Abstract
Patterns of nucleotide polymorphism within populations of Drosophila melanogaster suggest that insecticides have been the selective agents driving the strongest recent bouts of positive selection. However, there is a need to explicitly link selective sweeps to the particular insecticide phenotypes that could plausibly account for the drastic selective responses that are observed in these non-target insects. Here, we screen the Drosophila Genetic Reference Panel with two common insecticides; malathion (an organophosphate) and permethrin (a pyrethroid). Genome-wide association studies map survival on malathion to two of the largest sweeps in the D. melanogaster genome; Ace and Cyp6g1. Malathion survivorship also correlates with lines which have high levels of Cyp12d1, Jheh1 and Jheh2 transcript abundance. Permethrin phenotypes map to the largest cluster of P450 genes in the Drosophila genome, however in contrast to a selective sweep driven by insecticide use, the derived allele seems to be associated with susceptibility. These results underscore previous findings that highlight the importance of structural variation to insecticide phenotypes: Cyp6g1 exhibits copy number variation and transposable element insertions, Cyp12d1 is tandemly duplicated, the Jheh loci are associated with a Bari1 transposable element insertion, and a Cyp6a17 deletion is associated with susceptibility.
Keywords: Drosophila Genetic Reference Panel (DGRP), malathion, permethrin, acetylcholinesterase, Cyp6a17
Understanding the genetic basis of insecticide resistance is important, not only to inform the implementation of insecticides in agriculture and disease vector control, but also as an evolutionary case-study operating over observable periods of time. Utilizing genome-wide association studies (GWAS) to investigate insecticide resistance provides an unbiased way to identify multiple natural genetic variants associated with a phenotype, while the polymorphism data surrounding associated variants may provide clues to their evolutionary trajectory.
Since its introduction in 2012, the Drosophila Genetic Reference Panel (DGRP; Mackay et al. 2012) has proved to be a powerful tool for dissecting the genetic architecture of a range of Drosophila melanogaster phenotypes through the implementation of genome-wide association studies (GWAS) on the DGRP’s 205 inbred, sequenced lines. Insecticide-induced mortality has been among these phenotypes (Battlay et al. 2016; Denecke et al. 2017; Schmidt et al. 2017). In 2015, the DGRP’s utility was increased with the introduction of transcriptome data (Huang et al. 2015), allowing phenotypes to be tested for association directly with variation in individual transcripts across the D. melanogaster transcriptome.
The sequence data generated by the DGRP has also proved to be a valuable resource for the study of population genomics, and has allowed the identification of regions of strong, recent selection in the DGRP’s ancestral population (Garud et al. 2015). Two of the most pronounced of these signals, genome wide, come from insecticide resistance loci Cyp6g1 and Ace. Significant selective signals have also been identified around these loci in other D. melanogaster populations (Garud and Petrov 2016), and related species (Signor et al. 2017; D. simulans), as well as by targeted analyses in D. melanogaster of selection at Ace (Karasov, Messer and Petrov 2010) and Cyp6g1 (Catania et al. 2004; Schmidt et al. 2010).
The fact that insecticides appear to have played such an important role in the recent evolutionary history of the DGRP allows us the rare opportunity to study the quantitative genetics of a trait in the process of strong selection. It is unknown, however, what compound or compounds are causing this selection. Although natural variation in both Ace and Cyp6g1 has been demonstrated to confer resistance to various insecticides, attempts to detect these associations in the DGRP, and hence associate the selective sweeps at these loci with a particular compound, have departed from expectations.
Acetylcholineesterase (Ace) is the molecular target of organophosphate insecticides, and four non-synonymous substitutions in the enzyme’s active-site groove have been demonstrated to reduce the binding capacity of organophosphate insecticides (Menozzi et al. 2004). Battlay et al. (2016) were, however, unable to detect a significant effect of variation in Ace on resistance to the organophosphate azinphos-methyl in DGRP larvae, but instead detected a strong association with alleles that overexpressed Cyp6g1, a cytochrome P450 enzyme previously shown to confer metabolic resistance to DDT and imidacloprid when overexpressed (Daborn et al. 2001, Daborn et al. 2002, Joußen et al. 2008, Hoi et al. 2014). Although the link between natural alleles which overexpress Cyp6g1 and resistance to DDT has been demonstrated in a worldwide sample (Catania et al. 2004) and Australian populations (Schmidt et al. 2010), a similar result was not observed in the DGRP (Schmidt et al. 2017).
Aside from the recently reported association between azinphos-methyl resistance and Cyp6g1 in the DGRP (Battlay et al. 2016), previous investigations have mapped organophosphate resistance to a region including Cyp6g1 (Kikkawa 1961 [parathion]; Pyke et al. 2004 [diazinon]). Cross-resistance to the organophosphate malathion was reported at the mapping region on chromosome 2 by Kikkawa (1961), and Ogita’s (1958) mapping of DDT resistance. Le Goff et al. (2003) also reported malathion cross-resistance in DDT resistant lab lines (Hikone-R and Wisconsin), both of which showed heightened levels of Cyp6g1 transcript. Likewise, DDT-resistant 91-R (which carries a resistance allele at the Cyp6g1 locus [Schmidt et al. 2017] and overexpresses the enzyme [Pedra et al. 2004]), shows cross resistance to malathion (Misra et al. 2013).
In light of this evidence, resistance to organophosphates makes a compelling subject for study in the DGRP. Natural variants with the two strongest signals of selection in the population, Ace and Cyp6g1, may both confer resistance to these compounds, and the organophosphate class of insecticides has been employed widely over a long period of time, giving it opportunity to induce such selective pressures.
Pyrethroids have also been extensively utilized both spatially and temporally in insect control, however, natural variation contributing to pyrethroid insecticide class resistance in D. melanogaster is less well understood. Like Ace, resistance-causing mutations in the molecular target of pyrethroids and DDT, the voltage-gated sodium channel, are common in insect pest species (Dong et al. 2014). However, orthologous mutations have not been described as natural variants in D. melanogaster, although EMS mutagenesis has yielded mutations in para (the D. melanogaster voltage-gated sodium channel alpha subunit) that cause resistance to DDT and the pyrethroid deltamethrin (Pittendrigh et al. 1997). At least one D. melanogaster cytochrome P450 gene has been shown to be involved in pyrethroid biology. Cyp4e3 is both induced in response to permethrin exposure, and capable of increasing resistance to the insecticide when overexpressed (Terhzaz et al. 2015), however, once again natural variation in this gene has not been described, and any contribution to of this locus to pyrethroid resistance in wild populations is yet to be determined.
Organophosphates and pyrethroids are two of the oldest and most widely used insecticide classes in the world today. Here, we investigate the genetic basis of resistance in the DGRP to a representative of each of these classes; the organophosphate, malathion, and the pyrethroid, permethrin. We assess genomic and transcriptomic associations with both male and female adults at multiple doses and incorporate genotyping of structural variation and previously identified signatures of selective sweeps.
Materials and Methods
Fly lines
DGRP lines were generated by Mackay et al. (2012) and obtained from the Bloomington Drosophila stock center in Indiana. The 6g1HR-GAL4 driver line was generated by Chung et al. (2007). The RAL_517 Cyp6g1-KO line was generated by Denecke et al. (2017). The UAS-Cyp12d1 line was generated by Daborn et al. (2007). Cyp6a17KG04448 flies were generated by Bellen et al. (2011) and were obtained from the Bloomington Drosophila Stock Center. All fly stocks were maintained at 25° on rich medium containing, maltose (46g/L), dextrose (75g/L), yeast (35g/L), soy flour (20g/L), maize meal (73g/L), agar (6g/L), acid mix (14ml/L), and tegosept (16ml/L). The acid mix solution was made up of orthophosphoric acid (42ml/L), and propionic acid (412ml/L), while the tegosept solution was 50g tegosept dissolved in 950 ml of 95% EtOH.
Insect bioassays
Adult flies for bioassays were anesthetized with CO2 at 0-24 hr after eclosion and sorted by sex into holding vials containing rich media, where they were kept for 3-4 days, resulting in 3-5 day old adults for use in bioassays. Assays commenced between 11 am and 12 pm. 20mL glass scintillation vials were treated with 500μl of acetone/insecticide solution at the required concentration and rolled using a hotdog warmer (heat off) until the acetone had evaporated. ∼7 flies were transferred to each vial without the use of anesthesia, and cotton wool moistened with 10% sucrose solution was used to stopper the scintillation vials. DGRP lines were screened at a single dose.
Phenotype selection
Twenty-four-hour mortality in adult insects was selected as a phenotype as this is a standard assay in D. melanogaster insecticide resistance literature (Daborn et al. 2001; Schmidt et al. 2010; Schmidt et al. 2017), and DGRP lines show negligible control mortality in scintillation vial bioassays at this time point (Schmidt et al. 2017). ‘Knockdown”, a phenotype in which the fly lies paralyzed and twitching, or exhibits uncontrolled flight, is a field-relevant effect of pyrethroid insecticides. We therefore also measured incidence of this phenotype at three hours in permethrin-treated flies. As no pronounced knockdown effect was expected (or observed) in malathion-treated flies, we instead scored mortality at additional time points (3, 6 and 12 hr). Discriminant insecticide doses (1μg/vial for malathion, 10μg/vial for permethrin) were identified by screening 20 randomly selected DGRP lines on a range of concentrations. Transgenic lines were screened at multiple doses and scored for mortality at 24 hr. A minimum of three biological replicates were performed for each sex of each DGRP line, and for each sex at each dose for transgenic lines.
Calculation of LD50
For transgenic lines, linear models were fitted to dose-mortality data on a log-probit scale using ‘glm’ in R (R Core Team 2013) and scripts from Johnson et al. (2013). Median lethal dose (LD50) values and 95% confidence intervals were calculated using Fieller’s method from fitted linear models (Finney 1971).
Estimation of broad-sense heritability and sex effect
The broad-sense heritability of each phenotype was estimated for both sexes individually as σ2G / (σ2G + σ2E), using the variance components of a linear model of the form phenotype ∼1 + (1 | line) using ‘lme4’ in R (R Core Team 2013). Male sex effect was estimated from the male sex intercept of the model phenotype ∼ sex + (0 + sex | line) using ‘lme4’ in R (R Core Team 2013).
Genome-wide association studies
Phenotype files for 170 DGRP lines, consisting of mean mortality data for both males and females, were generated for all phenotypes and were submitted to the Mackay lab DGRP2 GWAS pipeline (http://dgrp2.gnets.ncsu.edu/; Huang et al. 2014). The genome wide significance threshold (1×10−5) was corrected for the number of phenotypes tested for each insecticide (8 and 4 for malathion and permethrin respectively) and applied to the Mackay lab pipeline ‘mixed p-value’ (association after correction for the effects of wolbachia and major chromosomal inversions). Bonferroni significance thresholds were calculated as 0.05 divided by the product of the number of genomic variants (1,877,810 and 1,876,330 for malathion and permethrin respectively) and phenotypes tested.
Assessment of variants within H12 peaks
Malathion associated variants were identified in four H12 selective sweep peaks. To test whether these variants may be the foci of these sweeps, distortions in candidate allele frequency in lines sharing the two most common haplotypes (H1 and H2) at each H12 peak were tested using Fisher’s exact test.
Transcriptome to phenotype associations
Transcriptome data for 1-3 day old adult flies from 185 DGRP lines were recovered from the DGRP website (http://dgrp2.gnets.ncsu.edu/data.html; Huang et al. 2015). Mean transcription level was calculated for each gene in each sex from two biological replicates, to give a mean level for each of the 18,139 transcripts measured by Huang et al. (2015) in each DGRP line, for both males and females. A linear model was fit between mean transcription level of each gene measured by Huang et al. (2015) for the relevant sex, and each malathion and permethrin phenotype individually. 1×10−3 (which roughly corresponds to the genome-wide significance threshold used in GWAS adjusted for the smaller number of tests performed against the transcriptome compared to the genome) was used as a base significance threshold for transcriptome associations after correction for the number of phenotypes tested for each insecticide (8 and 4 for malathion and permethrin respectively). Bonferroni significance thresholds were calculated as 0.05 divided by the product of the number of transcripts (18139) and phenotypes tested. Associated variants from GWAS were also tested for annotation as eQTL using data from Huang et al. 2015.
Genotyping of structural variation
BAM files containing alignments of DGRP line sequences from Illumina platforms to the y; cn bw sp; reference genome were recovered from the Baylor College of Medicine website (https://www.hgsc.bcm.edu/content/dgrp-lines; Mackay et al. 2012). Local alignments at candidate loci were visualized with IGV 2.0 software (Robinson et al. 2011) to manually score structural variation. Cyp6g1, Cyp6a17/23, and Cyp12d1 structural variants were previously genotyped in Good et al. (2014).
Genotyping of the Bari1 insertion presence upstream of Jheh1 and Jheh2 genes was provided by Josefa González derived from diagnostic PCR (33 lines; González, Macpherson and Petrov 2009) and T-lex software (119 lines; Fiston-Lavier et al. 2010). These datasets were supplemented with our own manual calling of the insertion using IGV 2.0 software (167 lines; Thorvaldsdóttir et al. 2013). This resulted in 80 DGRP lines with high confidence (at least two concurrent calls) Bari-Jheh genotypes that also had matching transcriptome and malathion phenotype data, which were used for further analysis.
Amplification events involving Cyp6g1 and Cyp6g2 were inferred from local read depth in DGRP BAM files. Read depth at each nucleotide position under consideration were recovered using the Genome Analysis Toolkit ‘DepthOfCoverage’ utility (McKenna et al. 2010). Regions interrogated were non-overlapping portions of the Cyp6g1 amplicon (2R:8072727-8074976), the Cyp6g1g2 amplicon (2R:8075688-8077656), and a control region of similar size just upstream of Cyp6g1 which does not exhibit structural variation in the DGRP (2R:8070657-8072656). Mean read depth for each amplicon was calculated for each DGRP line, and normalized to mean read depth of the control region.
Transgenic overexpression
Cyp12d1 was overexpressed using the GAL4/UAS system (Brand and Perrimon 1993) and the 6g1HR-GAL4 driver described by Chung et al. (2007). 6g1HR-GAL4 virgin females, in which GAL4 is regulated by Cyp6g1 upstream sequence originating from Hikone-R line flies, were crossed to males carrying an additional copy of Cyp12d1 under control of a UAS promoter. w1118 was used as a control (Daborn et al. 2007).
Frequencies of Ace and Cyp6g1 in the Drosophila Genome Nexus
FASTA files from the Drosophila Genome Nexus release 1.1 (Lack et al. 2016) were downloaded from http://www.johnpool.net/genomes.html. The provided scripts were used to mask data for identity by descent and population admixture. Variants were retrieved from the genomes using the provided dataslice.pl script. In the case of Cyp6g1, we used 2R:8072837, a SNP in complete linkage disequilibrium with derived alleles of Cyp6g1 in the DGRP, as a marker for derived Cyp6g1 alleles in the DGN data.
Data Availability
The DGRP strains are available from Bloomington Stock center. All data are reported in the manuscript or in the associated supplementary material. DGRP phenotypes and raw Mackay pipeline outputs for each phenotype are supplied in the supplementary data file. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6954443.
Results
Phenotypes
Male and female insecticide phenotypes (3, 6, 12 and 24-hour malathion mortality at 1μg/vial, permethrin 3-hour knockdown and permethrin 24-hour mortality at 10 μg/vial) were measured for 170 DGRP lines. Malathion broad-sense heritability (H2) ranged from 0.56-0.68, while permethrin H2 ranged from 0.56-0.61 (Table S1). For all insecticide phenotypes, males showed higher population mean susceptibility than females, and the effect of sex on phenotype was greater for permethrin (0.11-0.24) than malathion (0.02-0.04; Table S1).
Genome-wide association studies
Phenotypes were tested for associations with genomic variants using the DGRP2 pipeline, which corrects for the effects on phenotype of wolbachia infection status and five common chromosomal inversion genotypes in each DGRP line (Huang et al. 2014). Wolbachia infection significantly reduced insecticide susceptibility to both insecticides at multiple phenotypes (P < 0.05; Table S1). P-values arising from these mixed linear models are reported as ‘mixed p-values’, and two thresholds were utilized in the assessment of significant associations: The ‘genome-wide significance threshold’ (1×10−5), corrected for the number of phenotypes tested in each insecticide, and the Bonferroni significance threshold (0.05 corrected for the number of DGRP variants and phenotypes tested).
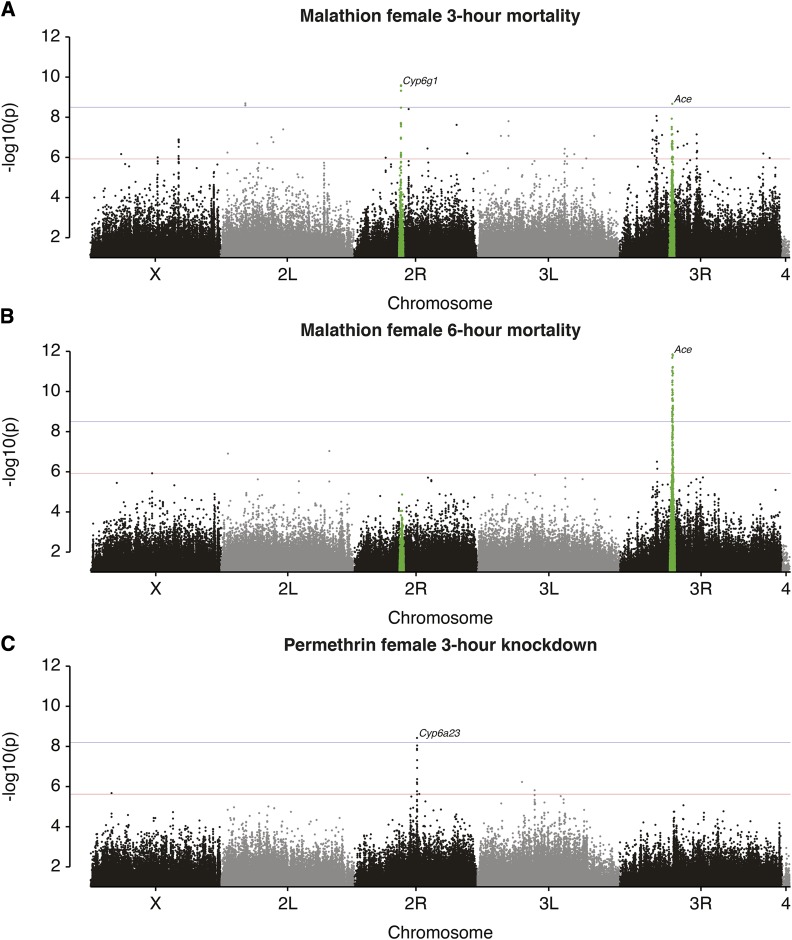
Across malathion phenotypes, 273 unique variants were identified with mixed p-values below the genome-wide significance threshold (1.25×10−6; Figure 1A, Figure 1B, Figure S1A, Table S2, Table S4), more than half of which (176) were only associated with a single sex. 12 nonsynonymous variants in 8 genes were associated with malathion phenotypes, including three in Ace, the molecular target of organophosphate insecticides. 62 variants were associated with mixed p-values below the Bonferroni-corrected significance threshold (3.33×10−9). Enrichment analyses using gene function, protein interactions and pathway relations failed to identify significant terms with the malathion phenotypes (Antonov 2011; Table S2).
Figure 1.
Most significant DGRP genomic variant associations with malathion and permethrin phenotypes. Manhattan plots (mixed p-value against genomic location) for two malathion phenotypes and one permethrin phenotype, showing strong associations around Cyp6g1, Ace, and members of a cluster of cytochrome P450s on chromosome 2R. Genome-wide significance thresholds are indicated in red, Bonferroni significance thresholds are indicated in blue. Green highlights on malathion Manhattan plots show variants within H12 selective sweep statistic peaks identified around Cyp6g1 and Ace (Garud et al. 2015).
For permethrin, 39 variants were associated with any phenotype with mixed p-values below the genome wide significance threshold (2.5×10−6; Figure 1C, Figure S1B, Table S5, Table S6), two of which achieved Bonferroni-corrected significance (P < 6.66×10−9). Eight of the nine variants common to both sexes, including the Bonferroni-significant associations, were identified in the cytochrome P450 cluster on the right arm of chromosome 2, and consequently P450 related terms were significant in gene ontology enrichment analyses (Antonov 2011; Table S2).
Selective sweeps at resistance loci
Variants that are indicative of previously described resistance haplotypes surrounding Ace and Cyp6g1 were among those strongly associated with malathion phenotypes (Figure 1A, Figure 1B, Figure S1A); these included the three Ace resistance substitutions that segregate in the DGRP (I199V [3R_9069054_SNP], G303A [3R_9069408_SNP] and F368Y [3R_9069721_SNP]; Mutero et al. 1994; Menozzi et al. 2004; Battlay et al. 2016) and 2R_8072884_INS, the Accord LTR insertion which differentiates the ancestral Cyp6g1-M allele from resistant Cyp6g1 alleles (Daborn et al. 2002; Schmidt et al. 2010). Selective sweeps involving Ace and Cyp6g1 have previously been described in the DGRP (Garud et al. 2015), and our malathion GWAS identified 128 and 18 associated variants from within boundaries of the Ace and Cyp6g1 sweeps respectively.
To ascertain whether any other selective sweep regions may associate with our phenotypes, we looked for localization of phenotype-associated variants within the most extreme signatures of selection in the DGRP identified by Garud et al. (2015) using the 10kb-windowed H12 statistic. Of 273 malathion variants, we observed a total of 151 associated variants within four of the 50 H12 selective sweep peaks. For the 39 permethrin variants, there were no overlaps with any H12 peaks. To assess whether malathion-associated variants were located on the haplotypes driving the selection signals at the four H12 peaks, we assessed their association with the two most common H12 test haplotypes (Figure S2). H1 and H2 (the two most common haplotypes at each sweep peak) are significantly more likely to contain resistant variants for all three Ace sites, Cyp6g1, and one variant in a third H12 peak (3R:13258656; Fisher’s exact test, P < 0.05). However, minor alleles at 3R:13258656 are only observed in H2 and absent from H1, making it unlikely that this variant is the major driver of haplotype structure at this locus.
Transcriptome associations
To determine the transcriptomic effects of our GWAS candidates, we interrogated datasets generated by Huang et al. (2015) for insecticide-associated variants which are also eQTL (genomic variants associated with variation in mean of a particular transcript level) and veQTL (genomic variants associated with variation in variance of a particular transcript level; Tables S3–S6). 53 unique malathion-associated variants (1.25×10−6) were eQTL, of which 16 within the Cyp6g1 sweep boundaries were eQTL of Cyp6g1 and its tandem paralog Cyp6g2. Almost all other malathion-associated eQTL (33), and 42 of the 45 malathion-associated veQTL, mapped to the Ace sweep region. Given the extended linkage disequilibrium within the sweep region, these are side-effects of the strong association between malathion phenotypes and Ace resistance mutations. Four permethrin-associated variants (2.5×10−6) were eQTL and veQTL, three of which were annotated to Cyp6a23 and were eQTL and veQTL of Cyp6a23’s tandem paralog, Cyp6a17.
We also tested for associations directly between insecticide phenotypes and Huang et al.’s (2015) DGRP transcriptome data. For malathion, 42 transcript associations were identified (P < 1.25×10−4; Table S7), one of which, Cyp6g1, was below the Bonferroni-corrected significance threshold (3.45×10−7), and was also the only transcript linked to malathion-associated genomic variants via eQTL. Notably, 10 of the 42 malathion candidate transcripts were among those found by Misra et al. (2011) to have their expression altered by twofold or more by ectopic expression of CncC. These include Cyp12d1, a cytochrome P450 enzyme that confers resistance to DDT and dicyclanil when overexpressed (Daborn et al. 2007), and Jheh1 and Jheh2, shown by Guio et al. (2014) to increase resistance to malathion when induced. In the case of permethrin, 11 transcript associations were identified (P < 2.5×10−4; Table S8), none of which were below the Bonferroni-corrected significance threshold (6.89×10−7). While Cyp6a17 transcript levels (of which three permethrin-associated variants are linked by eQTL and veQTL) do not reach significance at 2.5×10−4 in any of our transcriptome to phenotype association tests, they have a high rank in all phenotypes (male 3-hour knockdown rank = 26/18137, adjusted r2 = 0.060; female 3-hour knockdown rank = 4/18139, adjusted r2 = 0.064; male 24-hour mortality rank = 26/18137, adjusted r2 = 0.057; female 24-hour mortality rank = 26/18139, adjusted r2 = 0.066).
Structural variation in candidate genes
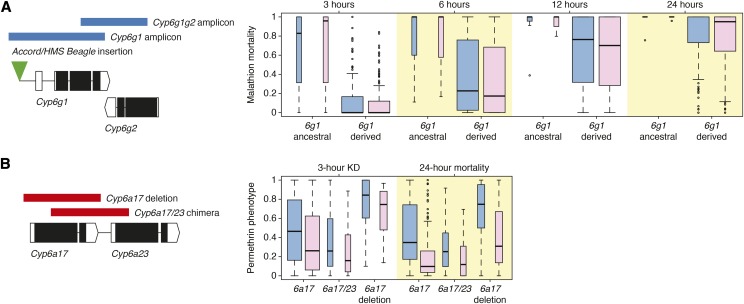
Structural variation among DGRP lines has previously been reported for association study candidates Cyp6g1, Cyp12d1, Jheh1/Jheh2 and Cyp6a17/Cyp6a23 (Zichner et al. 2013; Good et al. 2014; Guio et al. 2014; Figure 2, Figure S3). Of these structural variants, only Cyp6g1 is directly called in DGRP genotype data (2R_8072884_INS encodes the presence of the Accord transposable element insertion, present in all derived Cyp6g1 alleles). Therefore, the incidences of structural variants were manually tested for association against the relevant insecticide phenotype, and significant associations were found with Cyp6g1 derived alleles in the case of malathion, and the Cyp6a17 deletion allele in the case of permethrin (two tailed t-test assuming unequal variances, P < 0.05; Figure 2).
Figure 2.
Structural variation in candidate insecticide resistance genes. DGRP structural variation in insecticide resistance candidates (A) Cyp6g1 and (B) Cyp6a17 and Cyp6a23. Box plots show phenotype distributions among DGRP lines at each phenotype, grouped by structural variant allele. Blue plots represent males and pink plots represent females. Mean phenotypes for derived Cyp6g1 alleles and Cyp6a17 deletion alleles are significantly different from reference alleles in all relevant phenotypes (Table S9).
Discordant paired-end read mapping over the Cyp6g1 and Cyp6g2 loci shows that the two gene amplification events described in Schmidt et al. (2010) are present in the Cyp6g1-AA and Cyp6g1-BA alleles among DGRP lines. However, read depth across the region suggests substantial variation in copy number of both these amplicons, implying 1-5 copies of the Cyp6g1 amplicon, and 1-10 copies of the partial Cyp6g1g2 amplicon, which are correlated with transcript levels of both Cyp6g1 and Cyp6g2 (Figure S4).
Knockout of Cyp6g1 increases susceptibility to three organophosphate insecticides
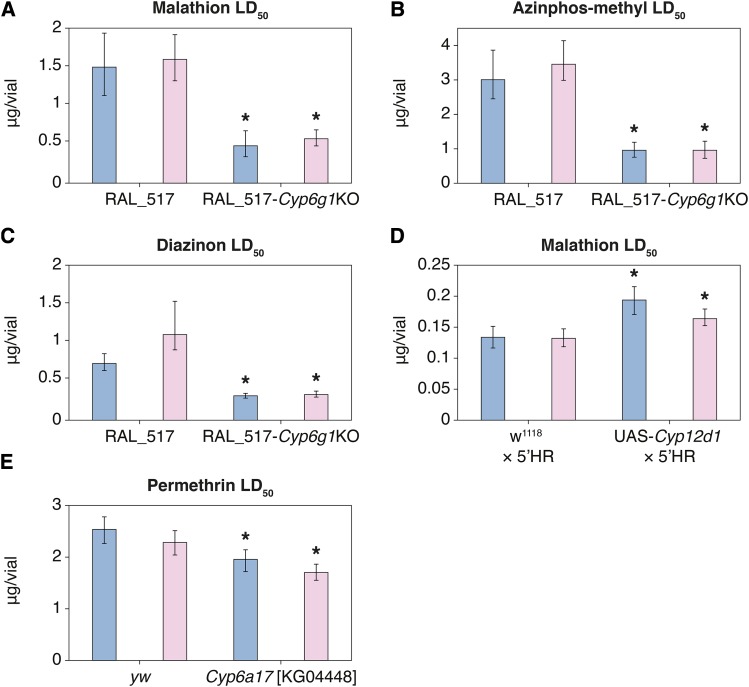
Natural variation at Cyp6g1 contributes to resistance to a range of insecticides including DDT (Schmidt et al. 2010), azinphos-methyl (Battlay et al. 2016) and imidacloprid (Denecke et al. 2017) and also ranks at the top of our genomic and transcriptomic association tests with malathion phenotypes. We verified Cyp6g1 involvement in malathion resistance using RAL_517-Cyp6g1-KO, a DGRP line in which the natural resistance allele Cyp6g1-BA is knocked out (Denecke et al. 2017). We observed a decrease in malathion median lethal dose (LD50) of approximately two thirds in both male and female RAL_517-Cyp6g1-KO flies when compared to unmodified RAL_517 flies (Figure 3A).
Figure 3.
Functional validation of insecticide resistance candidates. Insecticide LD50s for both male and female adults. Error bars represent 95% confidence interval. Blue bars represent males and pink bars represent females. CRISPR knockout of Cyp6g1 in the DGRP line RAL_517 background significantly increases susceptibility to organophosphate insecticides malathion, azinphos-methyl and diazinon. Transgenic overexpression of Cyp12d1 with the 6g1HR-GAL4 driver significantly increases resistance to malathion. Gene Disruption Project line Cyp6a17KG04448 shows increased permethrin susceptibility.
Battlay et al. (2016) previously demonstrated that Cyp6g1 overexpression, both transgenic (using GAL4-UAS overexpression) and among DGRP lines, was associated with resistance in larvae to the organophosphate azinphos-methyl. Pyke et al. (2004) mapped resistance to another organophosphate, diazinon, in an Australian natural population of D. melanogaster to a region containing Cyp6g1. Here we also present toxicological assays of RAL_517-Cyp6g1-KO and RAL_517 that demonstrate that the removal of the natural Cyp6g1-BA resistance allele from RAL_517 significantly reduces resistance to azinphos-methyl (Figure 3B) and diazinon (Figure 3C) in male and female adults.
Cyp12d1 overexpression increases malathion resistance
Increased expression of Cyp12d1 has previously been linked with resistance to the insecticides DDT and dicyclanil (Pedra et al. 2004; Daborn et al. 2007; Gellatly et al. 2015). In this study, we detected an association between male mortality at 24 hr and transcript level of Cyp12d1-p (adjusted r2 = 0.12; P = 4.80×10−6), one of the two copies of Cyp12d1 present in the y; cn bw sp; genome. Flies transgenically overexpressing Cyp12d1 had significantly higher malathion LD50s (∼30% and ∼20% increases for males and females respectively) than control crosses (Figure 3D).
Cyp6a17 disruption increases permethrin susceptibility
Top permethrin-associated genomic variants are eQTL of Cyp6a17, a locus at which two deletion variants exist in the DGRP. To test the hypothesis that Cyp6a17 contributes to permethrin resistance, we obtained a Gene Disruption Project line (Bellen et al. 2011), Cyp6a17KG04448, in which a P-element construct had been inserted into the coding region of Cyp6a17, early in the first exon. We found significantly reduced permethrin LD50s in both males and females from this line when compared to control flies (Figure 3E).
Discussion
In this study, the DGRP was assayed for resistance to malathion and permethrin, representatives of two of the most widely used insecticide classes, organophosphates and pyrethroids. Sexes were phenotyped separately and scored at multiple time points, increasing the resolution with which variants contributing to resistance could be identified. The majority of associations significant at the genome-wide association threshold are private to a single sex, including variants in and around a major malathion candidate, Cyp6g1, which were only significant in females at the three-hour mortality phenotype (Figure S1). Different sex and time point phenotypes may result in different associated candidates through two avenues: biological effects, like sexual dimorphism or a threshold exposure time required to elicit a response, (i.e., gene induction; Willoughby et al. 2006), and statistical effects that are consequences of skewed phenotypic distributions, whereby genes with low minor allele frequencies can be associated at more extreme phenotypes. Cyp6g1 appears to fall into the latter category, as the ancestral allele at this locus is rare (only present in nine DGRP lines; Battlay et al. 2016) and our transgenic results demonstrate that Cyp6g1 confers resistance to malathion and other organophosphates at a range of doses in both sexes (Figure 3A-C).
Evidence suggests that insecticides have played a large role in recent selection in D. melanogaster. Here, we find malathion-associated variants in four of the top 50 H12 selective sweep peaks identified in the DGRP by Garud et al. (2015). However only associations in the windows containing Cyp6g1 and Ace display distributions in haplotype structure congruent with the selective sweep windows containing them (Figure S2). There is strong evidence to support the hypothesis that both Cyp6g1 and Ace are the targets of selection in multiple populations including the ancestral population of the DGRP (Catania et al. 2004; Schmidt et al. 2010; Karasov et al. 2010; Garud et al. 2015; Garud and Petrov 2016), and the link to malathion resistance is now compelling.
Of the four substitutions in D. melanogaster Ace known to confer enzymatic insensitivity and hence resistance to organophosphate and carbamate insecticides (Menozzi et al. 2004), the three most common are present in DGRP lines (I199V, G303A and F368Y). Each of these three nonsynonymous sites independently achieved Bonferroni-significant associations with male and female malathion mortality at 6 hr. Only three combinations of these Ace alleles are present in the DGRP at moderate frequencies: The ancestral, susceptible Ace haplotype (Ace-IGF), and two resistant Ace substitution haplotypes (Ace-VGF (one substitution) and Ace-VAY (three substitutions). Ace-VGF and Ace-VAY enzymes have inhibitory constants of 6.4 and 32 to malaoxon (the activated form of malathion) respectively (Menozzi et al. 2004), and we find that DGRP population mean malathion mortalities for each haplotype corresponds to these relationships, suggesting that the previously characterized role of Ace resistance substitutions explains the strong malathion associations detected within Ace and the surrounding haplotype. In contrast, DGRP GWAS of resistance to the organophosphate azinphos-methyl did not detect strong associations with these alleles (Battlay et al. 2016). This is likely due to the lower inhibitory constants of these alleles to azinphos-methyl than malathion; Ace-VGF actually reduces the inhibitory constant to 0.92, and Ace-VAY only increases it to 4.8 (Menozzi et al. 2004).
Derived alleles of Cyp6g1 are associated, through both genomic and transcriptomic variation, with malathion resistance, and the link between malathion-associated genomic variants and Cyp6g1 expression is demonstrated by eQTL mapped by Huang et al. (2015; Table S4). This study adds to the mounting evidence that Cyp6g1 overexpression in wild populations confers resistance to multiple organophosphate insecticides (Kikkawa 1961; Pyke et al. 2004; Battlay et al. 2016), and that organophosphate selection may be a more likely explanation than DDT for the sweep observed at the Cyp6g1 locus (Schmidt et al. 2017). Moreover, we find that the DGRP harbors greater allelic diversity at Cyp6g1 than had previously been described at the locus, and that these additional structural variants, along with those previously characterized, are correlated with differences in transcription of Cyp6g1 and downstream Cyp6g2.
This work also implicates cytochrome P450s in resistance to permethrin. DGRP variants most strongly associated with permethrin map to a region on chromosome 2R containing nine P450 genes, with peaks over Cyp6a23 and Cyp317a1. Three of these variants are annotated by Huang et al. (2015) as eQTL and veQTL of Cyp6a23’s tandem paralog, Cyp6a17, and structural variation in the DGRP has previously been described involving Cyp6a17 and Cyp6a23 (Zichner et al. 2013; Good et al. 2014). Two deletions in this region are present among DGRP lines, one creates a single chimeric gene comprised of Cyp6a17 and Cyp6a23 sequence. In the other, Cyp6a17 is deleted, save for a small section which exists as a gene conversion in the otherwise intact Cyp6a23. Due to the homology between these genes, this gene conversion introduces only four nucleotide changes and a single non-synonymous substitution in Cyp6a23 (Good et al. 2014). Among DGRP lines, it is this deletion of Cyp6a17 that is associated with increased susceptibility to permethrin (Figure 2B). This is congruent with the finding that when Cyp6a17 is disrupted in Cyp6a17KG04448, it leads to relative susceptibility (Figure 3E). It is worth noting that the susceptible allele appears to be the derived state, as the duplication leading to the original divergence Cyp6a17 and Cyp6a23 seems to be at least as old as the divergence between D. melanogaster and D. ananassae (Good et al. 2014).
The transcription level of another P450, Cyp12d1-p, was associated with male 24-hour malathion mortality, and transgenic overexpression of the gene confers malathion resistance in both male and female adults (Figure 3D). Interestingly, we do not observe a strong association at either the genomic level (Table S9) or the transcriptome level (Table S6) with Cyp12d1-d, present as a duplicated paralog in 24% of DGRP lines. Coding (three amino acids differentiate Cyp12d1-p and Cyp12d1-d in the reference genome) or expression pattern differences between the genes may explain this observation. Alternatively, Cyp12d1-p’s significance may be inflated by its strong correlation with a group of co-regulated genes that are induced by oxidative stress. We found that ten of the top malathion-associated transcripts are among those known to be regulated by CncC (Misra et al. 2011).
Two more CncC-activated transcripts associated with malathion phenotypes are Jheh1 and Jheh2. Guio et al. (2014) demonstrated that the insertion of Bari1 upstream of Jheh1 and Jheh2 increases the inducibility of these genes in response to oxidative stress, which results in an increased resistance to malathion. In this study, we found associations between constitutive transcript levels of Jheh1 and Jheh2 and male malathion mortality at 24 hr (adjusted r2 = 0.10; P = 4.52×10−5) and 3 hr (adjusted r2 = 0.10; P = 4.62×10−5) respectively. However, we did not find that the presence of the Bari-Jheh insertion was significantly associated with malathion mortality at any of our four time points, for either sex (Figure S3A; Table S9). This is plausibly due to differences in the exposure times between our study (up to 24 hr) and the assays used in Guio et al. (2014; up to 214 hr). It makes sense that in our more acute assays, the baseline expression level of expression of Jheh1 and Jheh2 would be important, whereas over longer assay periods induction capacity would play a more important role.
An important question in DGRP insecticide resistance studies is the applicability of findings in this subset of variation to populations worldwide. Ace and Cyp6g1 resistance alleles have been identified in a range of populations, as have the footprints of their selection (Catania et al. 2004; Karasov, Messer and Petrov 2010; Schmidt et al. 2010; Garud and Petrov 2016). This is further reinforced by interrogation of the Drosophila Genome Nexus (DGN; Lack et al. 2016), which reveals Ace and Cyp6g1 allele frequencies comparable to the DGRP in many populations around the world (Table S10). The structural variants at Jheh1/2/3, Cyp12d1 and Cyp6a17/23 have also been described in populations outside the DGRP (González, Macpherson and Petrov 2009; Kolaczkowski et al. 2011; Chakraborty et al. 2018).
In this investigation of insecticide resistance in the DGRP using an organophosphate and a pyrethroid insecticide we saw stark differences in the genes involved as well as the evidence for a selective response to the compounds. The top candidates from malathion GWAS correlate with peaks of selection in the DGRP population, making organophosphate resistance a highly credible selective pressure on the population ancestral to the DGRP. Conversely, Cyp6a17, our top permethrin candidate, does not lie within a H12 selective sweep peak, nor would we expect the allele described to be the target of positive insecticide-based selection, given it increases susceptibility to permethrin. However, Cyp6a17 ranks fourth among D. melanogaster P450s (after Cyp6a13, Cyp6a2, and Cyp6a14) in similarity to malaria vector Anopheles funestus CYP6P9a and CYP6P9b. Naturally occurring duplications of each of these genes are associated with pyrethroid resistance in A. funestus (Wondji et al. 2009), and selective sweeps have been described at CYP6P9a in response to pyrethroid-based malaria interventions (Barnes et al. 2017).
An emerging picture of insecticide resistance, informed by results from DGRP studies as well as investigations in pest insect species (Joußen et al. 2012; Faucon et al. 2015) is that complex structural variation and high allelic diversity, along with selective sweep signatures, are common in genes contributing to resistance.
Acknowledgments
Josefa González, Phil Batterham/Batterham lab (stocks), Trudy Mackay, NECTAR computational services. We also thank Owain Edwards for discussion and the CSIRO for scholarship support of PB.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6954443.
Communicating editor: R. Kulathinal
Literature Cited
- Antonov A. V., 2011. BioProfiling. de: analytical web portal for high-throughput cell biology. Nucleic acids research, 39, W323–W327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K. G., Weedall G. D., Ndula M., Irving H., Mzihalowa T., et al. , 2017. Genomic footprints of selective sweeps from metabolic resistance to pyrethroids in African malaria vectors are driven by scale up of insecticide-based vector control. PLoS Genet. 13: e1006539 10.1371/journal.pgen.1006539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battlay P., Schmidt J. M., Fournier-Level A., Robin C., 2016. Genomic and Transcriptomic Associations Identify a New Insecticide Resistance Phenotype for the Selective Sweep at the Cyp6g1 Locus of Drosophila melanogaster. G3: Genes| Genomes| Genetics, 6(8), 2573–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743. 10.1534/genetics.111.126995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Catania F., Kauer M. O., Daborn P. J., Yen J. L., Ffrench-Constant R. H., et al. , 2004. World‐wide survey of an Accord insertion and its association with DDT resistance in Drosophila melanogaster. Mol. Ecol. 13: 2491–2504. 10.1111/j.1365-294X.2004.02263.x [DOI] [PubMed] [Google Scholar]
- Chakraborty M., VanKuren N. W., Zhao R., Zhang X., Kalsow S., et al. , 2018. Hidden genetic variation shapes the structure of functional elements in Drosophila. Nat. Genet. 50: 20–25. 10.1038/s41588-017-0010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Bogwitz M. R., McCart C., Andrianopoulos A., Batterham P., et al. , 2007. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175: 1071–1077. 10.1534/genetics.106.066597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P., Boundy S., Yen J., Pittendrigh B., ffrench-Constant R., 2001. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genomics 266: 556–563. 10.1007/s004380100531 [DOI] [PubMed] [Google Scholar]
- Daborn P. J., Yen J. L., Bogwitz M. R., Le Goff G., Feil E., 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. 10.1126/science.1074170 [DOI] [PubMed] [Google Scholar]
- Daborn P. J., Lumb C., Boey A., Wong W., Batterham P., 2007. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem. Mol. Biol. 37: 512–519. 10.1016/j.ibmb.2007.02.008 [DOI] [PubMed] [Google Scholar]
- Denecke S., Fusetto R., Martelli F., Giang A., Battlay P., et al. , 2017. Multiple P450s and Variation in Neuronal Genes Underpins the Response to the Insecticide Imidacloprid in a Population of Drosophila melanogaster. Sci. Rep. 7: 11338 10.1038/s41598-017-11092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K., Du Y., Rinkevich F., Nomura Y., Xu P., et al. , 2014. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 50: 1–17. 10.1016/j.ibmb.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucon F., Dusfour I., Gaude T., Navratil V., Boyer F., et al. , 2015. Unravelling genomic changes associated with insecticide resistance in the dengue mosquito Aedes aegypti by deep targeted sequencing. Genome Res. 25: 1347–1359. 10.1101/gr.189225.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney D. J., 1971. Probit Analysis, Ed. 3rd Cambridge University Press, Cambridge, UK. [Google Scholar]
- Fiston-Lavier A. S., Carrigan M., Petrov D. A., Gonzalez J., 2010. T-lex: a program for fast and accurate assessment of transposable element presence using next-generation sequencing data. Nucleic acids research 39: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud N. R., Petrov D. A., 2016. Elevated linkage disequilibrium and signatures of soft sweeps are common in Drosophila melanogaster. Genetics 203: 863–880. 10.1534/genetics.115.184002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly K. J., Yoon K. S., Doherty J. J., Sun W., Pittendrigh B. R., et al. , 2015. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic. Biochem. Physiol. 121: 107–115. 10.1016/j.pestbp.2015.01.001 [DOI] [PubMed] [Google Scholar]
- González J., Macpherson J. M., Petrov D. A., 2009. A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol. Biol. Evol. 26: 1949–1961. 10.1093/molbev/msp107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good R. T., Gramzow L., Battlay P., Sztal T., Batterham P., et al. , 2014. The molecular evolution of cytochrome P450 genes within and between Drosophila species. Genome Biol. Evol. 6: 1118–1134. 10.1093/gbe/evu083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guio L., Barrón M. G., González J., 2014. The transposable element Bari‐Jheh mediates oxidative stress response in Drosophila. Mol. Ecol. 23: 2020–2030. 10.1111/mec.12711 [DOI] [PubMed] [Google Scholar]
- Hoi K. K., Daborn P. J., Battlay P., Robin C., Batterham P., et al. , 2014. Dissecting the insect metabolic machinery using twin ion mass spectrometry: a single P450 enzyme metabolizing the insecticide imidacloprid in vivo. Anal. Chem. 86: 3525–3532. 10.1021/ac404188g [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R., et al, 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proceedings of the National Academy of Sciences, 112(44), E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. M., Dahlgren L., Siegfried B. D., Ellis M. D., 2013. Acaricide, fungicide and drug interactions in honey bees (Apis mellifera). PLoS One 8: e54092 10.1371/journal.pone.0054092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joußen N., Heckel D. G., Haas M., Schuphan I., Schmidt B., 2008. Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1‐overexpressing strains of Drosophila melanogaster, leading to resistance. Pest Manag. Sci. 64: 65–73. 10.1002/ps.1472 [DOI] [PubMed] [Google Scholar]
- Joußen N., Agnolet S., Lorenz S., Schöne S. E., Ellinger R., et al. , 2012. Resistance of Australian Helicoverpa armigera to fenvalerate is due to the chimeric P450 enzyme CYP337B3. Proceedings of the National Academy of Sciences, 201202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T., Messer P. W., Petrov D. A., 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6: e1000924 10.1371/journal.pgen.1000924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa H., 1961. Genetical studies on the resistance to parathion in Drosophila melanogaster. Annu Rep Sci Wks Osaka Univ 9: 1–20. [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack J. B., Lange J. D., Tang A. D., Corbett-Detig R. B., Pool J. E., 2016. A thousand fly genomes: an expanded drosophila genome nexus. Mol. Biol. Evol. 33: 3308–3313. 10.1093/molbev/msw195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff G., Boundy S., Daborn P. J., Yen J. L., Sofer L., et al. , 2003. Microarray analysis of cytochrome P450 mediated insecticide resistance in Drosophila. Insect Biochem. Mol. Biol. 33: 701–708. 10.1016/S0965-1748(03)00064-X [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi P., Shi M. A., Lougarre A., Tang Z. H., Fournier D., 2004. Mutations of acetylcholinesterase which confer insecticide resistance in Drosophila melanogaster populations. BMC Evol. Biol. 4: 4 10.1186/1471-2148-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R., Horner M. A., Lam G., Thummel C. S., 2011. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25: 1796–1806. 10.1101/gad.17280911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R., Lam G., Thummel C. S., 2013. Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect Biochem. Mol. Biol. 43: 1116–1124. 10.1016/j.ibmb.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutero A., Pralavorio M., Bride J. M., Fournier D., 1994. Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA 91: 5922–5926. 10.1073/pnas.91.13.5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita Z., 1958. A new type of insecticide. Nature 182: 1529–1530. 10.1038/1821529b0 [DOI] [PubMed] [Google Scholar]
- Pedra J. H. F., McIntyre L. M., Scharf M. E., Pittendrigh B. R., 2004. Genome-wide transcription profile of field-and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA 101: 7034–7039. 10.1073/pnas.0400580101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh B., Reenan R., Ganetzky B., 1997. Point mutations in the Drosophila sodium channel gene para associated with resistance to DDT and pyrethroid insecticides. Molecular and General Genetics MGG 256: 602–610. 10.1007/s004380050608 [DOI] [PubMed] [Google Scholar]
- Pyke F. M., Bogwitz M. R., Perry T., Monk A., Batterham P., et al. , 2004. The genetic basis of resistance to diazinon in natural populations of Drosophila melanogaster. Genetica 121: 13–24. 10.1023/B:GENE.0000019920.71944.2b [DOI] [PubMed] [Google Scholar]
- R Core Team 2013. R: A language and environment for statistical computing.
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. biotech., 29: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998 10.1371/journal.pgen.1000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Battlay P., Gledhill-Smith R. S., Good R. T., Lumb C., et al. , 2017. Insights into DDT Resistance from the Drosophila melanogaster Genetic Reference Panel. Genetics 207: 1181–1193. 10.1534/genetics.117.300310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor S. A., New F. N., Nuzhdin S., 2017. A large panel of Drosophila simulans reveals an abundance of common variants. Genome Biol. Evol. 10: 189–206. 10.1093/gbe/evx262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhzaz S., Cabrero P., Brinzer R. A., Halberg K. A., Dow J. A., et al. , 2015. A novel role of Drosophila cytochrome P450–4e3 in permethrin insecticide tolerance. Insect Biochem. Mol. Biol. 67: 38–46. 10.1016/j.ibmb.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby L., Chung H., Lumb C., Robin C., Batterham P., et al. , 2006. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem. Mol. Biol. 36: 934–942. 10.1016/j.ibmb.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Wondji C. S., Irving H., Morgan J., Lobo N. F., Collins F. H., et al. , 2009. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 19: 452–459. 10.1101/gr.087916.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zichner T., Garfield D. A., Rausch T., Stütz A. M., Cannavó E., et al. , 2013. Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res. 23: 568–579. 10.1101/gr.142646.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DGRP strains are available from Bloomington Stock center. All data are reported in the manuscript or in the associated supplementary material. DGRP phenotypes and raw Mackay pipeline outputs for each phenotype are supplied in the supplementary data file. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6954443.