Abstract
Halving of the genome during meiosis I is achieved as the homologous chromosomes move to the opposite spindle poles whereas the sister chromatids stay together and move to the same pole. This requires that the sister kinetochores should take a side-by-side orientation in order to connect to the microtubules emanating from the same pole. Factors that constrain sister kinetochores to adopt such orientation are therefore crucial to achieve reductional chromosome segregation in meiosis I. In budding yeast, a protein complex, known as monopolin, is involved in conjoining of the sister kinetochores and thus facilitates their binding to the microtubules from the same pole. In this study, we report Zip1, a synaptonemal complex component, as another factor that might help the sister kinetochores to take the side-by-side orientation and promote their mono-orientation on the meiosis I spindle. From our results, we propose that the localization of Zip1 at the centromere may provide an additional constraining factor that promotes monopolin to cross-link the sister kinetochores enabling them to mono-orient.
Keywords: Synaptonemal complex, Zip1, mono-orientation, meiosis, budding yeast
Conservation of ploidy during sexual reproduction depends on the successful generation of a gamete with genome content precisely half of its mother progenitor cell. The process through which this happens is meiosis which consists of one round of DNA replication followed by two rounds of chromosome segregations. During the first division of meiosis (meiosis I) the homologous chromosomes separate from each other and move to the opposite spindle poles. At the same time, the two sister chromatids that form each homologous chromosome, remain glued together by cohesin and move to the same pole. Meiosis I ends up with two nuclei each having ploidy half of the mother cell. The success of this reductional segregation of the chromosomes depends on three key meiosis I specific events. One, the pairing of the homologous chromosomes that culminates into recombination mediated physical linkage between the homologs. Second, the sister kinetochores attach to the microtubules emanating from the same spindle pole (mono-orientation) resulting in the sister chromatids to co-segregate and third, the cohesins at the centromere and pericentromere are protected from degradation.
Although the overall process of meiotic chromosome segregation is conserved from yeast to mammals, there are variations in the mechanism of how the sister kinetochores are mono-oriented on meiosis I spindle. For instance, Rec8-cohesin is the key factor for such mono-orientation in S. pombe and higher eukaryotes (Chelysheva et al. 2005; Yokobayashi and Watanabe 2005), whereas the same is not true in S. cerevisiae (Tóth et al. 2000). In this organism a four protein complex termed monopolin is responsible for holding the two sister kinetochores together enabling them to connect to a single microtubule and thus to become mono-oriented with respect to the spindle pole (Marston and Amon 2004; Monje-Casas et al. 2007). Mam1 is the first protein of this complex that was discovered and is expressed only during meiosis I (Tóth et al. 2000). The other two proteins, Csm1 and Lrs4, are nucleolar proteins and become targeted to the centromere at the beginning of meiosis I (Rabitsch et al. 2003). The lastly discovered component is Hrr25, a casein kinase whose kinase activity is required for mono-orientation (Petronczki et al. 2006).
It is believed that monopolin acts as a molecular clamp to keep the sister kinetochores together (Corbett et al. 2010; Corbett and Harrison 2012; Sarangapani et al. 2014). Therefore, in principle, any condition that would favor less rotational freedom for the sister kinetochores may facilitate a side-by-side geometry of the sister kinetochores which then, in turn, would promote localization of monopolin at the centromere to do the final task of clamping. In fact, the role of condensin on mono-orientation of the sister kinetochores in different organisms may follow this notion (Brito et al. 2010; Tada et al. 2011; Burrack et al. 2013).
In this study, we wished to address if there are other factors that might constrain the chromosomes to adopt a side-by-side geometry of the sister kinetochores and we assume such a factor should act upstream of the kinetochore-microtubule attachment. The pairing of the homologs at early prophase is such an upstream event. The assembly of the synaptonemal complex (SC) along the length of the homologous chromosomes reinforces the pairing. The SC is a tripartite structure with two lateral and one central elements, and it holds the two homologs in near vicinity that favors homolog pairing. In S. cerevisiae, the assembly of SC initiates through programmed double-strand break made by Spo11 endonuclease (Henderson and Keeney 2004). The major component of SC, Zip1 is necessary to tightly juxtapose the homologous chromosomes (Sym et al. 1993; Sym and Roeder 1995; Dong and Roeder 2000). Upon disassembly of SC toward the end of pachytene, the homologs remain joined to each other by reciprocal crossovers called chiasmata. Zip1 appears at the centromere before SC assembly starts and then spread along the chromosomes as SC forms. This early localization of Zip1 at the centromere causes a phenomenon called ‘centromere coupling’ where Zip1 promotes pairing of the non-homologous chromosomes in a homology-independent manner (Tsubouchi and Roeder 2005; Falk et al. 2010). Interestingly, Zip1 persists specifically at the centromere even after disassembly of SC until metaphase I, (Gladstone et al. 2009; Newnham et al. 2010), a time window that overlaps with the time when the sister kinetochores become attached unidirectionally to the microtubule (Miller et al. 2012). This extended localization of Zip1 at the centromere is believed to facilitate bi-orientation of the homologs on the meiosis I spindle (Gladstone et al. 2009; Newnham et al. 2010). It is proposed that apart from the physical linkage between the homologs as chiasmata, localization of Zip1 at the centromere constrains the homologous centromere pairs to take a back-to-back geometry so that each pair can attach to the microtubules from the opposite poles and become bi-oriented.
Given the facts that Zip1 is capable of bridging proteinaceous structure as it joins the lateral elements during SC formation and it can constrain the rotational freedom of the kinetochores by localizing specifically at the centromere, we wished to test if this protein has any similar role in cross-bridging sister kinetochores by constraining them to take a side-by-side geometry required for mono-orientation. To examine this, we analyzed the zip1 deletion mutant for its role in sister kinetochore mono-orientation. From the results presented here, we propose that retention of Zip1 at the centromere beyond SC disassembly and at the time of kinetochore-microtubule attachment, facilitates sister kinetochores staying together that in turn aids in Mam1 stability at the kinetochore and hence sister mono-orientation.
Materials and Methods
Yeast strains
All the yeast strains used in this study were of SK1 background. Detailed genotype for all the strains is mentioned in Table S1 of supplementary information. Deletion, tagging, and promoter shuffling of the genes were performed using PCR cassettes amplified from respective plasmids obtained from Euroscarf (Longtine et al. 1998; Janke et al. 2004).
Chromosome segregation assay
In order to fluorescently mark Chromosome V, a plasmid containing TetO repeats (224 copies) was integrated at 1.4 kb away from CENV, and GFP-TetR was expressed ectopically as described earlier (Michaelis et al. 1997). This marking could be either heterozygous (both the sisters of one homolog are marked) or homozygous (both the homologs are marked). Fluorescence microscope Zeiss Axio Observer.Z1 was used for live cell imaging as described earlier (Mehta et al. 2014; Prajapati et al. 2017). In all the cell biology experiments, cell counting was performed at least for two times, and the error bars represent the standard deviation from the mean.
Meiotic progression
Meiotic synchronization and progression of the cell cycle were performed as described earlier in details (Mehta et al. 2014). A single colony of respective yeast strain was inoculated in 5 ml of YPD (yeast extract 1%, peptone 2%, and dextrose 2%) broth and was grown for overnight. Cells were diluted to O.D.(600)=0.2 in 25 ml of YPA (1% yeast extract, 2% peptone, 1% potassium acetate) and were grown for 12 to 16 h. Further, cells of O.D.(600)=1.6-1.8, were transferred to 25 ml of sporulation media, SPM (potassium acetate 0.3%, raffinose 0.02%, supplemented with a 1/4th concentration of the auxotrophic amino acids). Cells were harvested at different time points for immunofluorescence and live cell imaging.
Sporulation efficiency and spore viability
As described above, cells were progressed into meiosis in the SPM media until sporulation (24 h). Samples were observed under a microscope for finding a total number of sporulated cells (having either 1,2,3 or 4 spores) which was divided with a total number of examined cells to calculate the sporulation efficiency as explained previously (Mehta et al. 2014). Spores were planted on YPD plate using Zeiss Scope.A1 microscope and calculation of germinated spores out of the total number of spores planted was denoted as percentage spore viability. Spore viability was calculated for two times with 30 tetrads dissected each time.
Indirect immunofluorescence and chromatin spread
Indirect immunofluorescence and Chromatin spread were performed as described previously in details (Mehta et al. 2014; Agarwal et al. 2015). Typically, the spheroplasted cells were incubated with blocking solution (5% skim milk prepared in 10 mg/ml BSA with Phosphate Buffered Saline) for 15 min, and after washing of the cells, primary antibodies (rat anti tubulin, YOL1/34, Serotech, UK, 1: 5000; rat anti HA 3F10, Roche, Germany, 1:200; mouse anti myc 9E10, Roche, Germany, 1:200; mouse anti GFP, Roche, Germany, 1:200) were added and kept for 1 h. After 3-4 washes with PBS, cells were incubated with appropriate secondary antibodies (TRITC-conjugated goat anti-rat, 112-025-167, Jackson ImmunoResearch Laboratories, USA, 1:200; Alexa flour 488-conjugated goat anti-mouse, 115-545-166, Jackson ImmunoResearch Laboratories, USA, 1:200). After 3-4 washes DAPI (1µg/ml) was added followed by mounting of the slides.
ChIP assay
Chromatin immunoprecipitation assay was performed as discussed earlier in details (Prajapati et al. 2017). Typically, the wild-type and zip1Δ strains were grown in YPA for around 14 to 18 h and were transferred to SPM for a synchronized meiotic progression. The samples were collected from 5 to 8 h in every one and a half hour interval for chromatin spread and Mam1-9Myc localization was examined. The Mam1 signal was clearly observed at 5 and 6.5 h in the wild-type and in the zip1Δ cells, respectively (Figure S4). Therefore, the cells were harvested at these respective time points to perform either ChIP assay (for the Mam1-9Myc association to the centromere) or co-localization study (for Mam1-9Myc and Ndc10-6HA) in the wild-type and zip1Δ strains. For ChIP assays, around 5x108 cells were fixed using 1% formaldehyde for 1 h. After lysis of the cells by glass beads, chromatin was sheared to around 200-500 bp using probe sonicator (21 s on, 1 min on ice, 12 cycles) in total 400 µl volume of lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM KCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, freshly added protease inhibitor cocktail from Roche, Germany). The lysate was cleared at 20000 g for 15-20 min; the supernatant was divided into three tubes as follows- 50 µl for Input/WCE (Whole Cell Extract), 150 µl each for +Ab and –Ab samples (250 µl lysis buffer was added to make up the volume till 400 µl). The anti myc (for Mam1-9Myc) antibodies (rabbit polyclonal, ab9106, 2 µgm/sample, Abcam) were added in +Ab samples. After 4 h incubation at 4° in rotating condition, protein A sepharose beads were added and kept in rotation for 1 h. Beads were washed and DNA was purified from all the samples after decrosslinking as previously described (Prajapati et al. 2017). The qPCR reaction was performed using Agilent Technologies MX3000P real-time PCR machine in SYBR green reaction mixture and the Ct values were imported from MxPro v4 10d Software (Agilent). To find out enrichment/input the ΔCT was calculated as mentioned earlier (Mehta et al. 2014; Verzijlbergen et al. 2014), ΔCT= Ct (ChIP) – [Ct (Input) – logE (Input dilution factor)], here E represents specific primer efficiency. The final enrichment/input value was obtained by E^- ΔCT. The ChIP experiment was performed for at least three times and the error bars represent the standard deviation from the mean. Primer sequences used for qPCR are mentioned in Table S2.
Other methods
For removal of the microtubules during the meiotic progression of spo11Δ, zip1Δ spo11Δ and mam1Δ spo11Δ strains, 100 µg/ml benomyl was added following 6 h of their release into SPM medium and the cultures were incubated for further 2 h before harvesting. Before the addition of benomyl, the cells were analyzed for the spindle length by immunofluorescence using anti tubulin antibodies to verify the metaphase I stage. The microtubules were depolymerized subsequently by addition of benomyl (Figure S3, Hochwagen et al. 2005).
To analyze the co-localization between Mam1 and Ndc10, the ‘Imaris coloc tool’ was utilized to calculate the Pearson’s correlation coefficient (Adler and Parmryd 2010). The detailed description of the method has been described elsewhere (Figure S9 of Prajapati et al. 2017).
In order to analyze the DAPI staining (Figure 2A) distributed in 3D space, the cell images were captured at multiple planes using ‘z-stack’ tool of Zeiss Axio-vision software using motorized Axio Observer.Z1 microscope from Zeiss. For each image, the appropriate planes (where the fluorescence is bright and focused) were selected and merged to get an image in 2D and that was used for the qualitative measurement of DAPI stain.
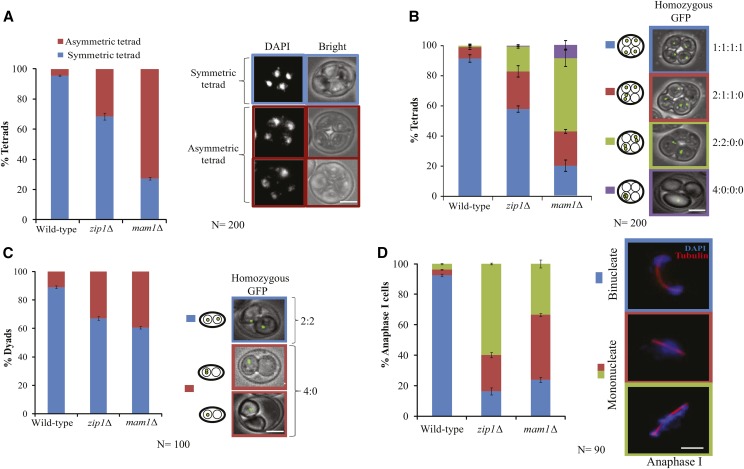
Figure 2.
zip1Δ mutant shows chromosome segregation defects similar to mam1Δ. (A) The histograms show the percentage of asymmetric and symmetric tetrads in wild-type (SGY116), mam1Δ (SGY223) and zip1Δ (SGY1127) strains. (B) Segregation pattern of homozygous GFP dots in the above strains. (C) Segregation pattern of homozygous GFP dots in the dyads of mam1Δ and zip1Δ strains. (D) Immunofluorescence of the above strains showing the percentage of anaphase I cells harboring mono- or bi-nucleates. DNA and spindle were visualized using DAPI and anti-tubulin antibodies, respectively. The absence of Zip1 gives an equal number of mono-nucleates like mam1Δ at the end of meiosis I. Bar, ∼2 µm. Two independent experiments were performed for each strain, and the average values are shown with a total number of cells represented by N. Error bars represent the standard deviation from the mean.
Data availability
Yeast strains are available upon request. The supplementary information file is available at FigShare. This file includes Tables S1, S2 and figures from S1 to S5. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7105220.
Results
zip1Δ mutant shows defect in meiosis with an increased percentage of dyads
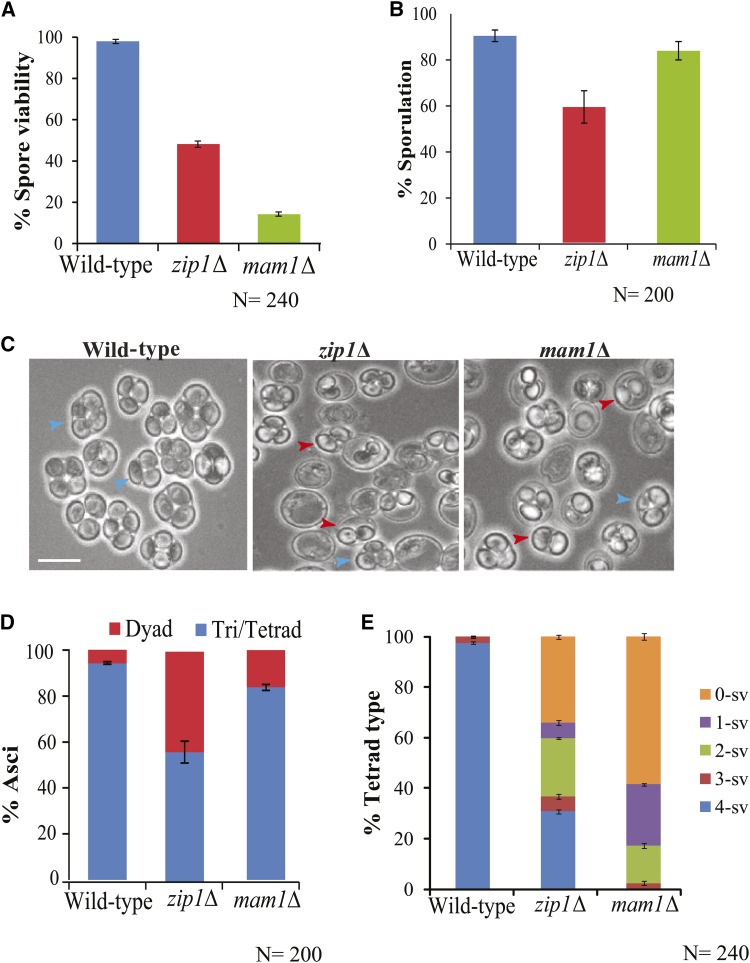
Earlier studies have demonstrated the roles of Zip1 in centromere coupling and the formation of SC (Sym et al. 1993; Dong and Roeder 2000; Tsubouchi and Roeder 2005; Falk et al. 2010). In this study, we wish to test its function in mono-orientation of sister chromatids during meiosis I. Therefore, as the first set of experiments we determined the spore viability of zip1Δ mutant and compared that with the wild-type and mam1Δ strains. While the wild-type strain showed 98%, zip1Δ and mam1Δ mutants showed ∼48% and ∼14% spore viability, respectively (Figure 1A) as reported earlier (Sym and Roeder 1994; Tóth et al. 2000; Bhuiyan and Schmekel 2004). Further, zip1Δ strain showed a reduced sporulation efficiency (59%) as reported earlier (Chen et al. 2015) as compared to both mam1Δ (84%) and wild-type (91%) strains (Figure 1B). Importantly, analysis of types of sporulation showed that zip1Δ gave more dyads asci (42%) than mam1Δ (15%) and wild-type strains (5%) (Figure 1C and D). Further, among the tetrads, we calculated the percentage of tetrads types on the basis of a number of viable spores from each tetrad (Figure 1E) and found 0- or 1-spore viable tetrads were more in the mam1Δ mutant. This result showed that generation of aneuploid spores was more in case of mam1Δ than zip1Δ mutant accounting for low spore viability for the former.
Figure 1.
Deletion of Zip1 causes reduced spore viability and increased dyads. (A) Spore viability assay in wild-type (SGY40), zip1Δ (SGY1066), and mam1Δ (SGY223). (B) Sporulation efficiency was determined by counting 200 cells for each strain in two independent experiments. (C) Field view of the sporulation cultures of the above strains after 24 h in SPM medium. Blue, and red arrowheads show tetrad/triad and dyads, respectively. Bar, ∼5 µm. (D) Distribution of types of asci in the above strains. Sporulation efficiency and type of asci were calculated after repetition of the experiments two times. (E) The percentage of tetrad type represents tetrads with 4, 3, 2, 1 and 0 spores viable (4-sv, 3-sv, 2-sv, 1-sv and 0-sv). % tetrad type was calculated from total 60 tetrads dissected for each strain. The spore viability and the percentage of tetrad type were calculated after tetrad dissection for two times (30 dissected in each time). Error bars represent the standard deviation from the mean.
Generation of dyads is a hallmark feature of a cell compromised in sister chromatid mono-orientation with intact cohesin where meiosis I division is abrogated, and the cell enters into meiosis II. The spores obtained from the dyads are likely to be diploids (non-maters), and they will be mostly inviable due to aneuploidy as all the sisters will not be bi-oriented. A similar occurrence of dyads was observed in spo13Δ, or in mam1Δ mutants harboring sister mono-orientation defect and where meiosis I division is bypassed (Rutkowski and Esposito 2000; Tóth et al. 2000). However, although we observed an increased occurrence of dyads in zip1∆, the spores of those dyads were found to be haploids as judged by the zygote formation after mating with the tester strains (the known MAT a and MAT α strains) and these were less viable than the spores obtained from mam1Δ (Figure S1A). In mam1Δ cells, in the absence of mono-orientation, bi-orientation of the sisters leads to abrogation of meiosis I. Whereas in zip1∆ cells, the defects in SC and cross-over formation (Sym et al. 1993) and in homolog bi-orientation (Gladstone et al. 2009; Newnham et al. 2010) may result in either meiosis I abrogation or meiosis I homolog non-disjunction. This following normal meiosis II can cause the genome to be packaged into two spores with high inviability of the spores. However, the possibility for an additional defect in mono-orientation of the sister chromatids in zip1∆ cells cannot be ruled out with these experiments.
In order to examine if the dyads formation in zip1Δ strain, is either due to the starvation condition and delayed prophase as described previously (Neiman 2005) or linked to kinetochore orientation defect, we used the zip1Δ spo11Δ double mutant which does not show arrest at prophase (Obeso and Dawson 2010; Thacker et al. 2014). After sporulation, we observed around 20% dyads (Figure S1B), which is approximately half of the dyad population observed in zip1Δ alone which indicates a possible role of Zip1 in kinetochore orientation.
The previous study has shown that the cells without Mam1 take a longer time in meiosis I due to sister chromatid orientation defect and the cells arrest transiently at metaphase I (Tóth et al. 2000). If the zip1∆ cells, like mam1∆, show an orientation defect, it is expected that the cells will spend more time in metaphase I. However, zip1∆ cells arrest at meiotic prophase before a kinetochore-microtubule attachment takes place and this arrest can be abrogated by removing chiasmata (Sym et al. 1993; Obeso and Dawson 2010; Thacker et al. 2014). Therefore, to investigate if the zip1∆ cells, like mam1∆, can halt transiently in metaphase I, we followed the kinetics of meiotic progression of the zip1∆ cells in presence or absence of Spo11 which is required to generate DSB, a prerequisite for chiasmata formation (Figure S1C). Both mam1Δ and zip1Δ mutants were found to progress more slowly through meiosis I than the wild-type. As expected, mam1Δ cells showed a transient arrest at metaphase I (Figure S1C, the arrow on the blue line) and a significant population of zip1Δ cells showed prolonged arrest at prophase. Notably, around 14% cells of zip1Δ alone showed arrest at metaphase I (Figure S1C, the arrow on the red line) while rest remained arrested at prophase. However, when we followed the progression of zip1Δ spo11Δ double mutant, we failed to observe any transient arrest at metaphase I. We speculate that even if the sister kinetochores may bi-orient in the absence of Zip1, the kinetochore-microtubule amphitelic connection may not be robust enough to elicit a transient metaphase I arrest.
zip1Δ cells show missegregation of the chromosomes similar to mam1Δ cells
Reduced spore viability in zip1Δ suggests that many of the generated spores obtaining from the tetrads population are aneuploids (Figure 1A and E). DAPI staining of the tetrads showed that this is true where around 31% of the tetrads were asymmetric in zip1Δ strain compared to only 5% in the wild-type (Figure 2A), whereas in mam1∆ around 72% cells showed asymmetric tetrads. In such tetrads, two nuclei had more DNA than the other two that might have caused due to a partial abrogation of meiosis I segregation (not for all chromosomes) due to sister chromatids bi-orientation and their non-disjunction due to retention of cohesion. The presence of less percentage of asymmetric tetrads in zip1∆ than in mam1∆, suggests that lack of Zip1 might cause fewer pairs of sister chromatids to bi-orient than in the cells lacking Mam1. However, the generation of asymmetric tetrads in zip1∆ cells may additionally occur due to perturbation in SC or crossover formation causing meiosis I non-disjunction. Nevertheless, these tetrads contain aneuploid spores resulting in less than 50% spore viability in zip1Δ cells (Figure 1A and E).
To further ascertain the fact that the generation of asymmetric tetrads is a consequence of the completion of meiosis II without any meiosis I, we followed the segregation of the homologs by labeling them with fluorescence. For this, we marked chromosome V with GFP-TetR/TetO system (Michaelis et al. 1997) in the homozygous condition in the wild-type, zip1Δ, and mam1Δ cells and analyzed the segregation of the four GFP dots each depicting one copy of chromosome V (Figure 2B). Faithful chromosome segregation during meiosis results in four nuclei each having one of the four marked sister chromatids (one GFP dot) and termed as 1:1:1:1 pattern of chromosome segregation (Figure S2A). Whereas, any defect due to lack of homolog pairing, homolog bi-orientation and/or mono-orientation of sister chromatids will generate tetrads with 2:1:1:0, 2:2:0:0 and 4:0:0:0 patterns (Figure S2B, C and D). Lack of homolog pairing will lead the homologs to segregate randomly in meiosis I that may result in 2:2 or 4:0 pattern in the bi-nucleates at the end of meiosis I which will turn into 1:1:1:1 or 2:2:0:0 pattern, respectively at the end of meiosis II. However, additional meiosis II non-disjunction will lead to 2:1:1:0 or 4:0:0:0 patterns, respectively. On the other hand, lack of mono-orientation in all the sister pairs will lead to no segregation of chromosomes in meiosis I followed by disjunction or non-disjunction in meiosis II to produce binucleates with 2:2 or 4:0 patterns, respectively. However, if only a subset of the sister pairs bi-orient, that will lead to an overall segregation of the chromosomes in meiosis I keeping the bi-oriented pairs at the middle and such cells upon entering into meiosis II will give 2:2:0:0 or 2:1:1:0 (if disjoined) and 4:0:0:0 (if non-disjoined) patterns (Figure S2C and D). Therefore, to judge the type of defect that zip1∆ strain might harbor in addition to known homolog pairing and bi-orientation defects, we compared the GFP dots distribution between mam1Δ and zip1Δ strains. Similar frequencies of 2:1:1:0 (∼25%) pattern was observed both in mam1Δ and zip1∆ mutants (Figure 2B) suggesting meiosis II non-disjunction occurs in both these strains. As expected mam1∆ showed a high percentage (48%) of 2:2:0:0 segregation accounting for lack of sister mono-orientation and abrogation of meiosis I. Interestingly, zip1Δ mutant also showed a moderate percentage (16%) of 2:2:0:0 segregation which may be due to both random segregation of the homologs in meiosis I due to failure in homolog pairing and bi-orientation as well as due to the abrogation of meiosis I upon bi-orientation of sisters on the meiosis I spindle. Wild-type cells, as expected, mostly showed 1:1:1:1 pattern of segregation.
If the observed percentage of dyads (Figure 1D) in zip1∆ and mam1∆ cells is due to complete abrogation of meiosis I, then normal meiosis II segregation will result in 2:2 segregation of homozygous GFP dots and any non-disjunction during the process will lead to 4:0 segregation. Remarkably, both mam1Δ and zip1Δ gave almost similar 2:2 and 4:0 patterns of GFP dot distribution among the dyads (Figure 2C) suggesting similar types of defects, presumably sister bi-orientation during meiosis I, occurring in these mutants.
zip1Δ mutant produces mono-nucleated cells similar to mam1Δ cells
From the appearance of dyads in the zip1Δ cells (Figure 1D), and the patterns of DAPI and GFP segregations in the dyads and the tetrads (Figure 2A-C), much like that of mam1Δ cells, it can be tempting to presume that Zip1 might have a role in mono-orienting sister chromatids. To test this further, we hypothesized that if mono-orientation is suppressed in the absence of Zip1, each pair of the sister chromatids will be pulled from opposite spindle poles. However, the cohesin present in between the sisters will restrict the separation resulting blockage of meiosis I chromosome segregation. Under this condition, the cells will remain as mono-nucleated but will proceed biochemically to elongate the spindle giving anaphase I spindle. This will subsequently proceed to complete meiosis II with the single nuclear division. This phenotype was previously reported in mam1∆ cells (Tóth et al. 2000).
To assess whether zip1∆ mutant can suppress mono-orientation and produce mono-nucleates, we analyzed the cells toward the end of meiosis I with anaphase I spindle. This excludes the zip1∆ cells that become arrested just after pachytene (Sym et al. 1993). Both zip1Δ and mam1Δ mutants showed ∼75% of mono-nucleated cells as compared to only 5% in the wild-type strain (Figure 2D). A high percentage of mono-nucleated cells in mam1∆ cells at the end of meiosis I, consistent with the earlier report (Tóth et al. 2000), occurs due to higher suppression of mono-orientation of the sister kinetochores. On the other hand, the similar high percentage of mono-nucleated cells in zip1∆ cells may result from lack of homolog bi-orientation (Gladstone et al. 2009) and/or suppression of mono-orientation of the sister kinetochores related to the function of this protein at the centromere. Lack of bi-orientation with poor SC formation can disperse the homologs at different positions between the two SPBs in metaphase I and upon anaphase I onset the homologs, and hence the DAPI can be found ‘distributed’ along the anaphase I spindle (Figure 2D, green shade). Whereas if the sisters are bi-oriented in metaphase I, they will remain at the middle of the two SPBs and can produce a ‘roundish’ DAPI mass on anaphase I spindle (Figure 2D, red shade). To distinguish these two categories, we observed the mono-nucleated cells arising in mam1∆ or zip1∆ mutants carefully. Out of the mono-nucleated cells in mam1∆, around 42% showed ‘roundish’ nuclei accounting for predominant suppression of mono-orientation. Whereas in zip1∆, the majority of the mono-nucleated cells (60%) showed ‘distributed’ DAPI (Figure 2D, green shade) indicating a defect in homolog bi-orientation and imperfect SC formation. Importantly, a sizable fraction of mono-nucleated cells (23%) also showed ‘roundish’ nuclei (Figure 2D, zip1∆ category, red shade) suggesting the occurrence of suppression of mono-orientation.
Zip1 facilitates mono-orientation of the sister kinetochores
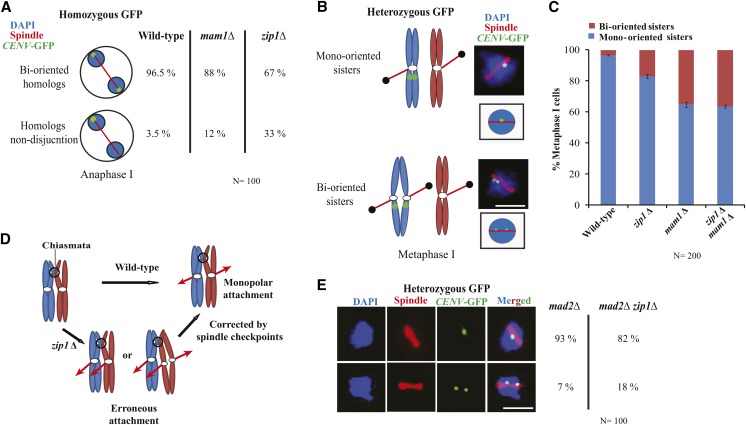
Previous studies have demonstrated that an early localization of Zip1 at the centromere is required for ‘centromere coupling’ (Tsubouchi and Roeder 2005; Falk et al. 2010). This is followed by its role in homolog pairing and SC formation. Interestingly, Zip1 remains at the centromere even after SC disassembly and this extended localization of Zip1 at the centromere promotes bipolar attachment of the homologous chromosomes (Gladstone et al. 2009) and the non-exchange chromosomes to the spindle (Newnham et al. 2010). To confirm this, we marked both the homologs of chromosome V with GFP-TetR/TetO (Michaelis et al. 1997) and observed the anaphase I cells of the wild-type, mam1Δ, and zip1Δ. As expected zip1Δ mutant showed an increased percentage of homolog non-disjunction (∼33%) as compared to the wild-type (3.5%) and mam1Δ (∼12%) strains (Figure 3A) suggesting, defects in bipolar attachment of the homologs, and our zip1Δ mutant conforms to the earlier report (Gladstone et al. 2009).
Figure 3.
The absence of Zip1 causes homologs non-disjunction and sister chromatid bi-orientation during meiosis I. (A) Immunofluorescence assay is showing the percentage of anaphase I cells of wild-type (SGY116), mam1Δ (SGY223) and zip1Δ (SGY1127) with homologs disjunction or non-disjunction. Around 100 cells were analyzed for each strain. (B) Schematic of mono or bi-orientation of sister chromatids along with the representative images during metaphase I (C) Immunofluorescence analysis showing the percentage of wild-type (SGY115), mam1Δ (SGY220), zip1Δ (SGY1427) and mam1Δ zip1Δ (SGY3190) cells at metaphase I with mono- or bi-oriented sister chromatids. The cells were counted from two independent experiments for each strain and the total number of cells are represented as N. (D) Schematic showing monopolar attachment of the sister chromatids during metaphase I and possible correction of erroneous attachment by spindle checkpoint proteins. (E) The mono-oriented and bi-oriented sisters were counted similarly as described in ‘B’ using mad2Δ (SGY1265) and zip1Δ mad2Δ (SGY1267) cells. Around 100 cells were observed for each strain. Bar, ∼5 µm. Error bars represent the standard deviation from the mean.
The role of Zip1 in bi-orientation of the homologs, perhaps by constraining the homologs (Gladstone et al. 2009; Newnham et al. 2010), may also principally facilitate the sister chromatids to mono-orient. Increased frequency of occurrence of dyads (Figure 1D), 2:2:0:0 segregation of the homozygous GFP dots (Figure 2B) and mononucleated cells with anaphase I spindle (Figure 2D) in the zip1Δ cells also suggest that Zip1 might possess a moderate role in mono-orientation of the sister kinetochores on metaphase I spindle. Therefore, to directly visualize the sister kinetochore orientation on metaphase I spindle, we marked only one copy of the homolog (only one pair of sister chromatids) with GFP-TetR/TetO at CENV in the wild type, mam1Δ, and zip1Δ cells. In the wild-type, sister chromatids remain cohesed and mono-oriented during metaphase I and appear as a single GFP dot on the spindle whereas, upon bi-orientation sister chromatids will be pulled from the opposite poles causing them to split and the sisters will appear as two split GFP dots on the spindle (Figure 3B). Therefore, to distinguish single vs. two split GFP dots, cells were released into synchronized meiosis. As discussed above, a majority of zip1Δ cells showed strong delay at pachytene/prophase (Figure S1C). However, a population of cells (14%) instead arrested at metaphase I. We counted the GFP dots in those metaphase I cells as judged by mono-nucleates with short spindles. A fraction of zip1Δ nuclei (17%) showed two split GFP dots which are more than thrice as compared to its isogenic wild-type (4%, Figure 3C). Whereas 35% of mam1Δ nuclei, used as a control, showed such splitting indicating bi-orientation of the sister chromatids on metaphase I spindle which is consistent with an earlier report (Kiburz et al. 2008, Figure 3C). Our observed frequency (17%) of two split GFP dots in zip1∆ mutant may be an underestimate. This is because earlier studies have shown that spindle checkpoint proteins can potentially correct erroneous attachment, if any, during metaphase and can lead the cell toward faithful chromosome segregation (Gillett et al. 2004). This correction becomes possible in a spindle checkpoint dependent way in the zip1Δ cells as they, besides prophase arrest, also arrest transiently at metaphase I when the kinetochore-microtubule attachment becomes established (Figure 3D, Sym et al. 1993; Bailis et al. 2000; Gladstone et al. 2009). Therefore, we thought that this correction could be the reason why the zip1Δ mutant did not show a higher frequency of sister bi-orientation. Hence, we wished to assess the mono-orientation defect in the zip1∆ cells devoid of any error correction mechanism. For this, we repeated the sister chromatid mono-orientation assay in zip1∆ cells deleted for spindle checkpoint gene MAD2. However, the zip1∆ mad2∆ double mutant showed a similar frequency of two split GFP dots (18%, Figure 3E) compared to zip1∆ (17%, Figure 3C) but more than mad2∆ alone (7%, Figure 3E).
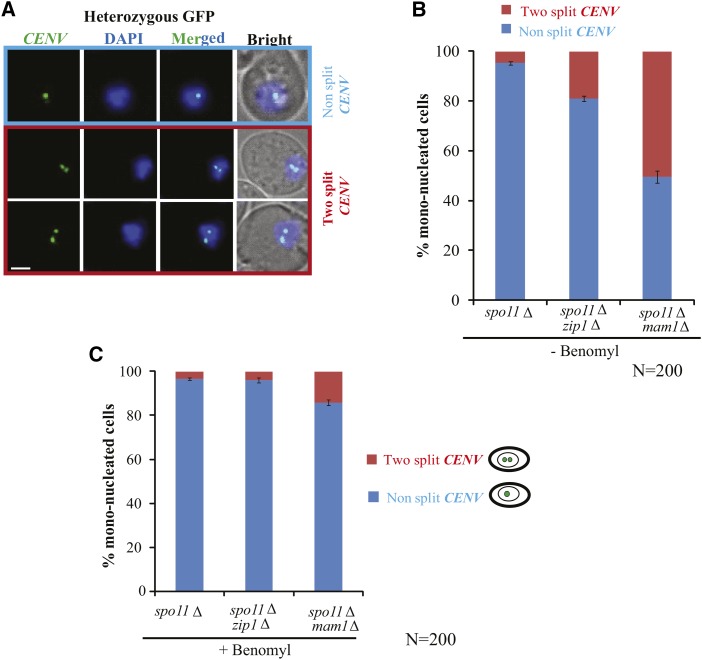
To negate the possibility that the increased two split GFP dots in the zip1∆ cells over the wild-type is due to a role of Zip1 in SC formation, we performed the above mono-orientation assay in the zip1∆ cells where SPO11 was also deleted. The spo11Δ alone and the mam1Δ spo11Δ double mutants were used as controls. We have analyzed the mono-nucleated cells after 8 h in SPM and examined the GFP dot segregation among all the strains (Figure 4A). We found that ∼50% mono-nucleated cells showed two split GFP dots which reflects the bi-orientation of sister chromatids in mam1Δ spo11Δ strain. However, around ∼18% of cells showed this phenomenon in the zip1Δ spo11Δ strain in comparison to 4–5% of spo11Δ cells (Figure 4B). This result indicates that the observed defect in mono-orientation is not due to lack of SC formation in the zip1∆ cells.
Figure 4.
Sister chromatids bi-orientation in zip1Δ is independent of SC formation but requires microtubule mediated pulling. (A and B) Live cell imaging showing two split vs. non split CENV-GFP dots in spo11Δ (SGY3180), zip1Δ spo11Δ (SGY3179) and mam1Δ spo11Δ (SGY3178) cells. The mono-nucleated cells were scored for the GFP dots after transferring the cultures into SPM for 8 h. (C) The same experiment was performed where the cells were treated with 100 µgm/ml benomyl for 2 h after 6 h of culturing into SPM and many of the cells were in metaphase I before addition of the drug. The cells were counted from two independent experiments for each strain and the total numbers of cells are represented as N. Error bars represent the standard deviation from the mean. Bar, ∼2 µm.
The observed splitting of two GFP dots in the zip1∆ cells may occur due to a cohesion defect between the two sisters. To examine this, the pulling force from microtubules was removed by depolymerization of microtubules so that any sister separation will account only for the cohesion defect but not for any bi-orientation. Removal of microtubules caused a large reduction in the frequency of two split GFP dots in all the strains (Figure 4C) indicating that the splitting of the sister centromeres occurs predominantly due to bi-orientation of the sisters. This also suggests that the data presented in figure 2B (the 2:2:0:0 segregation pattern) may not be due to cohesion defect between the sister chromatids. Depolymerization of the microtubules was confirmed by immunofluorescence using anti-tubulin antibodies (Figure S3).
Overall, the above results indicate that the lack of Zip1 causes bi-orientation of the sister chromatids, albeit at a moderate level but significantly at a frequency which is more than thrice to the wild-type. To address if the mono-orientation function of Zip1 is independent of Mam1, we investigated the percentage of two split GFP dots in zip1∆ mam1Δ double mutant strain. However, we found no synthetic enhancement of the defect in zip1∆ mam1Δ as opposed to mam1Δ (Figure 3C) cells suggesting a common pathway for the functioning of Zip1 and Mam1 in sister chromatid mono-orientation.
The localization of Mam1 at the centromere is reduced in the absence of Zip1
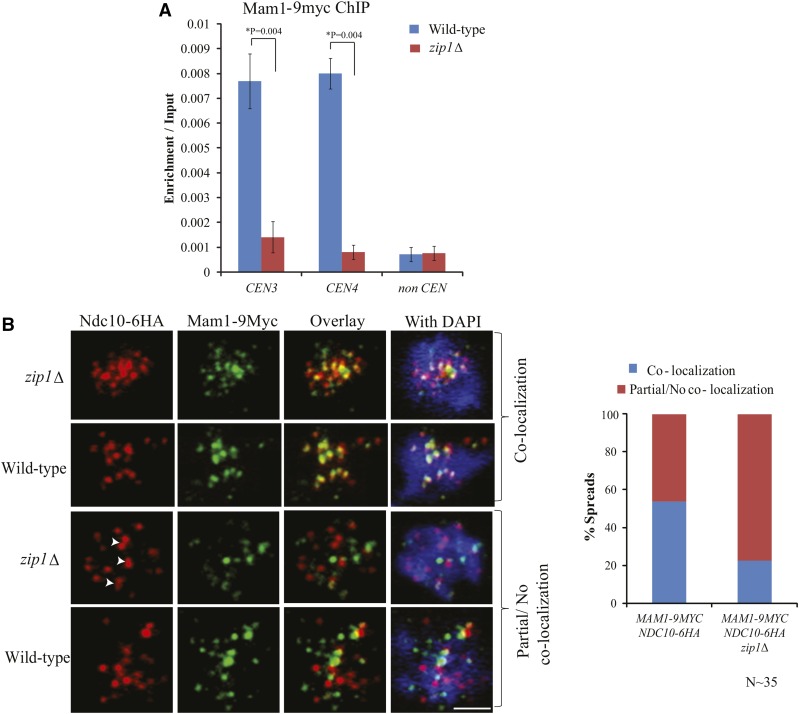
From the above results, it is evident that Zip1 facilitates the mono-orientation of the sister chromatids through the Mam1 pathway. We speculate that Zip1 does this by holding the sister kinetochores in the near vicinity which facilitates Mam1 to bridge the sisters by binding to the kinetochores through Dsn1 subunits (Sarkar et al. 2013). Therefore, we wished to test the localization of Mam1 at the centromere in the absence of Zip1 in cells arrested at metaphase I. We found a high perturbation in Mam1 localization at the centromere in the absence of Zip1 compared to the wild-type using ChIP assay (Figure 5A). Chromatin spreads were also performed from the same cells to investigate the localization of Mam1 at the centromeres that were marked by the inner kinetochore protein Ndc10 (Figure 5B and Figure S4). The Pearson’s correlation coefficient was measured using Imaris ‘coloc’ tool in order to find out the significant colocalization between Ndc10-6HA and Mam1-9Myc foci as previously described (Zinchuk and Zinchuk 2008; Adler and Parmryd 2010; Prajapati et al. 2017). We detected a significant mislocalization of Mam1 in the zip1∆ mutant as compared to the wild-type (Figure 5B, right panel). The dot plot showing the Pearson’s correlation coefficient for chromatin spread from the wild-type and the zip1∆ cells indicate most of the spreads in case of the mutant showed very less co-localization between Mam1 and Ndc10 (Figure S5). These results indicate that lack of Zip1 at the centromere limits Mam1 localization at the centromere which in turn causes mono-orientation defect.
Figure 5.
Mam1 is mislocalized in the absence of Zip1. (A) ChIP assay for quantifying the association of Mam1-9Myc at the centromeres in the presence or absence of Zip1. Anti myc antibodies were used to pull down Mam1-9Myc. The experiment was repeated for three times and error bars represent the standard deviation from the mean. *The P value was determined by using ‘two tailed t-test assuming unequal variances’ option from the data analysis tool of Microsoft Excel. (B) Chromatin spread showing localization of Mam1-9Myc and Ndc10-6HA in prophase/metaphase I cells of wild-type (SGY1444) and zip1Δ (SGY1410). Arrowhead shows doublet of Ndc10 foci that signifies kinetochores remained together by flanking crossover in the absence of Zip1 (Gladstone et al. 2009). On the basis of the localization of Mam1-9Myc with respect to Ndc10-6HA, the cells were divided into two categories: co-localization, partial or no co-localization. Bar, ∼5 µm.
Discussion
Mono-orientation of the sister kinetochores with respect to the meiotic spindle is of immense significance as this is one of the key events that lead to essential reductional division during meiosis I. The mechanism of this crucial event is poorly understood in higher systems. However, in budding yeast, a dedicated complex, called monopolin is directly involved in having the sister kinetochores co-oriented so that both can attach to the microtubules emanating from a single spindle pole (Rabitsch et al. 2003). The monopolin is believed to work as a clamp that puts the two sister kinetochores together constraining them to take side-by-side positions which facilitates their co-orientation (Corbett et al. 2010; Sarangapani et al. 2014). Therefore, any factor that can favor a side-by-side positioning would principally promote monopolin to achieve sister kinetochore mono-orientation. Such a factor might promote mono-orientation just by constraining the sister centromeres/kinetochores and favoring their side-by-side geometry congenial for unipolar spindle microtubule attachment. The function of condensin in sister mono-orientation is believed to follow this notion in a monopolin dependent way (Brito et al. 2010; Tada et al. 2011; Burrack et al. 2013). However, limiting rotation freedom could also impede proper orientation if the kinetochores are prevented from turning in the same direction. The ability of Zip1 to couple the centromeres (Tsubouchi and Roeder 2005), its role in cross-linking proteinaceous structure during SC assembly (Sym et al. 1993) and its extended localization at the centromere during kinetochore-microtubule attachment beyond SC disassembly (Gladstone et al. 2009), have encouraged us to hypothesize that Zip1 may be another factor that may constrain the sister centromeres to take side-by-side positions and thus Zip1 might facilitate mono-orientation of the sister kinetochores. In this work, we tested this hypothesis and found that Zip1 indeed has a moderate role in the process perhaps by stabilizing the monopolin at the centromere.
A reduction in sporulation efficiency (Figure 1B) and spore viability (Figure 1A and E) in zip1Δ is expected as SC formation is disrupted and crossing over frequency is reduced (Sym et al. 1993). Interestingly, we noticed more than 40% dyads in zip1Δ cells compared to less than 5% in the wild-type (Figure 1D) which was encouraging as the appearance of dyads is suggestive of abrogation of one nuclear division which may occur if meiosis I cannot take place due to suppression of mono-orientation. As expected dyads were generated in mam1Δ cells as well that were compromised in monopolin function (Figure 2C). However, the percentage of dyads was less in mam1Δ than in zip1Δ cells (Figure 1D). This is probably because although meiosis I is abrogated in most of the mam1Δ cells, some cells go ahead till the end of meiosis II producing four spores where two spores hardly contain any DNA (Tóth et al. 2000). Nevertheless, the formation of dyads gave a hint that in zip1∆ mutant mono-orientation may be affected.
It is important to address whether the observed role of Zip1 in mono-orientation of the sisters (Figure 2D, 3C) is independent of its function in SC formation. To answer this, bi-orientation frequency was measured in the zip1∆ and the mam1∆ cells in the absence of Spo11 and hence SC. The ratio of this frequency between zip1∆ and mam1∆ cells (1:2, Figure 3C) remains almost similar to what has been observed between spo11∆ zip1∆ and spo11∆ mam1∆ cells (1:2.6, Figure 4B). Further, the frequency was found more than three times higher in spo11∆ zip1∆ compared to spo11∆ alone (5% vs. 19%, Figure 4B) indicating that the role of Zip1 in mono-orientation is independent of SC formation. We believe that it is rather the localization and function of Zip1 at the centromere (Tsubouchi and Roeder 2005; Gladstone et al. 2009) that play a role in constraining the sister kinetochores to be co-oriented. In support of a targeted localization of Zip1 to the centromere, a physical interaction between Zip1 and the kinetochore protein, Nuf2 has been documented (Newman et al. 2000) which is reminiscent to the physical interaction of Mam1 with another kinetochore protein, Dsn1 in order to bind to the centromere (Newman et al. 2000; Sarkar et al. 2013).
To achieve mono-orientation, it is imperative that the rotational freedom of the sister kinetochores should be attenuated. They should be constrained such that they can form a side-by-side orientation that favors them to capture microtubules coming from the same spindle pole. The organisms with regional centromeres appear to use meiotic cohesin at the sister centromeres to conjoin them (Goldstein 1981; Li and Dawe 2009; Sakuno et al. 2009) in addition to other meiosis specific proteins such as Moa1 in fission yeast, functionally analogous to Spo13 of budding yeast (Yokobayashi and Watanabe 2005; Sakuno et al. 2009). In these organisms, a large heterochromatic pericentromere along with cohesin is believed to provide a structural rigidity defining the kinetochore architecture which is required for mono-orientation. Conversely, in budding yeast perhaps due to a point centromere and lack of pericentric heterochromatin, centromeric cohesin does not have any role in mono-orientation (Tóth et al. 2000); therefore, the necessary structural rigidity, in this organism, is provided by condensin that facilitates mono-orientation by augmenting monopolin function (Brito et al. 2010). Later similar observation was also made in S. pombe and in C. albicans, (Tada et al. 2011; Burrack et al. 2013). From these results, it is conceivable that any other factors besides cohesin and condensin that may physically constrain sister kinetochores in principle can aid in mono-orientation. In fission yeast, the linkage between the homologs in a bivalent (chiasmata) can resist bi-orientation of the sister kinetochores (Sakuno et al. 2011). Similarly, lack of chiasmata can lead to bi-orientation of the univalents in mouse and human (Kouznetsova et al. 2007; Nagaoka et al. 2011) whereas such a scenario has not been observed in budding yeast, worms or Arabidopsis (Klein et al. 1999; Chelysheva et al. 2005; Severson et al. 2009). Therefore, we speculated that there could be an additional factor present in the budding yeast that may provide geometric rigidity to the sister kinetochores required for their mono-orientation. The role of Zip1 in cross-linking axial elements on the homologs, its role in coupling centromeres and importantly its SC-independent presence at the centromere at the right time, i.e., during the kinetochore-microtubule attachment, makes us hypothesize that Zip1 could be another such factor helping in constraining sister kinetochores. Additionally, from its role in the pairing of the centromeres, it has been demonstrated that Zip1 promotes bi-orientation of the homologs on meiosis I spindle (Gladstone et al. 2009; Newnham et al. 2010). This function could potentially be a legacy of this protein’s role in mono-orientation of the sister kinetochores that we found, which is then eventually perceived as a role in bi-orientation of the homologs. It was important to address if Zip1 facilitates mono-orientation in addition to the monopolin mediated pathway. However, we found no enhancement of the mono-orientation defect in the zip1∆ mam1∆ double mutant cells over the mam1∆ single mutant (Figure 3C) indicating that in this aspect Zip1 works through Mam1. In support of this, we observed Mam1 localization at the centromere is perturbed in the absence of Zip1 (Figure 5A). Notably, if the mono-orientation defect in zip1∆ is due to mislocalization of Mam1 from the centromere, the erroneous bi-orientation frequency (two split GFP dots) is expected to be similar for zip1∆ and mam1∆ cells which we failed to observe (Figure 3C). This is perhaps due to a difference in the outcomes between zip1∆ and mam1∆ cells with respect to the levels of Mam1 activity available at the centromere. In zip1Δ cells, some Mam1 remains at the centromere which is possibly sufficient to rescue the mono-orientation defect to some level. During pachytene, Zip1 helps in the generation of a 100 nm wide structure (SC) at the chromosome axes to link the homologs, and it is reasonable to believe that this structure is maintained at the centromeres during metaphase I or at least at the time of kinetochore-microtubule attachment (Gladstone et al. 2009; Newnham et al. 2010). Here, the fact that the sister chromatids are closely held by condensin (Brito et al. 2010) and Zip1 is likely holding homologs apart enough, maybe a crucial requirement for cross-linking the sister kinetochores by monopolin complex. In this scenario, the absence of Zip1 will allow the homologs to come close enough so that monopolin may not be able to cross-link the sister centromeres leading to reduced Mam1 occupancy at the centromere (Figure 5A). However, mono-orientation of sister kinetochores maintained to some level (Figure 3C) may be via some feedback mechanisms which can allow the stabilized monopolin interaction at the centromere to some extent.
In this study, we provide evidence that Zip1 has a moderate but significant role in mono-orientation of the sister kinetochores, a crucial phenomenon to achieve faithful meiosis. It appears that Zip1 does this function by facilitating retention of Mam1 at the centromere. In budding yeast, localization of Mam1 at the centromere is a key event for sister chromatid mono-orientation. Given the importance of this process for the reductional division, it is not surprising that cells will adopt multiple mechanisms to reinforce this and thus Zip1 may serve as another receptor, besides Dsn1, for Mam1 at the centromere. Thus this work advances the knowledge by revealing another layer of regulation of sister mono-orientation in the budding yeast. Our findings have strong general implications as the SC proteins are also found in the higher eukaryotes and hence may have more roles to play particularly when these organisms lack monopolin like dedicated complex.
Acknowledgments
We would like to thank Gunjan Mehta for help in the construction of certain strains. This work is funded by BRNS and DBT (37(1)/14/30/2015/BRNS and BT/PR13909/BRB/10/1432/2015, respectively) Govt. of India. HKP, MA, and PM are supported by MHRD (10I30006), CSIR (Ref. no: EU-IV/2008/JUNE/327214) and UGC (Ref. no.: 17-06/2012(i) EU-V) fellowships, respectively.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7105220.
Communicating editor: J. Berman
Literature Cited
- Adler J., Parmryd I., 2010. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A 77: 733–742. 10.1002/cyto.a.20896 [DOI] [PubMed] [Google Scholar]
- Agarwal M., Mehta G., Ghosh S. K., 2015. Role of Ctf3 and COMA subcomplexes in meiosis: Implication in maintaining Cse4 at the centromere and numeric spindle poles. Biochim. Biophys. Acta 1853: 671–684. 10.1016/j.bbamcr.2014.12.032 [DOI] [PubMed] [Google Scholar]
- Bailis J. M., Smith A. V., Roeder G. S., 2000. Bypass of a Meiotic Checkpoint by Overproduction of Meiotic Chromosomal Proteins. Mol. Cell. Biol. 20: 4838–4848. 10.1128/MCB.20.13.4838-4848.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan H., Schmekel K., 2004. Meiotic chromosome synapsis in yeast can occur without spo11-induced DNA double-strand breaks. Genetics 168: 775–783. 10.1534/genetics.104.029660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito I. L., Yu H. G., Amon A., 2010. Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics 185: 55–64. 10.1534/genetics.110.115139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack L. S., Applen Clancey S. E., Chacon J. M., Gardner M. K., Berman J., 2013. Monopolin recruits condensin to organize centromere DNA and repetitive DNA sequences. Mol. Biol. Cell 24: 2807–2819. 10.1091/mbc.e13-05-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Diallo S., Vezon D., Gendrot G., Vrielynck N., et al. , 2005. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118: 4621–4632. 10.1242/jcs.02583 [DOI] [PubMed] [Google Scholar]
- Chen X., Suhandynata R. T., Sandhu R., Rockmill B., Mohibullah N., et al. , 2015. Phosphorylation of the Synaptonemal Complex Protein Zip1 Regulates the Crossover/Noncrossover Decision during Yeast Meiosis. PLoS Biol. 13: e1002329 10.1371/journal.pbio.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K. D., Harrison S. C., 2012. Molecular architecture of the yeast monopolin complex. Cell Reports 1: 583–589. 10.1016/j.celrep.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K. D., Yip C. K., Ee L. S., Walz T., Amon A., et al. , 2010. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142: 556–567. 10.1016/j.cell.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Roeder G. S., 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148: 417–426. 10.1083/jcb.148.3.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J. E., Chan A. C., Hoffmann E., Hochwagen A., 2010. A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev. Cell 19: 599–611. 10.1016/j.devcel.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Gillett E. S., Espelin C. W., Sorger P. K., 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164: 535–546. 10.1083/jcb.200308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstone M. N., Obeso D., Chuong H., Dawson D. S., 2009. The synaptonemal complex protein Zip1 promotes bi-orientation of centromeres at meiosis I. PLoS Genet. 5: e1000771 10.1371/journal.pgen.1000771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. S., 1981. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell 25: 591–602. 10.1016/0092-8674(81)90167-7 [DOI] [PubMed] [Google Scholar]
- Henderson K. A., Keeney S., 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 101: 4519–4524. 10.1073/pnas.0400843101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A., Wrobel G., Cartron M., Demougin P., Niederhauser-Wiederkehr C., et al. , 2005. Novel response to microtubule perturbation in meiosis. Mol. Cell. Biol. 25: 4767–4781. 10.1128/MCB.25.11.4767-4781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., et al. , 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kiburz B. M., Amon A., Marston A. L., 2008. Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 1199–1209. 10.1091/mbc.e07-06-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S. B., Michaelis C., et al. , 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. 10.1016/S0092-8674(00)80609-1 [DOI] [PubMed] [Google Scholar]
- Kouznetsova A., Lister L., Nordenskjold M., Herbert M., Hoog C., 2007. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat. Genet. 39: 966–968. 10.1038/ng2065 [DOI] [PubMed] [Google Scholar]
- Li X., Dawe R. K., 2009. Fused sister kinetochores initiate the reductional division in meiosis I. Nat. Cell Biol. 11: 1103–1108. 10.1038/ncb1923 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Marston A. L., Amon A., 2004. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5: 983–997. 10.1038/nrm1526 [DOI] [PubMed] [Google Scholar]
- Mehta G. D., Agarwal M., Ghosh S. K., 2014. Functional characterization of kinetochore protein, Ctf19 in meiosis I: an implication of differential impact of Ctf19 on the assembly of mitotic and meiotic kinetochores in Saccharomyces cerevisiae. Mol. Microbiol. 91: 1179–1199. 10.1111/mmi.12527 [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K., 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45. 10.1016/S0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- Miller M. P., Unal E., Brar G. A., Amon A., 2012. Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. eLife 1: e00117 10.7554/eLife.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Prabhu V. R., Lee B. H., Boselli M., Amon A., 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128: 477–490. 10.1016/j.cell.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S. I., Hodges C. A., Albertini D. F., Hunt P. A., 2011. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr. Biol. 21: 651–657. 10.1016/j.cub.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., 2005. Ascospore Formation in the Yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 565–584. 10.1128/MMBR.69.4.565-584.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. R., Wolf E., Kim P. S., 2000. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 13203–13208. 10.1073/pnas.97.24.13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnham L., Jordan P., Rockmill B., Roeder G. S., Hoffmann E., 2010. The synaptonemal complex protein, Zip1, promotes the segregation of nonexchange chromosomes at meiosis I. Proc. Natl. Acad. Sci. USA 107: 781–785. 10.1073/pnas.0913435107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeso D., Dawson D. S., 2010. Temporal characterization of homology-independent centromere coupling in meiotic prophase. PLoS One 5: e10336 10.1371/journal.pone.0010336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Matos J., Mori S., Gregan J., Bogdanova A., et al. , 2006. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 126: 1049–1064. 10.1016/j.cell.2006.07.029 [DOI] [PubMed] [Google Scholar]
- Prajapati H. K., Rizvi S. M., Rathore I., Ghosh S. K., 2017. Microtubule-associated proteins, Bik1 and Bim1, are required for faithful partitioning of the endogenous 2 micron plasmids in budding yeast. Mol. Microbiol. 103: 1046–1064. 10.1111/mmi.13608 [DOI] [PubMed] [Google Scholar]
- Rabitsch K. P., Petronczki M., Javerzat J. P., Genier S., Chwalla B., et al. , 2003. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4: 535–548. 10.1016/S1534-5807(03)00086-8 [DOI] [PubMed] [Google Scholar]
- Rutkowski L. H., Esposito R. E., 2000. Recombination can partially substitute for SPO13 in regulating meiosis I in budding yeast. Genetics 155: 1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T., Tada K., Watanabe Y., 2009. Kinetochore geometry defined by cohesion within the centromere. Nature 458: 852–858. 10.1038/nature07876 [DOI] [PubMed] [Google Scholar]
- Sakuno T., Tanaka K., Hauf S., Watanabe Y., 2011. Repositioning of Aurora B Promoted by Chiasmata Ensures Sister Chromatid Mono-Orientation in Meiosis I. Dev. Cell 21: 534–545. 10.1016/j.devcel.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Sarangapani K. K., Duro E., Deng Y., Alves F. L., Ye Q., et al. , 2014. Sister kinetochores are mechanically fused during meiosis I in yeast. Science 346: 248–251. 10.1126/science.1256729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Shenoy R. T., Dalgaard J. Z., Newnham L., Hoffmann E., et al. , 2013. Monopolin subunit Csm1 associates with MIND complex to establish monopolar attachment of sister kinetochores at meiosis I. PLoS Genet. 9: e1003610 10.1371/journal.pgen.1003610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Ling L., van Zuylen V., Meyer B. J., 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23: 1763–1778. 10.1101/gad.1808809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Engebrecht J. A., Roeder G. S., 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. 10.1016/0092-8674(93)90114-6 [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G. S., 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. 10.1016/0092-8674(94)90197-X [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G. S., 1995. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128: 455–466. 10.1083/jcb.128.4.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K., Susumu H., Sakuno T., Watanabe Y., 2011. Condensin association with histone H2A shapes mitotic chromosomes. Nature 474: 477–483. 10.1038/nature10179 [DOI] [PubMed] [Google Scholar]
- Thacker D., Mohibullah N., Zhu X., Keeney S., 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. 10.1038/nature13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A., Rabitsch K. P., Galova M., Schleiffer A., Buonomo S. B., et al. , 2000. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell 103: 1155–1168. 10.1016/S0092-8674(00)00217-8 [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Roeder G. S., 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308: 870–873. 10.1126/science.1108283 [DOI] [PubMed] [Google Scholar]
- Verzijlbergen K. F., Nerusheva O. O., Kelly D., Kerr A., Clift D., et al. , 2014. Shugoshin biases chromosomes for biorientation through condensin recruitment to the pericentromere. eLife 3: e01374 10.7554/eLife.01374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S., Watanabe Y., 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817. 10.1016/j.cell.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Zinchuk V., Zinchuk O., 2008. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr. Protoc. Cell Biol. Chapter 4: Unit 4.19 10.1002/0471143030.cb0419s39 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Yeast strains are available upon request. The supplementary information file is available at FigShare. This file includes Tables S1, S2 and figures from S1 to S5. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7105220.