Abstract
Dynamic control of gene expression is a hallmark of the circadian system. In mouse liver, approximately 5–20% of RNAs are expressed rhythmically, and over 50% of mouse genes are rhythmically expressed in at least one tissue. Recent genome-wide analyses unveiled that, in addition to rhythmic transcription, various post-transcriptional mechanisms play crucial roles in driving rhythmic gene expression. Alternative polyadenylation (APA) is an emerging post-transcriptional mechanism that changes the 3′-ends of transcripts by alternating poly(A) site usage. APA can thus result in changes in RNA processing, such as mRNA localization, stability, translation efficiency, and sometimes even in the localization of the encoded protein. It remains unclear, however, if and how APA is regulated by the circadian clock. To address this, we used an in silico approach and demonstrated in mouse liver that 57.4% of expressed genes undergo APA and each gene has 2.53 poly(A) sites on average. Among all expressed genes, 2.9% of genes alternate their poly(A) site usage with a circadian (i.e., approximately 24 hr) period. APA transcripts use distal sites with canonical poly(A) signals (PASs) more frequently; however, circadian APA transcripts exhibit less distinct usage preference between proximal and distal sites and use proximal sites more frequently. Circadian APA transcripts also harbor longer 3′UTRs, making them more susceptible to post-transcriptional regulation. Overall, our study serves as a platform to ultimately understand the mechanisms of circadian APA regulation.
Keywords: circadian rhythms, alternative polyadenylation, in silico, mouse liver
Post-transcriptional regulation is an integral part of controlling gene expression and can influence when, where, and how much protein will be generated (Keene 2010). The mammalian molecular clock system utilizes multiple post-transcriptional mechanisms, including alternative splicing, mRNA stability, translation, exosome, and miRNA regulation, to drive robust circadian gene expression (Kojima and Green 2015; Green 2017). Genome-wide studies demonstrated that approximately 5–20% of RNAs are expressed rhythmically in mouse liver, and over 50% of mouse genes are rhythmically expressed in at least one tissue, even though these percentages vary among studies due to the difference in experimental conditions and statistical analyses in detecting rhythmicity (Duffield 2003; Fang et al. 2014; Zhang et al. 2014; Janich et al. 2015; Kojima and Green 2015; Sato et al. 2017; Guan et al. 2018). These rhythmic gene expressions were thought to be driven primarily by rhythmic transcription; however, more recent transcriptome studies challenged the current belief and highlighted the importance of post-transcriptional regulation. For example, approximately 80% of all rhythmically expressed RNAs do not undergo rhythmic de novo transcription. Moreover, oscillating mRNA levels are not a prerequisite for rhythmic protein accumulation (Reddy et al. 2006; Koike et al. 2012; Menet et al. 2012; Mauvoisin et al. 2014; Robles et al. 2014). These studies clearly emphasize that it is not just transcription, but, rather, an intricate interplay between transcriptional and post-transcriptional regulation that is required to generate rhythmicity in gene expression.
One important mechanism of post-transcriptional regulation is the control of poly(A) tails. Despite the traditional view that poly(A) tail formation is a static and non-regulatory process, recent studies clearly demonstrated that poly(A) tails undergo dynamic regulation both in length and location to, ultimately, alter the fate of mRNAs. We and others identified mRNAs with rhythms in their poly(A) tail length (Robinson et al. 1988; Gerstner et al. 2012; Kojima et al. 2012) and demonstrated that this rhythmicity correlates with protein accumulation (Kojima et al. 2012). Differential usage of poly(A) sites, an event termed alternative polyadenylation (APA), yields transcripts differing in their 3′-ends, and can impact downstream RNA processing, such as mRNA stability (Mayr and Bartel 2009; Gupta et al. 2014), translation (Mayr and Bartel 2009; Lau et al. 2010; Pinto et al. 2011; Gruber et al. 2014; Masamha et al. 2014), subcellular localization of mRNAs (An et al. 2008; Djebali et al. 2012; Neve et al. 2016), or even localization of the encoded protein (Berkovits and Mayr 2015). APA regulation is widespread and prevalent, affecting ∼50% of genes in mammals as well as in other taxa, such as plants and flies (Tian et al. 2005; Tian and Manley 2016).
APA occurs in response to a variety of extracellular stimuli (An et al. 2008; Chang et al. 2015), changes in physiological conditions (Takagaki et al. 1996; Takagaki and Manley 1998; Sandberg et al. 2008; Wang et al. 2008; Ji et al. 2009; Ji and Tian 2009; Shepard et al. 2011), or pathological conditions (Mayr and Bartel 2009; Singh et al. 2009). It remains elusive, however, whether the circadian clock system also regulates poly(A) site locations. Interestingly, the protein levels of a few components of the 3′-end processing machinery, such as CSTF77 (gene name: CSTF3) or NUDT21 (gene name: CPSF5), are rhythmic in mouse liver (Mauvoisin et al. 2014; Robles et al. 2014), raising the possibility that APA is under circadian regulation. In fact, statistical analyses using circadian microarray datasets indicated that APA is under the circadian regulation in mouse white fat and liver (Liu et al. 2013; Ptitsyna et al. 2017). Although microarrays and RNA-seq both effectively quantify transcriptional levels and the results are reasonably correlated between the two technologies, RNA-Seq outperforms microarrays (even tiling arrays) in detecting transcripts with low abundance, distinguishing various isoforms, and enabling the detection of differentially expressed genes with higher fold-change (Agarwal et al. 2010; Zhao et al. 2014).
By using circadian RNA-seq datasets in mouse liver, here we report that 57.4% of all genes expressed undergo APA with, on average, 2.53 alternative polyadenylated transcripts per gene. Notably, 2.9% of all expressed genes (or 4.5% of genes undergoing APA regulation) alternate their poly(A) site usage with a circadian period. Distal poly(A) sites and canonical poly(A) signals (PASs) are more frequently used among all APA genes. This was also the case among circadian APA genes; however, rhythmic APA genes have less distinct differences in poly(A) site strength between proximal and distal sites and use proximal sites more frequently. In addition, circadian APA transcripts displayed longer 3′UTRs, which are generally rich in cis-regulatory elements, increasing the opportunity to be subjected to post-transcriptional regulation. These characteristics among circadian APA genes will ultimately help us understand the mechanisms of circadian APA regulation.
Materials and Methods
Alignment of short reads
Short read files from liver RNA-seq datasets were downloaded from the NCBI SRA database projects: SRP036186 in FASTQ format (Zhang et al. 2014), SRP014751 (Koike et al. 2012) and SRA025656 (Vollmers et al. 2012) in CSFASTA format with QUAL files. All reads were aligned to the mm9 genome using TopHat version 1.3.3 and Bowtie version 0.12.7 (Trapnell et al. 2009). To maximize the efficiency of mapping to the transcripts, rather than the genomes, we used a guide GFF file of M. musculus (-G option in TopHat) compromising the 3′-ends of all transcripts in mouse tissues (Hoque et al. 2013). Reads that were mapped to multiple regions were discarded. Single end SOLiD colorspace reads were aligned with the following options: -Q –C –-library-type fr-second strand –G TandemUTR_ALE.mm9.gff3. Paired-end Illumina reads were aligned with the following options: -r 200–library-type fr-firststrand –G TandemUTR_ALE.mm9.gff3. Read mapping rates were ∼61–65% for the dataset produced from the Illumina platform (Zhang et al. 2014), while these from the SOLiD platform (Koike et al. 2012; Vollmers et al. 2012) were considerably lower (∼21–30% and ∼16–20%, respectively). This may be due to high ribosomal RNA content in the library preparations and/or the inherent problems that arise from converting the SOLiD sequence data file from colorspace to basespace, which is known to have high error rates (Ondov et al. 2008). The resulting BAM files were separated into strand-specific files using SAMtools.
Quantification of poly(A) site usage frequency by MISO
We applied the Mixture-of-Isoforms probabilistic model (MISO) and calculated “ψ values” of each transcript (Katz et al. 2010). To this end, we first discarded genes with <20 reads in their 3′UTRs using the GTF 3′-end annotation files (Hoque et al. 2013) with the default settings. For both APAALE and APAUTR, we first eliminated isoforms whose average ψ values for all time points were < 0.1. We also eliminated isoforms whose ψ values were 0 at 2 or more time points for the datasets that have 2 circadian cycles, or at 1 or more time points for the dataset that has only1 circadian cycle. We also eliminated transcripts whose average Transcripts Per Million (TPM) of all time points were < 1 (see next section for RNA quantification methods).
To simplify the subsequent analyses, we then extracted the two transcripts isoforms with the highest ψ numbers (average of all time points) for each gene. If the second highest ψ value was identical among two or more isoforms, we treated each combination (of highest and second highest transcripts) as independent and kept all the combinations. We defined the “APA index” of these combinations by calculating = ψ proximal - ψ distal. Therefore, a negative APA index represents transcripts whose distal site usage was more frequent than proximal usage, while a positive APA index represents transcripts whose proximal site usage was more frequent than distal site usage.
Rhythmicity Detection by MetaCycle
All circadian parameters were detected using MetaCycle with the function meta2d (Wu et al. 2016). A period window was set to 20 to 28 hr. The datasets were then filtered for rhythmicity with meta2d output values of P < 0.05 and Meta2d rAmp > 0.1 (for APA index) or > 0.01 (for RNA abundance). A slightly lower threshold of P < 0.06 and Meta2d rAmp >0.09 was used to detect one transcript (Ccdc93) that was represented by all three datasets.
Gene ontology analysis
Gene ontology (GO) analyses were performed using DAVID (Huang da et al. 2009a; Huang da et al. 2009b). “Functional Annotation Clustering” feature was used, and the clusters with an Enrichment Score > 1.33 (i.e., equivalent of p-value< 0.05) were listed in Table S2.
3′RACE assay
3′- Rapid Amplification of cDNA Ends (3′RACE) assay was performed as previously described elsewhere (Scotto-Lavino et al. 2006). Briefly, total RNAs were extracted from mouse liver using TRIZOL (Life Tech), treated with TURBO DNase I (Life Tech) for 30min at 37C, and then 5ug of which was subjected to cDNA synthesis using SuperScript II (Life Tech). To specifically amplify target transcripts, nested PCR was performed for all of the genes. The first PCR was performed using primers Qo and Gene Specific Primer 1 (GSP1), followed by Qi and GSP2 with 1/200 diluted DNA samples from the first PCR. The images shown in Figure 2E are the results of the second PCR. Primer sequences are as follows: QTVN reverse transcription: 5′- CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTTVN-3′, 3RACE_Qo: 5′- CCAGTGAGCAGAGTGACG-3′, 3RACE_Qi: 5′-GAGGACTCGAGCTCAAGC-3′, Akr1c6_GSP1: 5′- ACTGGAGGTCCATTTTGTGC-3′, Akr1c6_GSP2: 5′- CTTGTGCCAGATGTCACTGC-3′, D930014E17Rik_GSP1: 5′- CCCAGGGACCTATTCTGTCA-3′, D930014E17Rik_GSP2: 5′- GCTCCAGT-GCCTTATCCAAG-3′, Sppl3_GSP1: 5′-AGTTTACCTGGCTTCCTCGC-3′, Sppl3_GSP2: 5′-TGATAGACTTGTGGGCTGTG-3′, Oit3_GSP1: 5′-CTGCAGAGCCAGACACTGAC-3′, Oit3_GSP2: 5′-CCCCTCTAGAACGACTTCCTG-3′
Figure 2.
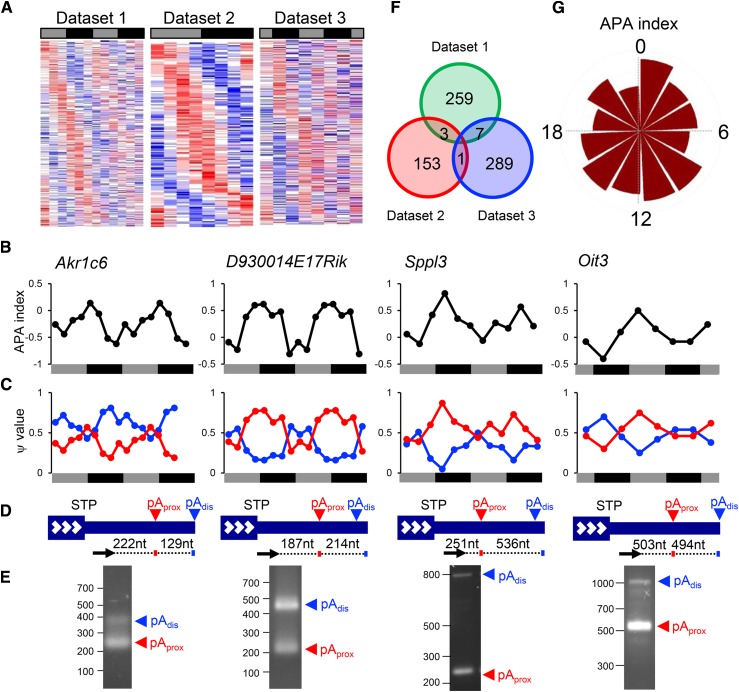
Identification of circadian APA genes. (A) Phase-sorted heat map of the APA index of circadian APA genes. Red and blue colors indicate higher or lower APA index, respectively. Each row represents one gene. (B-C) Examples of circadian APA gene. (B) The APA index profiles (=ψproximal - ψdistal) of Akr1c6 (Dataset 2), D930014E17Rik (Dataset 2), Sppl3 (Dataset 1), and Oit3 (Dataset 3) over the circadian cycles. (C) Individual ψ values for transcripts with either proximal (red) and distal (blue) poly(A) site usage over the circadian cycles. Data from Dataset 2 were double-plotted for easier visualization of rhythmicity. (D) 3′-end structures of APA example genes. STP: stop codon, pAprox: proximal poly(A) site, pAdis: distal poly(A) site, big navy box with white arrowheads: protein-coding region, navy line: 3′UTR. Black arrows represent the locations of the primers used in the 3′RACE assay in (E). (E) 3′RACE assay. Numbers on the left represent the DNA size makers. (F) Venn Diagram of the number of circadian APA transcripts among the three datasets. (G) Circular histogram plots of circadian APA index. Each wedge represents a 2-h bin. Gray or black bars indicate subjective day or night, respectively.
DATA AVAILABILITY
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6741623.
Results
Identification of circadian APA genes
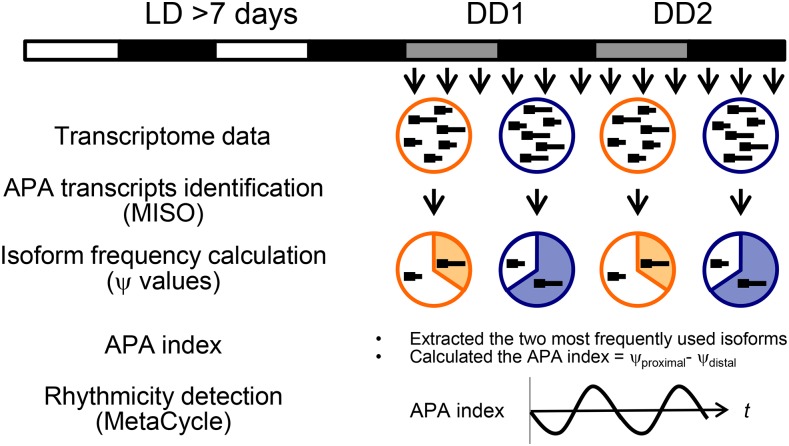
Our in silico strategy to identify transcripts undergoing circadian APA is described in Figure 1. First, we retrieved circadian transcriptome datasets from the Sequence Read Archive (SRA) at the National Institutes of Health (NIH) (https://www.ncbi.nlm.nih.gov/sra). Three datasets (Koike et al. 2012; Vollmers et al. 2012; Zhang et al. 2014) met our criteria that 1) the samples were harvested in a constant dark condition, 2) the datasets provided strand-specific information, which was critical for the downstream analyses because 3′-ends of transcripts sometimes overlap between genes, and 3) the samples were taken from mouse liver in which many other circadian genome-wide analyses have been performed (i.e., Nascent-seq, GRO-seq, ribosome profiling, proteomics, etc.).
Figure 1.
Schematic representation of our strategy to identify circadian APA genes. Briefly, we obtained three circadian transcriptome datasets from NCBI SRA. We then applied the Mixture-of-Isoforms probabilistic model (MISO) to detect alternatively polyadenylated transcripts. MISO further calculated “ψ values” of each transcript that represent the expression levels of alternatively polyadenylated isoforms relative to that of the total transcript within each sample (i.e., each time point). We subsequently extracted isoforms whose ψ numbers (average of all time points) were the two highest to simplify the analyses, and then designated the “APA index” of this combination for each time point, by calculating = ψ proximal - ψ distal. The rhythmicity of the APA index was assessed by MetaCycle. White/black bars and gray/black bars indicate light:dark (LD) = 12:12 or constant dark (DD) conditions, respectively, at which tissues were harvested.
In order to identify APA in the selected datasets, we applied the Mixture of Isoforms Model (MISO) algorithm (Katz et al. 2010), a probabilistic framework that identifies differentially regulated isoforms from transcriptome data. MISO was originally intended to detect alternative splicing, but later implemented to detect alternative polyadenylation by incorporating user-defined poly(A) site annotation data (Katz et al. 2010; Hooper 2014). We chose MISO over other algorithms, such as DaPars, QAPA, and APAtrap (Xia et al. 2014; Ha et al. 2018; Ye et al. 2018) that identify 3′-ends of transcripts de novo and thus require significantly higher read depth. The three circadian transcriptome datasets only had 10-30M mappable and usable reads. Using pre-defined poly(A) sites that had been detected in mouse tissues including liver (Hoque et al. 2013), MISO identifies two types of alternatively polyadenylated isoforms: alternative poly(A) sites within 3′UTRs (APAUTR), and alternative 3′-terminal exons (ALE) flanking the last intron (APAALE). MISO subsequently quantifies the relative expression levels of alternatively polyadenylated isoforms and returns “ψ values,” which represent the expression levels of alternatively polyadenylated isoforms relative to that of all transcripts at a given time point. The sum of ψ values, therefore, is 1 for each transcript at each time point. We used ψ values as the poly(A) site usage frequency of each alternatively polyadenylated isoform.
From the three mouse liver circadian transcriptomes, MISO detected approximately 5,000 APAALE and 16,000 APAUTR isoforms that met our filtering criteria (see Materials and Methods), consisting of approximately 8,000 unique genes (4,000 genes for APAALE and 7,500 genes for APAUTR) (Table 1). The average poly(A) sites per gene were 2.53 (Table 1), which is similar to what was observed in other mouse tissues, where it ranged from 1.5 to 4.0 (Tian et al. 2005; Hoque et al. 2013). Out of the approximately 8000 genes that were expressed, genes that had multiple poly(A) sites represented 57.4% (Table 2). This is also comparable to what has been reported in other mouse tissues, such as retina (49%), EST database (47%), fibroblasts (66%), and cell line mixtures (78.5%) (Tian et al. 2005; Hoque et al. 2013; Xiao et al. 2016; Hu et al. 2017).
Table 1. Numbers of APA isoforms and genes detected by MISO from mouse transcriptome data.
| Dataset | APAALE isoform | APAUTR isoform | Total isoforms | APAALE unique gene | APAUTR unique gene | Total unique gene | APAALE per gene | APAUTR per gene | PAS per gene |
|---|---|---|---|---|---|---|---|---|---|
| Dataset1 | 5,091 | 16,223 | 22,124 | 3,765 | 7,362 | 8,171 | 0.62 | 1.99 | 2.61 |
| Dataset2 | 4,416 | 13,501 | 17,917 | 3,306 | 6,848 | 7,668 | 0.58 | 1.76 | 2.34 |
| Dataset3 | 5,839 | 18,445 | 24,284 | 4,606 | 8,125 | 9,174 | 0.64 | 2.01 | 2.65 |
| Average | 5,115 | 16,056 | 21,172 | 3,892 | 7,445 | 8,338 | 0.61 | 1.93 | 2.53 |
Table 2. Numbers of genes/transcripts that possess single or multiple PASs.
| Dataset | Single PAS genes (ALE) | Single PAS genes (UTR) | Unique Single PAS genes (%) | Multiple PAS transcripts (ALE) | Multiple PAS transcripts (UTR) | Multiple PAS genes (ALE) | Multiple PAS genes (UTR) | Unique Multiple PAS genes (%) |
|---|---|---|---|---|---|---|---|---|
| Dataset1 | 2,499 | 1,657 | 3758 (41.3) | 3,402 | 14,566 | 1,262 | 4,737 | 5338 (58.7) |
| Dataset2 | 2,492 | 1,002 | 3228 (41.0) | 1,924 | 12,499 | 812 | 4,198 | 4645 (59.0) |
| Dataset3 | 3,694 | 2,036 | 5059 (44.4) | 2,145 | 16,409 | 912 | 5,970 | 6236 (55.6) |
| Average | 2,895 | 1,565 | 4,015 (42.6) | 2,490 | 14,491 | 995 | 4,968 | 5,406 (57.4) |
Using ψ values of each isoform at each time point, we next sought to identify genes that underwent circadian APA regulation. To simplify the statistical analyses and decrease complexity, we focused only on APAUTR, which comprised ∼75% of all the APA detected in our analyses (Table 1). We also focused on only the two most frequently used isoforms (i.e., the two transcripts with the highest average ψ values across all time points). This is justified, as the average number of poly(A) sites (including those with a single poly(A) site) was 2.53 (Table 1). We subsequently calculated the “APA index (=ψ proximal- ψ distal),” which represents the difference in the ψ values between the two APAUTR isoforms. Thus, genes with negative APA indexes use the distal poly(A) site (i.e., produce a longer transcript) more frequently, while genes with positive APA indexes use the proximal poly(A) site (i.e., produce a shorter transcript) more frequently. We then applied the rhythmicity detection algorithm MetaCycle (Wu et al. 2016) to detect rhythmic APA events (Figure 1).
We identified 269 (Dataset 1: 3.3%), 157 (Dataset 2: 2.0%), and 297 transcripts (Dataset 3: 3.2%), whose APA index was rhythmic (Figure 2A-C, Table S1). We experimentally validated the 3′-end sequences of both proximal and distal poly(A) sites for selected genes by 3′- Rapid Amplification of cDNA ends (RACE) (Figure 2D-E). The 3′-end sequences were further verified by Sanger sequence and these matched with annotated 3′-end in the bioinformatical analyses (Hoque et al. 2013) (data not shown).
Eleven genes were found to be rhythmic in at least two datasets (Ado, Cnot7, Cyhr1, Mapk8, Paqr9, Srsf10, and Tnfaip2 between Dataset 1 and 3, Ccdc93, Ctbs, and Sdccag3 between Dataset 1 and 2, and Hspa4 between Dataset 2 and 3) (Figure 2F). An additional 9 genes (Magi1, Mrps5, Oxr1, Pcf11, Samhd1, Ssh1, Ttpal, Vsp26a, Zfp259) had APA indexes that were rhythmic in two datasets; however, we did not categorize these genes as “overlapping” because, although one isoform was commonly detected in two datasets, the other isoform (typically the second highest ψ value) was not identical between the two datasets. Of note, we found one transcript (Ccdc93) that was represented by all three datasets when we slightly lowered the rhythmicity detection threshold (Table S1). Nevertheless, the small overlap between datasets is probably due to the differences in experimental conditions, such as the duration of sampling (24 hr for Dataset 2, and 48 hr for Dataset 1 and 3), the sampling interval (4 hr for Dataset 1, 3 hr for Dataset 2, and 6 hr for Dataset 3, the sampling start time (CT 0 for Datasets 1 and 2, and CT 22 for Dataset 3), and the source RNA used for library construction (ribosomal depleted RNA for Dataset 1 and poly(A)+ enriched RNA for Datasets 2 and 3), as well as technical noise over the course of experiments and analyses (Conesa et al. 2016).
One remarkable feature of APA is the global shortening or lengthening of transcripts (Flavell et al. 2008; Mayr and Bartel 2009). However, we did not detect any particular time points during which the APA index was disproportionally enriched (Figure 2G), although each dataset exhibited unique patterns (Supplemental Figure 1). This is in contrast to transcription and translation, both of which are under global regulation in mouse liver, peaking during early subjective night (Koike et al. 2012; Menet et al. 2012; Jouffe et al. 2013; Janich et al. 2015). This is also in stark contrast with circadian poly(A) tail length regulation, where the rhythmicity of poly(A) tail length peaks predominantly at subjective night (Kojima et al. 2012).
In order to shed light on the cellular processes that are regulated by circadian APA genes, we performed a gene ontology analyses using DAVID (Huang da et al. 2009a; Huang da et al. 2009b). The top 10 pathways/processes that were significantly enriched, were “Transcription,” “Nucleotide/ATP-binding,” “Metal(zinc)-binding,” “Endocytosis,” “Protein transport,” “Cell cycle,” “RNA-binding,” “Endoplasmic reticulum,” “Fibronectin type-II,” and “Biological rhythms” (Table S2). Notably, “Transcription” had the highest enrichment score in all three datasets (Table S2). It is possible that circadian APA indirectly regulates rhythmic transcription by affecting RNA stability or translation efficiency, for example, of those genes functioning in transcription.
Canonical PASs at distal sites are more frequently used in mouse liver, although proximal poly(A) sites more frequently used among rhythmic APA transcripts
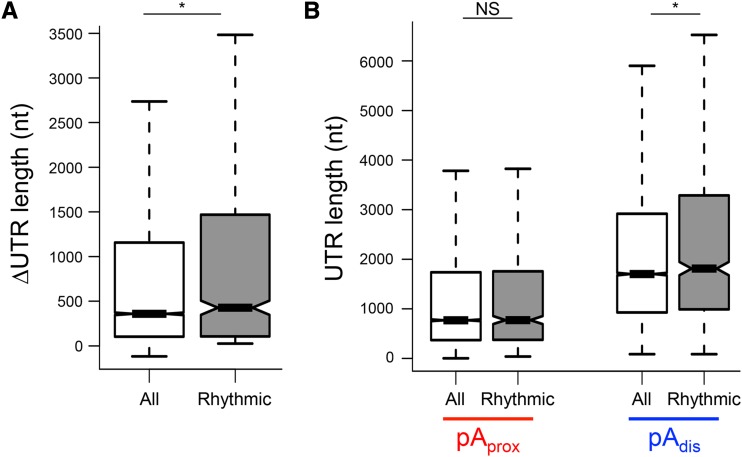
To shed light into the mechanism of circadian APA regulation, we next characterized the relative distance, location, sequence, and frequency of alternative poly(A) sites, and compared these characteristics between rhythmic and all APA transcripts (control). In mouse liver, we found that the relative 3′UTR length difference between alternatively polyadenylated transcripts is longer among rhythmic APA transcripts (median: 427 nt) compared to control APA transcripts (median: 359 nt) (Mann-Whitney-Wilcoxon test: P = 0.02806) (Figure 3A). Interestingly, the 3′UTR length of short isoforms was not different between all (median: 768.5 nt) and rhythmic APA transcripts (median: 772 nt) (Figure 3B). In contrast, the 3′UTR length of long isoforms was increased in rhythmic APA (median: 1811 nt) compared to all APA transcripts (median:1700.5 nt) (Mann-Whitney-Wilcoxon test: P = 0.02052) (Figure 3B). These data suggest that the long isoforms of rhythmic APA transcripts contain more cis-regulatory elements and are more likely subject to post-transcriptional regulation.
Figure 3.
Rhythmic APA genes have longer 3′UTRs. (A) Difference in the 3′UTR length between APA transcripts. (B) Distributions of 3′UTR length of the short (i.e., proximal poly(A) site usage) and long (i.e., distal poly(A) site usage) isoforms. All: n = 15246, and rhythmic: n = 723. Box-whisker plots with quartiles (box) and quartile ± 1.5 interquartile range (whiskers). *; P < 0.05. (Mann-Whitney-Wilcoxon Test). pAprox: proximal poly(A) site, pAdis: distal poly(A) site.
The most frequently detected PAS is AAUAAA, accounting for ∼60% of PASs among all expressed cDNAs/ESTs in human and mouse (Beaudoing et al. 2000; Tian et al. 2005). Another approximately 20 PAS variants have been observed, albeit less frequently (ex., AUUAAA: ∼15%), while some transcripts do not contain an obvious PAS at all (Beaudoing et al. 2000; Tian et al. 2005). Our analysis revealed that 64.2% of single PAS genes contained the canonical signal, AAUAAA, and 17.7% contained the variant AUUAAA within 40 nt from the 3′-end of each transcript (Table 3). In contrast, genes possessing more than one PAS (i.e., APA genes) had lower AAUAAA usage and only 43.2% possessed AAUAAA, whereas 17.4% contained AUUAAA (Table 3). The actual percentage may be even lower, since we counted the same motif twice if both poly(A) sites are within 40 nt of one another, which was the case for 7.9% of genes.
Table 3. APA transcripts and their PAS types.
| Dataset | Single PAS | Multiple PAS | ||
|---|---|---|---|---|
| AAUAAA (%) | AUUAAA (%) | AAUAAA (%) | AUUAAA (%) | |
| Dataset1 | 1004 (60.6) | 276 (16.7) | 6380 (46.8) | 2606 (17.9) |
| Dataset2 | 650 (64.8) | 186 (18.6) | 5330 (42.6) | 2096 (16.8) |
| Dataset3 | 1364 (67.0) | 363 (17.8) | 6985 (42.6) | 2762 (16.8) |
| Combined | 3018 (64.2) | 825 (17.6) | 18695 (43.0) | 7464 (17.2) |
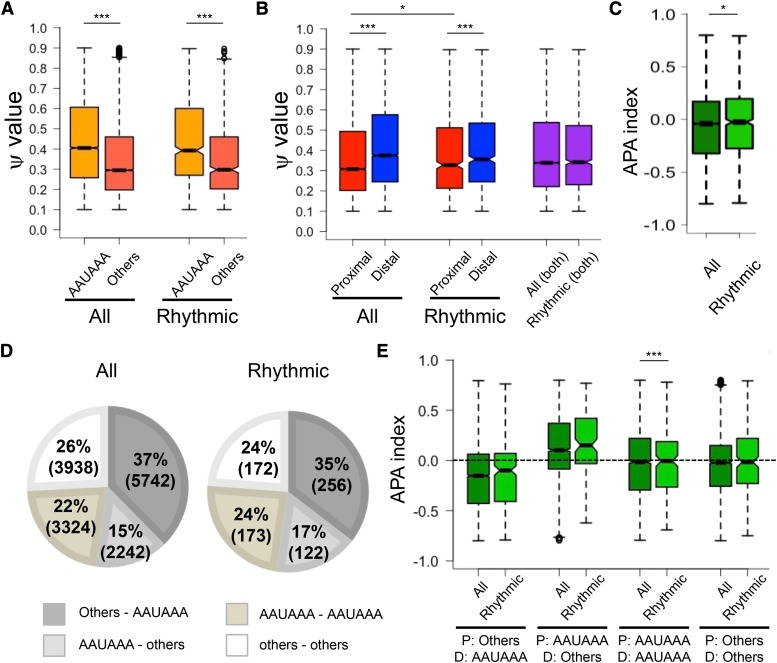
The canonical PAS (AAUAAA) has been considered strongest among other variants, due to its high prevalence among cDNAs/ESTs (Beaudoing et al. 2000; Tian et al. 2005). By using the average ψ values across all time points as an indicator of poly(A) site strength, we found that the isoforms with canonical PASs were indeed more frequent in mouse liver (Figure 4A, S2A). The median ψ value for isoforms with AAUAAA was 0.41, whereas that of other variants was 0.30 in all APA transcripts (Mann-Whitney-Wilcoxon test: P < 2.2e-16) (Figure 4A, S2A). This trend was also observed among rhythmic APA transcripts, in which the median ψ value for isoforms with AAUAAA was 0.39, whereas that of other variants was 0.30 (Mann-Whitney-Wilcoxon test: P < 2.2e-16) (Figure 4A, S2A).
Figure 4.
Hepatic APA genes use distal poly(A) sites and canonical PASs more frequently, but circadian APA genes have less distinct difference. (A) Distributions of ψ values in APA transcripts that have a canonical PAS (AAUAAA) or other PAS variants. All: AAUAAA; n = 14632, others; n = 15860, and rhythmic: AAUAAA; n = 724, others; n = 722. (B) Distributions of ψ values in transcripts that use distal or proximal poly(A) sites. All: proximal = 15246 and distal = 15246, Rhythmic: proximal = 723 and distal = 723. (C) Distribution of the APA index between all (N= 15246) and rhythmic (N= 723) APA transcripts. (D) Percentages of each PAS combination group. All: N = 15246, Rhythmic: N= 723. Numbers in parentheses indicate the number of transcripts that fall under each combination group. (E) Distribution of the APA index in each PAS group. All the box-whisker plots represent quartiles (box) and quartile ± 1.5 interquartile range (whiskers). *; P < 0.05. ***; P < 0.005 (Mann-Whitney-Wilcoxon Test).
Each tissue has a different frequency for the poly(A) site usage, with the distal sites generally being more frequently used in non-proliferating cells (Hoffman et al. 2016), such as hepatocytes that make up ∼80% of the liver. We found that distal sites were stronger than proximal sites in mouse liver, as the median ψ value of distal sites was 0.38, while that of proximal sites was 0.31 among all APA transcripts (Mann-Whitney-Wilcoxon test: P < 2.2e-16) (Figure 4B, S2B). Distal sites were also stronger among rhythmic APA transcripts, as the median ψ value of distal sites was 0.36, while that of proximal sites was 0.33 (Mann-Whitney-Wilcoxon test: P < 2.2e-16). Interestingly, however, the difference between proximal and distal sites was smaller in rhythmic APA transcripts (Δ median ψdistal-proximal = 0.03), compared to control APA transcripts (Δ median ψdistal-proximal = 0.07). This was not due to differences in the distribution of ψ values between all and rhythmic APA transcripts, as the median ψ value that includes both proximal and distal sites were the same (median ψ = 0.39) for both all and rhythmic APA transcripts (Figure 4B, S2B). Rather, this was because rhythmic APA transcripts use proximal poly(A) sites more frequently, as the median ψ values of proximal sites higher among rhythmic APA transcripts (median: 0.33) compared to all APA transcripts (median: 0.31) (Mann-Whitney-Wilcoxon test: P = 0.02134). This was further supported through comparison of the APA index, which represents the relative strength between proximal and distal poly(A) sites. We found that rhythmic APA transcripts had slightly, but significantly, higher APA index (median: -0.025) compared to all APA transcripts (median: -0.04) (Mann-Whitney-Wilcoxon test: P = 0.02811) (Figure 4C), indicating that rhythmic APA transcripts use proximal poly(A) sites more frequently.
Genes that have more than one poly(A) site often have canonical PASs at the most distal site, while variants tend to be found at more proximal sites, despite that promoter-proximal poly(A) sites have a natural advantage over distal ones to be transcribed first and can be used more frequently (Beaudoing et al. 2000; Tian et al. 2005; Davis and Shi 2014). This was also the case in mouse liver, and transcripts with a variant at a proximal site and AAUAAA at a distal site were most frequent among both all and rhythmic APA transcripts (Figure 4D, S3A). In addition, rhythmic APA transcripts tended to have higher APA indices relative to all transcripts, regardless of the combination of PASs at proximal and distal poly(A) sites (Figure 4E, S3B), consistent with our earlier observation (Figure 4C).
These results demonstrate that canonical PAS (AAUAAA) and distal sites are most commonly used in mouse liver. Rhythmic APA genes, in contrast, have less distinct differences in poly(A) site strength between proximal and distal sites and use proximal sites more frequently. Presumably, this allows the daily poly(A) site switch to be more amenable.
Discussion
In this study, we identified and characterized transcripts undergoing rhythmic alternative polyadenylation (APA) in mouse liver, using existing circadian transcriptome datasets combined with a probabilistic algorithm, MISO (Katz et al. 2010; Koike et al. 2012; Vollmers et al. 2012; Zhang et al. 2014). Identification of differentially expressed transcripts from transcriptome data has been a challenge, because distinguishing full-length isoforms from partially reconstructed fragments is not always possible and requires greater sequencing depth and more powerful statistical methods (Trapnell et al. 2009; Katz et al. 2010). MISO circumvents this issue by inverting the process by which reads are first produced and then inferring the underlying isoform abundances that best explain the observed reads. Our in silico analysis identified 712 transcripts (or 2.9% of all expressed genes) that undergo circadian APA regulation in mouse liver.
The number of circadian APA genes we detected in our analysis was smaller than a previous report, which identified 1153 circadian APA genes in mouse liver derived from microarray data (Liu et al. 2013). It is unclear how much our list of APA genes overlaps with the previous report, as the identity of these transcripts, except for a few, was not revealed (Liu et al. 2013). The difference in the number is most likely due to the frequency of tissue sampling, in which the liver samples were taken: every 1 hr (Hughes et al. 2009; Liu et al. 2013) or every 3-6 hr (this study). The other likely causes include the amplitude cutoff, the gene expression cutoff (i.e., presence vs. absence), the rhythmicity detection algorithm, and quantification method of the APA switch (i.e., “APA index” = ψ proximal - ψ distal vs. “the ext/com-ratio” (=ψ distal / ψ proximal)). Most notably, our use of transcriptome data constitutes a significant advantage over that of microarray data, as it allowed us to pinpoint the exact 3′-end locations of each transcript and characterize features commonly found in circadian APA genes. Tiling arrays have higher probe density than conventional microarrays, and yet do not have the sensitivity and accuracy to distinguish isoforms, due to their probe length (∼25-60mer) and spacing (∼400-10,000 nt on average) (Liu 2007; Agarwal et al. 2010; Zhao et al. 2014). In addition, our use of three independent datasets alleviated observing dataset-specific effects (for example, Figure 2G, Supplemental Figure 1-3).
Although the mechanisms of the circadian APA are not fully understood, we found several interesting characteristics that are unique to rhythmic APA transcripts. One such characteristic is that rhythmic APA genes have less distinct differences in poly(A) site strength between proximal and distal sites (Figure 4), possibly due to the lower frequency of canonical PAS usage among rhythmic APA genes (Table 3). This presumably allows the daily poly(A) site switch to be more feasible. The most likely mechanism is that the cis-elements specifically found in the long isoform (i.e., distal poly(A) site usage) either activate or repress one poly(A) site usage over another by interacting with trans-factors. It is also possible that these interactions affect the RNA processing (i.e., mRNA stability, translation, and localization) (Di Giammartino et al. 2011; Mayr 2016). To support this, circadian APA transcripts have a larger difference in the 3′UTR length between the APA isoforms (Figure 3). Identifying these cis-elements and understanding how their interaction with trans-factors affect the target poly(A) site choice in a circadian manner will, thus, help us better understand the mechanisms circadian APA regulation in the future.
Since components of the cleavage and polyadenylation machinery, e.g., CSTF77 and NUDT21, are expressed rhythmically in mouse liver (Mauvoisin et al. 2014; Robles et al. 2014), it is tempting to speculate that they are involved in regulating circadian APA at least for some genes. Changes in the levels of the other core components for cleavage and polyadenylation machinery, including CSTF-64/CSTG2, CSTF-64τ, CFI-68/CPSF6, CFI-25/CPSF5, FIP1L1, and PCF11 as well as auxiliary factors such as PABPN1, PTBP, RBBP6, and PAF1C, have all been shown to impact poly(A) site choices (Takagaki et al. 1996; Takagaki and Manley 1998; Kubo et al. 2006; de Klerk et al. 2012; Jenal et al. 2012; Martin et al. 2012; Yao et al. 2012; Yao et al. 2013; Di Giammartino et al. 2014; Lackford et al. 2014; Masamha et al. 2014; Gennarino et al. 2015; Li et al. 2015; Domingues et al. 2016; Yang et al. 2016). These proteins may also be involved in regulating circadian APA, although this remains to be investigated.
Despite the advances of sequencing technology and analytical tools that expand our ability to detect APA events, consequences of APA are still largely unknown. We measured the correlation between protein rhythmicity vs. APA rhythmicity utilizing three circadian proteome datasets from mouse liver (Reddy et al. 2006; Mauvoisin et al. 2014; Robles et al. 2014). We found that the percentage of rhythmic proteins was similar among circadian APA genes and control APA genes (data not shown). It is unclear whether this suggests that rhythmic APA regulation does not contribute to rhythmic protein production or the correlations are missed simply because the existing mammalian liver proteome data only cover a small percentage of the entire proteome. It is also possible that the impact of APA on translation is regulated by gene-specific, rather than global, mechanisms. Nevertheless, it would be of great interest in the future to examine the impact of circadian APA regulation on the proteome and/or transcriptome.
Obviously, experimental validation is key to underscoring the importance of our findings. Our attempts to measure circadian changes in APA by quantitative PCR were unsuccessful; however, quantitative PCR may not be an ideal method, since it is technically impossible to specifically detect shorter isoforms. Several genome-wide approaches are now available to detect 3′-end of transcripts quantitatively (Pelechano et al. 2012; Cornett and Lutz 2014; Hoque et al. 2014; Yao and Shi 2014; Xing and Li 2015; Zheng et al. 2016; Zheng and Tian 2017). These tools can be used in the future to quantify circadian APA, including de novo poly(A) sites.
Overall, our study provides evidence that the choice of poly(A) site and therefore mRNA 3′-end structure are under circadian regulation. Interesting research directions in the future would be to 1) experimentally validate the circadian poly(A) site switches, 2) identify cis-elements enriched in the circadian APA transcripts, and 3) delineate how these cis-elements confer rhythmicity in poly(A) site choices. By identifying unique characteristics of rhythmic APA genes and illustrating the landscape of poly(A) tails in mouse liver, our study serves as a platform to help us understand the mechanisms of circadian APA regulation.
Aknowledgments
The authors thank Ms. Naseem Maghzian and Ms. Connie Magarrelli for their technical assistance and Janet Webster for critical reading of the manuscript. This work was partially supported by a Grant for Basic Science Research Projects from the Sumitomo Foundation (to S.K.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6741623.
Communicating editor: J. Dunlap
Literature Cited
- Agarwal A., Koppstein D., Rozowsky J., Sboner A., Habegger L., et al. , 2010. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383 10.1186/1471-2164-11-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. J., Gharami K., Liao G. Y., Woo N. H., Lau A. G., et al. , 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187. 10.1016/j.cell.2008.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E., Freier S., Wyatt J. R., Claverie J. M., Gautheret D., 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10: 1001–1010. 10.1101/gr.10.7.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits B. D., Mayr C., 2015. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522: 363–367. 10.1038/nature14321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. W., Zhang W., Yeh H. S., de Jong E. P., Jun S., et al. , 2015. mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 6: 7218 10.1038/ncomms8218 [DOI] [PubMed] [Google Scholar]
- Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., et al. , 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17: 13 (erratum: Genome Biol. 17: 181) 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett A. L., Lutz C. S., 2014. RHAPA: a new method to quantify alternative polyadenylation. Methods Mol. Biol. 1125: 157–167. 10.1007/978-1-62703-971-0_14 [DOI] [PubMed] [Google Scholar]
- Davis R., Shi Y., 2014. The polyadenylation code: a unified model for the regulation of mRNA alternative polyadenylation. J. Zhejiang Univ. Sci. B 15: 429–437. 10.1631/jzus.B1400076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E., Venema A., Anvar S. Y., Goeman J. J., Hu O., et al. , 2012. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 40: 9089–9101. 10.1093/nar/gks655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino D. C., Nishida K., Manley J. L., 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., et al. , 2012. Landscape of transcription in human cells. Nature 489: 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues R. G., Lago-Baldaia I., Pereira-Castro I., Fachini J. M., Oliveira L., et al. , 2016. CD5 expression is regulated during human T-cell activation by alternative polyadenylation, PTBP1, and miR-204. Eur. J. Immunol. 46: 1490–1503. 10.1002/eji.201545663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield G. E., 2003. DNA microarray analyses of circadian timing: the genomic basis of biological time. J. Neuroendocrinol. 15: 991–1002. 10.1046/j.1365-2826.2003.01082.x [DOI] [PubMed] [Google Scholar]
- Fang B., Everett L. J., Jager J., Briggs E., Armour S. M., et al. , 2014. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159: 1140–1152. 10.1016/j.cell.2014.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S. W., Kim T. K., Gray J. M., Harmin D. A., Hemberg M., et al. , 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. 10.1016/j.neuron.2008.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino V. A., Alcott C. E., Chen C. A., Chaudhury A., Gillentine M. A., et al. , 2015. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. eLife 4 10.7554/eLife.10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Vanderheyden W. M., Lavaute T., Westmark C. J., Rouhana L., et al. , 2012. Time of day regulates subcellular trafficking, tripartite synaptic localization, and polyadenylation of the astrocytic fabp7 mRNA. J. Neurosci. 32: 1383–1394. 10.1523/JNEUROSCI.3228-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. B., 2017. Circadian Posttranscriptional Regulatory Mechanisms in Mammals. Cold Spring Harb. Perspect. Biol. 10: a030692 10.1101/cshperspect.a030692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R., Martin G., Muller P., Schmidt A., Gruber A. J., et al. , 2014. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun. 5: 5465 10.1038/ncomms6465 [DOI] [PubMed] [Google Scholar]
- Guan D, Xiong Y, Borck PC, Jang C, Doulias PT, Papazyan R, Fang B, Jiang C, Zhang Y, Briggs ER, et al. 2018. Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 174: 831–842 e812 10.1016/j.cell.2018.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta I., Clauder-Munster S., Klaus B., Jarvelin A. I., Aiyar R. S., et al. , 2014. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol. Syst. Biol. 10: 719 10.1002/msb.135068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K. C. H., Blencowe B. J., Morris Q., 2018. QAPA: a new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol. 19: 45 10.1186/s13059-018-1414-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Y, Bublik DR, PU A, Elkon R, Biniashvili T, Agami R, Oren M, Pilpel Y.2016. 3′UTR Shortening Potentiates MicroRNA-Based Repression of Pro-differentiation Genes in Proliferating Human Cells. PLoS Genet 12: e1005879 10.1371/journal.pgen.1005879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J. E., 2014. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Hum. Genomics 8: 3 10.1186/1479-7364-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M., Ji Z., Zheng D., Luo W., Li W., et al. , 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 10: 133–139. 10.1038/nmeth.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M., Li W., Tian B., 2014. Accurate mapping of cleavage and polyadenylation sites by 3′ region extraction and deep sequencing. Methods Mol. Biol. 1125: 119–129. 10.1007/978-1-62703-971-0_10 [DOI] [PubMed] [Google Scholar]
- Hu W., Li S., Park J. Y., Boppana S., Ni T., et al. , 2017. Dynamic landscape of alternative polyadenylation during retinal development. Cell. Mol. Life Sci. 74: 1721–1739. 10.1007/s00018-016-2429-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., et al. , 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet. 5: e1000442 10.1371/journal.pgen.1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P., Arpat A. B., Castelo-Szekely V., Lopes M., Gatfield D., 2015. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 25: 1848–1859. 10.1101/gr.195404.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kuhn U., et al. , 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553. 10.1016/j.cell.2012.03.022 [DOI] [PubMed] [Google Scholar]
- Ji Z., Lee J. Y., Pan Z., Jiang B., Tian B., 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA 106: 7028–7033. 10.1073/pnas.0900028106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Tian B., 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4: e8419 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C., Cretenet G., Symul L., Martin E., Atger F., et al. , 2013. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11: e1001455 10.1371/journal.pbio.1001455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E. T., Airoldi E. M., Burge C. B., 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7: 1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D., 2010. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology 151: 1391–1397. 10.1210/en.2009-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N., Yoo S. H., Huang H. C., Kumar V., Lee C., et al. , 2012. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338: 349–354. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Green C. B., 2015. Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry 54: 124–133. 10.1021/bi500707c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Sher-Chen E. L., Green C. B., 2012. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26: 2724–2736. 10.1101/gad.208306.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Wada T., Yamaguchi Y., Shimizu A., Handa H., 2006. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 34: 6264–6271. 10.1093/nar/gkl794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackford B., Yao C., Charles G. M., Weng L., Zheng X., et al. , 2014. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 33: 878–889. 10.1002/embj.201386537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. G., Irier H. A., Gu J., Tian D., Ku L., et al. , 2010. Distinct 3′UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF). Proc. Natl. Acad. Sci. USA 107: 15945–15950. 10.1073/pnas.1002929107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., You B., Hoque M., Zheng D., Luo W., et al. , 2015. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 11: e1005166 10.1371/journal.pgen.1005166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. S., 2007. Getting started in tiling microarray analysis. PLOS Comput. Biol. 3: 1842–1844. 10.1371/journal.pcbi.0030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu W., Murakawa Y., Yin J., Wang G., et al. , 2013. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 3: 2054 10.1038/srep02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Gruber A. R., Keller W., Zavolan M., 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports 1: 753–763. 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Masamha C. P., Xia Z., Yang J., Albrecht T. R., Li M., et al. , 2014. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510: 412–416. 10.1038/nature13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvoisin D., Wang J., Jouffe C., Martin E., Atger F., et al. , 2014. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA 111: 167–172. 10.1073/pnas.1314066111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., 2016. Evolution and Biological Roles of Alternative 3′UTRs. Trends in cell biology 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Bartel D. P., 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J. S., Rodriguez J., Abruzzi K. C., Rosbash M., 2012. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1: e00011 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve J., Burger K., Li W., Hoque M., Patel R., et al. , 2016. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 26: 24–35. 10.1101/gr.193995.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B. D., Varadarajan A., Passalacqua K. D., Bergman N. H., 2008. Efficient mapping of Applied Biosystems SOLiD sequence data to a reference genome for functional genomic applications. Bioinformatics 24: 2776–2777. 10.1093/bioinformatics/btn512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Wilkening S., Jarvelin A. I., Tekkedil M. M., Steinmetz L. M., 2012. Genome-wide polyadenylation site mapping. Methods Enzymol. 513: 271–296. 10.1016/B978-0-12-391938-0.00012-4 [DOI] [PubMed] [Google Scholar]
- Pinto P. A., Henriques T., Freitas M. O., Martins T., Domingues R. G., et al. , 2011. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 30: 2431–2444. 10.1038/emboj.2011.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyna N., Boughorbel S., El Anbari M., Ptitsyn A., 2017. The role of alternative Polyadenylation in regulation of rhythmic gene expression. BMC Genomics 18: 576 10.1186/s12864-017-3958-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., et al. , 2006. Circadian orchestration of the hepatic proteome. Curr. Biol. 16: 1107–1115. 10.1016/j.cub.2006.04.026 [DOI] [PubMed] [Google Scholar]
- Robinson B. G., Frim D. M., Schwartz W. J., Majzoub J. A., 1988. Vasopressin mRNA in the suprachiasmatic nuclei: daily regulation of polyadenylate tail length. Science 241: 342–344. 10.1126/science.3388044 [DOI] [PubMed] [Google Scholar]
- Robles M. S., Cox J., Mann M., 2014. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 10: e1004047 10.1371/journal.pgen.1004047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B., 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, Sassone-Corsi P. 2017. Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 170: 664–677 e611. 10.1016/j.cell.2017.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G., Frohman M. A., 2006. 3′ end cDNA amplification using classic RACE. Nat. Protoc. 1: 2742–2745. 10.1038/nprot.2006.481 [DOI] [PubMed] [Google Scholar]
- Shepard P. J., Choi E. A., Lu J., Flanagan L. A., Hertel K. J., et al. , 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. 10.1261/rna.2581711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Alley T. L., Wright S. M., Kamdar S., Schott W., et al. , 2009. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 69: 9422–9430. 10.1158/0008-5472.CAN-09-2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Manley J. L., 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2: 761–771. 10.1016/S1097-2765(00)80291-9 [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Seipelt R. L., Peterson M. L., Manley J. L., 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952. 10.1016/S0092-8674(00)82000-0 [DOI] [PubMed] [Google Scholar]
- Tian B., Hu J., Zhang H., Lutz C. S., 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33: 201–212. 10.1093/nar/gki158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Manley J. L., 2016. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C., Schmitz R. J., Nathanson J., Yeo G., Ecker J. R., et al. , 2012. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 16: 833–845. 10.1016/j.cmet.2012.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., et al. , 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Anafi R. C., Hughes M. E., Kornacker K., Hogenesch J. B., 2016. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32: 3351–3353. 10.1093/bioinformatics/btw405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., Donehower L. A., Cooper T. A., Neilson J. R., Wheeler D. A., et al. , 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 5: 5274 10.1038/ncomms6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M. S., Zhang B., Li Y. S., Gao Q., Sun W., et al. , 2016. Global analysis of regulatory divergence in the evolution of mouse alternative polyadenylation. Mol. Syst. Biol. 12: 890 10.15252/msb.20167375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Li Q. Q., 2015. RADPRE: a computational program for identification of differential mRNA processing including alternative polyadenylation. Methods Mol. Biol. 1255: 57–66. 10.1007/978-1-4939-2175-1_6 [DOI] [PubMed] [Google Scholar]
- Yang Y., Li W., Hoque M., Hou L., Shen S., et al. , 2016. PAF Complex Plays Novel Subunit-Specific Roles in Alternative Cleavage and Polyadenylation. PLoS Genet. 12: e1005794 (correction: PLoS Genet. 12: e1005883) 10.1371/journal.pgen.1005794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Biesinger J., Wan J., Weng L., Xing Y., et al. , 2012. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc. Natl. Acad. Sci. USA 109: 18773–18778. 10.1073/pnas.1211101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Choi E. A., Weng L., Xie X., Wan J., et al. , 2013. Overlapping and distinct functions of CstF64 and CstF64tau in mammalian mRNA 3′ processing. RNA 19: 1781–1790. 10.1261/rna.042317.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Shi Y., 2014. Global and quantitative profiling of polyadenylated RNAs using PAS-seq. Methods Mol. Biol. 1125: 179–185. 10.1007/978-1-62703-971-0_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Long Y., Ji G., Li Q. Q., Wu X., 2018. APAtrap: identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics 34: 1841–1849. 10.1093/bioinformatics/bty029 [DOI] [PubMed] [Google Scholar]
- Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B., 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA 111: 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Fung-Leung W. P., Bittner A., Ngo K., Liu X., 2014. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 9: e78644 10.1371/journal.pone.0078644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Liu X., Tian B., 2016. 3′READS+, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA 22: 1631–1639. 10.1261/rna.057075.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Tian B., 2017. Polyadenylation Site-Based Analysis of Transcript Expression by 3′READS. Methods Mol. Biol. 1648: 65–77. 10.1007/978-1-4939-7204-3_6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6741623.