can be categorized as plain or complex depending on whether they are made of one or multiple kinds of flagellin protein (6). In well-studied systems, such as peritrichously flagellated E. coli, there are multiple filaments composed of the FliC flagellin protein (7, 8). Similarly, the alphaproteobacterium Rhodobacter sphaeroides has one flagellum (laterally positioned on the cell) that is made up of a single flagellin protein (9, 10). On the other hand, examples of bacteria with filaments made of multiple flagellins include Caulobacter crescentus, which has a single polar flagellum but, remarkably, six flagellin proteins (11). Within the Rhizobiaceae family, S. meliloti, Rhizobium leguminosarum, Agrobacterium sp. strain H13-3, and A. tumefaciens all encode multiple flagellins. A. tumefaciens mutated in three of its four flagellin genes is reduced in virulence by about 38%, and Agrobacterium sp. H13-3, lacking all of its three flagellin genes, is resistant to flagellum-specific phage infection (12–16). Flagella with plain single flagellin filaments exhibit structural polymorphisms during the course of normal flagellar propulsion and rotational switching, whereas those with complex filaments do not exhibit these polymorphisms unless they are exposed to extreme conditions of pH and ionic strengths (13, 17). Studies in C. crescentus with six different flagellin genes suggest that multiple flagellins can have a certain level of redundancy (11). In some cases, there can be a single predominant flagellin required for flagellar function, such as those for S. meliloti, Agrobacterium sp. H13-3, and Rhizobium leguminosarum (12, 14). Multiple flagellins can also be differentially regulated, as in spirochetes, Vibrio cholerae, and C. crescentus (11, 18, 19).
A. tumefaciens has a lophotrichous arrangement of 5 to 6 flagellar filaments hypothesized to be composed of four flagellins. The flagellins all share significant sequence similarity with each other (see Table S1 and Fig. S1 in the supplemental material) and are closely related to the flagellins from S. meliloti and Agrobacterium sp. H13-3. The functions and coordination of the flagellins are, however, not well studied in A. tumefaciens. FlaA, FlaB, and FlaC are encoded by a single gene cluster and are of similar lengths (FlaA, 306 amino acids [aa]; FlaB, 320 aa; FlaC, 313 aa). These three flagellins share two domains with most other bacterial flagellins: an N-terminal domain (∼170 aa) and a C-terminal domain (∼70 aa), flanking a short internal nonconserved region (50 to 60 aa). In contrast, FlaD is the most dissimilar, located roughly 15 kb distal to the flaABC cluster, and it is oriented in the opposite direction relative to the other flagellins (Fig. S2). FlaD is 430 aa, with a large internal segment (∼170 aa) that is not shared with the other flagellins. FlaA is reported to be the major flagellin, as disruption of the flaA gene severely compromised motility, reportedly resulting in vestigial stubs instead of flagellar filaments (16, 20). Other single-flagellin mutants were shown to be attenuated in motility that produced filaments with some structural abnormalities. Mutants were generated either via transposon or antibiotic cassette insertions and thus are prone to polar effects on downstream genes and partial gene copies, confounding the results reported for these flagellins and making it difficult to evaluate individual gene function (16, 20).
S. meliloti and Agrobacterium sp. H13-3 have been studied for their multiple flagellins. Like A. tumefaciens, S. meliloti has four flagellins, and Agrobacterium sp. H13-3 has three flagellins, FlaA, FlaB, and FlaD (12). Mutational analysis suggested that FlaA along with another secondary flagellin is required to assemble a functional filament and render proficient motility. Although these three taxa are related, the amino acid sequence alignment of their flagellin proteins reveals that A. tumefaciens has a higher degree of variability in the less-conserved central region. Although the amino and carboxy termini are well conserved, the internal variability suggests the possibility that flagellar filaments of A. tumefaciens also have significant structural diversity (12).
In this study, we created nonpolar deletions of the four flagellin genes, flaA, flaB, flaC, and flaD, alone and in all possible combinations in order to genetically decipher their functions and determine the role(s) that each might play in establishing flagellar filaments in A. tumefaciens. We have found that the flagellins are not redundant in their functions and differ with respect to their abundance in flagella. By specific flagellin labeling, FlaA and FlaB can be visualized in otherwise normal flagellar filaments but adopt different distributions. FlaC and FlaD are present in demonstrably lower levels. Genetic suppressor analysis has revealed that existing flagellins and other regulatory genes that affect flagellin expression can compensate for the loss of the major flagellin FlaA in flagellin mutants, and that a single key residue in FlaA is a major determinant of the helical flagellar conformation.
RESULTS
FlaA is required for normal flagellar biogenesis and motility.
Mutational analysis of the A. tumefaciens fla genes has been reported, in which the flaA gene was identified as encoding the only flagellin absolutely required for synthesizing a functional filament to confer normal motility, and it was found that flaA mutants formed vestigial stubs (16). However, the interpretation of this work was confounded by the types of mutations generated, such as by marker integration, which has a propensity for problems, including polar effects on downstream genes. To circumvent these complications, we created a complete set of in-frame deletions for all flagellin genes (flaA Atu0545, flaB Atu0543, flaC Atu0542, and flaD Atu0567) in all combinations (see Fig. S2A and B in the supplemental material). The ΔflaA mutant produces straight, somewhat shortened filaments, as evaluated by transmission electron microscopy (TEM) (Fig. 1A), rather than producing stubs as reported in earlier studies (15). The ΔflaA mutant is also quite deficient in swimming through motility agar, although less so than an aflagellate ΔflgE hook mutant (Fig. 1B). Other flagellin single mutants, ΔflaB, ΔflaC, and ΔflaD mutants, make normal flagella and confer wild-type swim ring diameters (Fig. 1).
FIG 1.
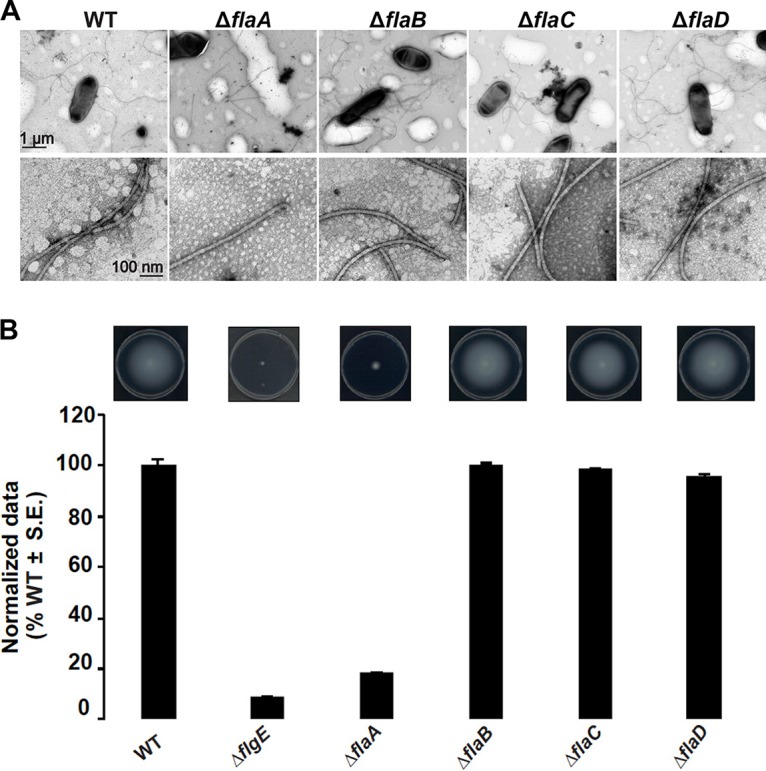
Filament morphology and swim phenotype of single-deletion flagellin mutants. (A) ΔflaA mutants exhibit straight filaments and all other mutants, ΔflaB, ΔflaC, and ΔflaD mutants, exhibit curved flagella similar to the wild type (some sheared flagella are also shown). The top row shows multiple filaments per mutant, and the bottom row shows zoomed-in views. Images were captured at magnifications of ×7,500 (top) and ×75,000 (bottom). The scale bar of 1 μm applies to all images in the top row, and the scale bar of 100 nm applies to all images in the bottom row. (B) Swimming motility phenotype of single-deletion flagellin mutants. The top images show the swim plates, indicating the expansion of the swim ring, and the graph below shows swim ring diameter data (7-day incubation) for the ΔflaA, ΔflaB, ΔflaC, and ΔflaD flagellin mutants, all normalized to the wild type. Values are the averages for three swim plates per strain. S.E., standard error of the mean.
All flagellin double and triple mutants with flaA have similar filament morphology and are equally compromised for motility (Fig. 2). The presence of flaB in the ΔflaA, ΔflaAC, ΔflaAD, and ΔflaACD flagellin mutants correlates with longer flagellar filaments than those which lack flaB (ΔflaAB, ΔflaABC, and ΔflaABD mutants) (Fig. 2A), suggesting that FlaB may have a specific role distinct from that of FlaC and FlaD. All mutants lacking flaA were complemented with plasmid-borne expression of flaA (expressed from its native promoter in combination with the E. coli lac promoter, Plac-PflaA), except for the ΔflaABCD quadruple mutant (Fig. 2B). Additionally, the ΔflaACD mutant harboring the flaA expression plasmid has roughly 50% wild-type motility, suggesting that together, the chromosomal flaB and the plasmid-borne flaA do not impart full motility.
FIG 2.
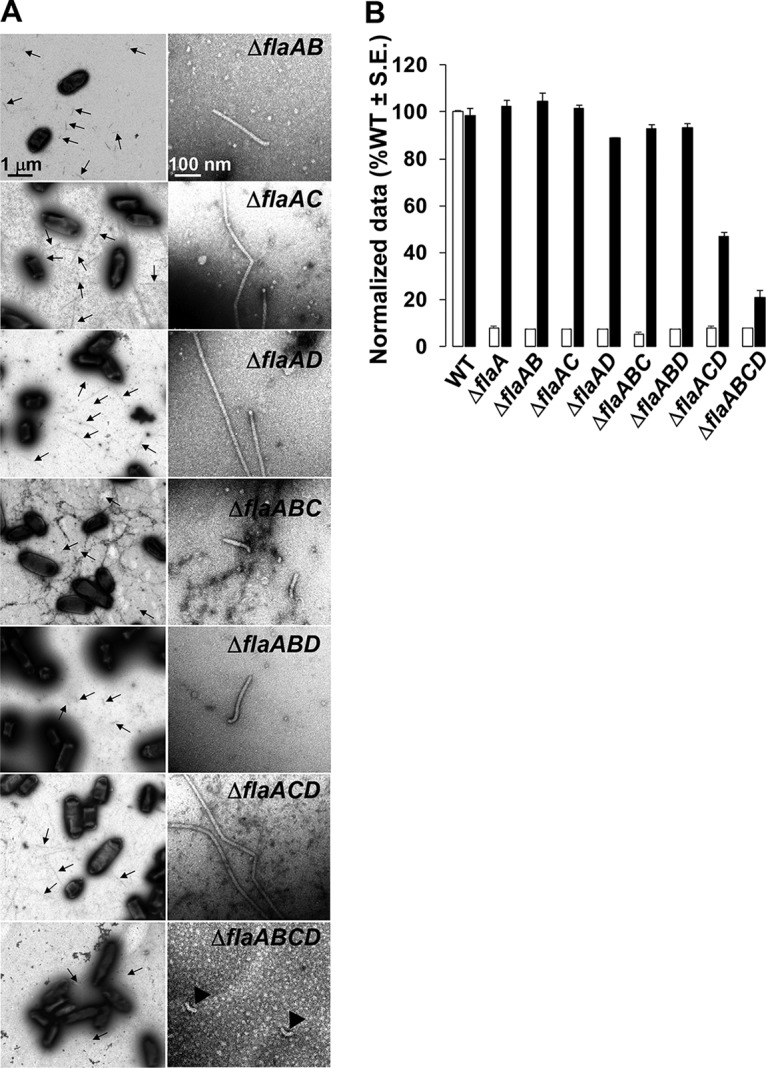
Filament morphology and swim phenotype of multiple-deletion flaA flagellin mutants. (A) All ΔflaA mutants exhibit straight filaments, and the ΔflaABCD mutant is aflagellate. The left column shows multiple filaments per mutant (indicated by arrows), and the right column shows zoomed-in images of 1 or 2 filaments. Images were captured at magnifications of ×6,000 (left) and ×60,000 (right). The ΔflaABCD mutants are aflagellate and show hooks. The scale bar of 1 μm applies to all images on the left, and the scale bar of 100 nm applies to all images on the right. (B) Swimming motility phenotype of all ΔflaA mutants harboring plasmid-borne flaA. Shown are swim ring diameter data (7-day incubation) for complementation of all ΔflaA mutants with plasmid-borne expression of flaA expressed (Plac-PflaA-flaA). Data are normalized to the wild type. White bars are the wild type or original mutants, and the black bars are wild type or the mutants carrying Plac-PflaA-flaA. Values are the averages for three swim plates per strain.
FlaA is required but not sufficient for effective motility.
FlaA appears to be absolutely required for biogenesis of normal flagella and motility; however, it was not clear if FlaA alone is sufficient for motility. The observation that the ΔflaABCD quadruple mutant with plasmid-borne flaA remained quite compromised in swimming indicated that on its own, flaA might not be sufficient for motility (Fig. 2B). In order to investigate this directly, an in-frame deletion of the three secondary flagellins, flaB, flaC, and flaD, was generated to obtain a ΔflaBCD triple mutant. The mutant exhibited very few flagellar filaments and was severely compromised in motility (Fig. 3). Although the few flagella produced by the ΔflaBCD mutant appeared to be curved, they were still insufficient to confer normal motility. Further evidence for the requirement of these flagellins is that the plasmid-borne flaA (Plac-PflaA-flaA) has little effect on this mutant (Fig. 3C). Dark-field microscopy of cells in liquid growth medium revealed some weakly swimming cells, in contrast to the completely nonmotile phenotype of the ΔflaBCD mutant alone (data not shown). This is also evident with microscopic observation of weak swimming behavior of a ΔflaABCD quadruple mutant with the plasmid-borne flaA. This suggests that flaA expression can independently confer residual basal motility and can create curved filaments, but FlaA alone does not confer normal motility.
FIG 3.
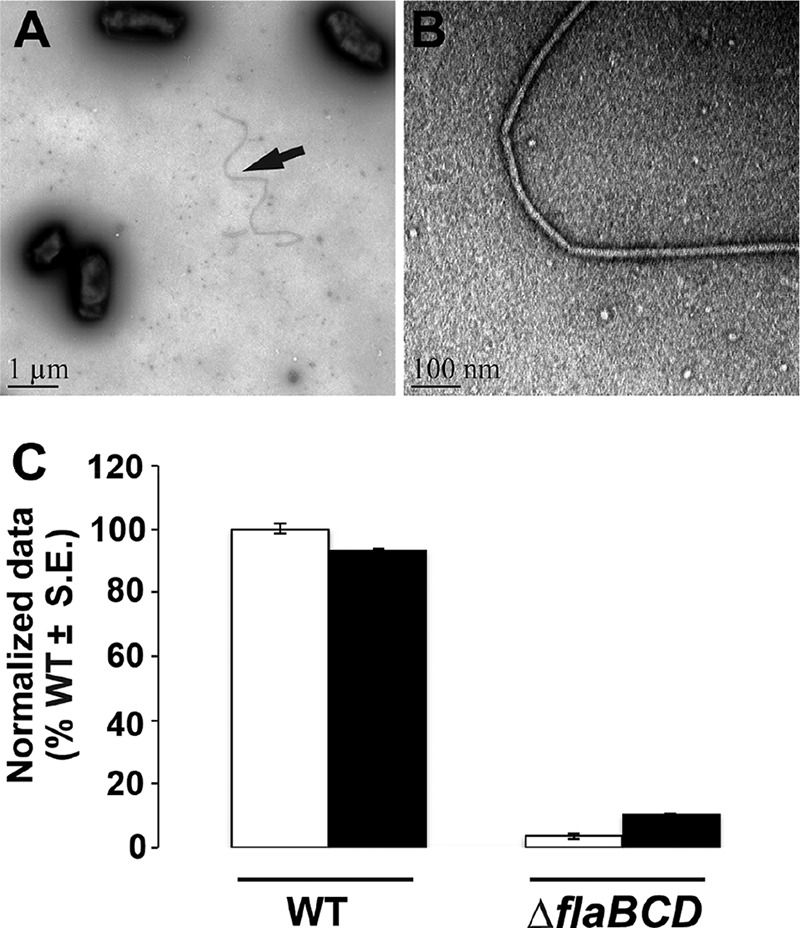
Filament morphology and swim phenotype of ΔflaBCD and motile flagellin mutants. (A and B) Curved filament morphology of the ΔflaBCD mutant. TEM images are captured at ×6,000 (A) and ×60,000 (B). (C) Day 7 swim ring diameter data for ΔflaBCD mutant alone and with plasmid-borne overexpression of flaA from Plac and native promoter of flaA (Plac-PflaA-flaA). Data are normalized to the wild type. White bars are wild type or the mutant, and the black bars are the strains with plasmid constructs. Values are the averages for three swim plates per strain.
FlaA and at least one other secondary flagellin are required for curved flagellar filaments and can confer wild-type motility.
In order to further investigate the requirement of flaBCD flagellins, we evaluated all flagellin double mutants that still retained a chromosomal copy of flaA along with any one of the secondary flagellins. Hence, we examined the following mutants: ΔflaBC mutant (retains flaA and flaD), ΔflaBD mutant (retains flaA and flaC), and ΔflaCD mutant (retains flaA and flaB). All these mutants exhibited normal flagella (Fig. 4A) and near-normal motility on swim agar (Fig. 4B). However, the ΔflaCD mutant appeared to show a significant reduction in motility in contrast to others, consistent with the observation of the ΔflaACD mutant with plasmid-borne flaA (Fig. 2B). Additionally, the ΔflaBCD mutant could be complemented with plasmid-borne expression of flaB, flaC, and flaD, indicating again that FlaA and any one secondary flagellin can confer significant levels of motility (Fig. 4B). These findings, along with the observation that the ΔflaABCD quadruple flagellin mutant could not be complemented with plasmid-borne flaA (Fig. 2B), emphasize the requirement for flaA and at least one of the three secondary flagellins genes flaB, flaC, and/or flaD).
FIG 4.
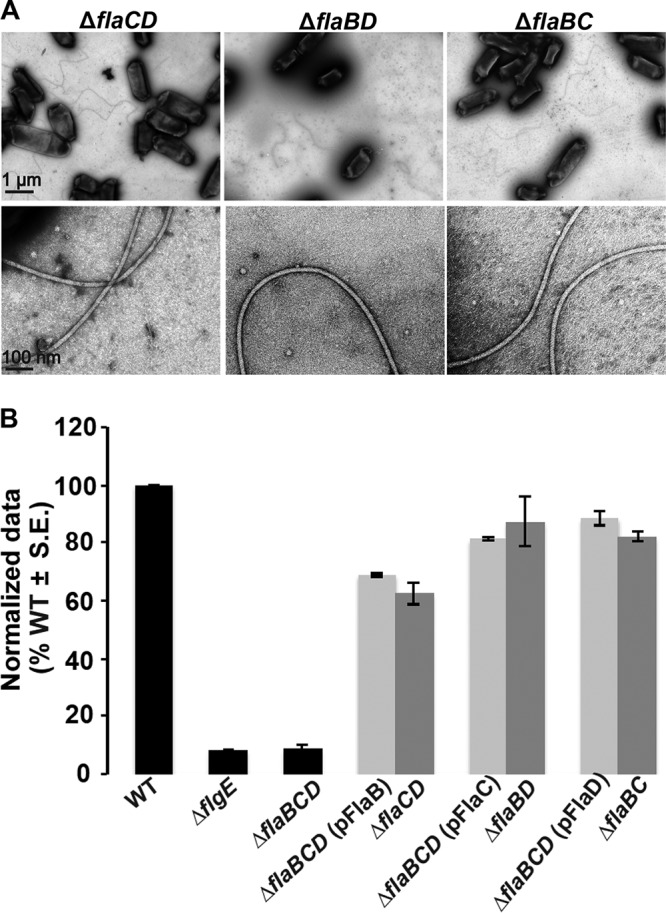
FlaA paired with any secondary flagellin restores curved flagella and motility. (A) TEM micrographs of ΔflaCD, ΔflaBD, and ΔflaBC mutants. The top row shows multiple filaments per mutant, and the bottom row shows zoomed-in views of 1 or 2 filaments. Images were captured at magnifications of ×6,000 (top) and ×60,000 (bottom). The scale bar of 1 μm applies to all images in top row, and the scale bar of 100 nm applies to all images in the bottom row. (B) Swim ring diameter data after 7 days for the ΔflaBCD mutant with plasmid-borne expression of either flaA, flaB, or flaC expressed from Plac (light-gray bars), induced with 400 μm IPTG. Data are normalized to the wild type. The double mutants are indicated for comparison (dark-gray bars). Wild type and other controls (ΔflgE and ΔflaBCD mutants) are shown as black bars. Values are the averages for three swim plates per strain.
FlaA and FlaB are the most abundant flagellins in wild-type filaments, but FlaC and FlaD are present at lower levels.
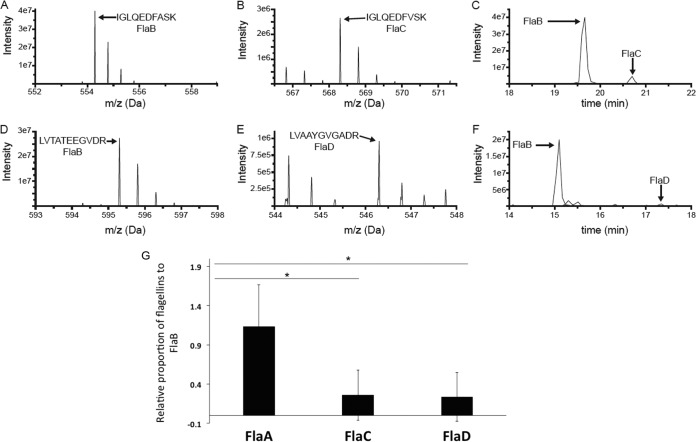
Flagellar filament proteins of the wild type and mutants had been observed by SDS-PAGE; however, the proportions of flagellins in wild-type filament preparations have not been reported (20). We quantified the relative amounts of the flagellins using liquid chromatography-mass spectroscopy (LC-MS). We first prepared wild-type filaments from three different replicates, as described in Materials and Methods. TEM analysis confirmed the presence of intact flagellar filaments in this preparation. The preparation was incubated with trypsin and analyzed by LC-MS. We identified signature peptides for all flagellins (FlaA, FlaB, FlaC, and FlaD) to be present in the wild-type filament preparation. The signal intensity observed in MS is not a direct indicator of molar amount, largely due to the fact that different peptides ionize with different efficiencies. Nevertheless, unique peptides derived from identical positions in each protein will possess similar, albeit not identical, amino acid residues, and hence will ionize to similar extents. Comparing these sets of diagnostic peptides, we determined the approximate molar ratio of each flagellin (normalized to FlaB) to be 1.3 (FlaA), 1 (FlaB), 0.2 (FlaC), and 0.2 (FlaD). Examples of MS survey scans of the peptides IGLQEDFASK and IGLQEDFVSK from FlaB and FlaC, respectively, are shown in Fig. 5A and B, and a plot of the signal intensities of these peptides as a function of LC elution time demonstrates that the signal from the FlaB-derived peptide is much more abundant (Fig. 5C). Similarly, the MS survey scans of LVTATEEGVDR and LVAAYGVGADR from FlaB and FlaD, respectively, are shown in Fig. 5D and E, and the plot of peptide intensity as a function of time demonstrates that the FlaB peptide is the more abundant of the two (Fig. 5F). The full data set of extracted peptide signals is reported in Table S2. Parallel analysis of the ΔflaABC mutant confirmed that these peptides were specific to the flagellins (data not shown).
FIG 5.
MS analysis data to determine the relative proportions of flagellin proteins in wild-type filament. (A to F) Representative examples of the MS-based relative quantification of flagellin isoforms. The experiment was conducted three times, and in the case of all peptides, the identity of the MS peak was confirmed at least once by MS/MS. (A and B) MS survey scans of two homologous doubly charged peptides from FlaB (IGLQEDFASK at 554.29 Da) and FlaC (IGLQEDFVSK at 568.30 Da) at the peak of their respective elution times. (C) Signal observed for those peptides as a function of LC elution time, with the peak for the FlaB peptide approximately 10 times more intense than that of the FlaC peptide. (D and E) MS survey scans of two homologous doubly charged peptides from FlaB (LVTATEEGVDR at 595.31 Da) and FlaC (LVAAYGVGADR at 546.30 Da) at the peak of their respective elution times. (F) Signal observed for those peptides as a function of LC elution time, with the peak for the FlaB peptide approximately 25 times more intense than that of the FlaD peptide. (G) Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis reveals the relative proportions of flagellin proteins in a wild-type filament preparation. The bar graph reports the relative abundances of FlaA, FlaC, and FlaD with respect to FlaB. FlaB was chosen as a reference, since the FlaB protein had the most peptides that were similar to peptides in the other three isoforms. The horizontal lines with an asterisk above the bars indicate that FlaA-to-FlaB ratio is relatively much higher than FlaC-to-FlaB and FlaD-to-FlaB ratios, via one-way ANOVA.
Comparing the ratios of different flagellin peptides and applying one-way analysis of variance (ANOVA), we found that quantities of FlaA and FlaB in a wild-type filament are not significantly different. After the removal of peptide ratio outliers, when normalized to FlaB levels, FlaA was significantly higher than either FlaC or FlaD. In contrast, FlaC levels were not significantly different from those of FlaD. This indicates that FlaA and FlaB are in higher proportions than FlaC and FlaD in a wild-type A. tumefaciens filament preparation and are in roughly equivalent amounts (Fig. 5G). However, observation of filament morphologies and swim assays of flagellin mutants revealed that FlaA is the functionally predominant flagellin and can confer proficient motility along with at least one of the other flagellins.
Cysteine labeling of FlaA and FlaB enables visualization of flagellar filaments.
In order to visualize flagellar filaments, we site-specifically mutated threonine residues in the flagellin coding sequences to cysteines, allowing them to be labeled with a cysteine-reactive Alexa Fluor 488 maleimide dye (21). Individual flagellins were mutated as follows: FlaA, T213C (FlaACys); FlaB, T190C (FlaBCys); FlaC, T216C (FlaCCys); and FlaD T276C (FlaDCys). All these sites were carefully chosen so that they were in a predicted exposed region of the flagellin structure, as determined by modeling on Salmonella flagellin FliC using the Phyre2 program (Fig. S3A). In order to confirm that these mutant alleles are functional, we tested complementation of the ΔflaA mutant with plasmid-borne expression of flaACys and the ΔflaBCD mutant with plasmid-borne expression of either flaBCys, flaCCys, or flaDCys, all expressed from the Plac promoter. All constructs complemented the corresponding mutants to the same level as their wild-type counterparts, indicating that these constructs are fully functional (Fig. S3B). The mutant FlaCys alleles were individually exchanged for the wild-type copy of the respective flagellin (Fla) gene in the chromosome. The site-specific mutation from threonine to cysteine does not hamper normal motility in these allelic replacement derivatives (Fig. S3C), although the flaB allele does exhibit a modest decrease in motility.
Wild-type cells encoding chromosomal FlaACys treated with Alexa Fluor 488 maleimide dye exhibited single curved long helical filaments and bundles. FlaA seems to be present uniformly along the entire filament. Flagella are often shed by the cell (Fig. 6), as was seen earlier in A. tumefaciens via transmission electron microscopy (22). In contrast to the FlaACys mutant, cells expressing FlaBCys revealed very short fluorescent filaments, many of which were not attached to cells (Fig. 6), despite full-length flagella in this mutant as indicated by TEM (data not shown) and by motility assays (Fig. S3C). This suggests that FlaB might not be present in the entire filament but rather comprise a discrete portion of it. We did not observe any fluorescent filaments with flagellated cells expressing either FlaCCys or FlaDCys, probably due to their low abundance, consistent with the findings from LC-MS analysis (Fig. 5G).
FIG 6.
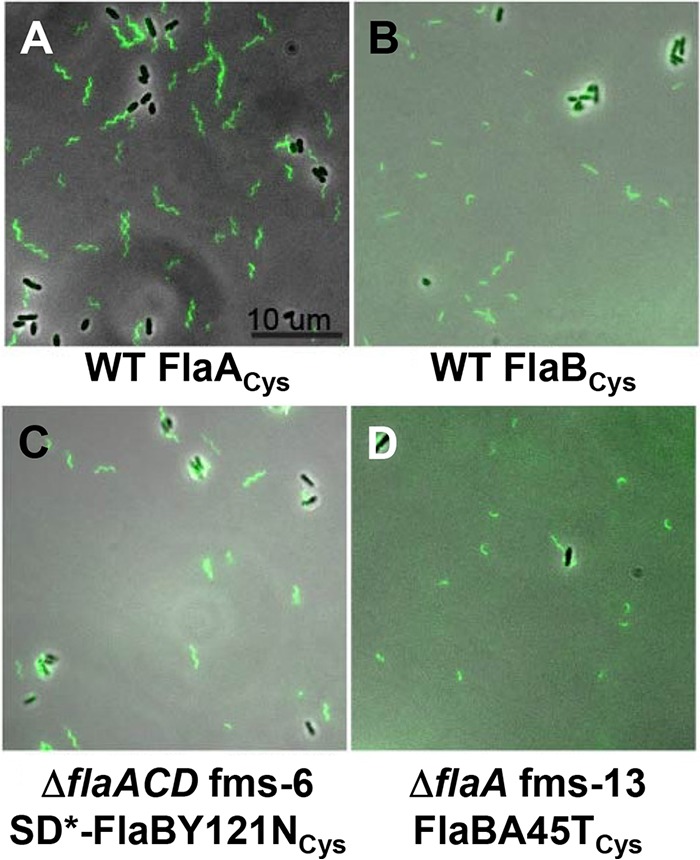
Filament morphology of wild-type and flagellin mutant suppressors that have chromosomal copies of FlaACys or FlaBCys. Flagella are visualized with fluorescence microscopy of cells labeled with Alexa Fluor maleimide dye linkage to cysteine residues in specific flagellins. The FlaCys residues were introduced into the flaA (A) or flaB (B to D) by allelic exchange of the resident gene with wild-type C58 (A and B), the ΔflaACD fms-6 mutant (C), and the ΔflaA fms-13 mutant. The scale bar of 10 μm applies to all images. Flagellar filaments with cysteine labeling of FlaC and FlaD could not be visualized, likely due to their low abundance. SD* refers to the mutated Shine-Dalgarno sequence in the suppressor mutant. Images were collected with a Nikon Ti microscope.
Isolation of suppressor mutants that retain a chromosomal flaB gene.
Migration through motility agar is severely compromised in flagellin mutants, including the ΔflaA, ΔflaAC, ΔflaAD, and ΔflaACD mutants. We observed, however, that extended incubation on motility agar gave rise to suppressors initially observed as flares emanating from the small swim rings of the parent mutants. Suppression only occurred in ΔflaA mutants having a chromosomal copy of flaB. The ΔflaAB, ΔflaABC, and ΔflaABD mutants did not produce any flares or suppressors. The isolated ΔflaA suppressor mutants exhibited significantly enhanced swim rings (Fig. S4). We designated these isolates flagellin mutation suppressor (fms) mutants.
Several candidate fms derivatives of the ΔflaA mutant were selected for further analysis, as follows: ΔflaA fms-13, ΔflaAC fms-5, ΔflaAD fms-12, ΔflaACD fms-1, and ΔflaACD fms-6. These flagellin suppressor mutants were first evaluated by sequencing for mutations in the flagellin regulators flaF and flbT, known to control flagellin expression in C. crescentus and Brucella melitensis (23–26); however, they were found to be wild type for these genes. We performed whole-genome sequencing (WGS) using an Illumina platform plus further targeted DNA sequencing to identify the mutations underlying the suppression phenotypes (Table 1 and Fig. 7).
TABLE 1.
Mutations and affected region(s) in fms derivatives
| Mutant | Parent mutant | WGS determined | Affected protein/region(s)a | Mutation(s) | Amino acid change(s) |
|---|---|---|---|---|---|
| fms-13 | ΔflaA | Yes | FlaB | G133A | A45T |
| fms-2 | ΔflaAC | No | SD-FlaB | G→A 13 bp US of flaB coding sequence | No change |
| fms-5 | ΔflaAC | Yes | FlaD, FlgE, US; SD-Atu2430 (SciP) | A311G (flaD), C1132G (flgE), G→T 9 bp US (Atu2430) | D104G (FlaD), Q378E (FlgE) |
| fms-12 | ΔflaAD | Yes | FlaB, Atu2430 (SciP) | G838C (flaB), C181T (Atu2430) | V280L (FlaB), L61F (Atu2430) |
| fms-1 | ΔflaACD | Yes | FlaB, Atu2430 (SciP) | T385A (flaB), G229C (Atu2430) | Y129N (FlaB), G77R (Atu2430) |
| fms-6 | ΔflaACD | Yes | FlaB, SD-FlaB | T385A (flaB), G→A 12 bp US of flaB start codon | Y129N |
SD, Shine-Dalgarno; US, upstream. Atu2430 is a homologue of SciP.
FIG 7.
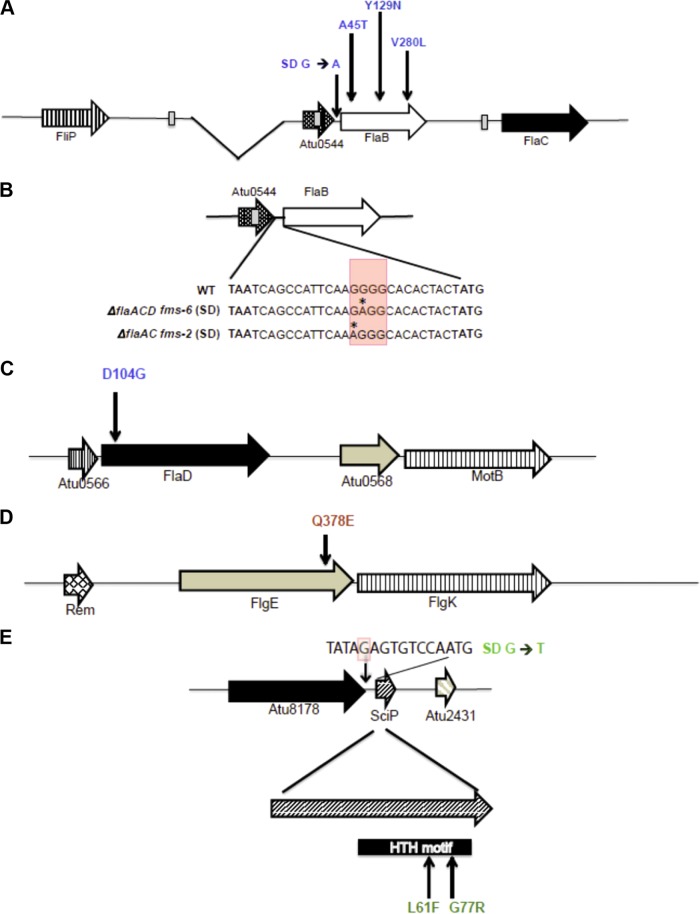
fms mutations mapped to flagellar genes and Atu2430 (SciP). The mutations are color-coded, with blue for flagellin mutations, red for the hook mutation, and green for SciP mutations. (A) Arrangement of flaB, flaC, and Atu0544 along with deletion in flaA indicated as the gap. Vertical arrows indicate fms mutations that are upstream of FlaB (Shine-Dalgarno) and within the flab coding region. (B) Arrangement of Atu0544 and FlaB and the genetic sequence starting from the Atu0544 stop codon (TAA) to the flaB start codon (ATG). Shown are details on the Shine-Dalgarno mutation at the genetic sequence level, with red shaded box indicating a predicted element and the asterisk indicating the mutated residue (SD G→A). (C and D) Arrangement of flaD (C) and flgE (D); vertical arrows indicate the fms mutations mapped to flaD and flgE. (E) Arrangement of SciP and with mutations mapped to Shine-Dalgarno (SD* G→T, shaded red in the sequence) and in the SciP coding region. FlaB, FlaD, FlgE, and SciP are 320, 430, 425, and 90 codons in length, respectively.
The most common mutations identified among the suppressors were either in the flaB coding sequence or its upstream sequences, often in combination with additional mutations affecting motility. We found that ΔflaA fms-13 had a single base substitution (G133A) in the flaB flagellin gene (Atu0543) producing a mutant FlaBA45T protein. We characterized two mutants isolated from the ΔflaAC parent. The ΔflaAC fms-2 mutant had a distinct G→A substitution located 13 bp upstream of the flaB coding sequence (Table 1) and thus had incurred a base substitution in its predicted Shine-Dalgarno sequence (Fig. 7A). However, this mutant was not analyzed by WGS, and hence, we are uncertain if there are other contributing mutations.
We performed WGS on the ΔflaAC fms-5 mutant, and in contrast to the two mutants described above, found it did not have mutations in or around flaB. Rather, fms-5 had three mutations (Table 1), including (i) a single base substitution (A311G) in the flaD flagellin gene (Fig. 7C), producing FlaDD104G mutant protein, (ii) a second mutation (C1132G) in the hook gene flgE (Fig. 7D) (Atu0574, 1,278 bp), producing a mutated FlgEQ378E protein, and (iii) a third mutation creating a single base substitution (G→T) located 9 bp upstream in the Shine-Dalgarno region of the predicted transcription factor sciP (Fig. 7E) (Atu2430, 276 bp). Originally identified in C. crescentus (27), SciP (small CtrA inhibitor protein) is an essential helix-turn-helix (HTH) transcription factor and has been shown to regulate several flagellar and chemotaxis genes (28).
The ΔflaAD fms-12 mutant was found to have incurred two mutations, a single base substitution (G838C) in the flaB flagellin gene producing FlaBV280L (Fig. 7A), and a second single base substitution also in Atu2430 (C181T), encoding the SciP regulator, generating a predicted SciPL61F mutant protein. The ΔflaACD triple mutant was particularly interesting, as it only has the single flagellin gene flaB; thus, we analyzed two independent suppressor mutants from this parent. The ΔflaACD fms-1 mutant had a single base substitution (T385A) in the flaB flagellin gene, producing FlaBY129N (Table 1), along with another single base substitution (G229C) also in the sciP gene (Atu2430), yielding SciPG77R (Fig. 7E). The ΔflaACD fms-6 mutant had the a Y129N FlaB flagellin mutation identical to that with fms-1, along with another single base substitution (G→A) located 12 bp upstream of the flaB coding sequence (Table 1) in its predicted Shine-Dalgarno sequence, which we designated SD*-FlaB (Fig. 7B).
All of the mutated flagellin residues are conserved between flagellins (FlaBA45, FlaBV280, and FlaDD104), with the exception of FlaBY129 (Fig. S1). Both SciP mutations at L61 and G77 mapped to its HTH motif and are conserved between SciP from A. tumefaciens and C. crescentus (Fig. S5).
Recreation of fms mutations to test sufficiency.
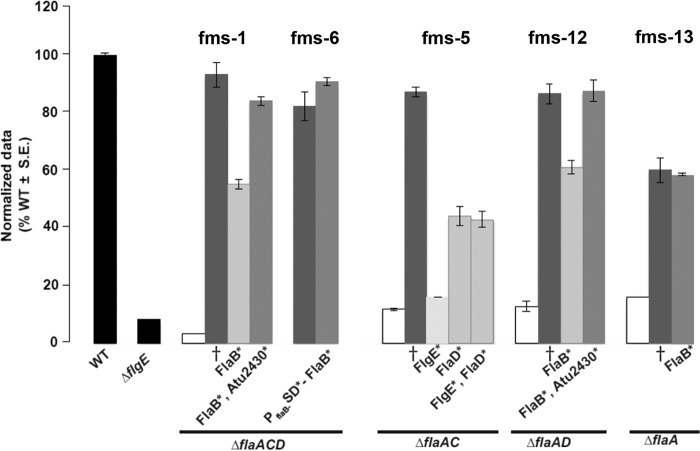
Allelic replacement was performed for the wild-type genes in the parent strains with the mutated copy of the implicated fms allele. The fms mutations were recreated either as a one-step allelic replacement as for ΔflaA fms-13, where only flaB was mutated, or in a multistep process as for ΔflaAD fms-12, where flaB and sciP were mutated (see Materials and Methods). Recreated ΔflaA fms-13 and ΔflaACD fms-6 mutants restored effective motility equivalent to that of the respective fms mutant, when either mutated flaB (for fms-13) or mutated flaB along with its Shine-Dalgarno mutation (for fms-6) was replaced in its parent background (Fig. 8). The recreated ΔflaAC fms-5, ΔflaAD fms-12, and ΔflaACD fms-1 mutants, however, could not completely restore the suppression swim phenotype with single allelic replacements of either mutated flaB (for fms-12 and fms-1) or flgE (for fms-5) alone. Both of the recreated ΔflaAD fms-12 and ΔflaACD fms-1 mutants also needed their respective sciP alleles in order to completely restore the suppression phenotype. Interestingly, recreated ΔflaAC fms-5 mutants with the mutated flgE allele alone did not rescue the nonmotile phenotype of the ΔflaAC mutant at all. Motility was partially restored to about the same degree with the mutated flaD allele alone or in combination with a flgE mutation. It is possible that the recreated ΔflaAC fms-5 mutant would be completely restored if the region upstream of its sciP is also replaced with the mutated allele, since in other cases (for fms-1 and fms-12), mutated sciP appears to play an important role in the suppression phenotype (Fig. 8). Based on studies of SciP in C. crescentus, we would predict that mutations that diminish its activity either through decreased expression levels or by inhibiting protein function would result in higher flagellin expression. Introduction of a plasmid-borne flaB-lacZ reporter fusion (pBM196) (Table S4) into three suppressor mutants, one with the mutation upstream of sciP (fms-5) and the other two with amino acid substitutions (fms-12 and fms-1) in the sciP product, revealed elevated expression levels in all of these suppressor mutants compared with their parent strains (Fig. S7).
FIG 8.
Swimming motility phenotype of fully and partially recreated fms mutants. Swim ring diameter data after 7 days showing the motility phenotype for originally isolated fms mutants and their recreated counterparts. The relevant fms mutant for each set of bars is indicated above them. Data are normalized to the wild type. The wild type and ΔflgE mutant are shown as black bars. White bars indicate the parent mutants. Dark-gray bars are the originally isolated fms mutants (†). Light-gray bars indicate the recreated suppressors that do not completely phenocopy the original suppressor. Medium-gray bars indicate recreated mutants that completely reproduce the original fms phenotype. Asterisks by the protein names indicate the mutated residues incorporated via allelic replacement in those strains. Values are the averages for three swim plates per strain.
FlaBY129N mutation can confer helicity to FlaB flagellar filaments.
FlaA and FlaB were found by LC-MS to be in similar proportions in a wild-type flagellar filament preparation (Fig. 5G). Since the fms mutations commonly mapped to FlaB, we further examined how the mutated FlaB might confer suppression and enhance motility. Also, all ΔflaA mutants make straighter, nonhelical, and nonfunctional flagella (Fig. 1A and 2A), but the suppressors have restored motility and thus either reverse or overcome the conformational deficiency. We introduced by allelic replacement the FlaBCys mutation into two suppressor mutants that had been confirmed to require only mutated FlaB (Fig. 8) to recreate efficient motility: ΔflaA fms-13, which was mutated for FlaBA45T, and ΔflaACD fms-6, which had both SD*-FlaB and FlaBY129N mutations (Fig. 7). The ΔflaA fms-13–FlaBCys mutant exhibited short curved flagellar filaments very similar to those of the wild type (WT) with FlaBCys, but interestingly, the ΔflaACD fms-6 FlaBCys exhibited helical flagellar filaments very similar to those observed for WT FlaACys (Fig. 6C and D). Hence, both of these suppressors made functional flagella but seemed to do so by different mechanisms.
However, it was unclear which mutation in the ΔflaACD fms-6 mutant (SD*-FlaB or FlaBY129N) contributed to the helical conformation of the FlaB-labeled flagellar filament. In order to ascertain which mutation was responsible, we separated these two mutations. We first compared the expression levels of a plasmid-borne wild-type PflaB-SD-lacZ translational fusion with that from a mutated PflaB-SD*-lacZ (with the fms-6 mutation) in the ΔflaACD mutant via a β-galactosidase assay. The expression for the mutated Shine-Dalgarno sequence of flaB is higher than that of the wild type (Fig. S6). It therefore seems likely that the SD*-FlaB mutation increases FlaB production.
We had observed repeatedly that plasmid-borne flaA expression driven from Plac in combination with the native promoter of flaA (PflaA) complements all ΔflaA mutants entirely, in contrast to expression from Plac alone in a ΔflaA mutant, presumably reflecting the strength of the PflaA promoter (compare Fig. 2 with S3B). We first expressed FlaBCys from the plasmid as a fusion to Plac-PflaA and examined this construct in the ΔflaA and ΔflaACD mutants. The ΔflaA mutant was included to examine the effect of the plasmid-encoded FlaB in a background where other flagellins were also present, and it was observed to partially rescue motility, whereas its expression in the ΔflaACD mutant did not (Fig. 9A). Both the ΔflaA and ΔflaACD mutants with Plac-PflaA-FlaBCys exhibited straight filaments (Fig. 10E and F) that correlate with their decreased swim ring diameters. We next expressed FlaBY129NCys from Plac-PflaA, and both the ΔflaA and ΔflaACD mutants were fully rescued for motility and exhibited helical flagella indistinguishable from those of the Plac-PflaA-FlaACys constructs in the same mutants (Fig. 9A and 10A, B, G, and H). This suggests that FlaBY129N is important for formation of helical flagella that impart proficient swimming in the suppressor mutants.
FIG 9.
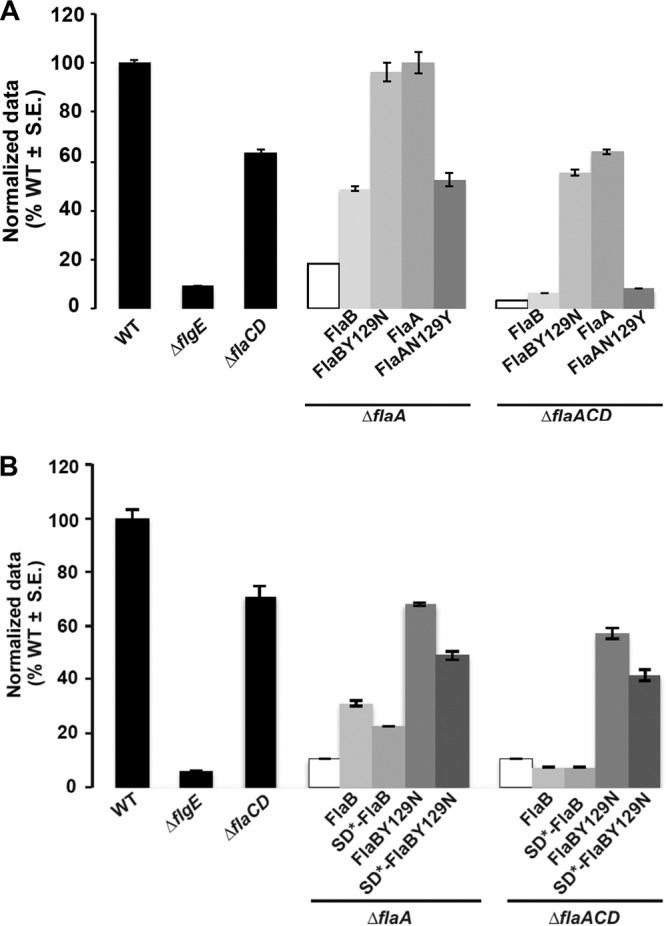
Swimming motility of ΔflaA and ΔflaACD mutants harboring plasmids carrying flaA and flaB alleles that impart altered helicity. Swim ring diameter of mutant A. tumefaciens derivatives. Results for the wild type, ΔflgE mutant, and ΔflaCD mutant are provided (black bars) for reference. Shown are the ΔflaA and ΔflaACD mutants (white bars), and derivatives harboring plasmids expressing FlaBCys or FlaBY129NCys and FlaACys or FlaAN129YCys expressed from Plac-PflaA (A) or from Plac-PflaB as well as the SD* mutation in the flaB Shine-Dalgarno sequence (B). Data are normalized to the wild type. Values are the averages for three swim plates per strain.
FIG 10.
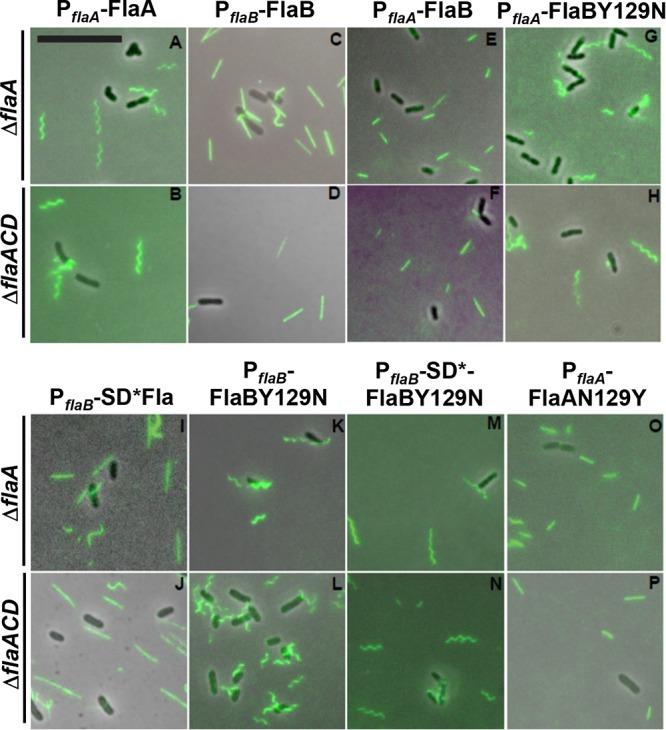
Filament morphology of plasmid-borne flagellin mutants visualized via fluorescence microscopy. Alexa Fluor 488 maleimide labeling of A. tumefaciens derivatives from ΔflaA (top row) and ΔflaACD (bottom row) strains. Flagellin constructs are expressed from Plac-PflaA or Plac-PflaB (as indicated). Mutants express either FlaACys or FlaBCys or SD*-FlaBCys and FlaAN129YCys or FlaBY129NCys, as indicated. Scale bar is 10 μm and applies to all images.
In order to examine the impact of the flaB Shine-Dalgarno mutation (from the fms-6 mutant), we expressed the wild-type FlaBCys allele and the mutant derivatives, SD*-FlaBCys, FlaBY129NCys, and SD*-FlaBY129NCys, from Plac-PflaB in the ΔflaA and ΔflaACD mutants and observed their flagella and swimming proficiency. Both mutants expressing plasmid-encoded FlaBCys constructs exhibited mostly straight flagella with occasional curvature similar to the WT FlaBCys (Fig. 10C and D), and only the ΔflaA mutant showed a modest improvement in motility (Fig. 9B). Cells with SD*-FlaBCys constructs also displayed a phenotype similar to that observed with FlaBCys (Fig. 10I and J). Interestingly, cells expressing FlaBCysY129N or SD*-FlaBY129NCys from Plac-PflaB exhibited helical flagellar filaments, with flagella for FlaBY129NCys (Fig. 10K and L) that are qualitatively shorter than those for the SD*-FlaBY129NCys (Fig. 10M and N). This suggests that FlaBY129N is independently contributing to helical flagella and that the Shine-Dalgarno mutation is contributing by increasing flagellin levels. Both ΔflaA and ΔflaACD mutants could be partially rescued with Plac-PflaB driven expression of either FlaBY129NCys or SD*-FlaBY129NCys (Fig. 9B). Here, we do observe a slight decrease in swim ring diameter for both the mutants harboring SD*-FlaBY129NCys (fms-6 mutation) constructs, consistent with a slight reduction in motility with the FlaBCys allelic replacement in WT (Fig. S3C), suggesting that this allele may manifest a minor deficiency.
Asparagine at position 129 in FlaA contributes to the formation of helical flagella.
Our results suggest that the FlaBY129N mutation is a major determinant for helical flagella in the ΔflaACD fms-6 suppressor and when this allele was expressed from a plasmid in both ΔflaA and ΔflaACD mutants. We therefore examined the function of this residue in the major flagellin FlaA. In FlaA, the residue at this position within the projected flagellin structure (Fig. S3A) is an asparagine (N129), which corresponds to the tyrosine (Y129) in FlaB (Fig. S1) that is mutated to an asparagine in several suppressor mutants. In order to investigate whether N129 in FlaA is important for rendering helical flagellar filaments, we site-specifically mutated FlaAN129Cys to FlaAY129Cys and expressed it from a plasmid-borne Plac-PflaA in the ΔflaA and ΔflaACD mutants. Interestingly, both the ΔflaA and the ΔflaACD mutants expressing Plac-PflaA-FlaAN129YCys exhibited straighter flagella (Fig. 10A, B, O, and P) and significantly weaker complementation (Fig. 9A and B) than the helical flagella and full complementation displayed by a mutant expressing FlaACys. This indicates that FlaAN129 is indeed important for the formation of helical flagella and thus an important contributor to motility.
DISCUSSION
Bacteria synthesize flagellar filaments from either one or multiple flagellins, and it is thought that these differences might have evolved in response to the environmental niche of each particular bacterial taxon (6, 29–31). Complex flagellar filaments composed of multiple flagellins are thought to have evolved in soil bacteria, like S. meliloti and Agrobacterium sp. H13-3, to provide stable filaments that enable efficient swimming and propulsion in highly viscous environments (6, 31). For host-associated bacteria, flagella are often recognized by the host defense response as pathogen-associated molecular patterns (PAMPs) (32). In fact, members of the Rhizobiaceae with complex flagella lack the recognized flagellin motif and are instead recognized by the host through perception of the translation factor Ef-Tu (33). It is clear that flagella are under multiple levels of strong selection in the environment for their abundance, distribution, and composition.
During flagellum assembly, flagellin proteins are transported through the narrow lumen of the filament and added at the growing tip. Recently, transport and assembly have been proposed to proceed via an injection-diffusion mechanism (34). Thus, the entry of flagellins into the export channel is a major step in the process. Given the proposed injection-diffusion assembly model, the mechanism by which bacteria coordinate flagellar biosynthesis with multiple different flagellins is even more intriguing.
Integration of multiple flagellins in A. tumefaciens motility.
In this study, we systematically examined the functions of the four flagellins that are thought to comprise the lophotrichous flagella tuft of A. tumefaciens. In certain bacteria, such as C. crescentus, multiple flagellins are thought to be redundant (11), and in others, they are differentially regulated or have unique functions (12, 14, 18, 19, 35, 36). In A. tumefaciens, FlaA is thought to be the major flagellin, with accessory roles for the secondary flagellins, FlaB, FlaC, and FlaD. The ΔflaA mutants were thought to form vestigial stubs and were impaired for motility (16, 20). However, our studies with carefully constructed in-frame mutations reveal that rather than forming stubs, ΔflaA mutants form filaments that lack the curvature found in wild-type filaments. The ΔflaA mutants do not appear to be completely immobilized, as for the hookless ΔflgE mutant (Fig. 2), and under the light microscope, very slow wiggling or traversing the field of view is observed (data not shown). Our findings suggest that FlaA is the major flagellin in part because it engenders helical shape to the filament.
The role of secondary flagellins was not clear from earlier studies. However, in the related Sinorhizobium meliloti and Agrobacterium sp. H13-3 organisms, secondary flagellins are also required for proficient swimming motility, as FlaA together with any one secondary flagellin were able to assemble a functional filament and thus enable the cells to swim (12). We performed detailed analysis in A. tumefaciens to ascertain the function of individual flagellins, finding that each flagellin can contribute to motility, and that the secondary flagellins FlaB, FlaC, and the most divergent FlaD, can combine with FlaA to drive efficient motility. However, there are discernible differences in motility, flagellar conformation, and length between specific flagellin mutants, suggesting distinct functionalities. In S. meliloti, feedback regulation has been reported to exist on flagellin synthesis, by which ΔflaA mutants have decreased expression of the secondary flagellins (12). We have not observed any significant effects on flagellar gene expression in the different A. tumefaciens flagellin mutants (B. Mohari and C. Fuqua, unpublished data).
Biophysical considerations of complex flagella.
What is the role for multiple flagellins in motile bacteria compared with those that have a single plain flagellin type? In plain flagella, flagellin monomers are organized in a helical pattern leaving an interior channel of 10 Å in radius; 11 monomers make up one turn of a helical element, with a rise of 52 Å (37). Hence, a flagellar filament usually consists of 11 protofilaments 10 to 20 μm in length, which can each transition between different degrees of helicity. Plain flagella can assume different forms, each with a distinct overall helical pitch (38). Transitions between these polymorphisms under hydrodynamic torque as a result of flagellar motor rotation reversal are responsible for flagellar bundle disruption, leading to tumbles in peritrichous multiflagellated bacteria. Each flagellin monomer can exist in one of two conformational states, L-type and R-type (that differ slightly in length), leading to L- or R-type protofilaments, given the high degree of cooperativity among subunits along each protofilament (39–41). When all protofilaments are L or R type, the flagellum is straight, while different proportions correspond to different ratios of L- and R-type monomers (38). Recent cryo-electron microscopy (cryo-EM) studies have provided the highest resolution analysis of plain filaments to date, allowing comprehensive demonstration of the bistable model using different flagellar structures obtained from mutants of Gram-positive and Gram-negative species (42).
A large proportion of bacteria produce complex flagellar filaments consisting of multiple flagellins, represented by monotrichous, peritrichous, and lophotrichous cellular distributions, as with A. tumefaciens. The complex filaments corresponding to monotrichous and lophotrichous arrangements are stiffer than the plain filaments of the better studied peritrichously flagellated bacteria, with consequences for the biophysical mechanisms underlying the generation of tumbles. Detailed experimental characterization of motility in Agrobacterium sp. H13-3 revealed that the rigid flagellar filaments do not undergo polymorphic transitions in the course of flagellar unbundling, triggered by changes in rotational speed of the flagella (13). Noting that the generation of supercoiling in simple flagella requires that monomeric subunits (and resulting protofilaments) exist in two different conformational states, we speculate that supercoiling in complex filaments instead arises from the existence of multiple flagellins with inherent structural differences. Rather than having different conformations of a single monomeric subunit, it is conceivable that packing multiple flagellins with inherent structural differences into filaments, perhaps in random distributions, also gives rise to mechanical strains that curve and twist the filament. This in turn may provide the different helical states that dictate propulsion with complex flagella, and in lophotrichous arrangements, such as A. tumefaciens, may also influence bundling. In A. tumefaciens, each type of flagellin and their distribution in filaments likely contributes in a different way, giving rise to some of the phenotypic changes we observe with the specific flagellin mutants.
Suppression of flagellin mutants by changes in relative expression and flagellum structure.
Given that we were able to observe suppressors of severely compromised flagellin mutants with the ΔflaA mutation, it seemed likely that in the appropriate contexts, secondary flagellins could compensate for the absence of FlaA. Suppression was observed only in mutants that retain a chromosomal copy of flaB, so we hypothesized that FlaA and FlaB might have somewhat similar functions. In the ΔflaACD fms-6 mutant, FlaBY129N in combination with base substitutions that alter the Shine-Dalgarno sequence reproduced the suppressor phenotype (Fig. 9B), but we discovered later that FlaBY129N is primarily responsible for rendering a helical shape to flagellar filaments in these mutants (Fig. 10L). It is interesting that this kind of suppression was only observed in ΔflaACD suppressors where FlaB is the only flagellin present. However, for the ΔflaACD fms-1 mutant, FlaBY129N in combination with SciP G77R were required to reproduce the suppressor phenotype (Fig. 8). Perhaps similarly, the increased expression from the SD*-FlaB mutation in the fms-6 mutant in part contributed to the improved motility in the ΔflaACD mutant. Overall, the above-mentioned observations suggest that the FlaBY129N mutation compensates for the absence of FlaA by rendering a helical shape to the flagella, and additional mutations can augment the impact of this change by increasing flaB expression.
The helicity of flagella is clearly an important attribute for their function. Extreme pH values of 3 and 11 can transform helical filaments to straight ones (13). We found that the mutation of a single tyrosine residue to an asparagine in FlaB (as in the fms-6 mutant) is sufficient to impart the helical shape of the mutant flagellar filament in our fms-6 mutant. Also, FlaA has an asparagine at the exact same position. Accordingly, we found that creating a FlaAN129Y (mutated) flagellin abolishes the helical flagellar conformation, transforming flagella to straight filaments that do not function in the context of a ΔflaACD mutant (Fig. 9A and 10O and P). Hence, it seems that FlaAN129 is a major contributor to the helical conformation of the flagella in A. tumefaciens. FlaC and FlaD have serines at the corresponding position, as do all of the flagellins from S. meliloti and Agrobacterium sp. H13-3. Of the six flagellins in C. crescentus at the equivalent position, five have an asparagine residue and one has a serine residue (11). In Shewanella oneidensis, two flagellins, FlaA and FlaB, comprise the filaments, and two key residues determine the activity of the FlaB flagellin; however, it is not known whether these residues determine helical shape (43). Here, we show that in A. tumefaciens, FlaAN129 is a critical residue that might contribute significantly to the role of FlaA as the major flagellin.
We observed that another frequent mechanism for suppression was via antagonism of SciP, a predicted inhibitor of flagellar gene expression. The suppressor mutations we isolated in sciP all resulted in elevated expression of flaB (and probably the other flagellins as well), one through mutation of the putative sciP Shine-Dalgarno sequence and the other two by changes in the SciP amino acid sequence. In C. crescentus, SciP (small CtrA inhibitor protein) is an essential HTH transcriptional factor that directly binds to CtrA (27), a two-component response regulator protein and the master regulator of flagellar gene control. As a class I flagellar gene at the top of the flagellar gene hierarchy, CtrA regulates the class II flagellar structural genes (28, 44). The SciP homologue from A. tumefaciens is 91% similar and 84% identical to that from C. crescentus, and aligning them reveals that the helix-turn-helix motif is well conserved (Fig. S5). SciP exerts its effects in swarmer cells of C. crescentus and thus controls the expression of many flagellar and chemotaxis genes by inhibiting CtrA, a function important for normal cell cycle progression. Several conserved residues, including R35 and R40 outside the HTH, are critical for SciP-CtrA interaction, and mutating these residues blocks the interaction, thus hampering the SciP overexpression phenotypes (27). The residues mutated in the A. tumefaciens fms mutants we isolated have not been previously identified as important; however, these are conserved and are clearly within the HTH, suggesting a role in DNA binding or for interactions with CtrA. The sciP homologue is not essential in A. tumefaciens, and thus its role in regulating CtrA, which is itself essential, remains speculative (45). Even so, it is possible that the mutations are to some extent relieving the inhibition of SciP on CtrA, thus allowing a greater transcription of flagellin genes, thus enhancing motility in the suppressors.
There thus seem to be multiple mechanisms to achieve suppression and thus overcome the absence of specific flagellins. An open question is whether flagellar filaments are composed of all four flagellins or whether they occur in specific combinations. There may be environmental conditions that lead to different proportions of the flagellins in filaments. We now know that the flagellins which comprise the flagellar filament of A. tumefaciens are not simply redundant in function, and we have gained insights into their specific roles and relative abundances.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Table S3 in the supplemental material lists all the bacterial strains and plasmids used in this study. Table S4 lists all oligonucleotides used. Buffers, reagents, antibiotics, and media were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma Chemical Co. (St. Louis, MO). Oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA). E.Z.N.A. plasmid miniprep kits (Omega Bio-tek, Norcross, GA) were used to purify DNA. All restriction enzymes and molecular biology reagents were obtained from New England BioLabs (Ipswich, MA). DNA was manipulated using standard techniques described earlier (46). Standard Sanger-type DNA sequencing was performed on an ABI 3730 sequencer at the Indiana Molecular Biology Institute, Bloomington, IN. Plasmids were introduced into A. tumefaciens via electroporation (47, 48). E. coli was grown in LB medium, and A. tumefaciens was grown in AT minimal medium (49) supplemented with 0.5% (wt/vol) glucose and 15 mM ammonium sulfate (ATGN). For allelic replacements, where ATSN medium was used, 5% (wt/vol) sucrose was used as the sole carbon source instead of glucose for sacB counterselection. The antibiotic concentrations used were as follows: for E. coli, 100 μg · ml−1 ampicillin (Amp) and 25 μg · ml−1 kanamycin (Km), and for A. tumefaciens, 300 μg · ml−1 Km and 50 μg · ml−1 spectinomycin (Sp). Isopropyl-thio-β-d-galactopyranoside (IPTG) was added to the media for Plac induction at the indicated concentrations, when appropriate.
Construction of complementation constructs.
Wild-type or site-specifically mutated flagellin coding sequences (wild-type flaA, flaB, flaC, and flaD and their corresponding mutated genes [flaAT213C, flaBT190C, flaCT216C, and flaDT276C]) were amplified with primers comp P1 and comp P2 from AtC58 genomic DNA using Phusion high-fidelity DNA polymerase. These fragments were gel purified and ligated to pGEM-T Easy, and their sequences were confirmed using appropriate restriction enzymes and then religated to pSRK-Km previously cleaved with corresponding restriction enzymes. For all complementation constructs except the ones with the native flagellin promoters, 5′ primers were designed to incorporate the NdeI site of pSRK-Km in frame with the lacZα start codon to be cloned into the IPTG-inducible expression vector pSRK-Km (50). The pSRK-Km constructs were electroporated into A. tumefaciens, as described previously (47). For the construction of Plac-PflaA-flaA and Plac-PflaA-flaAT213C, the entire promoter region upstream of flaA along with its coding region was amplified using flaA PflaA promoter comp P1 and flaA comp P2 primers. The 5′ flaA promoter comp P1 was designed with a stop codon, TGA, after the restriction site SpeI in order to terminate translation from lacZα to prevent occlusion of the native translation start site. Plac-Pnative-flaB was similarly constructed.
Construction of nonpolar deletion mutants.
Deletion mutants were constructed as described earlier (51). Primers for deleting the desired gene were carefully designed so that adjacent genes are not affected and to maintain potential translational coupling. DNA fragments were generated about 500 to 1,000 bp upstream and downstream flanking the desired gene via PCR using the P1 and P2 primers for the upstream sequence and the P3 and P4 primers for the downstream sequence. Primers P2 and P3 were designed so that they could overlap each other via 18 bp of complementarity (Table S4). The two flanking fragments were then joined together via a splicing by overlapping extension (SOE) PCR using Phusion high-fidelity DNA polymerase (NEB, Ipswich, MA), as described earlier (52–54). All single-deletion mutants were generated from the WT C58 strain. For generation of the ΔflaAB mutant, the ΔflaA mutant was used as the starting strain instead of the wild-type C58 in order to ensure that the Atu0544 open reading frame (ORF) is retained in the corresponding mutants (Fig. S2). FlaAP1 and FlaBP2 were used as the P1 and P2 primers, whereas FlaBP3 and FlaBP4 were used as the P3 and P4 primers. For subsequent generation of ΔflaABC and ΔflaABD mutants, the ΔflaAB mutant was used as the starting strain. The ΔflaABC mutant was generated with the use of FlaAP1 and FlaCP2 as the P1 and P2 primers and with FlaCP3 and FlaCP4 as the P3 and P4 primers. All multiple-flagellin-deletion mutants were generated starting from a single- or double-gene-deletion mutant. The flanking DNA fragments were gel purified, and another short PCR of 5 cycles was performed using this as the template and primer. In order to join the fragments, the above-described product was amplified via another amplification using primers P1 and P4. This final product of ≈1 kb was ligated to pGEM-T Easy (Promega, Madison, WI). Colonies were confirmed by DNA sequencing, digested with the appropriate restriction enzymes, and ligated into the suicide vector pNPTS138 cleaved with the same set of enzymes. The pNPTS138 plasmid (51; M. R. K. Alley, unpublished data) has the Km resistance (Kmr) gene, and the sacB gene imparts sucrose sensitivity (Sucs). Since it has a colE1 replication origin that does not function in A. tumefaciens, in order to form viable Kmr colonies, the plasmid must recombine into a stable endogenous replicon. Correct pNPTS138 derivatives were confirmed by digestion, and the miniprep was transformed into the E. coli mating strain S17-λpir. The mating strain harboring the deletion plasmid was conjugated with A. tumefaciens via mating on LB, and recombinant colonies were selected for growth on ATGN Km plates. In order to confirm that the desired deletion plasmid had integrated and the colonies are sucrose sensitive, Kmr colonies were restreaked on ATGN Km plates and ATSN Km plates. Colonies with normal growth on ATGN Km plates and very poor growth on ATSN Km plates were chosen for further processing. The next step was to select for the correct mutants that have lost the integrated plasmid due to a second recombination event and are sucrose resistant. To obtain these colonies, a single Kmr/Sucs colony was grown overnight in ATGN broth without Km selection and then plated on ATSN. The resultant Sucr colonies were patched on ATSN and ATGN Km to verify plasmid excision. Diagnostic colony PCR was performed using primers P1 and P4 that flank the desired gene to confirm that the target gene was deleted, and this amplification product was sequenced to confirm the deletion junction.
Allelic replacement of wild-type genes with the fms alleles.
Allelic replacement procedures were performed as described previously (55). Wild-type genes were replaced with either the fms or sciP allele in the parent flagellin mutant. The fragment containing the desired mutation in the middle was PCR amplified using primers P1 fms and P2 fms or using primers P1 sciP and P2 sciP from fms mutants, fms-1, fms-5, fms-6, fms-12, and fms-13, using Phusion high-fidelity DNA polymerase. Positioning the desired mutation in the center of the fragment increased the probability of recombination events on either side. The PCR fragments were gel purified and ligated to pGEM-T Easy. The remainder of the process was identical to the construction of nonpolar deletion mutants described above. This included incorporation of the pNPTS138::fms allele(s) in the parent mutant A. tumefaciens via mating. For allelic replacement of sciP alleles, the procedure was modified a bit due to the apparent toxicity of the gene when expressed in E. coli. We therefore used a triparental mating approach using the helper plasmid pRK2013 (56) or pRK2073 (57). After desired Kms/Sucr colonies were obtained either from the biparental or the triparental mating, colony PCR was performed to amplify the DNA, which was later gel purified, and the desired fms or sciP mutation was confirmed via DNA sequencing using internal sequencing primers. The procedure of allelic replacement was repeated sequentially when more than one mutation needed to be incorporated in a parent mutant.
Site-directed mutagenesis of flagellins.
Site-directed mutagenesis was performed as described earlier for FlaAT213C, FlaCT216C, and FlaDT276C, using the QuikChange protocol obtained from Stratagene Corp. (55). Long self-complementary mutagenic primers F1mut and R1mut (melting temperature [Tm], ∼68°C) were designed harboring the desired mutations individually in the middle for all flagellin genes (Table S4). Mutagenic primers and Phusion DNA polymerase were used to perform PCR on the following pGEM-T Easy plasmids: pBM139 (containing flaA), pBM147 (containing flaC), and pBM128 (containing flaD) (Table S4). DpnI digestion of the PCRs was performed in order to remove the parental wild-type and hemimethylated plasmid DNA. The newly synthesized uniformly nonmethylated mutated DNA should not be cleaved and will transform into E. coli, followed by sequencing to confirm the mutations and then introduction into the wild type by allelic replacement as described above. The mutated DNA fragment (either flaAT213C, flaCT216C, or flaDT276C) replaced its counterpart in the wild-type genome. Allelic replacement for the FlaBT190C strain was accomplished via an SOE PCR. Long self-complementary mutagenic primers FlaBT190CF1mut and FlaBT190CR1mut (Tm, ∼68°C) were designed harboring the desired mutation for T190C (flaB) (Table S4). As described above, about 500 to 1,000 bp of upstream and downstream DNA sequences flanking the desired gene were PCR amplified with the FlaB comp P1 and FlaBT190CR1mut primers (for the upstream sequence) and with FlaB comp P2 and FlaBT190CF1mut primers (for the downstream sequence). Owing to their self-complementarity, the two fragments were annealed, gel purified, and ligated to pGEM-T Easy, and the remaining procedure was identical to the protocol described above.
Genome resequencing of fms mutants.
After streak purifying the fms mutants on ATGN, a paired-end library was prepared from their total genomic DNA using the modified protocol of Lazinski and Camilli (http://tucf-genomics.tufts.edu/), as described earlier (54). The Quick Blunting kit (New England BioLabs) was used to blunt-end about 20 μg of sheared genomic DNA, followed by the addition of one deoxyadenosine to their 3′ ends using the Klenow fragment of DNA polymerase I. The Quick Ligation kit (New England BioLabs) was then used to ligate the DNA preparation with an adapter mix consisting of primers OLJ131 and OLJ137. The library was then amplified by PCR using the primers OLJ139 and OLJ140. Sequencing was performed on an Illumina MiSeq platform at the Center for Genomics and Bioinformatics at Indiana University.
Motility assays.
Petri dishes (100 mm) containing 0.25% ATGN motility agar were used to evaluate swimming proficiency. Swimming phenotypes were determined by inoculating fresh colonies into the center of the swim plates with 20 ml Bacto agar (BD, Sparks, MD) (58). Antibiotics and IPTG were added when necessary. The plates were incubated at 28°C for up to 7 days, and their swim ring diameters were measured daily.
TEM.
Transmission electron microscopy (TEM) was performed as described previously (22, 55). Three-hundred-mesh, 3-mm copper grids (Electron Microscopy Sciences, Hatfield, PA) were coated with carbon-Formvar films. The sample for TEM was A. tumefaciens cells grown in ATGN to an optical density at 600 nm (OD600) of ≈1.0 and diluted 1:10, 10 μl of which was applied to the Formvar-coated grids. After 5 min, each grid was dried with filter paper and then negatively stained with 2% uranyl acetate for another 5 min. The grids were again dried to remove excess stain and then examined with a JEOL JEM-1010 transmission electron microscope set to 80 kV in the Indiana University Electron Microscopy Center.
Visualization of flagellar filaments with Alexa Fluor 488 maleimide stain.
A. tumefaciens wild-type or mutant cells were grown overnight and then subcultured until they reached exponential phase. Alexa Fluor 488 maleimide dye was diluted to 25 μg/ml in ATGN minimal medium. Diluted dye (1 to 2 μl) was mixed with the same volume of cells. The mixture was then added on a slide and incubated for 5 to 10 min with a coverslip on it. The samples were then observed at ×60 magnification with a Nikon 90i microscope equipped with a Photometrics Cascade cooled charge-coupled-device (CCD) camera (excitation filter = 480/40 nm, dichromatic mirror = 505 nm, absorption filter = 535/50 nm).
Amino acid sequence alignment and structural analysis.
The FASTA A. tumefaciens FlaA, FlaB, FlaC, and FlaD amino acid sequences were imported from the PubMed Protein database to ClustalW2 and aligned as per the default settings (59). The FASTA sequences for SciP were aligned using Clustal Omega, with default settings (60). Phyre structures of the A. tumefaciens flagellins were individually generated using the Salmonella Typhimurium FliC flagellin structural template (PDB 3A5X). This structural analysis was important to create the site-specific mutations in the flagellins (see Fig. S1 and S3A for details) (61).
Mass spectrometry.
Flagellar filament preparation was done as described previously for C. crescentus (11). A. tumefaciens wild-type and ΔflaABC mutant cells were grown overnight, subcultured into 500 ml of ATGN minimal medium, and incubated until exponential phase. Cultures were then centrifuged at 10,000 × g for 15 min to remove cells. A fixed-angle Beckman 70 Ti rotor was used to ultracentrifuge the resultant supernatant at 74,800 × g for 30 min. Twenty milliliters of phosphate-buffered saline (PBS) was used to suspend the pellet, and the mixture was again centrifuged at 17,500 × g to remove any remaining cells. The supernatant was again ultracentrifuged at 74,800 × g for 30 min to concentrate the preparation. One hundred microliters of PBS was used to suspend the pellet, and the final filament preparation was examined for the presence of filaments via TEM.
Flagellar samples prepared as described above were analyzed via LC-tandem mass spectrometry (LC-MS/MS) in the Indiana University (IU) Laboratory for Biological Mass Spectrometry. These preparations were incubated for 45 min at 57°C with 2.1 mM dithiothreitol to reduce cysteine residue side chains. These side chains were then alkylated with 4.2 mM iodoacetamide for 1 h in the dark at 21°C. A solution containing 1 μg trypsin in 25 mM ammonium bicarbonate was added, and the samples were digested for 12 h at 37°C. The resulting peptides were desalted using a ZipTip (Millipore, Billerica, MA) The samples were dried down and injected into an Eksigent high-performance liquid chromatograph (HPLC) coupled to a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA) operating in “top five” data-dependent MS/MS selection. The peptides were separated using a 75-μm, 15-cm column packed in-house with C18 resin (Michrom Bioresources, Auburn, CA) at a flow rate of 300 nl/min. A 1-h gradient was run from buffer A (2% acetonitrile, 0.1% formic acid) to 60% buffer B (100% acetonitrile, 0.1% formic acid). Peptides shared between flagellin proteins as well as those specific to individual proteins were identified by MS database interpretation. For each peptide, LC peak areas were manually extracted using a ±0.1-Da mass window around the theoretical m/z value for the observed charge state. To determine the intensity of each flagellin protein relative to that of FlaB, MS-identified peptide pairs showing high sequence identity were compared. The ratio of the signal area for the peptide relative to its FlaB counterpart was determined. In the case of peptides containing a methionine residue, both the unmodified and oxidized versions of the peptide were observed, and in these cases, the total signal areas for both versions were calculated. One-way ANOVA was used to calculate if there were any significant differences between the ratios of peptides.
β-Galactosidase assays and lacZ fusions.
The lacZ reporter gene was fused to the start codons of flagellin genes, as described previously (62). Primers for lacZ fusion were designed so that the fragments had the ribosome-binding site and the gene start codon in-frame with the promoterless lacZ gene. The prom-1 and prom-2 primers (Table S4) were used to amplify promoter regions of flaB from A. tumefaciens wild-type and fms-6 genomic DNA with Phusion DNA polymerase, followed by gel purification and ligation to pGEM-T Easy. Their sequences were verified, followed by restriction digestion that cleaved them at the designed restriction sites, to be ligated to the vector pRA301, cleaved with appropriate restriction enzymes. Following another round of sequence verification, the constructs were electroporated into A. tumefaciens ΔflaACD mutants, as described earlier (47). β-Galactosidase assays were performed to determine the Miller units of at least three biological replicates, as described previously (63).
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by National Institutes of Health (NIH) grants GM080546 and GM120337 (to C.F.).
Electron microscopy was performed at the Indiana University (IU) Electron Microscopy Center. We thank Loubna Hammad for her contributions to the mass spectrometric analysis of flagellin preparations.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00327-18.
REFERENCES
- 1.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair DF. 2003. Flagellar movement driven by proton translocation. FEBS Lett 545:86–95. doi: 10.1016/S0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- 3.Turner L, Zhang R, Darnton NC, Berg HC. 2010. Visualization of flagella during bacterial swarming. J Bacteriol 192:3259–3267. doi: 10.1128/JB.00083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attmannspacher U, Scharf B, Schmitt R. 2005. Control of speed modulation (chemokinesis) in the unidirectional rotary motor of Sinorhizobium meliloti. Mol Microbiol 56:708–718. doi: 10.1111/j.1365-2958.2005.04565.x. [DOI] [PubMed] [Google Scholar]
- 5.Götz R, Schmitt R. 1987. Rhizobium meliloti swims by unidirectional, intermittent rotation of right-handed flagellar helices. J Bacteriol 169:3146–3150. doi: 10.1128/jb.169.7.3146-3150.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Götz R, Limmer N, Ober K, Schmitt R. 1982. Motility and chemotaxis in 2 strains of Rhizobium with complex flagella. J Gen Microbiol 128:789–798. [Google Scholar]
- 7.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 8.Brown MT, Steel BC, Silvestrin C, Wilkinson DA, Delalez NJ, Lumb CN, Obara B, Armitage JP, Berry RM. 2012. Flagellar hook flexibility is essential for bundle formation in swimming Escherichia coli cells. J Bacteriol 194:3495–3501. doi: 10.1128/JB.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson DA, Chacko SJ, Venien-Bryan C, Wadhams GH, Armitage JP. 2011. Regulation of flagellum number by FliA and FlgM and role in biofilm formation by Rhodobacter sphaeroides. J Bacteriol 193:4010–4014. doi: 10.1128/JB.00349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato S, Okamoto M, Asakura S. 1984. Polymorphic transition of the flagellar polyhook from Escherichia coli and Salmonella Typhimurium. J Mol Biol 173:463–476. doi: 10.1016/0022-2836(84)90391-7. [DOI] [PubMed] [Google Scholar]
- 11.Faulds-Pain A, Birchall C, Aldridge C, Smith WD, Grimaldi G, Nakamura S, Miyata T, Gray J, Li G, Tang J, Namba K, Minamino T, Aldridge PD. 2011. Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J Bacteriol 193:2695–2707. doi: 10.1128/JB.01172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharf B, Schuster-Wolff-Buhring H, Rachel R, Schmitt R. 2001. Mutational analysis of the Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J Bacteriol 183:5334–5342. doi: 10.1128/JB.183.18.5334-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharf B. 2002. Real-time imaging of fluorescent flagellar filaments of Rhizobium lupini H13-3: flagellar rotation and pH-induced polymorphic transitions. J Bacteriol 184:5979–5986. doi: 10.1128/JB.184.21.5979-5986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tambalo DD, Bustard DE, Del Bel KL, Koval SF, Khan MF, Hynes MF. 2010. Characterization and functional analysis of seven flagellin genes in Rhizobium leguminosarum bv. viciae. Characterization of R. leguminosarum flagellins. BMC Microbiol 10:219. doi: 10.1186/1471-2180-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yen JY, Broadway KM, Scharf BE. 2012. Minimum requirements of flagellation and motility for infection of Agrobacterium sp strain H13-3 by flagellotropic bacteriophage 7-7-1. Appl Environ Microbiol 78:7216–7222. doi: 10.1128/AEM.01082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chesnokova O, Coutinho JB, Khan IH, Mikhail MS, Kado CI. 1997. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol 23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi F, Tomaru H, Furukawa E, Ikeda K, Fukano H, Oosawa K. 2013. Key amino acid residues involved in the transitions of L- to R-type protofilaments of the Salmonella flagellar filament. J Bacteriol 195:3503–3513. doi: 10.1128/JB.02091-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klose KE, Mekalanos JJ. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol 180:303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Sal M, Marko M, Charon NW. 2010. Differential regulation of the multiple flagellins in Spirochetes. J Bacteriol 192:2596–2603. doi: 10.1128/JB.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deakin WJ, Parker VE, Wright EL, Ashcroft KJ, Loake GJ, Shaw CH. 1999. Agrobacterium tumefaciens possesses a fourth flagellin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology 145:1397–1407. doi: 10.1099/13500872-145-6-1397. [DOI] [PubMed] [Google Scholar]
- 21.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Haitjema CH, Fuqua C. 2014. The Ctp type IVb pilus locus of Agrobacterium tumefaciens directs formation of the common pili and contributes to reversible surface attachment. J Bacteriol 196:2979–2988. doi: 10.1128/JB.01670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llewellyn M, Dutton RJ, Easter J, O'Donnol D, Gober JW. 2005. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol Microbiol 57:1127–1142. doi: 10.1111/j.1365-2958.2005.04745.x. [DOI] [PubMed] [Google Scholar]
- 24.Anderson PE, Gober JW. 2000. FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol Microbiol 38:41–52. doi: 10.1046/j.1365-2958.2000.02108.x. [DOI] [PubMed] [Google Scholar]
- 25.Ferooz J, Lemaire J, Letesson JJ. 2011. Role of FlbT in flagellin production in Brucella melitensis. Microbiology 157:1253–1262. doi: 10.1099/mic.0.044867-0. [DOI] [PubMed] [Google Scholar]
- 26.Mangan EK, Malakooti J, Caballero A, Anderson P, Ely B, Gober JW. 1999. FlbT couples flagellum assembly to gene expression in Caulobacter crescentus. J Bacteriol 181:6160–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. 2010. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell 39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan MH, Kozdon JB, Shen X, Shapiro L, McAdams HH. 2010. An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc Natl Acad Sci U S A 107:18985–18990. doi: 10.1073/pnas.1014395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuwajima G, Asaka JI, Fujiwara T, Fujiwara T, Node K, Kondo E. 1986. Nucleotide-sequence of the hag gene encoding flagellin of Escherichia coli. J Bacteriol 168:1479–1483. doi: 10.1128/jb.168.3.1479-1483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enomoto M, Sakai A, Tominaga A. 1985. Expression of an Escherichia coli flagellin gene, hag48, in the presence of a Salmonella H1-repressor. Mol Gen Genet 201:133–135. doi: 10.1007/BF00397999. [DOI] [PubMed] [Google Scholar]
- 31.Cohen-Krausz S, Trachtenberg S. 1998. Helical perturbations of the flagellar filament: Rhizobium lupini H13-3 at 13 angstrom resolution. J Struct Biol 122:267–282. doi: 10.1006/jsbi.1998.4001. [DOI] [PubMed] [Google Scholar]
- 32.Felix G, Duran JD, Volko S, Boller T. 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276. doi: 10.1046/j.1365-313X.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 33.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. 2006. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 34.Renault TT, Abraham AO, Bergmiller T, Paradis G, Rainville S, Charpentier E, Guet CC, Tu Y, Namba K, Keener JP, Minamino T, Erhardt M. 2017. Bacterial flagella grow through an injection-diffusion mechanism. Elife 6:e23136. doi: 10.7554/eLife.23136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iida Y, Hobley L, Lambert C, Fenton AK, Sockett RE, Aizawa S. 2009. Roles of multiple flagellins in flagellar formation and flagellar growth post bdelloplast lysis in Bdellovibrio bacteriovorus. J Mol Biol 394:1011–1021. doi: 10.1016/j.jmb.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millikan DS, Ruby EG. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol 186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanner DE, Ma W, Chen Z, Schulten K. 2011. Theoretical and computational investigation of flagellin translocation and bacterial flagellum growth. Biophys J 100:2548–2556. doi: 10.1016/j.bpj.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauga E. 2016. Bacterial hydrodynamics. Annu Rev Fluid Mech 48:105–130. doi: 10.1146/annurev-fluid-122414-034606. [DOI] [Google Scholar]
- 39.Calladine CR. 1975. Construction of bacterial flagella. Nature 255:121–124. doi: 10.1038/255121a0. [DOI] [PubMed] [Google Scholar]
- 40.Calladine CR, Luisi BF, Pratap JVA. 2013. A “mechanistic” explanation of the multiple helical forms adopted by bacterial flagellar filaments. J Mol Biol 425:914–928. doi: 10.1016/j.jmb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namba K, Vonderviszt F. 1997. Molecular architecture of bacterial flagellum. Q Rev Biophys 30:1–65. doi: 10.1017/S0033583596003319. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Burrage AM, Postel S, Clark RE, Orlova A, Sundberg EJ, Kearns DB, Egelman EH. 2017. A structural model of flagellar filament switching across multiple bacterial species. Nat Commun 8:960. doi: 10.1038/s41467-017-01075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun L, Dong Y, Shi M, Jin M, Zhou Q, Luo Z-Q, Gao H. 2014. Two residues predominantly dictate functional difference in motility between Shewanella oneidensis flagellins FlaA and FlaB. J Biol Chem 289:14547–14559. doi: 10.1074/jbc.M114.552000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A 99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtis PD, Brun YV. 2014. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol 93:713–735. doi: 10.1111/mmi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 47.Mersereau M, Pazour GJ, Das A. 1990. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90:149–151. doi: 10.1016/0378-1119(90)90452-W. [DOI] [PubMed] [Google Scholar]
- 48.Fuqua WC, Winans SC. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tempé J, Petit A, Holsters M, Montagu MV, Schell J. 1977. Thermosensitive step associated with transfer of Ti plasmid during conjugation–possible relation to transformation in crown gall. Proc Natl Acad Sci U S A 74:2848–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morton ER, Fuqua C. 2012. Genetic manipulation of Agrobacterium. Curr Protoc Microbiol Chapter 3:Unit 3D.2. doi: 10.1002/9780471729259.mc03d02s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warrens AN, Jones MD, Lechler RI. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29–35. doi: 10.1016/S0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- 53.Merritt PM, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol 189:8005–8014. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Heindl JE, Fuqua C. 2013. Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens. PLoS One 8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohari B, Licata NA, Kysela DT, Merritt PM, Mukhopadhyay S, Brun YV, Setayeshgar S, Fuqua C. 2015. Novel pseudotaxis mechanisms improve migration of straight-swimming bacterial mutants through a porous environment. mBio 6:e00005-15. doi: 10.1128/mBio.00005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ditta G. 1986. Tn5 mapping of Rhizobium nitrogen-fixation genes. Methods Enzymol 118:519–528. doi: 10.1016/0076-6879(86)18098-0. [DOI] [Google Scholar]
- 58.Adler J. 1966. Chemotaxis in bacteria. Science 153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 59.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocol 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu J, Kim J, Koestler BJ, Choi J-H, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heckel BC, Tomlinson AD, Morton ER, Choi JH, Fuqua C. 2014. Agrobacterium tumefaciens exoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J Bacteriol 196:3221–3233. doi: 10.1128/JB.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.