Abstract
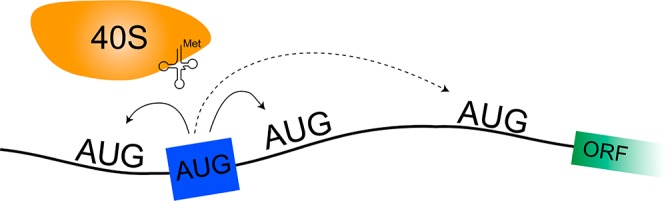
Cap-independent translation is believed to play an important role in eukaryotic protein synthesis, but the mechanisms of ribosomal recruitment and translation initiation remain largely unknown. Messenger RNA display was previously used to profile the human genome for RNA leader sequences that can enhance cap-independent translation. Surprisingly, many of the isolated sequences contain AUG triplets, suggesting a possible functional role for these motifs during translation initiation. Herein, we examine the sequence determinants of AUG triplets within a set of human translation enhancing elements (TEEs). Functional analyses performed in vitro and in cultured cells indicate that AUGs have the capacity to modulate mRNA translation either by serving as part of a larger ribosomal recruitment site or by directing the ribosome to defined initiation sites. These observations help constrain the functional role of AUG triplets in human TEEs and advance our understanding of this specific mechanism of cap-independent translation initiation.
Translation initiation is a critical process that requires recruitment of the ribosomal complex to the mRNA template and recognition of the initiation codon. The mechanism of initiation differs between prokaryotes and eukaryotes. In prokaryotes, ribosomal recruitment is facilitated by Watson–Crick base pairing between the ribosomal binding site in the mRNA template and a complementary region of the 16S rRNA.1,2 In eukaryotes, translation generally follows a cap-dependent mechanism in which the 43S ribosomal preinitiation complex (PIC) is recruited to a 7-methylguanosine cap located at the 5′ end of the RNA message.3,4 The ribosome then scans the 5′ leader region for an AUG codon that is recognized by the initiator tRNA bound to the ribosomal PIC. More recently, a growing body of evidence has highlighted the importance of an alternative method of initiation, termed cap-independent translation. In this noncanonical method of initiation, mRNA transcripts that contain translation enhancing elements (TEEs), cap-independent translation elements (CITEs), or internal ribosomal entry sites (IRESs) can bypass the requirement for a 5′ cap structure during ribosomal recruitment.5−7 Several studies have now demonstrated that cap-independent translation occurs during normal cellular processes, like mitosis and apoptosis, or when the cap-dependent translation machinery is compromised by viral infection or disease.8,9
Although the mechanism of cap-independent translation likely varies depending on the core RNA elements used to promote ribosomal initiation (e.g., TEEs, CITEs, and IRESs), common motifs that drive cap-independent translation activity remain elusive.10 Efforts to identify these core functional motifs have been hindered, in part, by the presence of upstream AUG (uAUG) triplets in the 5′ leader region of human genes. In a recent study, uAUGs were found in 40–50% of full-length human- and rodent-expressed mRNA transcripts.11 Many of these sites (20–30%) are conserved by evolution, suggesting mechanistic implications for distinguishing functional initiation codons from inactive AUG triplets.12 While sequence context is often used to predict the likelihood of AUG usage, only a fraction of human genes (∼35%) have a perfect Kozak sequence with a purine located at position −3 and a guanine located at position +4.13,14 Other factors that have made it difficult to identify the functional initiation codon include the length and structural stability of the 5′ leader sequence, the accessibility of the AUG codon to the ribosomal complex, and the potential for ribosomal initiation to occur at alternative non-AUG positions like ACG, CUG, and GUG.15−19
In a previous study, we used mRNA display to interrogate total human DNA for RNA sequences that have the capacity to mediate cellular cap-independent translation.7 By combining in vitro selection with next-generation deep sequencing, a catalog of >12000 TEE-bearing regions (TBRs), locations in the human genome that contain translation enhancing elements, was generated. Functional analysis studies performed in vitro and in cultured human cells indicate that many of the selected TEEs dramatically increase protein production levels when added to the 5′ leader region. These findings greatly increased the potential for cap-independent translation to occur in the human genome, which traditionally has been constrained to identified IRESs20, and supports the long-held belief that translation can proceed by different mechanisms.21
Herein, we explore the sequence determinants of human TEEs to better understand the mechanistic possibilities of TEE-associated cap-independent translation. Because many of the in vitro-selected TEEs contained multiple AUG triplets that could impede mRNA translation, we decided to investigate these sites as possible functional motifs in the mechanism of ribosomal recruitment and translation initiation. Our findings demonstrate that AUGs can have a dramatic but unpredictable effect on the efficiency of protein synthesis. Mutagenesis studies reveal a loss of function when AUGs are removed from high-activity TEEs and a failure to recapitulate activity when the same AUGs are inserted into an unrelated low-activity sequence that lacks AUGs. Protein sequencing confirmed that ribosomal initiation occurs at specific locations within the boundary of AUG-containing TEEs. Additionally, comparative functional genomics uncovered a short RNA motif that can modulate translational activity at a downstream initiation site. The presence or absence of this motif was shown to upregulate or downregulate activity, respectively, in multiple mammalian cell lines and sequence contexts. Furthermore, mutational studies identified the distance constraints of the motif from a well-defined downstream open reading frame (ORF). On the basis of these data, we postulate that AUG triplets play an important role in the mechanism of TEE-associated cap-independent translation initiation.
Materials and Methods
Cell Culture
HeLa, BSC40, RK13, and BHK cells were obtained from American Type Culture Collection (ATCC), while 129SV-MEF cells are fibroblasts taken from a 129 mouse embryo (a gift from Charles River Laboratories) that were then spontaneously transformed. HeLa, BHK, and RK13 cells were maintained in MEM (Invitrogen) with 5% fetal bovine serum (FBS, HyClone). BSC40 cells were maintained in DMEM (Invitrogen) supplemented with 5% FBS, while 129SV-MEF cells required DMEM with 10% FBS. All cell culture media contained 5 μg/mL gentamicin (Invitrogen). Cells were kept at 37 °C in a humidified atmosphere containing 5% CO2.
Luciferase Reporter Plasmids
Vectors for analyzing the effect of start codons (AUG) within the TEEs were created by modifying commercial vector pT7CFE1-CHis (Thermo Scientific). For studies requiring in vitro luciferase expression, the T7 promoter and viral EMCV IRES sequences were removed from the commercial vector using PvuII and BamHI restriction sites and replaced with a synthetically constructed T7 promoter. To insert luciferase into the vector, the TEE of interest and the luciferase gene were amplified out of a previously constructed reporter plasmid by polymerase chain reaction (PCR) and inserted using BamHI and NotI restriction sites, upstream of the poly(A) tail sequence and T7 termination site.7 For studies involving luciferase expression in cultured cells, the sequence of interest was inserted into a previously constructed reporter plasmid using BamHI and NcoI restriction sites. The insertion site was located downstream of the Vaccinia virus synthetic late promoter sequence and immediately upstream of the luciferase gene, which contained a poly(A) tail sequence at the 3′ end.7
Luciferase Reporter Assay
TEE sequences were functionally characterized by luciferase expression from reporter plasmids, both in vitro and within cells, as described previously.7 Cell-free characterization was performed using the Human In vitro Protein Expression Kit (Pierce), with 5′-capped or uncapped in vitro-transcribed RNA as a template. Luciferase expression was achieved following the manufacturer’s protocols using 500 ng of mRNA template and a 90 min translation at 30 °C. Luciferase activity was measured using the Promega Luciferase Assay System with a Glomax microplate luminometer (Promega). For characterization within cells, a transfect–infect assay was used. The desired cell type was seeded at a density of 15000 cells per well in white 96-well plates 18 h prior to transfection. Cells were transfected with a complex of the luciferase reporter plasmid (200 ng) and Lipofectamine 2000 (0.5 μL) in Opti-MEM (Invitrogen) and immediately infected with the Copenhagen strain (VC-2) of the wild-type Vaccinia virus at a multiplicity of infection (moi) of 5 PFU/cell. In this assay, the virus provides the RNA polymerase, thus facilitating mRNA production within the cytoplasm. Cellular ribosomes are then responsible for translation of the RNA message.22 Cells were lysed 6 h post-infection using 1× Reporter Lysis Buffer (Promega) in the 96-well plates and luciferase activity determined as described above. Reported luciferase values were additionally normalized to luciferase mRNA levels, as determined by real-time PCR. For both characterization methods, a minimum of three biological replicates were performed, in triplicate.
RNA Characterization
Messenger RNA used as a template for in vitro translation was generated using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs). Both 5′-capped mRNA and uncapped mRNA were generated following the manufacturer’s instructions, using 500 ng of linear DNA as a template and a 2 h incubation at 37 °C. To produce 5′-capped mRNA, 8 mM (final concentration) m7G(5′)ppp(5′)G RNA Cap Structure Analog (New England Biolabs) was added to the reaction mixture. Post-incubation, mRNA was purified using the Zymo Research RNA Clean and Concentrator (Fisher Scientific) following the manufacturer’s instructions. RNA integrity was confirmed by 1% agarose gel electrophoresis and a denaturing RNA loading dye (New England Biolabs). For cell-based assays, RNA was isolated 6 h post-infection via cell lysis using the RNeasy Micro kit (Qiagen) and following the manufacturer’s instructions, including the on-column DNase I treatment. The quantity of isolated RNA was determined using a NanoDrop ND-1000 instrument (Marshall Scientific), with only those samples containing an A260/A280 ratio between 1.9 and 2.1 deemed suitable for further use. The presence of clear 28S and 18S rRNA bands, as an indicator of RNA quality, was also confirmed by agarose gel electrophoresis as described above. Isolated RNA (200 ng) was reverse transcribed to generate complementary DNA (cDNA) using an oligo(dT22) primer and Superscript II reverse transcriptase (Fisher Scientific) at 42 °C for 1.5 h. Quantitative real-time PCR (qPCR) was used to measure mRNA levels and was conducted using the iQ SYBR Green Supermix (Bio-Rad) following the manufacturer’s protocol with 12 ng of cDNA as a template and each primer at a final concentration of 0.25 μM. Luciferase mRNA was amplified using primers RTlucF (5′ GCTGGGCGTTAATCAGAGAG) and RTlucR (5′ GTGTTCGTCTTCGTCCCAGT), while mRNA levels of the reference gene hypoxanthine-guanine phosphoribosyltransferase (HPRT, Entrez gene ID 3251) were determined using the primers RThprtF (5′ TGCTGAGGATTTGGAAAGGGTG) and RThprtR (5′ CCTTGAGCACACAGAGGGCTAC). Reaction mixtures were assembled in MicroAmp Fast optical 96-well plates (Applied Biosystems), which were then adhesively sealed using optical sealing tape (Bio-Rad). A StepOnePlus real-time PCR system (Applied Biosystems) was used to perform the qPCR, with the following cycling conditions: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. A preprogrammed melting curve was completed following cycling and confirmed uniform amplicon formation for each primer pair. Primer pair specificity was also confirmed through the observation of a single band by agarose gel electrophoresis following amplification. Each primer pair was validated in duplicate using a calibration curve with either plasmid DNA containing the luciferase gene or isolated HeLa cell DNA as a template. Analysis of these calibration curves yielded average slopes of −3.3596 and −3.2287 and y-intercepts of 7.4516 and 5.2165 for luciferase and HPRT, respectively, when the template concentration was plotted against the Ct value. For every qPCR performed, the reaction efficiency ranged from 0.97 to 1.10, with r2 values falling between 0.9949 and 0.9986 when amplification controls were analyzed. During analysis of translation activity driven by individual TEEs, luciferase mRNA levels were normalized to mRNA levels of HPRT using the ΔΔCt method.
Translation Initiation Analysis
Plasmids expressing the maltose binding protein (MBP) were constructed by replacing the luciferase gene with the MBP gene using NcoI and XhoI restriction sites within the reporter plasmids. Vectors expressing MBP were translated in the presence of [35S]methionine using the TnT Quick Coupled Transcription/Translation System (rabbit reticulocyte lysate, Promega) following the manufacturer’s protocol, without the presence of a cap structure. Templates (500 ng of plasmid) were translated using the TnT system for 60 min at 30 °C. Cell-free translation experiments performed with human ribosomes used the Human In vitro Protein Expression Kit (Pierce), lacking a cap structure in the reaction mix. Templates (500 ng of linear DNA) underwent transcription for 2 h at 32 °C followed by a 90 min translation at 30 °C in the presence of [35S]methionine. In both cases, MBP protein was purified on an amylose column (NEB) and recovered by being eluted in water containing 10 mM maltose. Recovery of radiolabeled MBP was monitored using a liquid scintillation counter (Beckman Coulter). On the basis of recovery counts, elution volumes were normalized, reduced, and loaded on a 4 to 12% gradient Bis-Tris polyacrylamide gel (Invitrogen) to generate bands of approximately equal intensity for analysis. To determine the exact translation initiation site, the MBP vectors were in vitro-transcribed using T7 RNA polymerase and expressed on a large scale (1 mL) using the Flexi Rabbit Reticulocyte Lysate system (Promega) with 100 mM KCl, 0.5 mM magnesium acetate, and 0.4 μM RNA template. The translation products were purified with an amylose column as described above and spotted onto a polyvinylidene fluoride membrane (0.2 μm, Bio-Rad). Samples were sequenced by researchers at Proseq Inc. (Boxford, MA).
Mutagenesis Studies
Deletion and insertion analysis vectors containing HGL6.877, HGL0.53, and various portions of the HGL6.985 sequence were constructed by Klenow DNA polymerase extension of overlapping oligonucleotides followed by a restriction enzyme digest with BamHI and NcoI. The digested fragments were ligated into the luciferase and MBP reporter plasmids. Similarly, the 13-nucleotide core motif was assayed for activity by constructing reporter vectors in which the 13-mer motif was either added to or deleted from the 5′ end of the desired TEEs. Expression from the reporter plasmids containing the modified sequences was performed in vitro and in cells as described above.
Results
Computational Analysis
We focused our analysis on a set of 225 human TEEs that were previously identified by mRNA display and assayed for function in cultured human cells using a luciferase reporter vector to assess translational activity.7 The sequences have an average length of 90 nucleotides and map with 100% identity to the human reference genome (hg18) (Table S1). Sequence analysis at the genome level revealed that our in vitro-selected TEEs are enriched with AUG triplets as compared to random samples of human DNA (average AUG density of 24 per kilobase vs 17 per kilobase; binomial test p value of <10–32). Furthermore, ∼95% of the TEEs contain at least one AUG triplet, with an average occurrence of three triplets per sequence (Figure S1a and Table S1). Sequences that contain only one AUG triplet prefer (93%) in-frame positions with respect to the downstream coding region (CDR) (Figure S1b), while those sequences that contain two or more AUGs have their triplets distributed between the in-frame and out-of-frame positions. Of the multi-AUG TEEs, 78% of the sequences have the first triplet in-frame with the CDR.
Ranking the sequences according to their translation enhancing activity failed to identify any correlations between activity and AUG abundance or activity and AUG position relative to the downstream CDR (Figure S2), indicating that AUG position and abundance in the 5′ leader region of uncapped mRNA transcripts are not determinants (positive or negative) of translation initiation activity. Surprisingly, the highest-activity sequence (HGL6.877) contains eight AUG triplets, while the lowest-activity sequence (HGL6.830) has none. Only one AUG from a total of 650 triplets in the data set of 225 TEEs resides in an optimal Kozak sequence context; however, that sequence exhibits only weak activity in our luciferase assay. Another 146 triplets are found in a variation of the optimal Kozak context, yet no clear correlation was observed between the Kozak AUG residing in-frame with the downstream CDR and sequence activity (Table S2). These results stand in contrast to conventional cap-dependent translation studies, where AUG triplets in the 5′ leader region tend to impede translation by serving as decoys to the authentic initiation site.
Functional Analysis
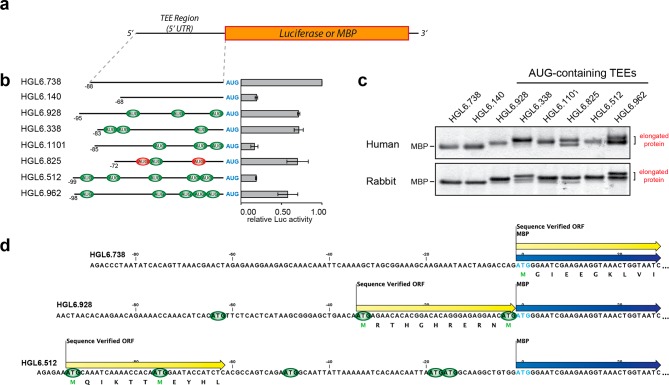
The observation that AUG abundance and location are not determinants of TEE-associated cap-independent translation initiation activity led us to speculate that differences in functional activity were caused by motifs within the selected TEEs that promoted the translation of uncapped mRNAs. To explore this possibility, we selected eight TEEs with a range of AUG triplet patterns (Table S3) for functional analysis. Some of the TEEs were shorter, and others longer. Some had no AUGs, and others many. Some were all in-frame, while others contained a mixture of in-frame and out-of-frame AUGs (Figure 1a). The set of eight TEEs was cloned into vectors that were used to evaluate translational activity. We used a luciferase reporter to measure translational activity and the maltose binding protein (MBP) to assay for initiation within the TEE sequences themselves. MBP was chosen for this assay because small differences in sequence length were not easily detected with luciferase, and affinity purification on an amylose resin ensured that the translated protein was correctly folded and functional.
Figure 1.
Functional analysis of a diverse set of human TEEs for translation initiation activity and early ribosomal initiation. (a) Schematic representation of the firefly luciferase reporter or maltose binding protein (MBP) constructs containing 5′ leader sequences that derive from human translation enhancing elements. (b) Translation initiation activity in HeLa cell lysate measured by luciferase expression: in-frame triplets (green), out-of-frame triplets (red), and the initiation codon at position +1 of the CDR (blue). Error bars designate the standard deviation. (c) Early ribosomal initiation on MBP constructs expressed in HeLa cell lysate and rabbit reticulocyte lysate. Protein expressed in the different lysate systems was purified, and recovery of radiolabeled MBP monitored by liquid scintillation counts. The amount of protein analyzed was normalized, based on the recovery counts, and gel electrophoretic mobility shifts indicate that human TEEs with AUG triplets in their sequence are prone to early ribosomal initiation. The slower-mobility bands correspond to MBP protein isoforms that contain N-terminal extensions. (d) Identification of authentic translation initiation sites by protein sequencing of in vitro-expressed MBP. The protein sequence is indicated below the nucleotide sequence of each TEE. Color scheme: MBP coding region (blue arrows), authentic translation initiation site (yellow arrows), and in-frame triplets relative to the MBP coding region (green).
We began by measuring the luciferase activity of each TEE in HeLa cell lysate, which provides a controlled medium for studying translational activity in a cap-independent environment, as no cap structure is present unless specifically added, and no RNA splicing occurs. To validate the use of HeLa cell lysate for evaluation of cap-independent translation, luciferase expression driven by both the encephalomyocarditis virus (EMCV) IRES and a short, unstructured sequence was assessed. The luciferase assay results confirm that only the EMCV control displayed translational activity when the cap structure was absent from the mRNA (Figure S3a). When the set of AUG-containing TEEs was analyzed, we noticed a number of interesting observations about the functional activity (Figure 1b and Table S4). The first observation is that TEEs lacking AUG triplets are not necessarily the best leader sequences for in vitro translation. For example, four of our AUG-containing TEEs are more efficient than a TEE (HGL6.140) that does not contain any AUG triplets. Second, the presence of out-of-frame AUG triplets is not an indicator of low translation enhancing activity. In this case, HGL6.825, which contains two out-of-frame AUG triplets, is equivalent to many high-activity TEEs that contain exclusively in-frame AUGs or no AUG triplets at all. Third, the effects of AUG triplet patterns appear to be context-dependent, as some sequences function with activity higher than that of others. For example, HGL6.512 and HGL6.962, which share a similar length, sequence composition, and secondary structure, have contrasting levels of activity even though both sequences contain five AUG triplets. Fourth, the AUG abundance is not a general indicator of TEE-associated cap-independent translation ability, as comparison of translational levels from capped and uncapped mRNA driven by the diverse set of TEEs reveals approximately equal expression in all cases (Figure S3b). The one exception is TEE HGL6.512, which displays higher translation levels from uncapped mRNA compared to those from the capped counterpart. This trend is not observed in HGL6.962 however, which contains the same number of AUGs as HGL6.512. Finally, the presence of upstream open reading frames (uORFs) is not an indicator of low sequence activity, as both HGL6.928 and HGL6.825 contain a uORF and display high translational activity (Table S5). Given the variation among sequence contexts and activity levels, these results suggest that the sequence determinants of TEE-associated cap-independent translation are more complex than what has been observed previously for conventional cap-dependent translation.
Mapping Translation Initiation Sites
To determine whether any of the sequences initiate translation within the 5′ leader region of their respective translation enhancing element, MBP vectors carrying the TEEs upstream of the CDR were expressed in HeLa cell lysate, purified by affinity chromatography, and analyzed for changes in protein length by gel electrophoresis. As the amount of MBP expressed from the different vectors varied in a manner similar to that of the level of luciferase expression (Table S4), an approximately equal amount of MBP protein was analyzed for each sample to ensure all samples could be visualized. Analysis of the resulting protein gel reveals a clear difference in band mobility between proteins produced from constructs that contain and lack AUG triplets (Figure 1c). All six of the TEEs that contain AUG triplets produce a protein band with mobility that is slower than that of the protein band produced from the two TEEs that lack AUG triplets. The shift in electrophoretic mobility is consistent with the synthesis of MBP isoforms that contain extended N-terminal tails due to early ribosomal initiation on the mRNA transcript.
To confirm that the shift in electrophoretic mobility was due to early initiation, we sequenced the first 10 amino acid residues of MBP samples produced from three of the eight TEEs. In each case, MBP was produced in HeLa cell lysate, purified by affinity chromatography, and sequenced by Edman degradation. For this analysis, we chose HGL6.738, which lacks AUG triplets and should initiate translation at position +1 of the CDR and two other human TEEs (HGL6.928 and HGL6.512) that contain multiple AUG triplets in their sequence. Protein sequencing reveals that HGL6.738 initiates translation at the designated start site (first codon in the CDR), while HGL6.928 and HGL6.512 initiate translation within the boundary of the TEE (Figure 1d). For HGL6.928, translation is initiated at the second AUG position, while that of HGL6.512 is initiated at the first AUG position near the 5′ terminus. This later observation was surprising given the proximity (six nucleotides) of the AUG to the 5′ terminus. In traditional cap-dependent translation, this position may be bypassed due to steric constraints caused by the cap binding complex.
To determine if the observed translation initiation pattern is specific to human ribosomes, we tested the set of eight human TEEs for activity in rabbit reticulocyte lysate. As with the HeLa cell lysate, the ability of the rabbit reticulocyte lysate to allow cap-independent translation was first validated using the EMCV IRES in the luciferase reporter (Figure S3a). Following validation, expression from the MBP-containing vectors was performed in vitro in a coupled transcription–translation reaction and purified, and a normalized amount of protein was analyzed by gel electrophoresis as described above (Table S4). The resulting electrophoretic mobility pattern observed in the rabbit reticulocyte lysate closely matched the pattern observed in the HeLa cell lysate (Figure 1c), suggesting that the initiation mechanism of TEE-associated cap-independent translation is not specific to ribosomes found in HeLa cells.
Local Sequence Context
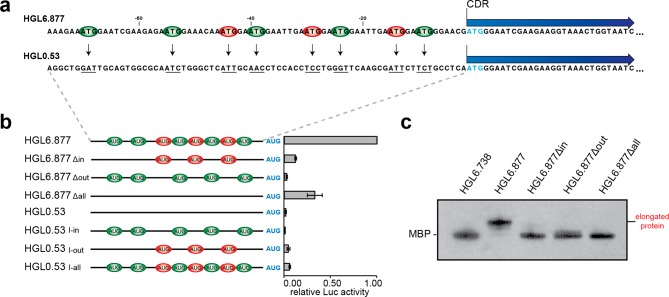
The observation that AUG-containing TEEs have the capacity to enhance mRNA translation activity led us to wonder whether AUG triplets alone are sufficient to confer activity or, instead, if translation is contingent upon the local sequence context of the translation enhancing element. To explore this question, we chose HGL6.877, the most active TEE previously identified in our screen of 225 human in vitro-selected TEEs (Table S1).7 HGL6.877 is an interesting sequence, because it contains eight AUG triplets that occur at in-frame and out-of-frame positions relative to the CDR and contains three ORFs (Table S5). We have previously shown that this sequence functions with high translational activity in vitro and in cultured human cells.7
To examine the role of sequence context in translational activity, we designed a deletion and insertion mutagenesis study that removed AUG triplets from HGL6.877 and inserted them back into an unrelated sequence of an identical length (Figure 2a). Uniform-length versions of HGL6.877 lacking in-frame, out-of-frame, and all AUG triplets were constructed by mutating the third position of each triplet from G to U. Likewise, similar mutations were used to insert the set of eight AUG triplets found in HGL6.877 into HGL0.53, an unselected sequence identified in our naïve human genome library.7 The engineered versions of HGL0.53 were constructed such that each AUG triplet was inserted at the precise location of its occurrence in HGL6.877 (Table S3). The wild-type and mutant constructs of HGL6.877 and HGL0.53 were then tested for translational activity in HeLa cell lysate. As shown in Figure 2b, none of the mutant versions of HGL6.877 or HGL0.53 exhibit any significant activity when compared to the wild-type HGL6.877 sequence (Table S6). Furthermore, removal of the AUGs from HGL6.877 greatly weakens the ability of the mRNA to be translated cap-independently, as compared to capped mRNA controls, while the insertion of AUGs into HGL0.53 does not confer cap-independent properties (Figure S3c). Even HGL6.877Δall, which is missing all eight AUG triplets, yields only modest levels of protein after mRNA translation. Intriguingly, HGL6.877Δin loses most of the activity of the original sequence, while the presence of the ORFs within the TEE remained unchanged, suggesting an important role for the triplets that were removed. In sequence HGL6.877Δout, ORFs were created during the removal of AUG triplets that may have impacted activity (Table S5), yet data from this study and others suggest that the presence of uORFs is not necessarily an indication of a decreased level of translation of the downstream CDR23,24 and ribosomal reinitiation can occur on nearby AUG triplets.25 Taken together, these observations imply that ribosomal recruitment sites have defined sequence motifs that are required for proper recruitment of the ribosomal machinery. While these sequences often contain AUG triplets, these motifs are not required for TEE-associated cap-independent translation initiation.
Figure 2.
Evaluating the local sequence context of AUG triplets in a human TEE with high translational activity. (a) Schematic of mutational analysis. In-frame (green) and out-of-frame (red) AUG triplets were removed from the 5′ leader sequence of a high-activity TEE (HGL6.877) and inserted into the 5′ leader sequence of an unselected genomic sequence (HGL0.53). Vectors were constructed with the luciferase or MBP coding regions (CDR). Initiation codon at position +1 of the CDR (blue). (b) Translation initiation activity of luciferase reporter vectors measured in HeLa cell lysate. The effect of insertion or deletion of in-frame (Δin), out-of-frame (Δout), or all (Δall) AUGs was determined. Error bars designate the standard deviation. (c) Early ribosomal initiation on MBP constructs expressed in HeLa cell lysate. Protein expressed in HeLa cell lysate was purified, and recovery monitored by liquid scintillation counts. On the basis of recovery counts, an equal amount of protein was analyzed by gel electrophoresis, with the resulting MBP protein from vectors containing mutations to HGL6.877 compared to those from vectors containing the wild-type sequence or a TEE that does not contain any AUGs (HGL6.738).
We were unable to identify the authentic initiation codon in HGL6.877 because of a translational modification that prevented sequencing of the N-terminal region of the MBP protein. However, gel electrophoresis, again performed with a normalized amount of protein per sample (Table S6), did confirm that HGL6.877 undergoes early initiation to produce an MBP isoform with an extended N-terminal tail (Figure 2c). Interestingly, engineered versions of HGL6.877 that lack some or all of the AUG triplets in the TEE region fail to undergo early initiation even though these sequences still contain several uAUGs.
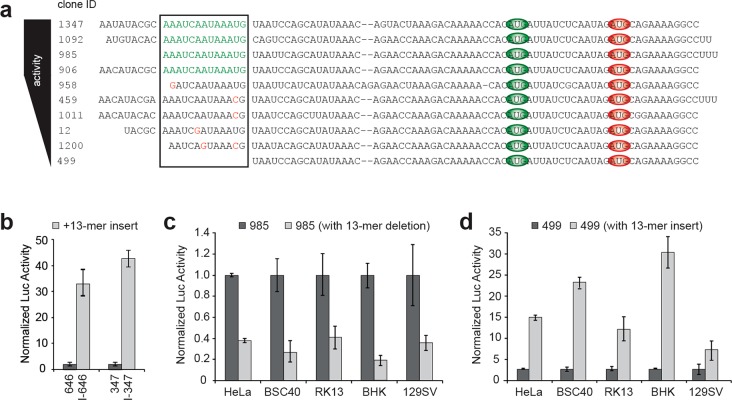
A Short High-Activity Motif
Identifying the core functional region of a TEE requires locating the motif within the 5′ leader sequence that is responsible for enhancing translation levels. While these sites are normally identified by end-mapping deletion analysis,22 our previous cellular data allowed us to use comparative functional genomics to identify a conserved 13-nucleotide motif (5′ AAAUCAAUAAAUG 3′) located at the 5′ end of a family of closely related sequences that was responsible for the functional activity of this sequence family (Figure 3a). The motif ends in an AUG triplet (underlined) that is important for activity because mutations in this region diminish translational activity in cells (Table S7). Interestingly, the presence of the AUG at the 3′ end of the motif creates several ORFs in combination with the downstream sequence (Table S5), yet activity levels of the sequences containing the 13-mer are some of the highest seen in our set of 225 TEEs (Table S1). Other portions of the 13-mer appear to be equally conserved, as mutations in this area lead to similar losses of activity. One member of this sequence family, HGL6.499, which lacks the 13-mer motif altogether, has very low translational activity in cultured HeLa cells, while the four highest-activity members (HGL6.1347, HGL6.1092, HGL6.985, and HGL6.906) all contain a perfect 13-mer motif and have activity that ranked in the top 10% of our previous cell-based screen of 225 human in vitro-selected TEEs.7
Figure 3.
Identification of a short 13-mer motif with strong translation enhancing activity. (a) Ten closely related human translation enhancing elements were identified and ranked according to their luciferase activity in cultured HeLa cells. The predicted ribosomal recruitment site (black box) was identified by a comparative functional genomic sequence alignment: functional 13-mer motif (green), point mutations (red), and AUG triplets within the selected sequence family (green and red ovals for in-frame and out-of-frame, respectively). (b–d) Functional analysis of sequences that contain and lack the 13-mer motif in cultured mammalian cells. Panel b shows the translational activity of two low-activity TEEs (HGL6.646 and HGL6.347) unrelated to the 13-mer sequence family in cultured HeLa cells. The effect of the 13-mer motif was determined by the addition of the motif to the 5′ end of each TEE. Panels c and d show the translational activity in cultured cells for one high-activity TEE (HGL6.985) and one low-activity TEE (HGL6.499), respectively, from the 13-mer sequence family. The effect of removing the 13-mer motif from HGL6.985 or adding the motif to the 5′ end of HGL6.499 was determined. Luciferase expression in all cases was normalized to luciferase mRNA levels. Error bars designate the standard deviation. Mammalian cell lines: HeLa (human), BSC40 (monkey), RK13 (rabbit), BHK (hamster), and 129SV (mouse).
To test the 13-mer as a small functional TEE, we examined the ability of the motif to modulate translational levels in different sequence contexts. We added the 13-mer to the 5′ end of HGL6.646 and HGL6.347, two low-activity sequences that are unrelated to each other and unrelated to the family of sequences from which the 13-mer was derived. When assayed for activity in cultured HeLa cells and HeLa cell lysates, the 13-mer motif was found to increase mRNA translation levels by up to 40-fold relative to those of the unmodified TEEs (Figure 3b and Figure S4b), indicating that the short 13-mer motif appears to function as a general enhancer of protein synthesis. Confirmation that nuclear expression does not contribute to our cell-based analysis provides confidence that false positives due to aberrant splicing did not occur (Figure S5). Moreover, when compared to those of capped mRNA controls in vitro, addition of the 13-mer increased cap-independent activity (Figure S3d), lending further support for its role as an enhancer.
Encouraged by this result, we decided to test the 13-mer for activity in different mammalian cell lines. For this assay, we chose HGL6.985 and HGL6.499, two sequences that differ in only the presence or absence of the 13-mer motif (Figure 3a). HGL6.985 contains the 13-mer motif and exhibits strong translational activity, while HGL6.499 lacks the motif and has very low activity. As a stringent test, we engineered vectors that removed the 13-mer from HGL6.985 and added the 13-mer to the 5′ terminus of HGL6.499. With this design format, we aimed to determine if translational activity could be inverted when the 13-mer motif was either removed from or added to the 5′ end of the RNA transcripts. When assayed in HeLa (human), BSC40 (monkey), RK13 (rabbit), BHK (hamster), and 129SV (mouse) cell lines, as well as in vitro using HeLa cell lysate, engineered versions of HGL6.985 lacking the 13-mer motif were 60–80% less active than the wild-type HGL6.985 sequence, while engineered versions of HGL6.499 containing the 13-mer motif were 600–3000% more active than the wild-type HGL6.499 sequence (Figure 3c,d and Figure S4a,b). Furthermore, the removal or addition of the 13-mer motif decreased or increased the translation activity, respectively, of the uncapped mRNA when compared to those of capped mRNA controls (Figure S3d). The strong concordance between cellular and cell-free expression provides confidence that the 13-mer functions with high translation initiation activity.
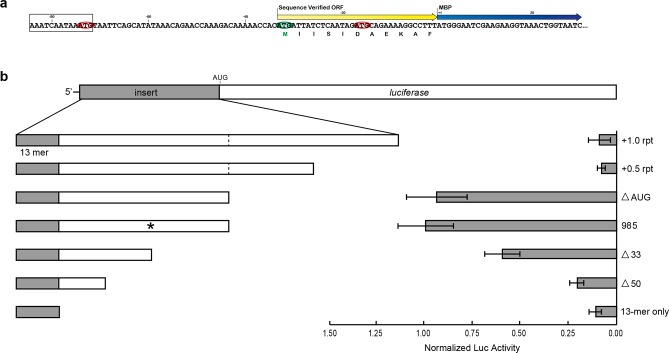
Analysis of the protein sequencing results of HGL6.985 indicated that the ribosome bypasses the AUG triplet in the 13-mer motif and initiates translation at position −33 of the TEE (Figure 4a). This result is consistent with a translation mechanism in which ribosomal recruitment and translation initiation proceed as two independent steps through recognition events that occur at separate locations on the mRNA template. To evaluate this process in greater detail, a series of insertion and deletion constructs were generated that increased and decreased the length of HGL6.985. Insertion constructs were designed by adding a repeat region at the 3′ end, which consisted of both the 5′-most half and the full sequence of HGL6.985. A repeating sequence was chosen so the overall nucleotide composition of the entire sequence would remain unchanged; no appreciable secondary structure was created (Table S8), and the HGL6.985 sequence alone (without the 13-mer) displayed minimal levels of translation enhancement (Figure 3c). In addition, a mutant version of HGL6.985 that mutated the authentic translation initiation site in the 5′ leader sequence was constructed. Analysis of luciferase expression levels in HeLa cells indicates that increasing the length of the leader sequence leads to a dramatic loss (80–90%) of activity, while shortening the length of the leader sequence leads to a progressive stepwise loss of activity (Figure 4b). Mutation of the authentic start site yields only a minimal change in activity. Interestingly, however, none of the modifications weakened the ability of the uncapped mRNAs containing the 13-mer motif to function cap-independently, when compared to capped RNA controls (Figure S3e). This suggests that the 13-mer motif is crucially important in cap-independent translation from our set of TEEs, yet the sequence flanking the motif dictates the overall strength of protein expression. Together, these results indicate that the selected version of HGL6.985 is the optimal length for this particular translation enhancing element and that ribosomal recruitment may involve other factors that require some distance from the initiation codon to function.
Figure 4.
Characterization of sequence constraints for a 13-mer motif. (a) Identification of authentic translation initiation sites by protein sequencing of in vitro-expressed MBP from the high-activity TEE HGL6.985. The protein sequence is listed below the nucleotide sequence. Color scheme: 13-mer motif (black box), MBP coding region (blue arrow), authentic translation initiation site (yellow arrow), and AUG triplets in-frame and out-of-frame with the MBP coding region (green and red ovals, respectively). (b) Mutational analysis of HGL6.985. Modifications were made to increase the 5′ leader length through addition of both a half-repeat and a full repeat (+0.5 rpt and +1.0 rpt, respectively) of the HGL6.985 sequence. A decrease in sequence length was achieved by removal of 33 (Δ33), 50 (Δ50), and all of the nucleotides downstream of the 13-mer motif (13-mer only). The authentic initiation site (*) within the TEE sequence was also deleted through mutation (ΔAUG). The functional impact of each mutation was assessed by measuring luciferase expression levels in HeLa cells relative to the unmodified HGL6.985 sequence and normalized to luciferase mRNA levels. Error bars designate the standard deviation.
Discussion
In traditional cap-dependent translation, the ribosome moves along the mRNA template toward the 3′ end until it encounters an initiation codon for translation.26 Mutagenesis studies indicate that introducing AUG codons upstream of the normal initiation site can dramatically inhibit translation when insertions are made at out-of-frame positions relative to the CDR, while insertions made at in-frame positions tend to supplant the original initiation site.27 Moreover, recognition of the AUG codon is often impaired when the initiation site lies close to the 5′ cap structure or is located in a region of strong secondary structure.28 In such cases, the ribosome will move to a downstream AUG codon that lies in an unobstructed region of the 5′ leader by processes termed leaky scanning and ribosomal shunting.26 When multiple AUG triplets are found in a 5′ leader region, mRNA translation can lead to the synthesis of multiple protein isoforms, but it is generally not known how and when specific AUG codons are utilized.
In some models of cap-independent translation, ribosomal recruitment sites are imbedded within a larger leader sequence and are thought to mimic the activity of a 5′ cap structure by recruiting the ribosome to the mRNA template.29 Apart from some initial studies by Mauro and others, which have demonstrated that certain mRNAs follow a prokaryotic model of mRNA–rRNA base pairing, very little is known about the mechanisms of cap-independent translation initiation in eukaryotic systems, as there are several possibilities.30,31 If this path is limited to only those mRNAs that can base pair to specific sites on the 18S rRNA, then protein-based regulation could provide a more universal solution to the problem of how eukaryotic ribosomes initiate translation on uncapped mRNA templates. However, it is also possible that nonhomologous mRNAs utilize adaptor molecules, such as noncoding RNAs, to link the mRNA template to the ribosomal complex. This later possibility is appealing, given the limited number of proteins that are known to interact with the 5′ leader of uncapped cellular transcripts32 and the preponderance of human noncoding RNAs.33
In this study, we examined the sequence determinants of AUG triplets in a defined set of naturally occurring human translation enhancing elements. This feature separates our study from others in which AUG triplets have been examined in synthetic constructs that are unrelated to natural genomes. Preliminary sequence analysis revealed that human TEEs contain an abundance of AUG triplets that would be expected to disrupt translational activity by diverting active ribosomes down unproductive pathways. However, we discovered that this was not the case, as many sequences with multiple AUGs were found to function with high activity in vitro and in cultured cells. Furthermore, we found that the absence of AUGs in the 5′ leader sequence does not automatically lead to higher translation levels. This result is the opposite of what would be expected on the basis of the linear scanning model for cap-dependent translation initiation.27 We showed that the stepwise removal of AUGs from a high-activity TEE led to significant losses of translational activity and that activity could not be increased by inserting the AUGs into a low-activity sequence. In all cases tested, AUG-containing TEEs were found to initiate translation within the boundary of the in vitro-selected sequence. Together, these results suggest that human TEEs contain specific regions that can modulate translational activity and that AUGs can be a critical component of these regions.
We discovered one example in which a short AUG-containing 13-mer motif is able to modulate translational activity in multiple sequence contexts and mammalian cell lines. We found that translational levels increase by as much as 40-fold when the motif is added to the 5′ end of two unrelated low-activity sequences. We also showed that the presence or absence of the motif at the 5′ end of a family of related sequences could lead to gains and losses of activity in five different types of cultured mammalian cells. Interestingly, the effect of the 13-mer motif on TEE-associated cap-independent translation appears to be influenced heavily by the surrounding sequence context, as evidenced by our analysis comparing capped and uncapped transcripts. A striking example of this lies in TEE HGL6.985. This TEE, which contains the 13-mer motif, drives high levels of translation when compared to other TEEs yet is only weakly cap-independent when compared to its capped counterpart. However, when the motif is added to HGL6.499, which lacks the 13-mer but is very similar across the remaining sequence, a dramatic increase in cap-independent activity was observed. This trend was also noticed when the motif was inserted into sequences with differing compositions; however, the overall effect varied. These findings together indicate that differences in the sequence downstream of the 13-mer motif, however subtle, influence the ability of the motif to drive cap-independent translation. Furthermore, protein sequencing of HGL6.985 revealed that ribosomal initiation occurs at an AUG codon downstream from the 13-mer motif. Altogether, this example suggests that TEE-associated cap-independent translation involving this 13-mer likely occurs by a two-step mechanism in which ribosomal recruitment and translation initiation proceed as separate recognition events and is conserved across closely related mammals.
The pronounced ability of the 13-mer motif to regulate mRNA translation levels in multiple cell lines led us to consider its molecular mechanism inside the cell. Recognizing the importance of the mouse Gtx element as a model for ribosomal recruitment,34−36 we searched the human 18S rRNA sequence for regions that might be complementary to the 13-mer motif. This analysis revealed a putative base pairing region at the base of expansion segment 3 that is complementary to a seven-nucleotide site at the 3′ end of the motif (Figure S6a). Further inspection of our set of human TEEs revealed that the TEEs containing the intact 13-mer motif share complementary with this same region of the 18S rRNA (Figure S6b). We mapped this putative mRNA binding site onto the three-dimensional structure of the eukaryotic ribosome to determine whether this region of the globular structure could be accessible to an mRNA template.37 The site is located, however, on the surface of the small ribosomal subunit some distance from the entrance tunnel of the ribosome (Figure S6c). These findings make it unlikely that mRNA is directly fed into the active site of the ribosome immediately following interaction with the 13-mer motif.38 This, coupled with the findings that the 13-mer appears to require a distance of at least 38 nucleotides from the initiation site and the composition of those nucleotides can have a profound effect on the overall activity of the motif, suggests that other factors may be involved in the translation driven by the 13-mer. It is entirely possible that the 13-mer motif or any of the other TEEs serve as interaction sites between mRNA and other cellular factors, much like IRES trans-activating factors (ITAFs) have been implicated in cellular IRES function.39 Moreover, these sites may facilitate an increased level of reinitiation of recycling ribosomes, through either cellular factors or direct interaction with rRNA, thereby increasing the efficiency of translation.40 While several possibilities exist, the exact mechanisms by which this AUG-containing 13-mer motif facilitates translation require further exploration.
One implication of our study is that human translation enhancing elements may contribute to the diversity of the human proteome by allowing translation to occur at different positions in primary transcripts. This possibility is particularly striking when considering the prevalence of human TEEs in our genomes.7 Translation at these sites could lead to the synthesis of protein isoforms or novel proteins from short open reading frames (ORFs). Indeed, recent computational and experimental studies suggest that thousands of short ORFs are translated in mammalian cells.41,42 Many of these gene products have been confirmed by proteomics analysis, indicating that the human proteome is very diverse.43−45 While the biological functions of most human peptides remain uncharacterized, several non-human examples have been studied in detail. The peptides (tal-1A, tal-2A, tal-3A, and tal-AA) that encode the polycistronic tarsel-less (tal) gene in Drosophila, for example, have been shown to modulate the activity of the shavenbaby transcription factor.46 Determining the extent to which human TEEs contribute to the total diversity of the human proteome is an interesting question that warrants further study.
In summary, we provide evidence that AUG triplets play an important yet often unpredictable role in TEE-associated cap-independent translation initiation. While the overabundance of AUGs within out set of TEEs hints at the importance of these triplets, their exact role appears to vary between sequences. Our results demonstrate that in some instances AUGs contribute by serving as part of a larger ribosomal recruitment motif. In other cases, they act as a start site for translation initiation. As other studies analyzing sequences that can facilitate cellular cap-independent translation have concluded, a common, general mechanism for this mode of translation initiation remains elusive, and it is likely that multiple mechanisms exist.47,48 Nonetheless, our observations provide a new opportunity to explore the mechanistic details of human translation enhancing elements and their contribution to the proteome.
Conclusions
Cap-independent protein translation has been shown to occur during normal cellular processes like mitosis and during times of cellular stress, such as a viral infection. Despite the observed importance of this method of protein production, the underlying mechanisms are not well understood. To address this issue, we analyzed a set of cap-independent translation enhancing elements from the human genome and uncovered an abundance of AUG triplets within their sequences. We experimentally demonstrated these AUGs can serve as part of a larger sequence context that can recruit the ribosome to the RNA message and that these triplets may or may not serve as an initiation site for translation to begin. The results of our study highlight the importance of these triplets in stimulating TEE-associated cap-independent protein translation and how these sequences can lead to the production of novel proteins, thereby contributing to the protein diversity encoded by the human genome.
Acknowledgments
The authors thank S. Kumar and K. Kukurba for help with the sequence analysis, A. Hansen for assistance with molecular cloning, M. Flores and M. Huynh for assistance with in vitro protein expression, B. Jacobs for help with cell culture, and members of the Chaput and Wellensiek laboratories for discussions about and comments on the manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00785.
Upstream AUG triplet patterns observed for 225 human TEEs (Figure S1), AUG content versus activity for 225 human TEEs (Figure S2), translation levels in the presence and absence of a cap structure (Figure S3), activity of modified TEEs in cell lysates (Figure S4), nuclear expression controls for the cell-based translation assay (Figure S5), proposed ribosomal interaction of the 13-mer motif (Figure S6), length, activity rank in cells, and AUG frame determination for 225 identified TEEs (Table S1), Kozak sequence analysis (Table S2), sequence details of TEEs used for analysis of upstream AUG triplets (Table S3), translational activity of eight selected TEEs with varying AUG patterns (Table S4), upstream open reading frame analysis (Table S5), activity of constructs used to evaluate the contributions of AUG to TEE function (Table S6), activity of 11 closely related TEEs used to identify a ribosomal recruitment site (Table S7), and details of modifications to TEE HGL6.985 (Table S8) (PDF)
This work was supported by a National Institutes of Health Eureka Award (GM085530) to J.C.C. and Midwestern University intramural funds granted to B.P.W.
The authors declare no competing financial interest.
Supplementary Material
References
- Shine J.; Dalgarno L. (1975) Determinant of cistron specificity in bacterial ribosomes. Nature 254, 34–38. 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Kaminishi T.; Wilson D. N.; Takemoto C.; Harms J. M.; Kawazoe M.; Schluenzen F.; Hanawa-Suetsugu K.; Shirouzu M.; Fucini P.; Yokoyama S. (2007) A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure 15, 289–297. 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Sonenberg N.; Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J.; Hellen C. U. T.; Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C. U. T.; Sarnow P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612. 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Terenin I. M.; Andreev D. E.; Dmitriev S. E.; Shatsky I. N. (2013) A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 41, 1807–1816. 10.1093/nar/gks1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellensiek B. P.; Larsen A. C.; Stephens B.; Kukurba K.; Waern K.; Briones N.; Liu L.; Snyder M.; Jacobs B. L.; Kumar S.; Chaput J. C. (2013) Genome-wide profiling of human cap-independent translation-enhancing elements. Nat. Methods 10, 747–750. 10.1038/nmeth.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G.; Carter M. S.; Eisen M. B.; Brown P. O.; Sarnow P. (1999) Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. U. S. A. 96, 13118–13123. 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs K. A.; Stoneley M.; Bushell M.; Willis A. E. (2008) Re-programming of translation following cell stress allows IRES-mediated transaltion to predominate. Biol. Cell 100, 27–38. 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S.; Elias-Kirma S.; Nir R.; Gritsenko A. A.; Stern-Ginossar N.; Yakhini Z.; Weinberger A.; Segal E. (2016) Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351 (pii), aad4939. 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- Iacono M.; Mignone F.; Pesole G. (2005) uAUG and uORFs in human and rodent 5′ untranslated mRNAs. Gene 349, 97–105. 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Churbanov A.; Rogozin I. B.; Babenko V. N.; Ali H.; Koonin E. V. (2005) Evolutionary conservation suggests a regulatory function of AUG triplets in 5 ’-UTRs of eukaryotic genes. Nucleic Acids Res. 33, 5512–5520. 10.1093/nar/gki847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener D.; Ray S. (1991) Eukaryotic start and stop translation sites. Nucleic Acids Res. 19, 3185–3192. 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S.; Niimura Y.; Gojobori T.; Tanaka H.; Miura K. (2007) Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 36, 861–871. 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D.; Mauro V. P. (2010) Determinants of initiation codon selection during translation in mammalian cells. PLoS One 5, e15057 10.1371/journal.pone.0015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J.; Sonenberg N. (1985) Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526. 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Peabody D. S. (1989) Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264, 5031–5035. [PubMed] [Google Scholar]
- Touriol C.; Bornes S.; Bonnal S.; Audigier S.; Prats H.; Prats A.-C.; Vagner S. (2003) Geration of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biology of the Cell 95, 169–178. 10.1016/S0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Ingolia N. T.; Lareau L. F.; Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrejs M.; Masek T.; Vopalensky V.; Hlubucek P.; Delbos P.; Pospisek M. (2010) IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 38, D131–D136. 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malys N.; McCarthy J. E. G. (2011) Translation initiation: variations in the mechanism can be anticipated, Cell. Cell. Mol. Life Sci. 68, 991–1003. 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellensiek B. P.; Larsen A. C.; Flores J.; Jacobs B. L.; Chaput J. C. (2013) A leader sequence capable of enhancing RNA expression and protein synthesis in mammalian cells. Protein Sci. 22, 1392–1398. 10.1002/pro.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen H.; Schumperli D.; Rosenberg M. (1984) Affecting gene expression by altering the length and sequence of the 5′ leader. Proc. Natl. Acad. Sci. U. S. A. 81, 7698–7702. 10.1073/pnas.81.24.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S.; Subramani S.; Berg P. (1986) Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol. Cell. Biol. 6, 2704–2711. 10.1128/MCB.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S.; Berg P. (1986) Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol. Cell. Biol. 6, 2695–2703. 10.1128/MCB.6.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiology and Molecular Biology Reviews 75, 434–467. 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol. 108, 229–241. 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867–19870. [PubMed] [Google Scholar]
- Chappell S. A.; Edelman G. M.; Mauro V. P. (2006) Ribosomal tethering and clustering as mechanisms for translation initiation. Proc. Natl. Acad. Sci. U. S. A. 103, 18077–18082. 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresios J.; Chappell S. A.; Zhou W.; Mauro V. P. (2006) An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 13, 30–34. 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- Meng Z.; Jackson N. L.; Shcherbakov O. D.; Choi H.; Blume S. W. (2010) The human IGF1R IRES likely operates through a Shine-Dalgarno-like interaction with the G961 loop (E-site) of the 18S rRNA and is kinetically modulated by a naturally polymorphic polyU loop. J. Cell. Biochem. 110, 531–544. 10.1002/jcb.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elroy-Stein O., and Merrick W. C. (2007) Translation inititiation via cellular internal ribosome entry sites. In Translational Control in Biology and Medicine (Mathews M., Sonenberg N., and Hershey J. W. B., Eds.) Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Bertone P.; Stolc V.; Royce T. E.; Rozowsky J. S.; Urban A. E.; Zhu X. W.; Rinn J. L.; Tongprasit W.; Samanta M.; Weissman S.; Gerstein M.; Snyder M. (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246. 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Matveeva O. V.; Shabalina S. A. (1993) Intermolecular mRNA-rRNA hybridization and the distribution of potential interaction regions in murine 18S rRNA. Nucleic Acids Res. 21, 1007–1011. 10.1093/nar/21.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro V. P.; Edelman G. M. (1997) rRNA-like sequences occur in diverse primary transcripts: Implications for the control of gene expression. Proc. Natl. Acad. Sci. U. S. A. 94, 422–427. 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro V. P.; Edelman G. M. (2002) The ribosome filter hypothesis. Proc. Natl. Acad. Sci. U. S. A. 99, 12031–12036. 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. J.; Devkota B.; Huang A. D.; Topf M.; Narayanan E.; Sali A.; Harvey S. C.; Frank J. (2009) Comprehensive molecular structure of the eukaryotic ribosome. Structure 17, 1591–1604. 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A. V.; Kolupaeva V. G.; Yusupov M. M.; Hellen C. U.; Pestova T. V. (2008) Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 27, 1609–1621. 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A. A.; Hatzoglou M. (2011) Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240. 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin M. A.; Skabkina O. V.; Hellen C. U.; Pestova T. V. (2013) Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell 51, 249–264. 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C.; Forrest A. R.; Nourbakhsh E.; Pang K. C.; Kai C.; Kawai J.; Carninci P.; Hayashizaki Y.; Bailey T. L.; Grimmond S. M. (2006) The abundance of short proteins in the mammalian proteome. PLoS Genet. 2, e52. 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T.; Ghaemmaghami S.; Newman J. R. S.; Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M.; Itagaki C.; Hata H.; Suzuki Y.; Izumi T.; Natsume T.; Isobe T.; Sugano S. (2004) Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs. Genome Res. 14, 2048–2052. 10.1101/gr.2384604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S. A.; Mitchell A. J.; Schwaid A. G.; Cabili M. N.; Ma J.; Levin J. Z.; Karger A. D.; Budnik B. A.; Rinn J. L.; Saghatelian A. (2013) Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64. 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M.; Kozuka-Hata H.; Suzuki Y.; Semba K.; Yamamoto T.; Sugano S. (2007) Diversity of translation start sites may defien increased complexity of the human short ORFeome. Mol. Cell. Proteomics 6, 1000–1006. 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- Kondo T.; Plaza S.; Zanet J.; Benrabah E.; Valenti P.; Hashimoto Y.; Kobayashi S.; Payre F.; Kageyama Y. (2010) Small peptides switch the transcriptional activity of shavenbaby during drosophila embryogenesis. Science 329, 336–339. 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- Baird S. D.; Turcotte M.; Korneluk R. G.; Holcik M. (2006) Searching for IRES. RNA 12, 1755–1785. 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsenko A. A.; Weingarten-Gabbay S.; Elias-Kirma S.; Nir R.; de Ridder D.; Segal E. (2017) Sequence features of viral and human Internal Ribosome Entry Sites predictive of their activity. PLoS Comput. Biol. 13, e1005734 10.1371/journal.pcbi.1005734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.