Abstract
Recently, studies have explored the use of probiotics like the Weissella cibaria strain, CMU (oraCMU), for use as preventive dental medicine instead of chemical oral care methods. The present study was conducted to investigate the antibacterial properties of the cell-free supernatant (CFS) from this bacterium. Cell morphology using the scanning electron microscope, and the antibacterial effect of CFS under various growth conditions were evaluated. The production of hydrogen peroxide, organic acids, fatty acids, and secretory proteins was also studied. Most of the antibacterial effects of oraCMU against periodontal pathogens were found to be acid- and hydrogen peroxide-dose-dependent effects. Lactic acid, acetic acid, and citric acid were the most common organic acids. Among the 37 fatty acids, only 0.02% of oleic acid (C18:1n-9, cis) was detected. Proteomic analysis of the oraCMU secretome identified a total of 19 secreted proteins, including N-acetylmuramidase. This protein may be a potential anti-microbial agent effective against Porphyromonas gingivalis.
Keywords: Weissella cibaria, antibacterial, cell-free supernatant, organic acid, secretome, probiotic
1. Introduction
Oral health can have a great impact on our general health and quality of life. Several oral care products are commonly and widely used for oral hygiene. However, the use of several antibacterial agent-containing oral care products may disrupt the natural microbial ecology of the mouth. This could lead to an overgrowth of opportunistic pathogens or exogenous microorganisms. Recently, the potential application of probiotics for oral health has been reported by many researchers [1,2,3]. Oral care probiotics have been commercialized as substitutes for the conventional treatment for oral diseases, including halitosis, dental caries, gingivitis, and periodontitis [4,5,6,7,8].
Probiotics are live organisms that confer health benefits to the host when administered in adequate amounts [9]. They produce functionally valuable substances, such as exopolysaccharides, folate, and antioxidants [10]. They also fight pathogens by altering the surrounding environment through production of antibacterial compounds and by competition for space and nutrients [11], and they modulate immune and inflammatory responses as well [12]. While several strains of lactobacilli 41 are known to have probiotic effects against oral pathogens, Weissella cibaria has largely been ignored.
A lactic acid bacterium, W. cibaria can be found in a multitude of places—not only in human saliva, but also in traditional fermented foods, such as kimchi, tarhana, and sourdough [13,14,15,16]. W. cibaria CMU, CMS1, CMS2, and CMS3 were isolated from the saliva of healthy infants that were caries-free [13] and patented as strains to suppress halitosis (US7250162B2). W. cibaria CMU (oraCMU) was recently commercialized as an oral care probiotic in South Korea owing to its beneficial effect on oral cavities [4]. Since the early 2000s, our research group has been studying the effects of W. cibaria on oral health through clinical studies [17,18]. Our previous studies have shown that W. cibaria can function in several ways: By directly inhibiting the growth of harmful bacteria [17] through competitive adhesion to the oral cavity [19]; through immune-regulatory actions to prevent inflammatory cytokines [20]; and by inhibiting Streptococcus mutans biofilm formation by converting water-insoluble glucans into water-soluble ones [18]. W. cibaria has been known to produce antibacterial substances, such as hydrogen peroxide, that can inhibit the proliferation of malodor-inducing bacteria [17]. However, it is unknown whether W. cibaria can produce other compounds that can act against periodontopathogens, and whether the notable bactericidal properties are due to pH changes or secretory components.
Therefore, the aim of this study was to identify the antibacterial component of the cell-free supernatant (CFS) from oraCMU. Our investigation was carried out with a focus on estimating hydrogen peroxide production capabilities and identifying organic and fatty acids through high-performance liquid chromatography (HPLC) and gas chromatography (GC). Furthermore, the potential secretory proteins responsible for the antibacterial activity were analyzed using 2D gel electrophoresis, as well as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis.
2. Results
2.1. Cell Morphology of oraCMU
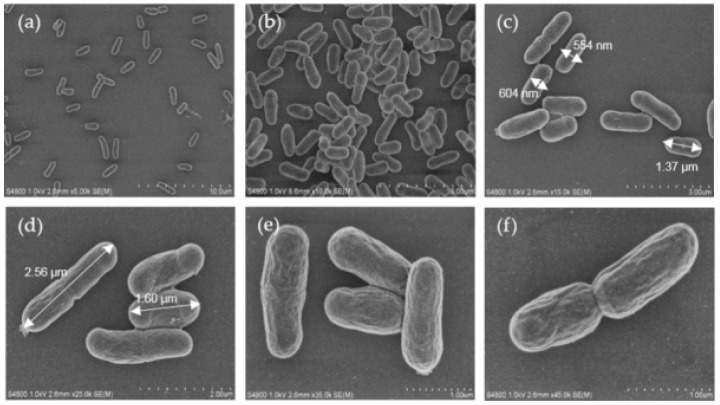
In order to observe the cell morphology of oraCMU, scanning electron microscopic (SEM) analysis was used at various magnifications, such as 5000×, 10,000×, 15,000×, 25,000×, 35,000×, and 45,000×. The bacteria were grown in MRS broth for 16 h, as shown in Figure 1. Cells were observed as short rods growing in pairs and were 0.5–0.7 µm wide and 1.3–2.5 µm long.
Figure 1.
Scanning electron microscopic (SEM) images of oraCMU at various magnifications: 5000× (a), 10,000× (b), 15,000× (c), 25,000× (d), 35,000× (e), and 45,000× (f).
2.2. Characterization of Antibacterial Substances
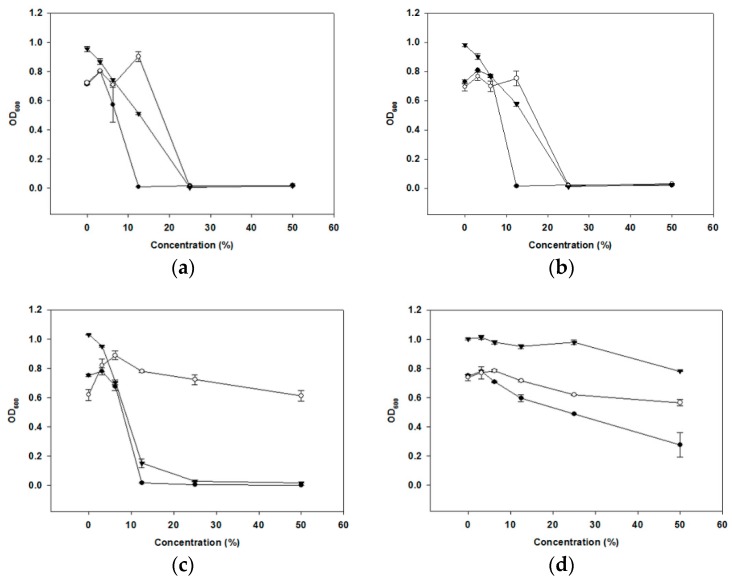
The CFS was treated in different ways to find out the components of cell-free supernatants (CFS) (organic acids, hydrogen peroxide (H2O2) or bacteriocin-like compounds (BLC)) that may be responsible for the anti-periodontopathogenic activity of oraCMU. As shown in Figure 2, the untreated CFS inhibited the growth of all periodontopathogens in a dose-dependent manner. The inhibitory effect of oraCMU was not affected by proteinase K and catalase treatment (Figure 2b). The H2O2-dependent effect of CFS was found to act against Porphyromonas gingivalis and Prevotella intermedia. On the other hand, BLC-dependent antimicrobial action of CFS was effective only against P. gingivalis. However, most of the antibacterial activity of CFS disappeared after neutralization.
Figure 2.
Dose-dependent effects of organic acid, hydrogen peroxide (H2O2), and a bacteriocin-like compound (BLC) in the cell-free supernatants (CFS) of oraCMU against periodontopathogenic bacteria. Optical density at 600 nm (OD600) of the cell suspension was measured. (a) Untreated CFS effect; (b) organic acid-dependent effect was measured using proteinase K and catalase-treated CFS; (c) H2O2-dependent effect was measured by the neutralized and proteinase K-treated CFS; (d) BLC-dependent effect was evaluated by the neutralized and catalase-treated CFS. P. gingivalis (●); Fusobacterium nucleatum (○); P. intermedia (▼).
2.3. Kinetics of H2O2 Production
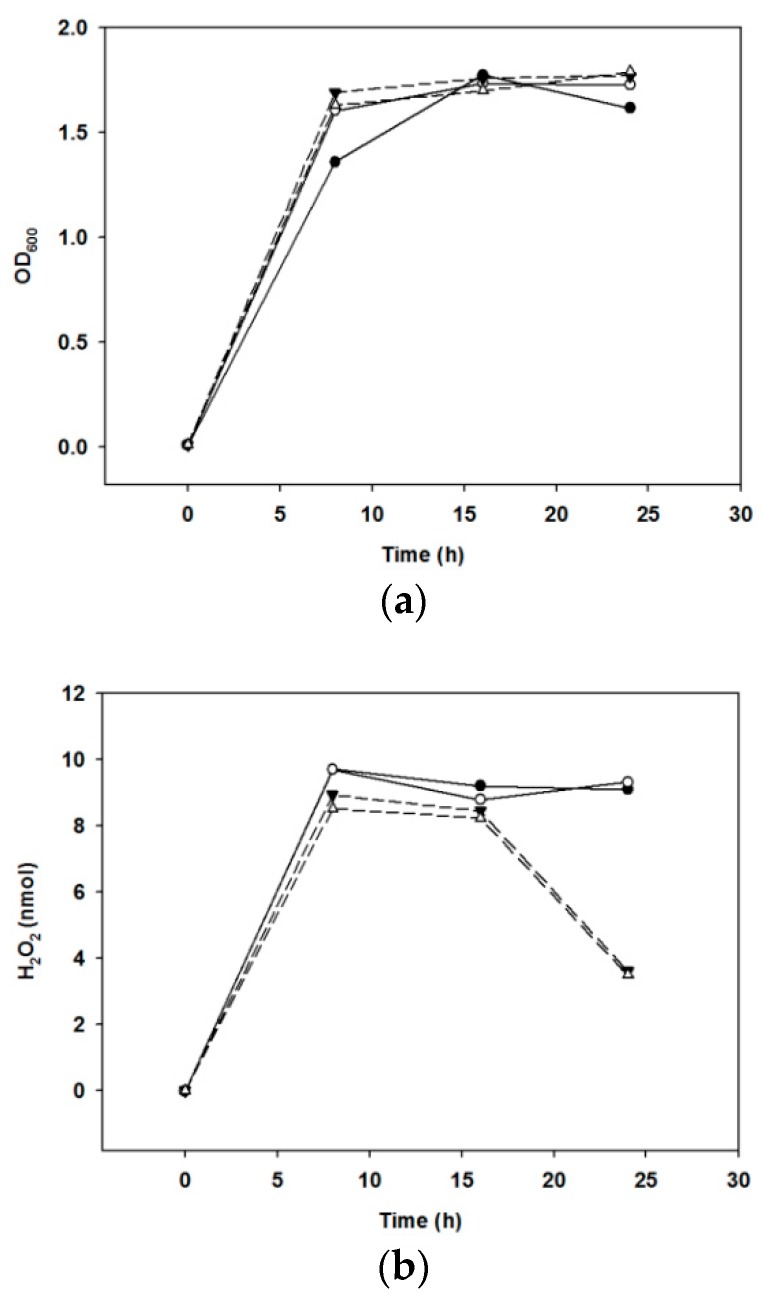
The amount of H2O2 produced by the growth of bacteria was compared when cultivated under various culture conditions. We found that the optical density at 600 nm increased under anaerobic culture conditions until 24 h, but the production of H2O2 decreased (Figure 3).
Figure 3.
Growth and H2O2 production under various conditions. (a) Growth was monitored spectrophotometrically at 600 nm. (b) Production of H2O2, determined as mentioned in Section 4. Solid lines, aerobic incubation at 30 °C (●) and at 37 °C (○); broken lines, anaerobic incubation at 30 °C (▼) and at 37 °C (△).
2.4. Determination of Organic and Fatty Acid Production
We examined the presence and concentration of five different organic acids (viz. oxalic acid, citric acid, succinic acid, lactic acid, and acetic acid) in the CFS by using the HPLC technique. The concentration of lactic acid was found to be highest at 6.42 mg/mL, followed by acetic acid and citric acid, which showed concentrations of 2.78 mg/mL and 0.88 mg/mL, respectively. Oxalic acid and succinic acid were not detected. When the presence and concentration of 37 fatty acids in the CFS were examined by GC, only oleic acid (C18:1n-9, cis) and palmitic acid (C16:0) were detected in the CFS, while the other fatty acids could not be detected (Table 1).
Table 1.
Quantitative analysis of organic and fatty acids in the CFS of oraCMU after 16 h incubation at 37 °C.
| Acids | mg/mL | Acids | mg/mL | Acids | mg/mL |
|---|---|---|---|---|---|
| Oxalic acid | ND | Pentadecanoic acid (C15:0) | ND | cis-11-Eicosenoic acid (C20:1) | ND |
| Citric acid | 0.88 a | cis-10-Peptadecenoic acid (C15:1) | ND | cis-11,14-Eicosadienoic acid (C20:2) | ND |
| Succinic acid | ND b | Palmitic acid (C16:0) | 0.00 | cis-11,14,17-Eicosatrienoic acid (C20:3n-3) | ND |
| Lactic acid | 6.42 | Palmitoleic acid (C16:1) | ND | cis-8,11,14-Eicosatrienoic acid (C20:3n-6) | ND |
| Acetic acid | 2.78 | Heptadecanoic acid (C17:0) | ND | Arachidonic acid (C20:4n-6) | ND |
| Butyric acid (C4:0) | ND | cis-10-Heptadecenoic acid (C17:1) | ND | cis-5,8,11,14,17-Eicosapentaenoic acid (C20:5n-3) | ND |
| Caproic acid (C6:0) | ND | Stearic acid (C18:0) | ND | Heneicosanoic acid (C21:0) | ND |
| Caprylic acid (C8:0) | ND | Elaidic acid (C18:1n-9, trans) | ND | Behenic acid (C22:0) | ND |
| Capric acid (C10:0) | ND | Oleic acid (C18:1n-9, cis) | 0.2 | Erucic acid (C22:1n-9) | ND |
| Undecanoic acid (C11:0) | ND | Linoleiaidic acid (C18:2n-6, trans) | ND | cis-13,16-Docosadienoic acid (C22:2) | ND |
| Lauric acid (C12:0) | ND | Linoleic acid (C18:2n-6, cis) | ND | cis-4,7,10,13,16,19-Docosahexaenoic acid (C22:6n-3) | ND |
| Tridecanoic acid (C13:0) | ND | α-Linolenic acid (C18:3n-3) | ND | Tricosanoic acid (C23:0) | ND |
| Myristic acid (C14:0) | ND | γ-Linolenic acid (C18:3n-6) | ND | Lignoceric acid (C24:0) | ND |
| Myristoleic acid (C14:1) | ND | Arachidic acid (C20:0) | ND | Nervonic acid (C24:1) | ND |
a Data expressed as the mean. b ND: not detected.
2.5. Secretome Analysis
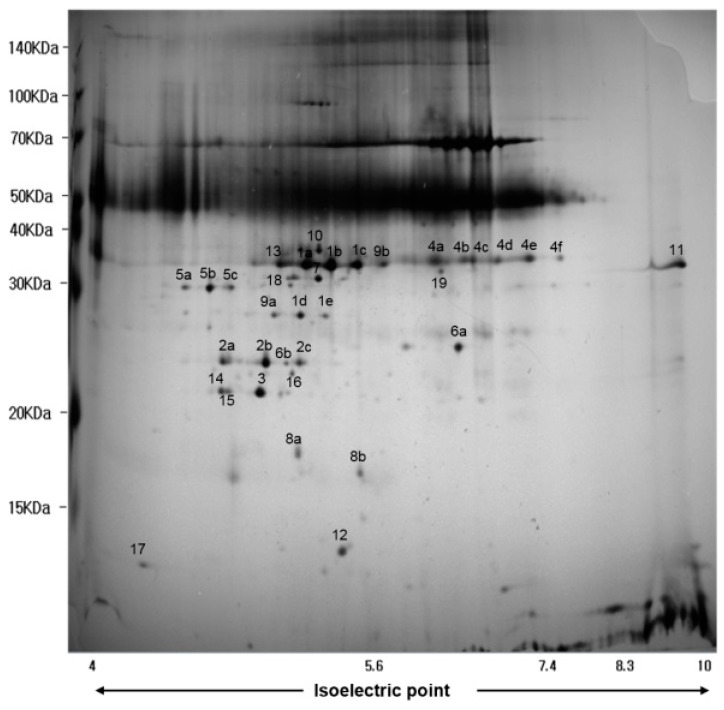
In order to identify secreted proteins in the CFS of oraCMU, proteins were separated by 2D gel electrophoresis (Figure 4). Using MALDI-TOF MS, a total of 19 secreted proteins were identified (Table 2). Amongst the identified proteins, the high-intensity spot proteins found were as follows: type 1 glyceraldehyde-3-phosphate dehydrogenase; N-acetylmuramidase; TIGR00266 family protein; leukocyte elastase inhibitor; l-lactate dehydrogenase; and a hypothetical protein. Spot numbers 1, 5, 9, 10, 16, and 19 were involved in energy production and conversion; spot number 2 in the hydrolysis of the peptidoglycan layer; spot numbers 3 and 6 were of unknown function; spot number 4 was a leukocyte elastase inhibitor; spot number 7 was involved in NADPH-dependent reductase; spot number 8 was related to DNA recombination and repair; spot number 11 in the outer membrane; spot numbers 12, 13, and 14 in nucleotide metabolism; and spot numbers 15, 17, and 18 in carbohydrate transport and metabolism.
Figure 4.
2D-gel electrophoresis of secreted proteins in the CFS of oraCMU after 16 h incubation at 37 °C.
Table 2.
Secreted proteins identified in the CFS of oraCMU.
| Spot No. a | Protein Description b | Accession Number c | MW (kDa) | PI | Protein Score CI (%) d |
|---|---|---|---|---|---|
| 1a–e | Type 1 glyceraldehyde-3-phosphate dehydrogenase | WP_010369259 | 35.756 | 5.03 | 100.00 |
| 2a–c | N-acetylmuramidase | WP_010373609 | 29.343 | 5.40 | 100.00 |
| 3 | TIGR00266 family protein | WP_043941068 | 24.976 | 4.91 | 99.19 |
| 4a–f | Leukocyte elastase inhibitor | XP_003482167 | 42.658 | 6.13 | 100.00 |
| 5a–c | l-lactate dehydrogenase | WP_010372268 | 33.942 | 4.75 | 100.00 |
| 6a,b | Hypothetical protein | WP_043711225 | 23.526 | 5.60 | 99.99 |
| WP_063083480 | 23.318 | 5.04 | 100.00 | ||
| 7 | Aldo/keto reductase | WP_063083131 | 36.627 | 5.08 | 100.00 |
| 8a,b | Single-stranded DNA-binding protein | WP_056973096 | 17.991 | 4.99 | 100.00 |
| WP_043709699 | 15.326 | 5.17 | 99.99 | ||
| 9a,b | NAD-dependent glyceraldehyde-3-phosphate dehydrogenase | KKU03010 | 41.843 | 5.54 | 100.00 |
| 10 | N-acetylglucosamin-6-phosphate deacetylase | WP_0432710743 | 42.715 | 5.17 | 100.00 |
| 11 | BMP family ABC transporter substrate-binding protein | WP_060655478 | 39.514 | 9.18 | 100.00 |
| 12 | Nucleoside-diphosphate kinase | WP_043710743 | 42.715 | 5.17 | 100.00 |
| 13 | Glyceraldehyde 3-phosphate reductase | WP_010370341 | 36.629 | 5.00 | 100.00 |
| 14 | 16S rRNA (cytosine(1402)-N(4))-methyltransferase RsmH | WP_004044730 | 35.71 | 8.73 | 99.64 |
| 15 | HTH-type transcriptional activator RhaR | WP_008787014 | 33.031 | 6.59 | 99.94 |
| 16 | 2,3-diphosphoglycerate-dependent phosphoglycerate mutase | WP_010371458 | 26.717 | 4.94 | 100.00 |
| 17 | Phosphocarrier protein HPr | WP_010370017 | 9.191 | 4.57 | 99.19 |
| 18 | PTS mannose transporter subunit EIIAB | WP_043709678 | 35.417 | 4.97 | 100.00 |
| 19 | Zinc-dependent alcohol dehydrogenase | XP_003482167 | 42.658 | 6.13 | 99.99 |
a Spot numbers refer to the proteins labeled in Figure 4. b Protein description derived from the MASCOT software (www.matrixscience.com). c Information obtained from the NCBI database (www.ncbi.nih.gov). d CI, confidence interval.
3. Discussion
Although a number of treatment approaches aim to inhibit the growth of pathogenic bacteria, probiotics have been applied as a novel way for the improvement of oral health [1,2,3]. They have been used to replace conventional antibiotic therapy, which has been increasingly ineffective due to increased bacterial resistance [4,5,6,7,8]. Probiotics not only have antimicrobial activity, but also inhibit the reappearance of oral pathogenic bacteria. In order to choose the best oral care probiotics, it is important to select bacteria that originate from the oral cavity which can withstand poor oral conditions, have low acid production, have antimicrobial activity, and are capable of inhibiting biofilm formation. The most common probiotic bacteria belong to the Lactobacillus species. Contrary to conventional belief, most lactobacilli are highly acidic and not suitable as oral care probiotics, as they have the potential to cause tooth decay. Certain strains of W. cibaria have been studied as optimal oral care probiotics [4,13,17,18]. Previous studies have shown that oraCMU strains produce less acid and more hydrogen peroxide than other commercial bacteria, and thereby can prevent halitosis or dental caries [4].
The surface morphology of the dextran of W. cibaria, examined using SEM, has been reported [21]. However, as far as we are aware, this is the first report to analyze the morphology of W. cibaria using SEM. In this study, the cells of W. cibaria were found to be short rods, 0.5–0.6 µm wide and 1.2–2.5 µm long (Figure 1). This result is consistent with the known fact that the cells of W. cibaria are short rods growing in pairs. However, the size determined was slightly different from the previously reported sizes of them being 0.8–1.2 µm in width and 1.5–2.0 µm in length [22].
Dental plaques are complex biofilms that accumulate on tooth enamel surfaces. Among over 700 bacterial species that form biofilms in the oral cavity, pioneer bacteria which initiate the colonization of the enamel salivary pellicle in a continuous fashion, followed by secondary bacterial colonization. Oral streptococci are the predominant species in the initial colonization stage. They produce antibacterial substances, such as H2O2, as a byproduct of aerobic metabolism. H2O2 acts as a protective mechanism for the initial colonizers against competing species [23], and is one of the typical antimicrobial substances produced by lactic acid bacteria (LAB) [10]. Previous studies have shown that oraCMU has the best ability to produce H2O2 than other commercial oral care probiotics [4]. A high degree of H2O2 could therefore be sufficient to inhibit the growth of bad breath-causing bacteria [18]. In the present study, oraCMU showed H2O2–dependent inhibition of the periodontopathogenic bacteria P. gingivalis and P. intermedia (Figure 2).
Hydroxyl radicals react with nucleic acids to damage genes, increase membrane permeability, limit membrane transport, and denature proteins within cells [24]. H2O2 is a strong antimicrobial substance produced by oraCMU. We aimed to determine whether H2O2 production was highest when oraCMU was cultivated under optimal conditions. In our present study, although no difference was observed in the growth of oraCMU under aerobic or anaerobic conditions, the H2O2 production under anaerobic conditions after 16 h decreased faster than when under aerobic conditions. This result is similar to a previous report that showed a higher H2O2 secretion by lactobacilli under aerobic conditions than under anaerobic conditions [25].
A large number of LAB inhibits the growth of bacterial pathogens by producing metabolites, such as acetic acid and lactic acid, and by lowering the pH [10]. In the present study, the antibacterial activity of CFS from oraCMU was not lost after a catalase and proteinase K treatment. Additionally, CFS, whose pH was adjusted to 7 and treated with proteinase K, still showed antibacterial activity, except on F. nucleatum. This suggests that the antibacterial activity of oraCMU might principally be attributed to some acids and H2O2.
In this study, HPLC analysis confirmed the presence of lactic, acetic, and citric acids in the CFS. The growth of P. gingivalis, P. intermedia, and F. nucleatum strains were also inhibited in the presence of lactic acid, acetic acid, and citric acid. The inhibitory effect of these acids has been reported to occur due to diffusion across the cell membrane towards the more alkaline cytosol, which interferes with the essential metabolic functions of the cell. The toxic effects of these acids include a decrease in intracellular pH and dissipation of membrane potential [26].
Oleic acid has been reported to be bactericidal against group A streptococci [27]. Wong et al. [28] also identified oleic acid to be the major component of the lipid fraction in the CFS of LAB strains that exhibited a strong inhibitory effect against Staphylococcus aureus. Nuñez de Kairuz et al. [29] reported that the H2O2-generating system was specifically induced by one of the saturated fatty acids from 4:0 to 16:0, or oleic acid. The bacterial membranes of lactobacilli typically consist of linear saturated, unsaturated, and cyclopropane fatty acids [29]. Oleic acid was incorporated into the membranes of lactobacilli when grown in an MRS broth supplemented with Tween 80, which consists of up to 90% oleic acid. It has been reported that only oleic acid protected L. rhamnosus GG when exposed to an acidic environment [30]. In the present study, we used GC to identify the presence of potential anti-microbial fatty acids in the CFS, and our results identified only oleic acid in the CFS, suggesting that oleic acid from the cell wall components might contribute significantly to the antibacterial activity of oraCMU.
The present study showed that there was a significant decrease in antibacterial activity in the neutralized and catalase-treated CFS. However, oraCMU still showed growth inhibition against P. gingivalis in this condition. This suggests the presence of another antimicrobial substance, such as BLC. To date, there have been no reports regarding the proteins secreted by W. cibaria, meaning that this is the first paper to report on the proteomic analysis of secretory proteins of the W. cibaria strain. In our study, type 1 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected most frequently among the identified proteins. GAPDH is known to be an essential enzyme involved in the production and conversion of intracellular energy, and is found not only in bacteria but also in fungi and eukaryotes [31]. These cytoplasmic enzymes are commonly known as housekeeping molecules. Recently, it was also reported that cell wall-associated GAPDH, found on the surface of non-pathogenic lactobacilli, was involved in the adherence [32].
In addition to GAPDH, other proteins which are known to be involved in energy production and conversion, such as l-lactate dehydrogenase, NAD-dependent GAPDH, N-acetylglucosamine-6-phosphate deacetylase, 2,3-diphosphoglycerate-dependent phosphoglycerate mutase, and zinc-dependent alcohol dehydrogenase, were identified. In the present study, several other proteins were also identified, including the BMP family transporter substrate-binding proteins in the outer membrane, enzymes involved in nucleotide metabolism, proteins involved in DNA recombination and repair, and proteins involved in carbon transport and metabolism. Proteins of the TIGR00266 family and hypothetical proteins with unknown function were also detected.
In addition, we identified several proteins secreted by the oraCMU strain of which some were related to bacterial cell wall damage and the modulation of inflammation. Among the identified proteins, N-acetylmuramidase may be a specific candidate as an antibacterial substance. N-acetylmuramidase, also called lysozyme, has an enzymatic activity that cleaves the 1,4-linkage between N-acetylmuramic acid and N-acetylglucosamine in the peptidoglycan found in the bacterial cell wall [33]. Interestingly, the result of this study showed that this protein may cause lysis of P. gingivalis after binding to its cell wall, but does not affect P. intermedia and F. nucleatum.
P. gingivalis, as a major periodontopathogen, plays a role in the reduction of the secreted leukocyte protease inhibitor [34]. It has been suggested that the concentration of secreted leukocyte protease inhibitors, including the leukocyte elastase inhibitor (LEI) are likely to reduce periodontitis [34,35]. LEI, also known as a porcine serine protease inhibitor (serpin B1), belongs to the protein clade founded by ovalbumin [36]. On the other hand, MRS broth is known to contain abundant amounts of serpin B1 [37]. Sánchez et al. [38] reported that the same serpin is secreted by L. rhamnosus GG, and it is presumed to be a proteolytic product cleaved by L. rhamnosus GG protease. It is important to note that these are crude extracts that also include proteins derived from the MRS broth, meaning that the identified LEI may already be present in the broth. Interestingly, only the substance BLC of CFS in the oraCMU showed any antibacterial effect in this study. Furthermore, our previous study showed that oraCMU has the capability of inhibiting F. nucleatum-induced interleukin-6 and interleukin-8 production [20]. Therefore, our hypothesis is that the secretory proteins of oraCMU strains are likely to be involved in immunomodulatory processes.
In summary, our study examined the antibacterial properties of oraCMU on periodontopathogenic bacteria. Our results suggest that there are at least three different mechanisms by which oraCMU suppresses the growth of periodontopathogenic bacteria: an acidic pH, production of hydroxyl radicals, and the secretion of specific proteins with antimicrobial activity. Lactic acid, acetic acid and citric acid as organic acids, and oleic acid as fatty acids, were identified as potential antibacterial compounds. The H2O2-secretion ability of oraCMU under aerobic conditions was better than that under anaerobic conditions. Specific proteins, such as N-acetylmuramidase, secreted by the probiotic bacterium oraCMU was identified, which could contribute to its antimicrobial properties. Additionally, LEI may be responsible for the beneficial effects of oraCMU through their interaction with host cells. These secreted proteins could be involved in the probiotic effects exerted by oraCMU. Further studies are needed to evaluate the potential of the identified proteins.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
W. cibaria CMU (oraCMU; Oradentics Inc., Seoul, South Korea), which has oral care probiotic properties, was used as a model microorganism [4,17,18,19,20]. W. cibaria was grown aerobically in MRS broth (Difco, Detroit, MI, USA) at 37 °C for 16 h. To prepare the CFS of bacterium, the cells were removed by centrifugation (4000× g, 20 m, 4 °C) and the supernatant was filter-sterilized (0.45 μm pore size; Millipore, Burlington, MA, USA). P. gingivalis KCTC 5352 and F. nucleatum KCTC 2488 were purchased from the Korean Culture Collection for Type Cultures (Daejun, South Korea). P. intermedia ATCC 25,611 was kindly provided by the Chonnam National University (Gwangju, Korea) F. nucleatum and P. intermedia were grown anaerobically in brain heart infusion broth (BHI broth, Difco) supplemented with 1% yeast extract (Difco), 0.1% cysteine (Sigma, St. Louis, MO, USA), 5 μg/mL hemin (Kisan Bio Co., Ltd., Seoul, South Korea), and 0.5 μg/mL menadione (Kisan Bio) at 37 °C for 48 h. P. gingivalis was grown anaerobically in tryptic soy broth (Kisan Bio) supplemented with 5 μg/mL hemin and 0.5 μg/mL menadione at 37 °C for 48 h.
4.2. Growth Inhibition of Periodontopathogenic Bacteria
The CFS of probiotics was tested against F. nucleatum, P. gingivalis, and P. intermedia to determine its dose-dependent inhibitory activity, reflecting the activity of the organic acids, H2O2, or BLC, according to a previous method [39]. The inhibitory effects of organic acids on growth were determined after mixing CFS with proteinase K (0.1 mg/mL; Sigma) and catalase (0.05 mg/mL; Sigma). The hydrogen peroxide-dependent activity was evaluated using neutralized and proteinase K-treated CFS. BLC-dependent activity was analyzed with neutralized and catalase-treated CFS. CFS was 2-fold serial diluted and added to each well. Cultures of periodontopathogenic bacteria were adjusted to OD600 = 0.05 with each growth medium. The same quantity of CFS of each probiotic was inoculated in a 96-well plate containing periodontopathogenic bacteria. Each well was read at 600 nm with a microplate reader (VersaMax, Molecular devices, San Jose, CA, USA) after anaerobic incubation for 48 h at 37 °C.
4.3. SEM Analysis
SEM analysis was performed as described by Sorrentino et al. [40]. Briefly, bacterial culture grown for 16 h was centrifuged (4000× g, 20 min, 4 °C), pellet-washed 2 times with a phosphate buffer (PB), and fixed overnight at 4 °C in 2.5% glutaraldehyde in 0.1 M PB. After fixing, samples were rinsed 3 times with PB and 1% OsO4, dehydrated for 1 h, and rinsed 2 times (0.1 M PB and distilled water each) in a graded ethanol series (50%, 60%, 70%, 80%, 90%, and 100%) for 15 min each. For substitution, hexamethyldisilazane (Samchun Pure Chemical, Seoul, South Korea) was treated 2 times for 10 min. After drying it overnight, samples were sputter-coated with gold and observed through SEM (Hitachi, Model: S-4800, Tokyo, Japan) at an accelerating voltage of 1.0 kV.
4.4. H2O2 Determination
In order to determine the kinetics of H2O2 production from the CFS under aerobic or anaerobic conditions, growth (OD600) of bacterial culture at 30 °C or 37 °C after 8, 16, and 24 h were measured simultaneously before centrifugation. CFS were then neutralized to pH 7.0, filter-sterilized (0.45 μm), and assayed for hydrogen peroxide according to the protocol (ab102500, Abcam, Cambridge, MA, USA) [4]. A 0.1-mL sample of supernatant placed in a microplate was read at 570 nm with a microplate reader.
4.5. Analysis of Organic Acids by HPLC
Analysis of organic acids in the CFS was carried out according to the method described by Yeo and Liong [41] with some modifications. Briefly, filtered samples were injected (10 μL) in triplicate into a HPLC system (Shiseido, Tokyo, Japan) equipped with Prevail Organic Acid column (250 × 4.6-mm i.d.), maintained at 40 °C. Organic acids (oxalic, citric, succinic, lactic, and acetic acids) were quantified by using a PDA detector (Shiseido Co., Ltd., Tokyo, Japan) monitoring the absorbance at 214 nm. Under these selected chromatographic conditions, the chromatogram of the standard mixture of all organic acids was obtained. A degassed mobile phase (0.25 M KH2PO4 with pH = 2.4) was used at a flow rate of 0.4 mL/min. Standards (Sigma) were prepared in deionized filter-sterilized water.
4.6. Analysis of Fatty Acids by GC
The crude lipid fraction was converted to fatty acid methyl esters (FAME) through sodium methoxide catalysis [28]. The FAMEs were analyzed on SP-2560 columns (100 m × 0.25 mm × 0.20 µm) using an Agilent Model 6850 gas chromatograph equipped with an FID (Agilent Technologies, Little Falls, PA, USA). Operating conditions were as follows: injector, 225 °C; detector, 285 °C; initial, 100 °C (hold 4 min); ramp, 208 °C (3 °C/3 min); final, 244 °C (hold 15 min); carrier gas, helium; flow rate, 0.75 mL/min; linear velocity, 18 cm/s; and split ratio, 200:1. The injection volume was 10 μL. Supelco 37 Component FAME Mix (Sigma) was used as a standard.
4.7. Analysis of Secretory Proteins by MALDI-TOF MS
All chemicals used in this study were purchased from Sigma. To identify secretory proteins of oraCMU, CFS was mixed with 7 M urea, 2 M Thiourea containing 4% (w/v) 3-[(3-cholamidopropy) dimethyammonio]-1-propanesulfonate (CHAPS), 1% (w/v) dithiothreitol (DTT), 2% (v/v) pharmalyte, and 1 mM benzamidine for 1 h. After centrifugation at 15,000× g for 1 h at 15 °C, soluble fractions (200 μg) were separated on 2D gel electrophoresis and then silver-stained, as described by Oakley et al. [42]. Quantitative analysis of digitized images was performed using the PDQuest (version 7.0; Bio-Rad Laboratories, Hercules, CA, USA) software according to the manufacturer’s protocols. Protein spots were cut from the gel for protein identification by peptide mass fingerprinting, digested with trypsin (Promega, Madison, WI, USA), resuspended in α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid, and analyzed by MALDI-TOF MS analysis (Microflex LRF 20, Bruker Daltonics, Billerica, MA, USA) as described by Fernandez et al. [43]. Spectra were collected at 300 shots per spectrum in the range of 600–3000 Da and calibrated with two-point internal calibration using trypsin auto-digestion peaks. The peak list was generated using Flex Analysis 3.0 (Bruker Daltonics). The data were analyzed with MASCOT software.
Author Contributions
M.-S.K. designed the experiments and wrote the manuscript. H.-S.L., J.-E.Y., and S.-P.H. performed the experiments and analyzed the data. All authors reviewed the manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1D1A1B03030952) and by Ministry of Science and ICT (MSIT) in Korean government and Korea Industrial Technology Association (KOITA) as “A study on the programs to support follow-up R&D of corporate R&D centers with academia and research institutes”.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Samot J., Badet C. Antibacterial activity of probiotic candidates for oral health. Anaerobe. 2013;19:34–38. doi: 10.1016/j.anaerobe.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Terai T., Okumura T., Imai S., Nakao M., Yamaji K., Ito M., Nagata T., Kaneko K., Miyazaki K., Okada A., et al. Screening of probiotic candidates in human oral bacteria for the prevention of dental disease. PLoS ONE. 2015;8:e0128657. doi: 10.1371/journal.pone.0128657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-López A., Camelo-Castillo A., Ferrer M.D., Simon-Soro Á., Mira A. Health-associated niche inhabitants as oral probiotics: The Case of Streptococcus dentisani. Front. Microbiol. 2017;10:379. doi: 10.3389/fmicb.2017.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jang H.J., Kang M.S., Yi S.H., Hong J.Y., Hong S.P. Comparative study on the characteristics of Weissella cibaria CMU and probiotic strains for oral care. Molecules. 2016;20:1752. doi: 10.3390/molecules21121752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flichy-Fernández A.J., Ata-Ali J., Alegre-Domingo T., Candel-Martí E., Ata-Ali F., Palacio J.R., Peñarrocha-Diago M. The effect of orally administered probiotic Lactobacillus reuteri-containing tablets in peri-implant mucositis: A double-blind randomized controlled trial. J. Period. Res. 2015;50:775–785. doi: 10.1111/jre.12264. [DOI] [PubMed] [Google Scholar]
- 6.Nishihara T., Suzuki N., Yoneda M., Hirofuji T. Effects of Lactobacillus salivarius-containing tablets on caries risk factors: A randomized open-label clinical trial. BMC Oral Health. 2014;14:110. doi: 10.1186/1472-6831-14-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masdea L., Kulik E.M., Hauser-Gerspach I., Ramseier A.M., Filippi A., Waltimo T. Antimicrobial activity of Streptococcus salivarius K12 on bacteria involved in oral malodour. Arch. Oral Biol. 2012;57:1041–1047. doi: 10.1016/j.archoralbio.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Maekawa T., Hajishengallis G. Topical treatment with probiotic Lactobacillus brevis CD2 inhibits experimental periodontal inflammation and bone loss. J. Period. Res. 2014;49:785–791. doi: 10.1111/jre.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Food and Agriculture Organization of the United Nations (FAO) World Health Organization (WHO) Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria. Food and Agriculture Organization of the United Nations (FAO); Rome, Italy: World Health Organization (WHO); Geneva, Switzerland: 2001. [Google Scholar]
- 10.Kanmani P., Satish Kumar R., Yuvaraj N., Paari K.A., Pattukumar V., Arul V. Probiotics and its functionally valuable products-a review. Crit. Rev. Food Sci. Nutr. 2013;53:641–658. doi: 10.1080/10408398.2011.553752. [DOI] [PubMed] [Google Scholar]
- 11.Popova M., Molimard P., Courau S., Crociani J., Dufour C., Le Vacon F., Carton T. Beneficial effects of probiotics in upper respiratory tract infections and their mechanical actions to antagonize pathogens. J. Appl. Microbiol. 2012;113:1305–1318. doi: 10.1111/j.1365-2672.2012.05394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller F., Duchmann R. Intestinal flora and mucosal immune responses. Int. J. Med. Microbiol. 2003;293:77–86. doi: 10.1078/1438-4221-00246. [DOI] [PubMed] [Google Scholar]
- 13.Kang M.S., Yeu J.E., Oh J.S., Shin B.A., Kim J.H. Complete Genome Sequences of Weissella cibaria Strains CMU, CMS1, CMS2, and CMS3 isolated from infant saliva in South Korea. Genome Announc. 2017;5:e01103–e01117. doi: 10.1128/genomeA.01103-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang B.K., Cho M.S., Park D.S. Red pepper powder is a crucial factor that influences the ontogeny of Weissella cibaria during kimchi fermentation. Sci. Rep. 2016;6:28232. doi: 10.1038/srep28232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sengun I.Y., Nielsen D.S., Karapinar M., Jakobsen M. Identification of lactic acid bacteria isolated from Tarhana, a traditional Turkish fermented food. Int. J. Food Microbiol. 2009;135:105–111. doi: 10.1016/j.ijfoodmicro.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 16.Bounaix M.S., Robert H., Gabriel V., Morel S., Remaud-Siméon M., Gabriel B., Fontagné-Faucher C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010;311:18–26. doi: 10.1111/j.1574-6968.2010.02067.x. [DOI] [PubMed] [Google Scholar]
- 17.Kang M.S., Kim B.G., Chung J., Lee H.C., Oh J.S. Inhibitory effect of Weissella cibaria isolates on the production of volatile sulphur compounds. J. Clin. Periodontol. 2006;33:226–232. doi: 10.1111/j.1600-051X.2006.00893.x. [DOI] [PubMed] [Google Scholar]
- 18.Kang M.S., Chung J., Kim S.M., Yang K.H., Oh J.S. Effect of Weissella cibaria isolates on the formation of Streptococcus mutans biofilm. Caries Res. 2006;40:418–425. doi: 10.1159/000094288. [DOI] [PubMed] [Google Scholar]
- 19.Kang M.S., Na H.S., Oh J.S. Coaggregation ability of Weissella cibaria isolates with Fusobacterium nucleatum and their adhesiveness to epithelial cells. FEMS Microbiol. Lett. 2005;253:323–329. doi: 10.1016/j.femsle.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Kang M.S., Lim H.S., Kim S.M., Lee H.C., Oh J.S. Effect of Weissella cibaria on Fusobacterium nucleatum-induced Interleukin-6 and Interleukin-8 Production in KB Cells. J. Bacteriol. Virol. 2011;41:9–18. doi: 10.4167/jbv.2011.41.1.9. [DOI] [Google Scholar]
- 21.Tingirikari J.M., Kothari D., Shukla R., Goyal A. Structural and biocompatibility properties of dextran from Weissella cibaria JAG8 as food additive. Int. J. Food Sci. Nutr. 2014;65:686–691. doi: 10.3109/09637486.2014.917147. [DOI] [PubMed] [Google Scholar]
- 22.Björkroth K.J., Schillinger U., Geisen R., Weiss N., Hoste B., Holzapfel W.H., Korkeala H.J., Vandamme P. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 2002;52:141–148. doi: 10.1099/00207713-52-1-141. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L., Kreth J. The Role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell Longev. 2012;2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas E.L. Myeloperoxidase-hydrogen peroxide-chloride antimicrobial system: Effect of exogenous amines on antibacterial action against Escherichia coli. Infect. Immun. 1979;25:110–116. doi: 10.1128/iai.25.1.110-116.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung J., Cooper D., Turner M.S., Walsh T., Giffard P.M. Cystine uptake prevents production of hydrogen peroxide by Lactobacillus fermentum BR11. FEMS Microbiol. Lett. 2003;227:93–99. doi: 10.1016/S0378-1097(03)00653-0. [DOI] [PubMed] [Google Scholar]
- 26.Konings W.N., Poolman B., Driessen A.J. Bioenergetics and solute transport in lactococci. CRC Crit. Rev. Microbiol. 1989;16:419–476. doi: 10.3109/10408418909104474. [DOI] [PubMed] [Google Scholar]
- 27.Speert D.P., Wannamaker L.W., Gray E.D., Clawson C.C. Bactericidal effect of oleic acid on group A streptococci: Mechanism of action. Infect. Immun. 1979;26:1202–1210. doi: 10.1128/iai.26.3.1202-1210.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong C.B., Khoo B.Y., Sasidharan S., Piyawattanametha W., Kim S.H., Khemthongcharoen N., Ang M.Y., Chuah L.O., Liong M.T. Inhibition of Staphylococcus aureus by crude and fractionated extract from lactic acid bacteria. Benef. Microbes. 2015;6:129–139. doi: 10.3920/BM2014.0021. [DOI] [PubMed] [Google Scholar]
- 29.Nuñez de Kairuz M.S., Olazabal M.E., Oliver G., Pesce de Ruiz Holgado A.A., Massa E., Farías R.N. Fatty acid dependent hydrogen peroxide production in Lactobacillus. Biochem. Biophys. Res. Commun. 1988;152:113–121. doi: 10.1016/S0006-291X(88)80687-9. [DOI] [PubMed] [Google Scholar]
- 30.Corcoran B.M., Stanton C., Fitzgerald G.F., Ross R.P. Growth of probiotic lactobacilli in the presence of oleic acid enhances subsequent survival in gastric juice. Microbiology. 2007;153:291–299. doi: 10.1099/mic.0.28966-0. [DOI] [PubMed] [Google Scholar]
- 31.Tristan C., Shahani N., Sedlak T.W., Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell. Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurmalainen V., Edelman S., Antikainen J., Baumann M., Lähteenmäki K., Korhonen T.K. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology. 2007;153:1112–1122. doi: 10.1099/mic.0.2006/000901-0. [DOI] [PubMed] [Google Scholar]
- 33.Striker R., Kline M.E., Haak R.A., Rest R.F., Rosenthal R.S. Degradation of gonococcal peptidoglycan by granule extract from human neutrophils: Demonstration of N-acetylglucosaminidase activity that utilizes peptidoglycan substrates. Infect. Immun. 1987;55:2579–2584. doi: 10.1128/iai.55.11.2579-2584.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretschmar S., Yin L., Roberts F., London R., Flemmig T.T., Arushanov D., Kaiyala K., Chung W.O. Protease inhibitor levels in periodontal health and disease. J. Period. Res. 2012;47:228–235. doi: 10.1111/j.1600-0765.2011.01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laugisch O., Schacht M., Guentsch A., Kantyka T., Sroka A., Stennicke H.R., Pfister W., Sculean A., Potempa J., Eick S. Periodontal pathogens affect the level of protease inhibitors in gingival crevicular fluid. Mol. Oral Microbiol. 2012;27:45–56. doi: 10.1111/j.2041-1014.2011.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remold-O’Donnell E. The ovalbumin family of serpin proteins. FEBS Lett. 1993;315:105–108. doi: 10.1016/0014-5793(93)81143-N. [DOI] [PubMed] [Google Scholar]
- 37.Remold-O’Donnell E., Chin J., Alberts M. Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor. Proc. Natl. Acad. Sci. USA. 1992;89:5635–5639. doi: 10.1073/pnas.89.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez B., Schmitter J.M., Urdaci M.C. Identification of novel proteins secreted by Lactobacillus rhamnosus GG grown in de Mann-Rogosa-Sharpe broth. Lett. Appl. Microbiol. 2009;48:618–622. doi: 10.1111/j.1472-765X.2009.02579.x. [DOI] [PubMed] [Google Scholar]
- 39.Kang M.S., Oh J.S., Lee H.C., Lim H.S., Lee S.W., Yang K.H., Choi N.K., Kim S.M. Inhibitory effect of Lactobacillus reuteri on periodontopathic and cariogenic bacteria. J. Microbiol. 2011;49:193–199. doi: 10.1007/s12275-011-0252-9. [DOI] [PubMed] [Google Scholar]
- 40.Sorrentino E., Reale A., Tremonte P., Maiuro L., Succi M., Tipaldi L., Di Renzo T., Pannella G., Coppola R. Lactobacillus plantarum 29 inhibits Penicillium spp. involved in the spoilage of black truffles (Tuber aestivum) J. Food Sci. 2013;78:M1188–M1194. doi: 10.1111/1750-3841.12171. [DOI] [PubMed] [Google Scholar]
- 41.Yeo S.K., Liong M.T. Effect of prebiotics on viability and growth characteristics of probiotics in soymilk. J. Sci. Food Agric. 2010;90:267–275. doi: 10.1002/jsfa.3808. [DOI] [PubMed] [Google Scholar]
- 42.Oakley B.R., Kirsch D.R., Morris N.R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez J., Gharahdaghi F., Mische S.M. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS) Electrophoresis. 1998;19:1036–1045. doi: 10.1002/elps.1150190619. [DOI] [PubMed] [Google Scholar]