Abstract
A majority of traits are determined by multiple quantitative trait loci (QTL) that can have pleiotropic effects. A multi-parent advanced generation inter-cross (MAGIC) population is well suited for genetically analyzing the effects of multiple QTL on traits of interest because it contains a higher number of QTL alleles than a biparental population. We previously produced the JAPAN-MAGIC (JAM) population, derived from eight rice (Oryza sativa L.) cultivars with high yield and biomass in Japan, and developed the method of genome-wide association study (GWAS) using haplotype information on the JAM lines. This method was effective for identifying major genes such as Waxy for eating quality and Sd1 for culm length. Here, we show that haplotype-based GWAS is also effective for the evaluation of multiple QTL with small effects on rice grain shape in the JAM lines. Although both the haplotype- and SNP-based GWAS identified multiple QTL for grain length and width, the sum of the estimated trait values of each allele for the QTL detected by haplotype-based GWAS had higher correlation with observed values than those detected by SNP-based GWAS, indicating high-accuracy QTL detection in the haplotype-based GWAS. Furthermore, the study revealed pleiotropic effects of some QTL regions in regulation of grain shape, suggesting that the haplotype-based GWAS using the JAM lines is an effective means to evaluate the main and side effects of haplotypes at each QTL. Information on the pleiotropic effects of haplotypes on various traits will be useful for designing ideal lines in a breeding program.
Keywords: GWAS, haplotype, QTL, SNP, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
Rice is a major crop, especially in Asia, and is a staple food for billions of people in the world. Rice cultivars have been improved by breeding for thousands of years, today with the aid of DNA marker-assisted selection (Collard et al. 2008). To identify desirable genes for introduction, researchers have searched for DNA markers linked to the causative genes of agriculturally important traits, e.g., Waxy for eating and cooking quality (Zhou et al. 2003), SUB1A for submergence tolerance (Xu et al. 2006), Xa1 for bacterial leaf blight resistance (Yoshimura et al. 1998), and pi21 for blast resistance (Fukuoka et al. 2009). These DNA markers have been developed through genetic analysis using biparental populations owing to their simple genetic structure.
Genome-wide association study (GWAS) is a powerful method to find QTL associated with target traits. Currently, GWAS is being performed using groups of cultivars and accessions as populations that are more genetically diverse than the biparental populations typically used in animal and plant breeding (Han and Huang 2013; Visscher et al. 2017). In rice, GWAS using a selected population consisting of 176 cultivars uncovered genes controlling agronomic traits such as days to heading and panicle length and number (Yano et al. 2016). However, GWAS is often problematic because complicated population structures hamper accurate detection of allelic effects on traits. One of the reasons for the success of Yano et al.’s study was careful selection of rice varieties that were not genetically highly structured or interrelated but had phenotypic diversity. Thus, there are bottlenecks restricting the elucidation of QTL alleles in GWAS when using genetically diverse populations, which we call “uncontrolled populations” in this paper.
Multi-parental inter-mated populations were first shown in mouse to enable analysis of higher numbers of genetic variations, which in turn give rise to wider phenotypic distributions (Churchill et al. 2004). In plants, the multi-parental inter-mated lines are generally referred to as multi-parent advanced generation inter-cross (MAGIC) lines. MAGIC lines have been developed in a number of plants including Arabidopsis thaliana (Kover et al. 2009), tomato (Pascual et al. 2015), faba (Sallam and Martsch 2015), cotton (Islam et al. 2016), wheat (Huang et al. 2012; Mackay et al. 2014), barley (Sannemann et al. 2015), maize (Dell’Acqua et al. 2015), sorghum (Ongom and Ejeta 2018), and rice (Bandillo et al. 2013; Meng et al. 2016a; Meng et al. 2016b; Ogawa et al. 2018). MAGIC populations are intermediate between biparental populations and uncontrolled populations in terms of genetic complexity and diversity. The population structure of MAGIC lines is not as complicated as that of uncontrolled populations because there are fewer alleles and recombination events. Genetic analyses of the MAGIC populations require a high density of DNA polymorphisms to distinguish chromosomal segments with low linkage disequilibrium derived from multiple parents, but recent progress in DNA sequencing technology facilitates the identification of high numbers of SNPs and insertion/deletion mutations. A MAGIC population derived from 19 Arabidopsis thaliana accessions was analyzed at the level of haplotype, which is defined as a chromosomal segment from a founder (Kover et al. 2009). The program for haplotype estimation is freely available. These tools facilitate GWAS using newly developed MAGIC populations to examine genetic variations for traits of interest.
We produced the JAPAN-MAGIC (JAM) population by using four japonica cultivars and four indica cultivars that have high grain yield and biomass when cultivated in Japan (Ogawa et al. 2018). In the process of developing the JAM population using the single-seed descent (SSD) method, lines were randomly selected from among those that produced any amount of seed. Sterility caused by hybridization between japonica and indica cultivars can lead to distortion of the founder genome proportions in the resulting population, but in this study the proportions of haplotypes among the eight founders were almost equal, at least at the whole-genome level, to their proportions in the JAM lines (Ogawa et al. 2018). This finding indicates that the JAM lines are suitable for evaluation of alleles from the eight founders. The JAM population shows higher phenotypic diversity than a set of four biparental recombinant inbred lines (RILs) produced from the same eight parents used to generate the JAM lines. GWAS using haplotype data enabled us to determine precise positions of QTL and to discriminate multiple alleles for major genes such as Waxy and Sd1 (Ogawa et al. 2018). This result indicated that haplotype-based GWAS can be used to detect genes with major effects. However, it was still unknown whether haplotype-based GWAS was effective for analyzing traits controlled by multiple factors.
Genetic studies of QTL using various populations mainly focus on effects of QTL on expression of specific target traits, but QTL often have pleiotropic effects. In rice, qPE9-1, detected as a QTL for panicle erectness, also affects plant height and panicle length (Yan et al. 2007). GHD8/DTH8, detected as a QTL for heading date, affects the CO2 assimilation rate in photosynthesis, plant height, and grain yield (Adachi et al. 2017; Yan et al. 2011). To produce ideal lines through DNA marker-assisted selection using QTL information, we have to know not only the main effects of QTL for target traits but also their side effects, which may be detrimental to crop performance. From this aspect, the pleiotropic effects of QTL should be paid more attention in molecular breeding.
In this study, we applied the haplotype-based GWAS method to the JAM lines to identify QTL associated with grain shape, which is known to be regulated by many genetic factors (Li et al. 2018; Nagata et al. 2015; Zuo and Li 2014). By comparing this method with ordinary SNP-based GWAS, we validated the accuracy of the haplotype-based GWAS method. Furthermore, we focused on the pleiotropic effects (both main and side effects) of the QTL for grain shape, which were detected in the haplotype-based GWAS using the JAM lines. This analysis shed light on the different effects of the QTL in the regulation of grain shape.
Materials and Methods
Development of the JAM population
As described in a previous paper (Ogawa et al. 2018), the JAM population was developed from eight founders: Ruriaoba (RU), Hokuriku 193 (HO), Takanari (TK), Suweon 258 (SU), Mizuhochikara (MI), Bekogonomi (BE), Tachiaoba (TC), and Akidawara (AK). On the basis of variant analysis of all annotated genes using SnpEff software (Cingolani et al. 2012), the first four listed cultivars were identified as indica and the second four as japonica (Ogawa et al. 2018). This result is consistent with a phylogenetic analysis of 126 rice accessions, including five of the founders of the JAM population, which was performed using 1046 genome-wide SNP markers in our previous paper (Yonemaru et al. 2014). We first crossed each indica cultivar with a different japonica cultivar to produce four types of hybrid seed. These hybrids were crossed to produce four-way recombinants and then crossed again to produce eight-way recombinants. We originally planned to produce 1,000 JAM lines (F2) by developing 10 lines from each of 100 eight-way recombinants. In fact, we obtained F6 seeds from 981 JAM lines through the SSD method and used 372 JAM lines (F5) for this study.
Cultivation of JAM lines
Germinated seeds were sown in trays filled with soil on May 9, 2016 and incubated at 30° in the dark for 2 days. Seedlings were grown in a paddy field (Kannondai District, Tsukuba, Japan) covered with thin plastic film to protect the plants from the cold. Individual seedlings were transferred to another paddy field on June 8, 2016 and cultivated according to standard procedures at the National Agricultural Research Organization (NARO) in Tsukuba.
Analysis of grain shape
To collect rice grains from panicles, the panicle part containing the grains was put into a glass Petri dish, with the panicle base remaining outside of the dish. The grains were removed by using the lid of the Petri dish to compress the panicle against the bottom edge of the dish and pulling the panicle base away from the dish, thus leaving the grains inside the dish. Awns and pedicels were removed by rubbing the grains in a bag and then by vacuum. Any awns and pedicels that remained were cut off with scissors. We put grains of each JAM line directly on the glass of a scanner (Seiko Epson, Suwa City, Japan) and aligned them to avoid contact between adjacent grains. We scanned the grains (600 dpi) and analyzed the images (0.042433 mm/pixel) using the SmartGrain software program (Tanabata et al. 2012), which can automatically detect every single grain in an image and evaluate its length, width, and area. The number of grains per plant ranged from 11 to 628 in the 372 JAM lines. The averages of grain length, width, and area of the JAM lines were used for GWAS.
GWAS and Sum of the Additive effects of QTL (SAQ)
SNP and haplotype data for the JAM lines were obtained in our previous work (Ogawa et al. 2018). A total of 16,345 SNP locations were detected by GBS analysis of the 376 JAM lines. The haplotype data for the JAM lines were predicted by using the method proposed for haplotype prediction in an Arabidopsis MAGIC population (Kover et al. 2009). All parameters were set to their defaults, except “-p”, which was set to 2.
SNP-based GWAS was performed using R software (https://www.r-project.org/). Associations of the imputed genotype data with phenotype data for grain length and width were analyzed with the General Linear Model (GLM) and Mixed Linear Model (MLM). The GLM was applied for the naïve (population structure not considered) and Q (population structure results included as fixed effects) analyses. Population structure was determined by principal component analysis (PCA) using the R function “prcomp”. The number of principal components (PCs) was set as 6 because the variance of PC score decreased gradually thereafter (Figure S1). The K (kinship results included as fixed effects) and QK models were analyzed with MLM (Yu et al. 2006) implemented in the “GWAS” function of the “rrBLUP” package (Endelman 2011). The kinship data were computed by using the “A.mat” function of “rrBLUP.”
Haplotype-based GWAS was performed by using haplotype information at each of the SNPs. To distinguish the newly identified QTL from previously identified QTL for rice grain shape, we named the newly identified QTL as JAM-GL for grain length and as JAM-GW for grain width. Both JAM-GL and JAM-GW, which were identified by using haplotype-based GWAS, were analyzed with non-parametric ANOVA (Kruskal–Wallis rank sum test) using the R function “kruskal.test”.
Peaks of the 5, 10, 20, and 30 highest −log10 P values in the results of both SNP-based and haplotype-based GWAS analyses were detected using the “findpeaks” function (minpeakdistance = 100) in the “pracma” package, and the SNPs associated with the peaks were defined as the QTL. Estimates of the additive effects of these QTL were calculated by the means of grain length and width for each SNP allele or haplotype using the R function “tapply”, and the sum of the additive effects of QTL (SAQ) was calculated.
Scatter plots were generated using the “ggplot2” package in the R software. These plots were used to show correlations between SAQ and observed values, and to illustrate the effects of haplotype at each QTL position on grain length and width.
Sequences of GW2 alleles
The sequences of GW2 alleles in the founders were determined using short-read Illumina resequencing data (Ogawa et al. 2018). The mutations in these alleles were analyzed for their effects on gene function in SnpEff software (Cingolani et al. 2012), which provides the number of variations that belong to one of four categories (variants_impact_high, _moderate, _low, or _modifier) according to their putative effect. Allele classification was performed as reported previously (Ogawa et al. 2018).
Data availability
Germplasm of JAM lines and founders, and their haplotypes and phenotypes, are available by request. They are offered through a material transfer agreement (MTA) from NARO via Daisuke.Ogawa@affrc.go.jp. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7027991.
Results
The JAM lines have phenotypic diversity for grain shape
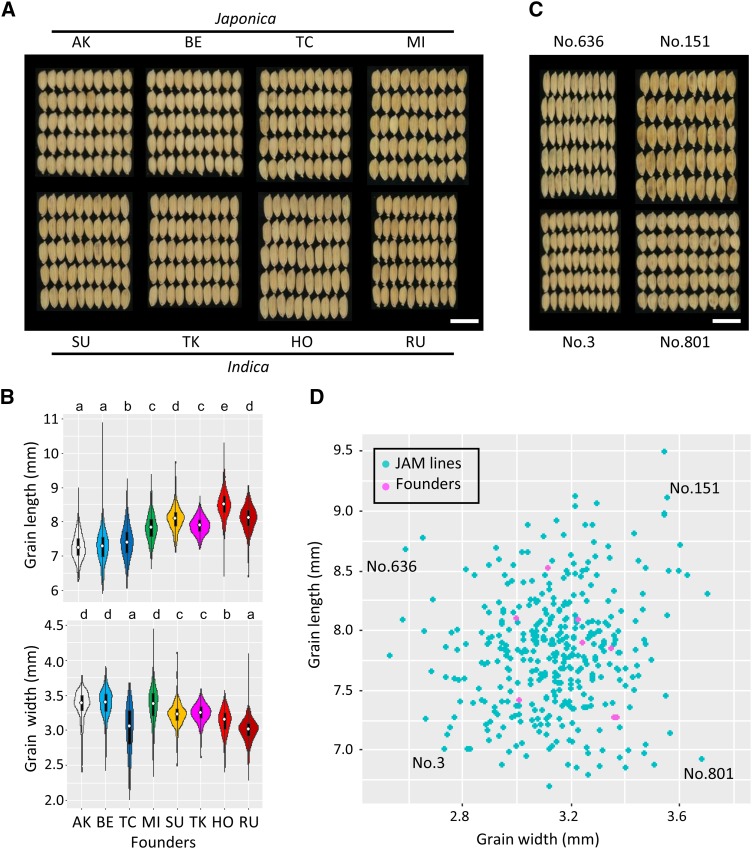
The eight founders of the JAM lines include four japonica cultivars and four indica cultivars. The grain of japonica cultivars is generally more round and that of indica more slender (Misra et al. 2017; Nagata et al. 2015). This trend could be seen in the founders: for example, the grain shape of japonica founders AK and BE was significantly shorter and wider than that of the indica founders, whereas the grain of indica founders HO, SU, and RU was significantly longer and narrower than all but one of the japonica founders (Figure 1A and B).
Figure 1.
Grain length and width of the eight founders and JAM lines. (A) Alignments of 50 grains of each of the four japonica cultivars (Akidawara [AK], Bekogonomi [BE], Tachiaoba [TC], and Mizuhochikara [MI]) and four indica cultivars (Suweon 258 [SU], Takanari [TK], Hokuriku 193 [HO], and Ruriaoba [RU]) used as founders. Pictures of the grain of each founder were taken individually and then merged. (B) Violin plots of the grain length and width of the founders. They include information on kernel density estimation, quantiles (black boxes), and median values (white circles). Different letters indicate significant differences (Tukey method, P < 0.05). (C) Alignments of 50 grains each of four JAM lines showing characteristic features. (D) Scatter plot of average grain length and width for each JAM line and the eight founders. White bars indicate 1 cm.
To understand the phenotypic distribution of grain shape in the 372 JAM lines, we evaluated their grain length and width (Figure 1C and D). The grain length ranged from 6.7 to 9.5 mm and grain width from 2.5 to 3.7 mm. The distributions of both grain length and grain width were continuous, and the two characteristics were not obviously correlated. Compared to the eight founders, JAM lines with longer, shorter, narrower, and wider grain were found, indicating transgressive segregation. This result is consistent with the involvement of multiple genetic factors in grain shape. We also checked the grain areas of the JAM lines. Grain area was highly correlated with both grain length and grain width (Figure S2), suggesting that grain length and width contribute to grain area almost equally.
Haplotype-based GWAS using the JAM lines is useful for detection of QTL for grain shape
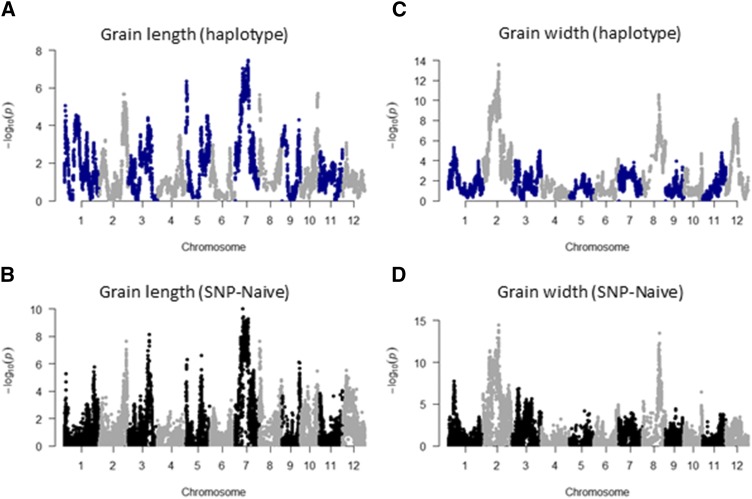
To clarify what factors regulate grain shape, we carried out haplotype-based GWAS and SNP-based GWAS with a naïve model (Figures 2 and S3). Both analyses identified several QTL for which the −log10 P value was more than 5, indicating that grain shape is a complex trait regulated by several factors. We also performed SNP-based GWAS with a Q model for considering population structure, K model for considering kinship, and QK model for considering both (Figure S4). The plot pattern of the SNP-based GWAS with the Q, K, and QK models was substantially different from that of SNP-based GWAS with the naïve model and the haplotype-based GWAS, confirming that the result of GWAS is changed by consideration of the population structure and kinship. However, what method is proper for the GWAS to detect QTL for the trait of grain shape is unknown.
Figure 2.
Manhattan plots of GWAS on grain length and width measurements. GWAS was carried out for (A, B) grain length and (C, D) width using (A, C) haplotype data and (B, D) SNP data. The naïve model was used for the SNP-based GWAS.
To answer the question, we investigated the accuracy of QTL detection by checking the correlation between SAQ on grain length and width and the actual values in each JAM line. We determined the SNP positions of QTL using the software program “findpeaks” and chose the top 5, 10, 20, and 30 QTL positions (Tables S1 and S2). For both traits, the correlations between QTL additive effects and observed values in haplotype-based GWAS tended to be higher than those for SNP-based GWAS with the naïve, Q, K, and QK models (Tables 1 and 2). Especially for the top 5 QTL, the pattern of correlation in the haplotype-based GWAS was remarkably different from the others, i.e., the data points were aligned less vertically (Figures S5 and S6). That might be attributable to the difference in the number of possibilities between haplotype-based and SNP-based GWAS: for 5 QTL, the former is 32,768 (85) because there are 8 possible haplotypes per locus and the latter is 32 (25) because there are 2 possible SNP alleles per locus. These results suggest that haplotype-based GWAS accurately detects the positions of QTL for grain shape.
Table 1. Pearson correlations between the observed values of grain length in the JAM lines and the sum of the additive effects of QTL (SAQ) obtained from the five models tested.
| Number of QTL | Haplotype-based model | Naïve model | Q model | K model | QK model |
|---|---|---|---|---|---|
| 5 | 0.53 | 0.46 | 0.39 | 0.44 | 0.45 |
| 10 | 0.60 | 0.61 | 0.49 | 0.59 | 0.54 |
| 20 | 0.71 | 0.66 | 0.62 | 0.70 | 0.65 |
| 30 | 0.76 | 0.69 | 0.72 | 0.75 | 0.74 |
All of the correlation values in the five models are significant (P < 0.001).
Table 2. Pearson correlations between the observed values of grain width in the JAM lines and the sum of the additive effects of QTL (SAQ) obtained from the five models tested.
| Number of QTL | Haplotype-based model | Naïve model | Q model | K model | QK model |
|---|---|---|---|---|---|
| 5 | 0.56 | 0.49 | 0.50 | 0.55 | 0.47 |
| 10 | 0.66 | 0.58 | 0.60 | 0.63 | 0.50 |
| 20 | 0.72 | 0.68 | 0.65 | 0.72 | 0.67 |
| 30 | 0.76 | 0.70 | 0.70 | 0.76 | 0.73 |
All of the correlation values in the five models are significant (P < 0.001).
The haplotype-based GWAS of grain length and width uncovered the main and side effects of the QTL
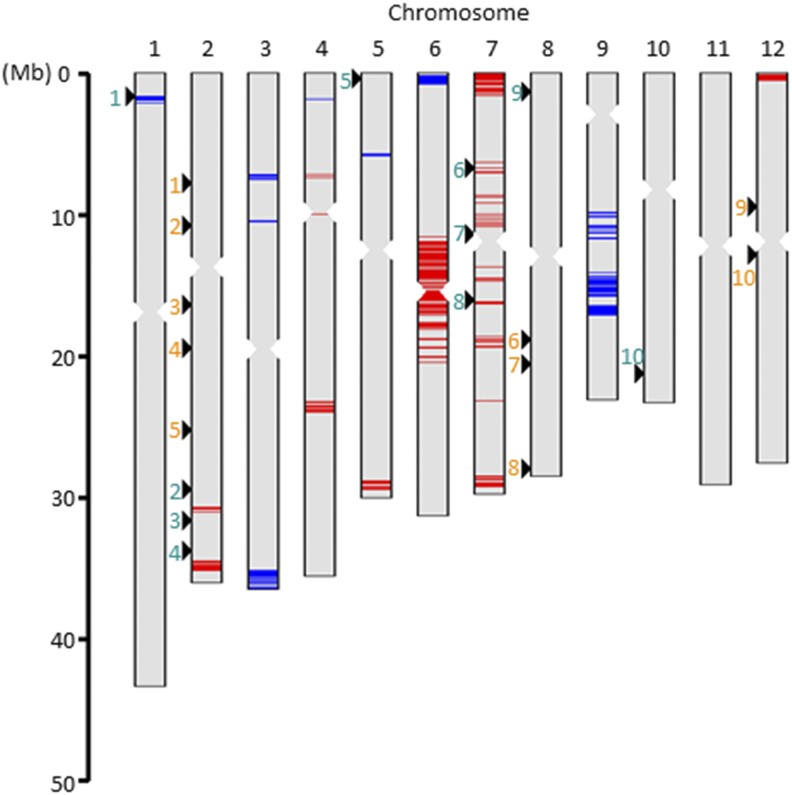
Genetic studies generally focus on the main effects of QTL (i.e., those affecting only the target trait) but not their influence on other traits (here called side effects), despite the known existence of pleiotropic effects of QTL. We asked whether QTL for grain length affect grain width and vice versa. We plotted averages of grain length and width in haplotypes at the positions of the top 10 QTL for grain length and width detected in the haplotype-based GWAS (designated JAM-GL1 to -GL10 and JAM-GW1 to -GW10; Figures S3, S7, and S8; Tables S3 and S4). In addition, we calculated the Pearson correlation values between grain length and width for the entire set of SNPs (Figure 3). This analysis revealed both negative and positive correlations between grain length and width among haplotype effects at the position of JAM-GL1 and in the regions of JAM-GL6 to -GL8, respectively. In other words, some genome positions with major QTL affecting one grain shape trait also had effects on the other trait. This result implies that haplotype-based GWAS using the JAM lines has the ability to reveal haplotype effects both on the target trait and on other traits at the location of each QTL.
Figure 3.
Chromosomal positions of QTL and haplotypes with effects on grain length and width. Arrowheads indicate the QTL positions of JAM-GL1 to -GL10 (cyan) and JAM-GW1 to -GW10 (orange). Horizontal lines on the chromosomes indicate regions where haplotypes have correlated effects on grain length and width. Red and blue indicate positive and negative Pearson correlations (P < 0.05), respectively.
The regions of JAM-GLs and JAM-GWs identified by haplotype-based GWAS partially overlapped those of previously detected QTL
We examined whether the QTL identified in this study overlapped with those previously detected in a genetic study of grain shape using biparental RILs of japonica and indica cultivars (Nagata et al. 2015). A total of 6 out of the top 20 QTL identified in the present study overlapped with those previously detected by Nagata et al. (Table S5). In that study, QTL were detected in a BC4F2 population produced by crosses between japonica cultivar Koshihikari and indica cultivar IR-64 (Nagata et al. 2015). This finding indicates that these QTL may be positioned as common QTL for variation of grain shape in japonica and indica.
We searched for candidate genes for the QTL by using haplotype data and sequence information for the eight founders. Intriguingly, most of the QTL detected in this study did not seem to correspond to the previously identified genes listed in Table S6 (Li et al. 2018; Zuo and Li 2014): either the allelic pattern was inconsistent with the effect of haplotype on grain shape or the QTL were located more than 1 Mbp away from the position of known grain shape regulators. Among known genes, only GW2 is located near a QTL identified in this study, JAM-GW1. Analysis of resequencing data (Ogawa et al. 2018) of GW2 alleles of the founders revealed 26 mutations including SNPs and 1-bp insertion/deletion mutations (Figure S9). Four GW2 alleles were found in the eight founders but mutations changing amino acid sequence of GW2 and detected as functional SNPs (FNPs) in a previous report (Song et al. 2007) did not exist in the JAM founders.
Discussion
Here, we showed that haplotype-based GWAS of the JAM lines produced higher correlation values between “sum of the additive haplotype effects of QTLs on the grain shape” and “actual values of grain shape” than did ordinary SNP-based GWAS with various models. This indicates that the peaks in the haplotype-based analysis can accurately detect QTL for grain shape, which is controlled by multiple QTL with small effects. These results suggest that haplotype-based GWAS using the JAM lines is effective for detecting QTL associated with complex traits.
Although GWAS using MAGIC and uncontrolled populations have revealed effective alleles for various traits, the ordinary method of GWAS based on biallelic SNPs often results in inconsistency between SNP alleles and functional alleles because of low coverage of SNPs and multi-alleles, often leading to imprecise detection of QTL. In this study, we showed that genome-wide haplotype information on the JAM lines overcomes this problem, as illustrated by the clearer peaks in the Manhattan plots. As suggested by our findings with the JAM lines, haplotype information rather than biallelic SNP information could be also useful in GWAS of uncontrolled populations, although the difficulty of haplotype estimation depends on the population. Recently, the ever-increasing availability of rich genome sequence information and improved algorithms for haplotype prediction have increased the accuracy of genetic analyses. In cassava, the haplotype map identified a deleterious mutation that concomitantly altered starch and ketone metabolism pathways during domestication (Ramu et al. 2017). In sweet potato, a haplotype-based analysis uncovered the evolutional history of chromosomal regions and polyploidy (Yang et al. 2017). Thus, genetic approaches using haplotype information are becoming standard for the detection of QTL.
The haplotype-based GWAS of grain length and width using the JAM lines revealed both the main and side effects of haplotypes on grain shape at each QTL. This demonstrates the importance of genetic analysis to understand trait–trait interaction because the scatter plots of grain length and width in the JAM lines did not show significant positive or negative relationships between these traits. Grain shape is regulated through signaling pathways such as G proteins, proteasomes, phytohormones, and MAP kinases (Li et al. 2018; Zuo and Li 2014). These signaling molecules affect grain shape in different ways; as shown in Table S6, some genes affect either length or width independently, whereas others affect grain area or length/width ratio. From this knowledge, it is understandable that the JAM lines contain genomic regions exhibiting haplotype effects on more than one grain shape trait. Although studies of trait–trait interactions are possible using biparental populations, MAGIC populations may be more advantageous because of the higher number of haplotypes. The haplotype-based GWAS method for genetic analysis using JAM lines should also be applicable to traits other than grain shape.
In summary, our results showed that the haplotype-based GWAS in the JAM lines was effective for detecting QTL associated with grain shape, which is an important trait for grain yield. The haplotype-based GWAS method can provide information on both main and side effects of QTL for agronomic traits, and the obtained information will contribute to producing ideal lines.
Acknowledgments
We gratefully acknowledge Emi Abe, Yukari Shimazu, Chiharu Kikuchi, Megumi Suzuki and Midori Nakajima for field support and scanning rice grain of the JAM lines. This work was supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (grant numbers 27007B and 29002A). Seeds of the founders were provided by Tohoku Agricultural Research Centre (BE), Central Region Agricultural Research Centre (HO), Kyushu Okinawa Agricultural Research Centre (RU, TC, MI) and Genetic Resources Centre (SU), NARO. The authors declare no competing interests.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7027991.
Communicating editor: E. Huang
Literature Cited
- Adachi S., Yoshikawa K., Yamanouchi U., Tanabata T., Sun J., et al. , 2017. Fine mapping of carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Front. Plant Sci. 8: 60 10.3389/fpls.2017.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A., Lobina I. T., et al. , 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice (N. Y.) 6: 11 10.1186/1939-8433-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. 10.1038/ng1104-1133 [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain W1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard B. C., Vera Cruz C. M., McNally K. L., Virk P. S., Mackill D. J., 2008. Rice molecular breeding laboratories in the genomics era: Current status and future considerations. Int. J. Plant Genomics 2008: 524847 10.1155/2008/524847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua M., Gatti D. M., Pea G., Cattonaro F., Coppens F., et al. , 2015. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biol. 16: 167 10.1186/s13059-015-0716-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. 10.3835/plantgenome2011.08.0024 [DOI] [Google Scholar]
- Fukuoka S., Saka N., Koga H., Ono K., Shimizu T., et al. , 2009. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001. 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- Han B., Huang X., 2013. Sequencing-based genome-wide association study in rice. Curr. Opin. Plant Biol. 16: 133–138. 10.1016/j.pbi.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Huang B. E., George A. W., Forrest K. L., Kilian A., Hayden M. J., et al. , 2012. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 10: 826–839. 10.1111/j.1467-7652.2012.00702.x [DOI] [PubMed] [Google Scholar]
- Islam M. S., Thyssen G. N., Jenkins J. N., Zeng L., Delhom C. D., et al. , 2016. A MAGIC population-based genome-wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genomics 17: 903 10.1186/s12864-016-3249-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Xu R., Duan P., Li Y., 2018. Control of grain size in rice. Plant Reprod. 31: 237–251. 10.1007/s00497-018-0333-6 [DOI] [PubMed] [Google Scholar]
- Mackay I. J., Bansept-Basler P., Barber T., Bentley A. R., Cockram J., et al. , 2014. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 (Bethesda) 4: 1603–1610. 10.1534/g3.114.012963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Zhao X., Ponce K., Ye G., Leung H., 2016a QTL mapping for agronomic traits using multi-parent advanced generation inter-cross (MAGIC) populations derived from diverse elite indica rice lines. Field Crops Res. 189: 19–42. 10.1016/j.fcr.2016.02.004 [DOI] [Google Scholar]
- Meng L. J., Guo L. B., Ponce K., Zhao X. Q., Ye G. Y., 2016b Characterization of three rice multiparent advanced generation intercross (MAGIC) populations for quantitative trait loci identification. Plant Genome 9: 1–14. 10.3835/plantgenome2015.10.0109 [DOI] [PubMed] [Google Scholar]
- Misra G., Badoni S., Anacleto R., Graner A., Alexandrov N., et al. , 2017. Whole genome sequencing-based association study to unravel genetic architecture of cooked grain width and length traits in rice. Sci. Rep. 7: 12478 10.1038/s41598-017-12778-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Ando T., Nonoue Y., Mizubayashi T., Kitazawa N., et al. , 2015. Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica x indica cross. Breed. Sci. 65: 308–318. 10.1270/jsbbs.65.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Yamamoto E., Ohtani T., Kanno N., Tsunematsu H., et al. , 2018. Haplotype-based allele mining in the Japan-MAGIC rice population. Sci. Rep. 8: 4379 10.1038/s41598-018-22657-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongom P. O., Ejeta G., 2018. Mating design and genetic structure of a multi-parent advanced generation intercross (MAGIC) population of sorghum (Sorghum bicolor (L.) Moench). G3 (Bethesda) 8: 331–341. 10.1534/g3.117.300248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L., Desplat N., Huang B. E., Desgroux A., Bruguier L., et al. , 2015. Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13: 565–577. 10.1111/pbi.12282 [DOI] [PubMed] [Google Scholar]
- Ramu P., Esuma W., Kawuki R., Rabbi I. Y., Egesi C., et al. , 2017. Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49: 959–963. 10.1038/ng.3845 [DOI] [PubMed] [Google Scholar]
- Sallam A., Martsch R., 2015. Association mapping for frost tolerance using multi-parent advanced generation inter-cross (MAGIC) population in faba bean (Vicia faba L.). Genetica 143: 501–514. 10.1007/s10709-015-9848-z [DOI] [PubMed] [Google Scholar]
- Sannemann W., Huang B. E., Mathew B., Leon J., 2015. Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 35: 86 10.1007/s11032-015-0284-7 [DOI] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X., 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Tanabata T., Shibaya T., Hori K., Ebana K., Yano M., 2012. SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160: 1871–1880. 10.1104/pp.112.205120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Wray N. R., Zhang Q., Sklar P., McCarthy M. I., et al. , 2017. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., et al. , 2006. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708. 10.1038/nature04920 [DOI] [PubMed] [Google Scholar]
- Yan C. J., Zhou J. H., Yan S., Chen F., Yeboah M., et al. , 2007. Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor. Appl. Genet. 115: 1093–1100. 10.1007/s00122-007-0635-9 [DOI] [PubMed] [Google Scholar]
- Yan W. H., Wang P., Chen H. X., Zhou H. J., Li Q. P., et al. , 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4: 319–330. 10.1093/mp/ssq070 [DOI] [PubMed] [Google Scholar]
- Yang J., Moeinzadeh M. H., Kuhl H., Helmuth J., Xiao P., et al. , 2017. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 3: 696–703. 10.1038/s41477-017-0002-z [DOI] [PubMed] [Google Scholar]
- Yano K., Yamamoto E., Aya K., Takeuchi H., Lo P. C., et al. , 2016. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 48: 927–934. 10.1038/ng.3596 [DOI] [PubMed] [Google Scholar]
- Yonemaru J., Mizobuchi R., Kato H., Yamamoto T., Yamamoto E., et al. , 2014. Genomic regions involved in yield potential detected by genome-wide association analysis in Japanese high-yielding rice cultivars. BMC Genomics 15: 346 10.1186/1471-2164-15-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Yamanouchi U., Katayose Y., Toki S., Wang Z. X., et al. , 1998. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 95: 1663–1668. 10.1073/pnas.95.4.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- Zhou P. H., Tan Y. F., He Y. Q., Xu C. G., Zhang Q., 2003. Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor. Appl. Genet. 106: 326–331. 10.1007/s00122-002-1023-0 [DOI] [PubMed] [Google Scholar]
- Zuo J., Li J., 2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48: 99–118. 10.1146/annurev-genet-120213-092138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Germplasm of JAM lines and founders, and their haplotypes and phenotypes, are available by request. They are offered through a material transfer agreement (MTA) from NARO via Daisuke.Ogawa@affrc.go.jp. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7027991.