Abstract
The Saccharomyces cerevisiae strain JAY270/PE2 is a highly efficient biocatalyst used in the production of bioethanol from sugarcane feedstock. This strain is heterothallic and diploid, and its genome is characterized by abundant structural and nucleotide polymorphisms between homologous chromosomes. One of the reasons it is favored by many distilleries is that its cells do not normally aggregate, a trait that facilitates cell recycling during batch-fed fermentations. However, long-term propagation makes the yeast population vulnerable to the effects of genomic instability, which may trigger the appearance of undesirable phenotypes such as cellular aggregation. In pure cultures of JAY270, we identified the recurrent appearance of mutants displaying a mother-daughter cell separation defect resulting in rough colonies in agar media and fast sedimentation in liquid culture. We investigated the genetic basis of the colony morphology phenotype and found that JAY270 is heterozygous for a frameshift mutation in the ACE2 gene (ACE2/ace2-A7), which encodes a transcriptional regulator of mother-daughter cell separation. All spontaneous rough colony JAY270-derived isolates analyzed carried copy-neutral loss-of-heterozygosity (LOH) at the region of chromosome XII where ACE2 is located (ace2-A7/ace2-A7). We specifically measured LOH rates at the ACE2 locus, and at three additional chromosomal regions in JAY270 and in a conventional homozygous diploid laboratory strain. This direct comparison showed that LOH rates at all sites were quite similar between the two strain backgrounds. In this case study of genomic instability in an industrial strain, we showed that the JAY270 genome is dynamic and that structural changes to its chromosomes can lead to new phenotypes. However, our analysis also indicated that the inherent level of genomic instability in this industrial strain is normal relative to a laboratory strain. Our work provides an important frame of reference to contextualize the interpretation of instability processes observed in the complex genomes of industrial yeast strains.
Keywords: Saccharomyces cerevisiae, Loss-of-heterozygosity, ACE2, Fermentation, Colony morphology, Mitotic recombination
Over the last decade, the genomic characterization of many diverged diploid isolates of the yeast Saccharomyces cerevisiae has revealed the existence of substantial intra-strain variability in nucleotide sequence and chromosome structure (Argueso et al. 2009; Borneman et al. 2011; Magwene et al. 2011; Peter et al. 2018). While most conventional laboratory strains, when diploidized, harbor nearly completely homozygous genomes and display stable phenotypes, the genome architecture of wild strains, including clinical and industrial isolates, has often been found to be more complex, with abundant inter-homolog nucleotide heterozygosity and structural variation.
In several species, widespread heterozygosity is often associated with beneficial phenotypes. It is suspected that this genomic configuration may also contribute to the adaptation of wild yeasts to their respective habitats, as well as to the robust growth, stress tolerance, and other desirable traits characteristic of industrial yeasts (Muller and McCusker 2009; Plech et al. 2014). Interestingly, the maintenance of heterozygosity by these single-celled organisms is challenging because allelic mitotic recombination events between homologous chromosomes occurs at relatively high rates (∼10−4/cell division; (St Charles and Petes 2013)), and can lead to immediate fixation of tracts of loss-of-heterozygosity (LOH) in the clonal cell lineage. Accordingly, in JAY270/PE2 and other wild strains, heterozygous regions are interspersed with stretches of homozygosity indicating the occurrence of LOH events in clonal ancestors (Magwene et al. 2011). In addition, multiple studies have characterized the emergence of chromosomal rearrangements in cells clonally derived from industrial strains (Nadal et al. 1999; Carro and Pina 2001; Basso et al. 2008; Querol and Bond 2009; de Amorim et al. 2017). Those detailed studies, combined with informal field observations occasionally reported by operators of wine, beer and biofuel fermentations, have earned industrial yeast strains a reputation for having unstable genomes. This genomic instability could, and likely does, make fermentations that use such strains vulnerable to process disruption due to the loss of selected traits or appearance of undesirable phenotypes.
One of the first heterozygous S. cerevisiae strains to have its genome characterized was JAY270/PE2, widely used in sugarcane bioethanol production (Argueso et al. 2009). This heterothallic diploid was originally isolated at a Brazilian distillery as an aggressive, yet highly productive wild contaminant (Basso et al. 2008). One of its most attractive features is its suitability to continuous cell recycling in batch-fed fermentations. JAY270/PE2 offers distillery operators predictable fermentation conditions during the 8-9 month sugarcane harvest season, because it is often able to outcompete wild yeast contaminants that are steadily introduced through the feedstock into the recycling system.
Here, we report a case study of two easily discernible and related mutant phenotypes that result from a specific rearrangement of the JAY270/PE2 genome. We characterized the spontaneous appearance of clones displaying a mother-daughter cell separation defect among pure cultures of this strain. This cell separation defect resulted in both rough colony morphology in agar plates and fast sedimentation in liquid culture. We found that these phenotypes were recurrently caused by an LOH event spanning the ACE2 locus. We measured the quantitative rate and characterized the qualitative spectrum of mitotic recombination at the region of chromosome XII (Chr12) where ACE2 is located. In order to provide reference for a broader comparison of genomic instability levels, we also measured the rates of LOH at three additional independent chromosomes, using both JAY270/PE2 and a conventional diploid homozygous laboratory strain. We found that the rates of mitotic recombination were quite similar overall between the four genomic regions and between the two strain backgrounds. These results showed that, while chromosomal rearrangements did occur at detectable rates and could lead to new phenotypes in JAY270/PE2, its inherent level of genomic instability was not elevated relative to the reference strain. Instead, we found that the genome of the conventional laboratory strain was just as dynamic as to that of JAY270/PE2.
Materials and Methods
Yeast genetic backgrounds and culture procedures
Saccharomyces cerevisiae strains used in this study descended from either the JAY270 or CG379 strain backgrounds (Table S1). JAY270 is a heterothallic diploid single colony isolate derived from the industrial bioethanol strain PE2 (Argueso et al. 2009). CG379 diploid is related to the S288c laboratory strain background (Morrison et al. 1991; Argueso et al. 2008).
Yeast cells were grown in YPD (20 g/L glucose, 20 g/L peptone, 10 g/L yeast extract, 20 g/L bacteriological agar for solid media) and synthetic media (20 g/L glucose, 5 g/L ammonium sulfate, 1.7 g/L yeast nitrogen base without amino acids, 1.4 g/L complete drop-out mix, 20 g/L bacteriological agar). Standard procedures for yeast transformation, crossing and sporulation were followed (Ausubel et al. 1998). Counter-selections against URA3 were performed in synthetic media supplemented with 1g/L of 5-Fluoroorotic Acid (5-FOA).
Allele replacement and complementation tests
A two-step allele replacement plasmid was constructed by cloning a segment of the wild type ACE2 allele containing the 8-adenine homopolymer into the URA3 pRS306 integrative vector (Sikorski and Hieter 1989). First, pRS306 was digested with BamHI and KpnI and ligated to a BglII- and KpnI-digested segment of the wild type ACE2 allele (PCR amplified from JAY289 genomic DNA using primers JAO904 and JAO905). The resulting plasmid, pAR1, was linearized with EcoRI and transformed into 5-FOA resistant (ura3 mutant) derivatives of JAY291 and JAY292 haploids (both mutant ace2-A7). Pop-in Ura+ integration transformants were selected, and pop-out events were then selected for resistance to 5-FOA. 5-FOA resistant candidates were screened by PCR and Sanger sequencing to verify the presence of the wild type ACE2 allele. This procedure resulted in the ACE2 strains JAY1051 and JAY1039, isogenic to JAY291 and JAY292 respectively. Complementation tests using these strains were performed as shown in Figure 2C.
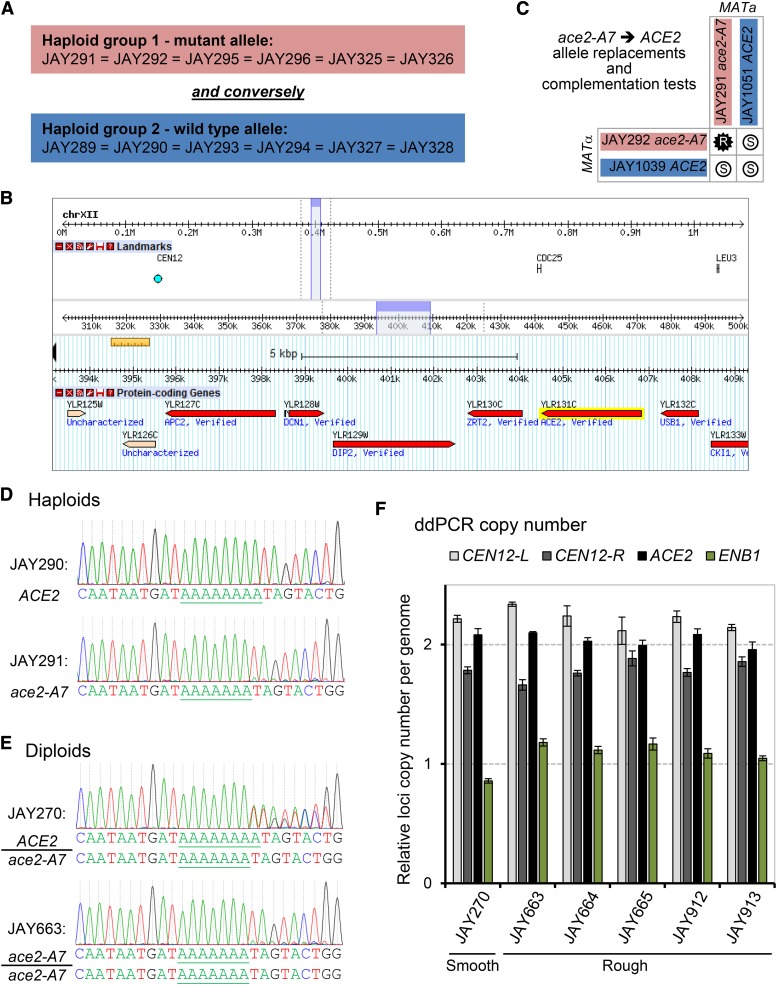
Figure 2.
Mapping and characterization of the ace2-A7 mutation. A The genetic mapping approach consisted of identifying HetSNP linkage regions in which one allele (M or P) co-segregated in all six haploids inferred to carry the rough mutant allele (red), while the other allele (P or M) co-segregated in all six haploids inferred to contain the smooth wild type allele (blue) (see Fig. 1K). B Genome Browser view of a Chr12 candidate region that satisfied the strict co-segregation criterion. This region spans 13 Kb and contains 9 genes (from YLR125W to CKI1). Review of the functional annotations of the genes in this region identified the ACE2 locus (highlighted in yellow) as a likely candidate, and further sequence review uncovered the ace2-A7 mutation. A second candidate region on Chr11 (not shown) was also identified through co-segregation analysis, but was not pursued further. C Allele replacements and complementation tests. The ace2-A7 allele from two haploid strains JAY291 (MATa) and JAY292 (MATα) was replaced with the ACE2 allele, generating JAY1051 and JAY1039, respectively. Matings between the ace2-A7 and ACE2 haploids confirmed the restoration of the smooth phenotype in diploid strains derived from at least one haploid parent containing the ACE2 allele. D and E Sanger DNA sequencing analysis of ACE2 locus. D shows segments of PCR-Sanger sequencing chromatograms for the ACE2 locus from haploids with the wild type ACE2 (JAY290) and mutant ace2-A7 (JAY291) alleles. Note the difference in the A-homopolymer runs, with eight consecutive peaks in ACE2 and seven consecutive peaks in ace2-A7. E shows PCR-Sanger chromatograms from diploids (primer extension was from left to right in all cases). The JAY270 chromatogram is a mixture of the two alleles (heterozygous), while the rough colony clone JAY663 only had the ace2-A7 pattern. The inferred DNA sequences in D and E are shown below the chromatograms. The chromatograms from rough clones JAY664, JAY665, JAY912 and JAY913 were indistinguishable from JAY663 (not shown). F Gene copy number analysis of the ACE2 locus. Plots show the calculated copy number for the four genomic regions probed, relative to the average of the signal between the control probes flanking the Chr12 centromere (CEN12-L and CEN12-R), both known to be present in two copies in the JAY270 genome. The ENB1 region was another control probe, known to be present as one copy in JAY270.
Construction of strains used in LOH assays
The JAY270 strains used in the LOH assays were constructed from a homozygous ura3Δ/ura3Δ derivative of JAY270 (JAY585; gift from F. Galzerani). The CORE2 cassette containing the Kluveromyces lactis URA3 gene, the S. cerevisiae URA3 gene and the KanMX4 geneticin resistance marker (KlURA3-ScURA3-KanMX4) was amplified from pJA40 (Zhang et al. 2013) with primers targeting integration at four different genomic regions (Table S2). Integration in the maternal or paternal Chr12 homolog was checked by PCR and Sanger genotyping of linked HetSNPs. The same procedures and primers were used integrate CORE2 in the CG379 background.
Quantitative LOH rate assays
Diploid yeast cells hemizygous for CORE2 insertions were streaked to single colonies on solid YPD medium and incubated at 30° for two days. Single colonies were inoculated into 5 ml liquid YPD, and incubated for 24 hr at 30° in a rotating drum. The cultures were serially diluted and plated on YPD (permissive) or 5-FOA (selective) media. Colonies were counted after 2 days of growth on permissive and 4 days on selective plates, and colony counts were used to calculate recombination rates and 95% confidence intervals using the Lea & Coulson method of the median within the FALCOR web application (http://www.keshavsingh.org/protocols/FALCOR.html) (Lea and Coulson 1949; Hall et al. 2009). Statistical analyses of pairwise comparisons between recombination rates were performed using a two-sided nonparametric Mann Whitney test in GraphPad Prism software.
Genotyping specific HetSNPs Through PCR-RFLP and molecular karyotyping
Genotyping was performed by PCR-amplification of regions containing the HetSNPs followed by restriction digestion and agarose gel analysis fragment length polymorphisms. In cases of markers where the HetSNPs were not associated with an RFLP, PCR products were Sanger-sequenced. A complete list of the HetSNPs coordinates, primers and restriction enzymes used is provided in Table S3.
Genome Sequencing Analyses
The genomes of the JAY270 parent strain and of 56 haploids derived from 14 complete JAY270 tetrads were sequenced using the Illumina short read whole genome sequencing platform. The sequencing reads were used to create a phased draft map of high confidence HetSNPs in JAY270’s genome (detailed information about the pipeline used are provided in the Supplemental Material). Genome sequencing data associated with this study is available in the Sequence Read Archive (SRA) database under study number SRP082524.
Quantitative digital droplet PCR
Genomic DNA was prepared from PFGE agarose plugs as previously described (Zhang et al. 2013). Sheared DNA was then digested with MfeI-HF, a restriction enzyme that cuts the genome frequently, but does not cut within the ddPCR amplicons. Digested DNA was diluted to 0.05 ng/μl and 2 μl of this dilution was used as a template for each reaction using BioRad’s QX200 EvaGreen Supermix. The internal genomic DNA template normalizer was the average between signal from DNM1 and YLR001C (regions of known two copy number), divided by 2.
Data and reagent availability
Strains and plasmids are available upon request. Supplemental files available at FigShare. Sequence data are available at the Sequence Read Archive (SRA) database under study number SRP082524. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6406598.
Results
Spontaneous JAY270 derivatives display a recurrent cell separation defect
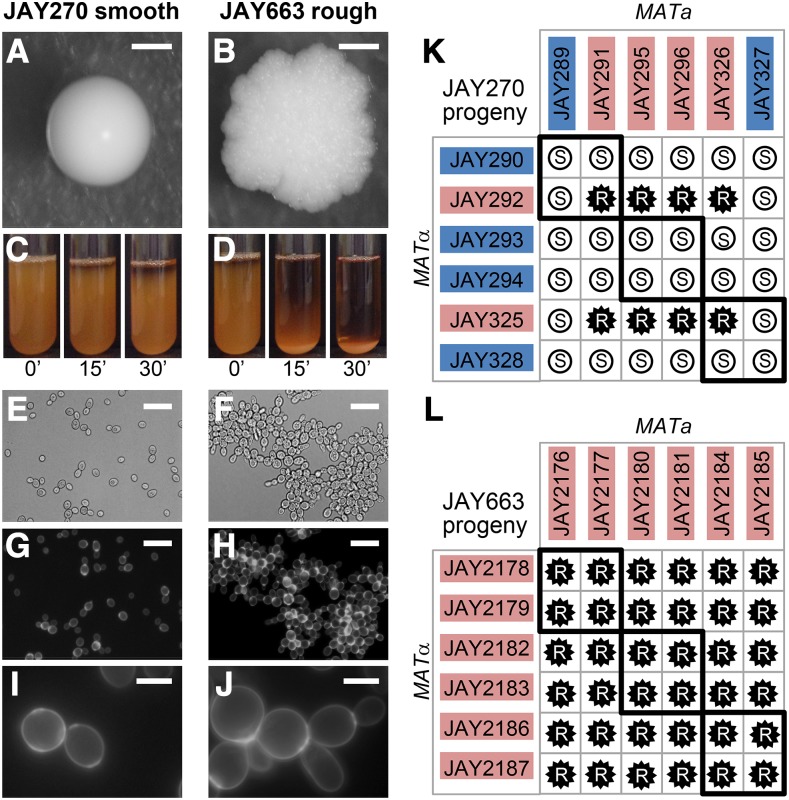
Yeast cellular aggregation can be a problem in batch-fed sugarcane bioethanol production because it reduces fermentation kinetics and leads to clogging of the centrifuges and pipes used to recycle the yeast biomass from one tank to the next (Della-Bianca et al. 2013; Paulino de Souza et al. 2018). One of the desirable industrial traits of the JAY270/PE2 strain (hereafter referred to simply as JAY270) is that its cells do not usually aggregate (Basso et al. 2008). Accordingly, JAY270 produces normal hemispherical colonies with smooth surfaces and edges when grown in solid agar medium, and remains in dispersed suspension in liquid culture for prolonged periods (Figure 1A and 1C). While these are the phenotypes typically observed, over the course of our studies using this strain we noticed the sporadic occurrence of colonies clonally derived from JAY270 that displayed altered morphology: relatively flat-growing colonies with rough surfaces and edges (Figure 1B). In addition, when grown in liquid media, these clones sedimented at a faster rate compared to JAY270 (Figure 1D). Under bright field microscopic examination, yeast cells derived from such rough colonies appeared to grow in chains, showing a budding pattern consistent with a defect in the separation of the daughter cells from their mother (Figure 1E-F). We stained these cells with calcofluor white to visualize the chitin-rich ring septa, confirming the persistent attachment of mother and daughter cells at the budding neck site (Figure 1G-J).
Figure 1.
Smooth and rough colony morphologies, liquid sedimentation, mother-daughter cell attachment, and phenotypes of diploids derived from mating specific haploids. A-J show images of the JAY270 smooth parent diploid strain (left panels) and its spontaneous rough derivative JAY663 (right panels). A and B, colony morphologies on YPD agar after 3 days growth at 30C. C and D, cell sedimentation kinetics. 5 ml liquid YPD cultures were grown overnight at 30C in a rotating drum. Test tubes were vortexed vigorously for 10 sec to fully resuspend the cells, then were left to rest and photographed at 15 min intervals (the 0’ pictures were taken immediately after vortexing). E and F, bright field, and G-J, fluorescence microscopy of cells stained with calcofluor white to highlight chitin septa and the mother-daughter cell attachment. Scale bars are 1mm (A-B), 20µm (E-F) and 5µm (G-J). K and L, Smooth (S, white circles) and rough (R, black stars) phenotypes of diploids formed by crossing the indicated MATa and MATα haploids isolated from three tetrads of each JAY270 (K) and JAY663 (L). Thick black lines indicate the four diploids derived from matings of intra-tetrad sibling haploids. The colored backgrounds for each haploid correspond to their inferred genotype (Blue, dominant wild type allele; Red, recessive mutant allele). All 12 haploids from panel K had their whole genomes sequenced. Co-segregation analysis with JAY270 HetSNPs (Fig. S1) was used for identification of the causal mutation at the ACE2 locus (Fig. 2).
We initially isolated five independent examples of such rough colonies for genetic characterization (JAY663, JAY664, JAY665, JAY912 and JAY913), all of which were derived either directly from JAY270 or from JAY270-isogenic strains. The rough colony phenotype of these isolates was stably maintained and was not reversible over several clonal generations, suggesting that it was likely encoded genetically and not caused by a transient transcriptional or post-transcriptional state. We estimated that these five isolates appeared spontaneously from a pool of ∼5 × 104 smooth colonies. Assuming a genetic origin and based on this high frequency of occurrence in diploid cells, we reasoned that this phenotype was unlikely to be caused by a rare dominant de novo nucleotide point mutation, which might be expected to occur in the 10−9-10−8/cell division rate range. Instead, a more plausible mutational mechanism was mitotic recombination leading to loss-of-heterozygosity (LOH), because it occurs at a compatible rate (∼10−4/cell division; (St Charles and Petes 2013)) and would not require a dominant phenotypic effect.
Investigation of the genetic basis of the rough colony phenotype
In a parallel project, we observed that crossing two specific haploid descendants of JAY270 (JAY291 MATa and JAY292 MATα; (Argueso et al. 2009)) resulted in diploid cells that displayed rough colony morphology and fast sedimentation phenotypes indistinguishable from those exhibited by the five isolates described above. Analysis of the JAY291 and JAY292 haploids showed that both displayed a mother-daughter cell separation defect and fast sedimentation, but neither had the rough colony phenotype (data not shown). JAY291, JAY292 and all other haploid derivatives of JAY270 display the normal smooth colony morphology.
The ability to consistently reproduce the mutant colony morphology phenotype in controlled crosses between specific haploids opened an avenue to investigate its genetic basis. We pursued this approach by carrying out crosses between twelve haploid descendants of JAY270 (3 tetrads; Figure 1K), all of which had their whole genomes sequenced (see below). All possible MATa x MATα crosses were performed producing 36 different diploids. Among them, we found eight with rough and 28 with smooth colony surfaces, in a pattern that was consistent with recessive inheritance of a trait controlled by a single gene. Even though the rough colony phenotype was not observed in any of the parents, the phenotypes of the respective diploids allowed us to infer which allele was present in the haploids: either the wild type dominant allele or the recessive mutant allele.
In addition, for comparison, we conducted a similar tetrad dissection and mating analysis with one of the spontaneous rough-colony isolates, JAY663. We examined the phenotypes of its haploid derivatives and found that none displayed the rough colony phenotype; they were all smooth (∼100 examined). We then took twelve of these haploids comprising three full tetrads (JAY2176 through JAY2187), determined their mating types, and conducted all possible mating combinations between them (Figure 1L). In this case, all 36 crosses resulted in rough colony diploids, indicating that all twelve haploid parents carried the causal recessive mutant allele, and therefore, JAY663 must have been homozygous for it. This was consistent with the hypothesis that copy-neutral LOH might be responsible for the sporadic appearance of the mutant morphology phenotype in JAY270 clonal derivatives.
Rough colony phenotype results from loss of the wild type ACE2 allele in diploids
The results described above suggested that the rough colony phenotype was diploid-specific, and caused by a recessive mutation present in heterozygous state in JAY270. Based on this premise, we divided the twelve sequenced JAY270-derived haploids from Figure 1K into two groups according to their inferred genotype (Figure 2A). Group 1 included the six haploids inferred to carry the mutant recessive allele of the causal gene, whereas group 2 included the six haploids with the wild type dominant allele.
We previously reported the whole genome sequence of the JAY291 haploid (Argueso et al. 2009). Since then we have sequenced the genomes of 55 additional JAY270-derived haploids, including the haploids used in the crosses shown in Figure 1K. This genome sequence dataset, comprising fourteen sets of four-spore tetrads, was generated in a project to characterize the abundance, distribution and phasing of heterozygous loci in the JAY270 genome, the full results of which will be described elsewhere. These haploid genomic sequences were used to create a draft map of phased heterozygous single nucleotide polymorphisms (HetSNPs) containing 12,023 loci unevenly distributed across the genome (Fig. S1).
In order to map the gene underlying the colony morphology trait, we searched for the JAY270 HetSNPs that co-segregated among the haploids from groups 1 or 2. We interrogated each of the genome-wide HetSNPs, and determined which had alleles that co-segregated in all six individuals within group 1, and conversely, had the other allele co-segregating in all six individuals within group 2. This analysis identified only two candidate regions that fit the strict co-segregation criterion. One of the candidate regions corresponded to ∼30 Kb on Chr11, including thirteen genes (not shown); and the other spanned ∼15 Kb containing nine genes on the right arm of Chr12 (Figure 2B), located ∼50 Kb centromere proximal to the ribosomal DNA tandem repeats (rDNA).
We reviewed the annotations of the 22 candidate genes, and identified a gene located in the Chr12 region, ACE2, which encodes a transcription factor that controls the expression of genes involved in the mother-daughter cell separation process (Weiss 2012). In cells lacking Ace2p, the daughter cell remains attached to the mother cell at the bud neck, resulting in the accumulation of multicellular clusters. These clusters lead to faster sedimentation in liquid culture compared to dispersed cells. Importantly, while both haploid ace2 and diploid ace2/ace2 mutants form multicellular clusters and display faster sedimentation in liquid culture, only the diploids form rough colonies in agar media (Voth et al. 2005). Haploid ace2 mutants form normal smooth colonies. This ploidy-dependent colony morphology phenotype is a result of the interaction between the defect in mother-daughter cell separation and the difference in bud site selection between haploids (axial budding pattern) and diploids (bipolar-pattern) (Voth et al. 2005).
We inspected the genomic sequence of the ACE2 gene in the haploid derivative JAY291 (Argueso et al. 2009), and compared it to the sequence in the S288c reference genome. Only one difference was identified: The wild type ACE2 allele in S288c contains a homopolymer run of 8 adenine nucleotides, while the mutant allele in JAY291 has 7 adenines in this region, resulting in a -1 frameshift mutation and a stop codon shortly downstream. Hence, we named the mutant allele ace2-A7. We then conducted reciprocal complementation tests to formally demonstrate that ace2-A7 was the causal mutation responsible for the rough colony phenotype. The mutant alleles in haploids JAY291 and JAY292 were replaced with the wild type allele, resulting respectively in the isogenic ACE2 strains JAY1051 and JAY1039. When these allele replacement strains were crossed to ace2-A7 strains (Figure 2C), the resulting diploids displayed the smooth colony phenotype. This result confirmed that the wild type ACE2 allele fully complemented the ace2-A7 mutation and restored normal colony morphology in heterozygous diploids in this genetic background.
LOH is the primary mechanism leading to the appearance of spontaneous rough colony isolates
After identifying the causal relationship between a mutation at the ACE2 locus and the rough colony phenotype, we determined its DNA sequence in the five spontaneous rough colony derivatives isolated earlier in the study. We PCR-amplified and Sanger-sequenced the region containing the adenine homopolymer run in ACE2 from JAY270, from the haploid derivatives JAY290 and JAY291, and from the rough colony isolates (Figure 2D-E). This analysis confirmed the presence of a run of 8 adenines in ACE2 (JAY290) and 7 adenines in ace2-A7 (JAY291). The chromatogram in the JAY270 heterozygous diploid was consistent with a mixture of ACE2 and ace2-A7 DNA templates being present in the sequencing reaction: single nucleotide peaks were observed at positions primer-proximal to the homopolymer run, and out-of-register double peaks were seen downstream of the seventh adenine nucleotide. The chromatograms for JAY663 (Figure 2E) and all four additional rough colony isolates (data not shown) were consistent with the presence of the ace2-A7 frameshift mutation and absence of the ACE2 allele. The loss of the ACE2 allele in the diploid rough colony isolates may be explained by either copy-neutral LOH mechanisms, such as inter-homolog mitotic recombination and de novo -1 contraction in the adenine homopolymer run of the ACE2 allele, or by copy-negative mechanisms such as segmental deletion spanning ACE2 or Chr12 monosomy. To distinguish between these scenarios, we conducted quantitative digital droplet PCR analysis to determine the copy number status of the ACE2 locus relative to genes immediately to the left (DNM1) and right (YLR001C) of the Chr12 centromere (CEN12-L and CEN12-R ddPCR probes, respectively), and relative to the ENB1 gene, which is known to be present as single hemizygous copy in JAY270’s Chr15 (Argueso et al. 2009). This analysis showed that all five rough colony isolates contained two copies of ACE2 locus DNA material (Figure 2F), thus ruling out the segmental deletion and monosomy mechanisms.
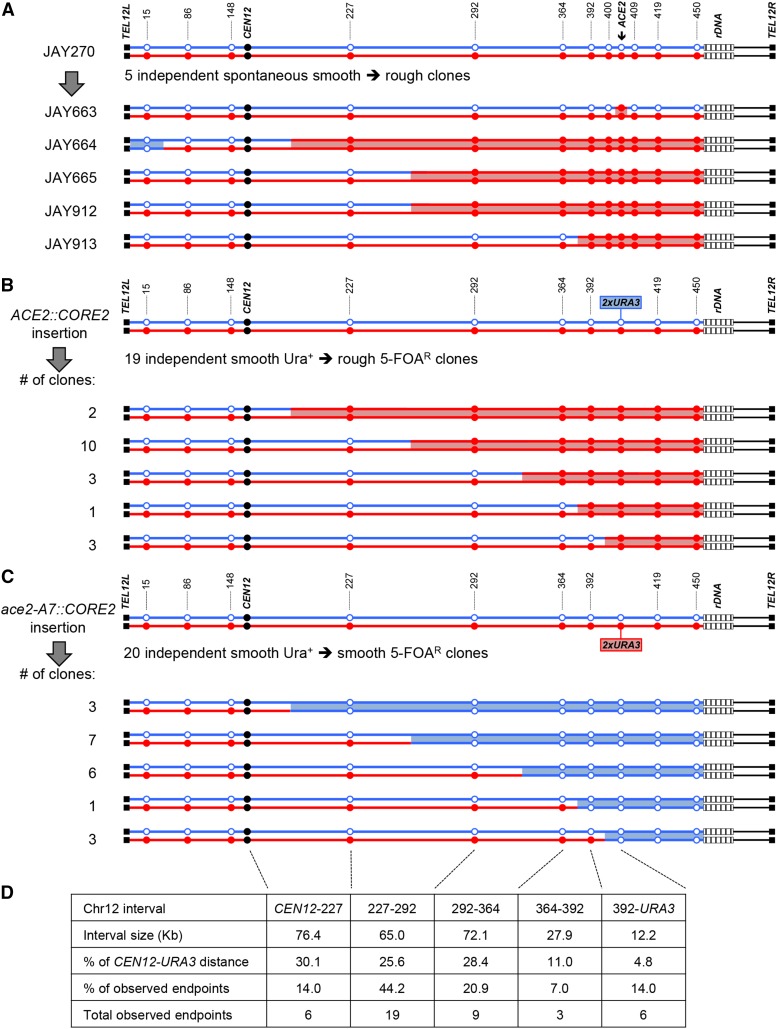
LOH due to interhomolog recombination is typically a regional mutational mechanism. Interstitial tracts of homozygosity can span tens of kilobases, and terminal tracts are even longer, extending all the way to the telomeres (St Charles and Petes 2013). Therefore, in addition to being homozygous for ace2-A7, the rough colony isolates, if derived from interhomolog recombination, might also be homozygous for flanking HetSNPs. In contrast, copy neutral LOH caused by a de novo -1 contraction should be restricted to the adenine homopolymer run. We distinguished these mechanisms by determining the genotypes at eleven Chr12 HetSNPs flanking the ACE2 locus using PCR followed by Sanger sequencing or restriction fragment length polymorphism analysis (RFLP; Table S3). The results of this analysis were compiled to produce the LOH tract maps shown in Figure 3A. As expected, JAY270 was heterozygous for all eleven markers tested. Notably, Chr12 in this strain is only heterozygous for positions to the left of the rDNA cluster (Fig. S1). This pattern is similar to that described previously for other heterozygous diploid S. cerevisiae genomes and is suggestive of ancestral LOH events mediated by rDNA instability (Magwene et al. 2011).
Figure 3.
LOH tract maps of the ACE2 and Chr12 heterozygous regions. A The genotypes at twelve phased JAY270 Chr12 HetSNP marker loci were determined using PCR and RFLP or Sanger sequencing analyses (Table S3). The approximate coordinates of the markers are shown in Kb. The Chr12 homolog containing the ace2-A7 allele was arbitrarily designated as maternal (Chr12-M, red) and the homolog containing the wild type ACE2 allele as paternal (Chr12-P, blue). JAY270 was heterozygous at all markers, and all rough colony isolates were homozygous for the ace2-A7 allele. White boxes distal to the 450 Kb HetSNP represent ∼1,500 Kb of ribosomal DNA repeats (rDNA). Chr12 right arm regions distal to the rDNA do not contain any heterozygous markers in JAY270. The red or blue shading between homologs corresponds to the directions (M or P, respectively) and approximate endpoint positions of the LOH tracts (midpoint between HetSNP markers). Panels B and C show the patterns of LOH found in independent 5-FOA resistant clones derived from JAY270 with hemizygous insertions of the CORE2 cassette (KlURA3-ScURA3-KanMX4) either adjacent to the ACE2 allele in the paternal Chr12 homolog (B) or adjacent to the ace2-A7 allele in the maternal Chr12 homolog (C). The clones selected in B were ace2-A7/ace2-A7 and formed rough colonies, whereas the clones selected in C were ACE2/ACE2 and formed smooth colonies. The genotypes at the nine HetSNP loci at the indicated positions were determined by PCR-RFLP. The clones showing continuous LOH tracts were grouped according to the endpoint interval between HetSNPs, and the number of clones in each group is indicated to the left. Panel D shows a representation of the endpoint distribution among clones with Chr12 LOH. These clones were arranged in groups according to the PCR-RFLP HetSNPs marker positions used in A, B and C. The table shows the analysis of absolute interval size, relative size compared to the CEN12 to CORE2 distance, and the number and frequency of endpoints (combined from A, B and C) for each Chr12 interval. Note that only clones with uninterrupted tracts are shown in B-C. Two clones with complex tracts (non-contiguous or bidirectional) were omitted from this figure and the endpoint distribution analysis.
Analysis of the JAY663 isolate showed that, while it was homozygous for the ace2-A7 mutation, it remained heterozygous at all other flanking markers, including those immediately proximal and immediately distal to the ACE2 locus. Therefore, in this particular case, we were unable to distinguish between a short gene conversion tract limited to the 8.7 Kb region between the immediate flanking HetSNPs vs. a de novo -1 contraction. Despite this limitation, we favor the gene conversion mechanism, since the rate of mitotic recombination (∼10−4; (St Charles and Petes 2013)) is ∼10,000 fold higher than the estimated rate of single nucleotide contractions within A homopolymer runs (∼10−8); (Tran et al. 1997)).
The four remaining isolates (JAY664, JAY665, JAY912, and JAY913) were homozygous for regions well beyond the ACE2 locus. All four LOH tracts were uninterrupted with homozygosity for nucleotides present in the Chr12 homolog that contained the ace2-A7 allele, which we arbitrarily designated the maternal homolog (Chr12-M; red in all figures). The centromere-proximal endpoints of the LOH tracts were roughly mapped to positions ranging from 39 Kb (JAY913) to 184 Kb (JAY664) from the ACE2 locus. On the distal side, these isolates had an additional 45 Kb of LOH that extended to the HetSNP at position 450 Kb, located 1.4 Kb proximal to the rDNA repeats. From this point, Chr12 contains ∼1,500 Kb of rDNA repeats plus another ∼600 Kb of distal homozygous single copy sequences. Since the 450 Kb HetSNP was the most distal marker in Chr12, we could not distinguish if these LOH tracts were generated as very long interstitial gene conversion events, or if they extended to the right telomere. This analysis also revealed an unexpected unselected LOH event on the left arm of Chr12 in JAY664, but in this case, it was associated with homozygosity for the nucleotide from the paternal homolog (Chr12-P, blue in all figures).
JAY270 does not show inherently higher rates of mitotic recombination
Taken together, the results described above showed that the majority of the isolates with altered colony morphology were homozygous not only at the ACE2 locus, but also at surrounding regions, indicating that interhomolog mitotic recombination was readily detectable in JAY270 and that it likely had a substantial role in shaping the genome of this strain. This was consistent with the uneven genomic distribution of HetSNPs (Fig. S1), with long tracts of homozygosity, suggesting that multiple LOH events accumulated in the ancestral lineage before JAY270 was isolated (Basso et al. 2008).
To gain a deeper understanding of the degree of genomic instability in JAY270, we conducted experiments to directly measure the rate of LOH in this strain (Figure 4A-B). Starting with a homozygous ura3Δ/ura3Δ derivative of JAY270 (FGY50; gift from F. Galzerani), we introduced one copy of the KlURA3-ScURA3-KanMX4 CORE2 counter-selectable cassette (Zhang et al. 2013) at a position immediately proximal to the ACE2 locus (1.3 Kb from the adenine homopolymer run) in either homolog. We grew cultures of strains carrying this insertion and plated the cells in media containing 5-FOA to select for clones that had lost the cassette. We performed these quantitative LOH rate measurements using the derivative carrying the hemizygous CORE2 insertion at Chr12-M, in which selected LOH produces cells with an ACE2/ACE2 genotype that remain dispersed in liquid culture. The reciprocal strain, with the CORE2 insertion at Chr12-P, could not be used for this quantitative application, since the selected LOH events produces cells with the ace2-A7/ace2-A7 genotype that aggregate, and thus the number of colony forming units observed in 5-FOA plates may not precisely correspond to the number of LOH-carrying cells in the population.
Figure 4.
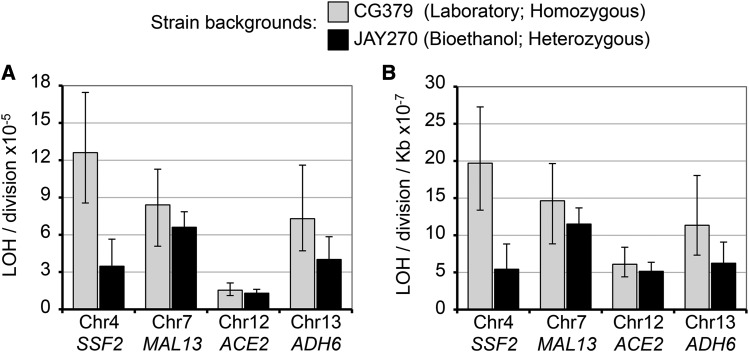
Quantitative analyses of LOH. The median absolute rates of LOH events (A) and normalized LOH rates per kilobase (respective CEN to CORE2 distance) (B) at four different genomic positions were measured in the CG379 laboratory strain background (gray bars) and in the JAY270 bioethanol strain background (black bars). CG379-derived diploids were homozygous at all genomic positions, except at the MAT locus and at the indicated marker positions. JAY270-derived diploids were heterozygous at multiple sites across the genome (Fig. S1), and at the indicated marker positions. In the X axis, Chr4, Chr7, Chr12, and Chr13 indicate diploids with hemizygous insertions of the CORE2 cassette (KlURA3-ScURA3-KanMX4) at each of those chromosomes. The LOH rate shown for Chr12 in JAY270 was derived from the strain carrying the ACE2/ace2-A7::CORE2 insertion (smooth Ura+ to smooth 5-FOAR transition). Error bars indicate 95% confidence intervals (CI) for each rate measurement.
The median frequency of selected homozygosity for the ACE2 allele was 1.2 × 10−4, and the corresponding LOH rate was calculated to be 1.5 × 10−5/cell division (Figure 4A). These measurements were in agreement with the unselected frequency of ace2-A7 homozygosity estimated earlier in the study (5 rough isolates from ∼50,000 colonies). We also used hemizygous CORE2 insertions to measure LOH rates at three other positions in the genome (Chr4 near SSF2, Chr7 near MAL13, and Chr13 near ADH6). In order to provide a reference for comparison of LOH rates measured in JAY270, we created these same four CORE2 insertions in a conventional homozygous diploid laboratory strain background routinely used to study genomic instability mechanisms, including LOH (strain CG379; Figure 4A-B) (Morrison et al. 1991; Rosen et al. 2013; Cornelio et al. 2017). The measured LOH rates (Figure 4A) were either not significantly different between the two strain backgrounds at Chr12 (P = 0.225) and at Chr7 (P = 0.307), or when different, they were somewhat lower in JAY270 at Chr13 (P = 0.033) and Chr4 (P < 0.001). Overall, these direct quantitative comparisons indicated that the two strain backgrounds have similar LOH rates, and therefore their respective inherent levels of genomic instability appear to be quite comparable (i.e., JAY270 is as stable as a conventional laboratory strain).
In addition, we asked whether LOH events occur more frequently at the Chr12 interval that spans ACE2 relative to other regions of the genome. Given that most LOH tracts were suggestive of crossover or break-induced replication (BIR) recombination mechanisms (see below), we calculated the normalized LOH rate per Kb for each interval, by dividing the absolute LOH rate by the distance between the centromeres and the respective CORE2 insertion sites. This analysis (Figure 4B) indicated that all four intervals have relatively similar rates of LOH, and that recombination near the ACE2 locus does not appear to be elevated by the presence of the rDNA repeats on its distal side.
We also used the 5-FOAR selection approach to characterize the qualitative nature of Chr12 LOH tracts in a larger set of independently selected clones. In contrast to the quantitative rate assays above, the difference in cell aggregation phenotype between LOH leading to ACE2/ACE2 vs. ace2-A7/ace2-A7 genotypes was not a concern. Thus, we used nine PCR-RFLP markers to map LOH tracts from 5-FOAR clones derived from both CORE2 insertions, at Chr12-P (Figure 3B) or at Chr12-M (Figure 3C). The LOH tracts selected from both strains resembled each other’s, and the tracts observed in the five initial spontaneous rough colony isolates (Figure 3A). All 41 selected Chr12 LOH clones were heterozygous for the left arm, and 39 of them had uninterrupted LOH tracts going from M/P to P/P or M/M on the right arm, starting at positions between CEN12 and ACE2, and extending up to the 450 Kb HetSNP. The two remaining clones had more complex tracts, including interruptions or homozygosity for the homolog opposite to the direction of selection (data not shown). The predominant LOH tract patterns were consistent with interhomolog mitotic crossover or BIR mechanisms.
Finally, we also investigated the distribution of LOH endpoints on Chr12. The region between CEN12 and ACE2 was divided in five intervals delimited by PCR-RFLP HetSNP markers. The distribution of endpoints found at these intervals was not significantly different between the strains carrying the CORE2 insertion at Chr12-P and Chr12-M (χ2 = 0.855; P = 0.93), suggesting that both homologs shared similar mitotic recombination properties. This similarity allowed us to pool the endpoint distribution data from the 19 5-FOAR clones derived from the Chr12-P insertion to the endpoints from 20 5-FOAR clones derived from the Chr12-M, and four of the original spontaneous rough clones (JAY664, JAY665, JAY912, JAY913). We compared the total number of endpoints leading to LOH observed within each interval to the expected distribution if endpoints were allocated purely as a function of the sizes of the physical intervals. This analysis indicated that the observed endpoint distribution was significantly different from this simple model (χ2 = 18.51; P = 0.001) (Figure 3D), suggesting the possible presence of a mild hotspot for initiation of mitotic recombination, particularly between the 227 and 292 Kb HetSNP markers.
Discussion
Rough colony phenotype and mutation in ACE2
We showed that a frequent colony morphology transition in JAY270 resulted from mitotic recombination events at a region of Chr12 heterozygous for a mutation in the ACE2 gene. Importantly, ace2-A7/ace2-A7 homozygosity also significantly accelerated the sedimentation of yeast cells in liquid culture, an undesirable trait that is typically associated with decreased fermentation kinetics and clogging of centrifuges and pipes at distilleries that use yeast recycling in fed-batch sugarcane bioethanol production (Paulino de Souza et al. 2018; Della-Bianca et al. 2013).
The ACE2 gene encodes a transcription factor that controls the expression of at least 20 genes involved in septum destruction and mother-daughter cell separation (Weiss 2012; Doolin et al. 2001). The rough colony phenotype was easily discernible in ace2-A7/ace2-A7 diploids, but was not observed in ace2-A7 haploids. This ploidy-specificity of the colony morphology phenotype caused by loss of ACE2 function has been previously attributed to the difference in budding patterns between diploid and haploid cells (Voth et al. 2005). Diploid cells typically display a bipolar pattern of budding, which when combined with cell-separation defects result in textured colony morphology. In contrast, haploid cells follow an axial budding program, during which each new bud emerges in a position adjacent to the previous bud scar. Mutation of the BUD4 gene gives rise to haploids that display a bipolar budding pattern typical of diploid cells. Voth et al. 2005 showed that ace2bud4 double mutant haploids display bipolar budding and a rough colony appearance indistinguishable from ace2/ace2 diploids. Additionally, mutations in ACE2 regulators, such as CBK1, HYM1, KIC1, MOB2, cause similar phenotypes including defects in cell separation and altered colony morphology.
The ace2-A7 −1 frameshift allele found in JAY270 leads to a premature stop codon resulting in a truncated, likely inactive, protein that lacks three zinc finger domains and a nuclear localization sequence (McBride et al. 1999). Another heterozygous diploid industrial strain, FostersB used in brewing (Borneman et al. 2011), has a +1 frameshift variant in the same adenine homopolymer region of ACE2 (ace2-A9 allele), suggesting that it is also vulnerable to phenotypic changes caused by LOH spanning this locus.
Interestingly, de novo mutations in ACE2 have been previously identified in multiple bioreactor and chemostat experimental evolution studies using S. cerevisiae haploids (Oud et al. 2013; Koschwanez et al. 2013; Ratcliff et al. 2015; Hope et al. 2017). The ace2-A7, ace2-A9, and additional loss-of-function alleles were identified in aggregating, fast-sedimenting adapted clones. This acquired multi-cellularity trait is thought to have provided them with enhanced fitness under the specific laboratory conditions where they evolved. In the specific case of JAY270 in the bioethanol distillery environment, it is not known if or how the switch between dispersed to aggregated cell growth states has an effect on fitness. However, it is possible that JAY270’s heterozygous ACE2/ace-A7 genotype may offer it the benefit of versatility, by giving individuals in the population the ability to quickly access the aggregated state through LOH. This may provide a short-term adaptive solution as cells encounter various environmental challenges during industrial fermentation (Basso et al. 2008). Individuals in the population could then later return to the dispersed state through expansion or contraction of the adenine run (Tran et al. 1997), or through sporulation and mating to an ACE2 haploid. Interestingly, a recent microsatellite-based phylogenetic study of yeast strains isolated from sugarcane bioethanol distilleries reported that within a cluster of isolates that were genetically similar to strain PE2 (the parent of JAY270), some of the isolates formed rough colonies while others had smooth colonies (Reis et al. 2017). This observation suggests the possibility that smooth-to-rough and perhaps also rough-back-to-smooth, transitions could actually be occurring within PE2 populations in distillery environments.
Analyses of spontaneous and selected LOH clones
Mapping of Chr12 heterozygosity in 44 spontaneous and selected clones revealed long tracts of LOH extending for several Kb and spanning hundreds of HetSNPs in addition to the ACE2 locus (Figure 3). Overall, the pattern of recombination observed was simple and consistent with the majority of events being the result of allelic mitotic crossovers or BIR (Symington et al. 2014; Kramara et al. 2018). However, we disfavor the BIR mechanism in this case, because the efficiency of BIR is greatly reduced (<40%) when the segment of DNA to be synthesized is longer than ∼100 Kb (Donnianni and Symington 2013). In the clones characterized here, BIR would require DNA synthesis across extremely long Chr12 DNA segments, including regions proximal to the 450 Kb HetSNP, across ∼1,500 Kb of the highly repetitive rDNA region, and additional ∼600 Kb to reach the right telomere. Thus, completing BIR through this specific region would be particularly challenging, while resolving the recombination intermediates toward the crossover outcome would not.
The ACE2 gene is positioned fairly close (∼50 Kb proximal) to the most unstable region of the yeast genome, the rDNA repeats (Ganley and Kobayashi 2014). We asked whether this relative proximity to the rDNA repeats at Chr12 could be leading to an elevated incidence of recombination involving the ACE2 locus. The normalized rate of mitotic recombination measured at ACE2 was very similar to or lower than the rates measured at other regions of the genome (Figure 4B), thus we conclude that the rDNA does not destabilize the centromere-proximal regions of the right arm of Chr12. This result was consistent with previous studies that showed a ∼10 fold higher rate of recombination on the distal side of the rDNA compared to the proximal side (Casper et al. 2008), and with Chr12 distal tracts of LOH detected in several wild S. cerevisiae isolates (Magwene et al. 2011).
Surprisingly, our analysis uncovered one rough clone (JAY664) that carried two coincident LOH events in each arm of Chr12 and in opposite directions (the selected [ace2-A7/ace2-A7] M/M LOH in the right arm, and an unselected P/P LOH in the left arm). The rate of LOH that we measured at the right arm of Chr12 was approximately 1.5 × 10−5, thus the rate for two independent events on this chromosome is predicted to be extremely rare (∼10−10). Intriguing observations of elevated rates of coincident mitotic recombination in yeast have been reported previously and generally interpreted as linked to a subpopulation of recombination-prone cells (Fogel and Hurst 1963; Minet et al. 1980; Montelone et al. 1981; Golin and Esposito 1984; Golin and Tampe 1988; Freeman and Hoffmann 2007; Forche et al. 2009). We speculate that JAY664 may have been derived from an analogous genome-wide instability mechanism. Further work offering supporting evidence in this regard is in progress and will be described elsewhere.
Genomic instability in JAY270 relative to a conventional laboratory strain background
Finally, the rates of LOH measured at all four genomic regions in JAY270 were comparable to the rates at the same positions in the CG379 laboratory strain background (Figure 4). The rates of LOH observed in this study in both strains were also compatible with the rates of allelic mitotic recombination reported in the literature (St Charles and Petes 2013). This result is significant because industrial yeast strains, including PE2, generally have a reputation for having inherently unstable genomes (de Amorim et al. 2017). The example characterized in our study suggests that, while karyotype instability is indeed frequently detected in yeast clones isolated from different industrial settings, the genomes of this group of strains may not necessarily be inherently less stable than those of conventional laboratory strains. This misconception may stem from the structural complexity of the genomes of wild and industrial isolates that make recombination between pairs of polymorphic homologs detectable through a simple karyotype analysis. These same recombination events, however, produce no net genetic change when occurring between perfectly identical pairs of homologs in homozygous laboratory diploids. Since allelic interhomolog mitotic recombination is not detectable in most conventional laboratory experimental settings, these strains may appear to be relatively stable, when in reality, recombination is also happening in them at similar rates. We believe that, at least in the case of JAY270, the high genomic instability reputation is not deserved, and the same might also be true for other industrial strains.
Acknowledgments
We thank Tom Petes and Michael McMurray for valuable insights and comments on the manuscript. We are also grateful to the anonymous reviewers who offered very thoughtful feedback on our manuscript. NMVS received a pre-doctoral fellowship from Brazil’s CAPES (0316/13-0). PC and AVP were supported by a UGC fellowship. KTN was supported by a Wellcome Trust-DBT India Alliance Intermediate fellowship- (IA/I/11/2500268) and IISER-TVM intramural funds. Research reported here was supported by the NIH award number R35GM119788 to JLA.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6406598.
Communicating editor: J. Hesselberth
Literature Cited
- Argueso J. L., Carazzolle M. F., Mieczkowski P. A., Duarte F. M., Netto O. V. C., et al. , 2009. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res. 19: 2258–2270. 10.1101/gr.091777.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850. 10.1073/pnas.0804529105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al. , 1998. Current Protocols in Molecular Biology, John Wiley & Sons, New York. [Google Scholar]
- Basso L. C., de Amorim H. V., de Oliveira A. J., Lopes M. L., 2008. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 8: 1155–1163. 10.1111/j.1567-1364.2008.00428.x [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Desany B. A., Riches D., Affourtit J. P., Forgan A. H., et al. , 2011. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 7: e1001287 10.1371/journal.pgen.1001287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro D., Pina B., 2001. Genetic analysis of the karyotype instability in natural wine yeast strains. Yeast 18: 1457–1470. 10.1002/yea.789 [DOI] [PubMed] [Google Scholar]
- Casper A. M., Mieczkowski P. A., Gawel M., Petes T. D., 2008. Low levels of DNA polymerase alpha induce mitotic and meiotic instability in the ribosomal DNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 4: e1000105 10.1371/journal.pgen.1000105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelio D. A., Sedam H. N., Ferrarezi J. A., Sampaio N. M., Argueso J. L., 2017. Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability. DNA Repair (Amst.) 52: 110–114. 10.1016/j.dnarep.2017.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Amorim H. V., de Amorim Neto H. B., Lopes M. L., de Lima Paulillo S. C., 2017. Evolution of Yeast Selection for Fuel Ethanol: Breaking Paradigms, Diversity and Benefits of Microorganisms from the Tropics, edited by de Azevedo J., Quecine M. Springer, New York: 10.1007/978-3-319-55804-2_17 [DOI] [Google Scholar]
- Della-Bianca B. E., Basso T. O., Stambuk B. U., Basso L. C., Gombert A. K., 2013. What do we know about the yeast strains from the Brazilian fuel ethanol industry? Appl. Microbiol. Biotechnol. 97: 979–991. 10.1007/s00253-012-4631-x [DOI] [PubMed] [Google Scholar]
- Donnianni R. A., Symington L. S., 2013. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 110: 13475–13480. 10.1073/pnas.1309800110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolin M. T., Johnson A. L., Johnston L. H., Butler G., 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40: 422–432. 10.1046/j.1365-2958.2001.02388.x [DOI] [PubMed] [Google Scholar]
- Fogel S., Hurst D. D., 1963. Coincidence relations between gene conversion and mitotic recombination in Saccharomyces. Genetics 48: 321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Magee P. T., Selmecki A., Berman J., May G., 2009. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182: 799–811. 10.1534/genetics.109.103325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman K. M., Hoffmann G. R., 2007. Frequencies of mutagen-induced coincident mitotic recombination at unlinked loci in Saccharomyces cerevisiae. Mutat. Res. 616: 119–132. 10.1016/j.mrfmmm.2006.11.014 [DOI] [PubMed] [Google Scholar]
- Ganley A. R., Kobayashi T., 2014. Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 14: 49–59. 10.1111/1567-1364.12133 [DOI] [PubMed] [Google Scholar]
- Golin J. E., Esposito M. S., 1984. Coincident gene conversion during mitosis in saccharomyces. Genetics 107: 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golin J. E., Tampe H., 1988. Coincident recombination during mitosis in Saccharomyces: Distance-dependent and -independent components. Genetics 119: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. M., Ma C. X., Liang P., Singh K. K., 2009. Fluctuation analysis calculator: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565. 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope E. A., Amorosi C. J., Miller A. W., Dang K., Heil C. S., et al. , 2017. Experimental Evolution Reveals Favored Adaptive Routes to Cell Aggregation in Yeast. Genetics 206: 1153–1167. 10.1534/genetics.116.198895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschwanez J. H., Foster K. R., Murray A. W., 2013. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife 2: e00367 10.7554/eLife.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramara J., Osia B., Malkova A., 2018. Break-Induced Replication: The Where, The Why, and The How. Trends Genet. 34: 518–531. 10.1016/j.tig.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 28: 491–511. [DOI] [PubMed] [Google Scholar]
- Magwene P. M., Kayikci O., Granek J. A., Reininga J. M., Scholl Z., et al. , 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 1987–1992. 10.1073/pnas.1012544108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H. J., Yu Y., Stillman D. J., 1999. Distinct regions of the Swi5 and Ace2 transcription factors are required for specific gene activation. J. Biol. Chem. 274: 21029–21036. 10.1074/jbc.274.30.21029 [DOI] [PubMed] [Google Scholar]
- Minet M., Grossenbacher-Grunder A. M., Thuriaux P., 1980. The origin of a centromere effect on mitotic recombination: A study in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 2: 53–60. 10.1007/BF00445694 [DOI] [PubMed] [Google Scholar]
- Montelone B. A., Prakash S., Prakash L., 1981. Spontaneous mitotic recombination in mms8–1, an allele of the CDC9 gene of Saccharomyces cerevisiae. J. Bacteriol. 147: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A., 1991. Eukaryotic DNA polymerase amino acid sequence required for 3′-5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 88: 9473–9477. 10.1073/pnas.88.21.9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L. A., McCusker J. H., 2009. Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol. Ecol. 18: 2779–2786. 10.1111/j.1365-294X.2009.04234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal D., Carro D., Fernandez-Larrea J., Pina B., 1999. Analysis and dynamics of the chromosomal complements of wild sparkling-wine yeast strains. Appl. Environ. Microbiol. 65: 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oud B., Guadalupe-Medina V., Nijkamp J. F., de Ridder D., Pronk J. T., et al. , 2013. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 110: E4223–E4231. 10.1073/pnas.1305949110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J., De Chiara M., Friedrich A., Yue J. X., Pflieger D., et al. , 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plech M., de Visser J. A., Korona R., 2014. Heterosis is prevalent among domesticated but not wild strains of Saccharomyces cerevisiae. G3 (Bethesda) 4: 315–323. 10.1534/g3.113.009381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol A., Bond U., 2009. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol. Lett. 293: 1–10. 10.1111/j.1574-6968.2008.01480.x [DOI] [PubMed] [Google Scholar]
- Ratcliff W. C., Fankhauser J. D., Rogers D. W., Greig D., Travisano M., 2015. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 6: 6102 10.1038/ncomms7102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis V. R., Antonangelo A. T. B. F., Bassi A. P. G., Colombi D., Ceccato-Antonini S. R., 2017. Bioethanol strains of Saccharomyces cerevisiae characterised by microsatellite and stress resistance. Braz. J. Microbiol. 48: 268–274. 10.1016/j.bjm.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D. M., Younkin E. M., Miller S. D., Casper A. M., 2013. Fragile Site Instability in Saccharomyces cerevisiae Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication. PLoS Genet. 9: e1003817 10.1371/journal.pgen.1003817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino de Souza J., Prado C. D., Eleutherio E. C. A., Bonatto D., Malavazi I., et al. , 2018. Improvement of Brazilian bioethanol production – Challenges and perspectives on the identification and genetic modification of new strains of Saccharomyces cerevisiae yeasts isolated during ethanol process. Fungal Biol. 122: 583–591. 10.1016/j.funbio.2017.12.006 [DOI] [PubMed] [Google Scholar]
- St Charles J., Petes T. D., 2013. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet. 9: e1003434 10.1371/journal.pgen.1003434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Rothstein R., Lisby M., 2014. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198: 795–835. 10.1534/genetics.114.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. T., Keen J. D., Kricker M., Resnick M. A., Gordenin D. A., 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17: 2859–2865. 10.1128/MCB.17.5.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth W. P., Olsen A. E., Sbia M., Freedman K. H., Stillman D. J., 2005. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot. Cell 4: 1018–1028. 10.1128/EC.4.6.1018-1028.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. L., 2012. Mitotic exit and separation of mother and daughter cells. Genetics 192: 1165–1202. 10.1534/genetics.112.145516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zeidler A. F., Song W., Puccia C. M., Malc E., et al. , 2013. Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics 193: 785–801. 10.1534/genetics.112.146522 [DOI] [PMC free article] [PubMed] [Google Scholar]