Abstract
Background
Age‐related changes in blood pressure are associated with a variety of poor health outcomes. Genetic factors are proposed contributors to age‐related increases in blood pressure, but few genetic loci have been identified. We examined the role of mitochondrial genomic variation in blood pressure by sequencing the mitochondrial genome.
Methods and Results
Mitochondrial DNA (mtDNA) data from 1755 participants from the LIFE (Lifestyle Interventions and Independence for Elders) studies and 788 participants from the Health ABC (Health, Aging, and Body Composition) study were evaluated using replication analysis followed by meta‐analysis. Participants were aged ≥69 years, of diverse racial backgrounds, and assessed for systolic blood pressure (SBP), diastolic blood pressure, and mean arterial pressure. After meta‐analysis across the LIFE and Health ABC studies, statistically significant associations of mtDNA variants with higher SBP (m.3197T>C, 16S rRNA; P=0.0005) and mean arterial pressure (m.15924A>G, t‐RNA‐thr; P=0.004) were identified in white participants. Among black participants, statistically significant associations with higher SBP (m.93A>G, HVII; m.16183A>C, HVI; both P=0.0001) and mean arterial pressure (m.16172T>C, HVI; m.16183A>C, HVI; m.16189T>C, HVI; m.12705C>T; all P's<0.0004) were observed. Significant pooled effects on SBP were observed across all transfer RNA regions (P=0.0056) in white participants. The individual and aggregate variant results are statistically significant after multiple comparisons adjustment for the number of mtDNA variants and mitochondrial regions examined.
Conclusions
These results suggest that mtDNA‐encoded variants are associated with variation in SBP and mean arterial pressure among older adults. These results may help identify mitochondrial activities to explain differences in blood pressure in older adults and generate new hypotheses surrounding mtDNA variation and the regulation of blood pressure.
Clinical Trial Registration
URL: http://www.ClinicalTrials.gov. Unique identifiers: NCT01072500 and NCT00116194.
Keywords: aging, blood pressure, DNA sequencing, mitochondria
Subject Categories: Genetics; Genetic, Association Studies; Hypertension; Aging; Blood Pressure
Clinical Perspective
What Is New?
These data provide evidence that specific mitochondrial DNA variants are associated with elevated blood pressures in meta‐analyses involving participants sampled from 2 well‐characterized cohorts of older adults.
What Are the Clinical Implications?
Observed associations of mitochondrial DNA variants with blood pressure suggest that mitochondrial variants may have an important role in blood pressure variation and may ultimately contribute to improved diagnosis or treatment of hypertension among older adults.
Introduction
Life expectancy continues to increase in developed countries, leading to ever‐increasing representation of older adults within the population.1 Given the disproportionate utilization of healthcare resources by older adults—for example, nearly three quarters of cardiovascular disease–related expenditures2—the maintenance of health and well‐being among older adults is a critical scientific and public health priority. Among the potential targets for improving health among older adults, hypertension represents one of the most prevalent and potentially modifiable risk factors. Hypertension causes >7 million premature deaths per year and contributes to 4.5% of the total disease burden worldwide,3 and the majority of hypertension‐related morbidity and mortality is observed among older adults.4 Despite tremendous advances in the understanding of hypertension pathophysiology and treatment, a need remains to further elucidate factors that regulate blood pressure among older adults.
Blood pressure regulation is considered a multifactorial trait involving interactions among genetic and environmental and lifestyle factors. Genetic variants account for 19% to 56% of systolic blood pressure (SBP) variation,5 37% to 52% of diastolic blood pressure (DBP) variation,5 and up to 80% of mean arterial pressure (MAP) variation.6, 7 To date, however, the majority of genetic variants associated with blood pressure have been identified in the nuclear genome.8, 9, 10 In contrast, less is known regarding the relative contribution of the mitochondrial genome to blood pressure variation. This gap in knowledge is important, given the well‐known contributions of mitochondria to meeting cellular energy requirements, particularly within the heart11 and vascular endothelium.12 Although a small number of studies suggest that mitochondrial DNA (mtDNA) contribute to hypertension, possibly due to cardiac and/or endothelial oxidative stress (reviewed by Ding et al13), larger and more comprehensive studies are needed to evaluate the potential contribution of mtDNA to blood pressure variation in various clinically relevant populations.
Energy from mitochondrial oxidative phosphorylation is dependent on the coordinated expression and interaction of genes encoded in the nuclear DNA and mtDNA. The majority of genes encoded by the mtDNA are crucial for the machinery that converts metabolic energy into ATP, including 37 genes encoding polypeptides, transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs). Collections of rare variants within genes or genomic regions influence phenotypic associations in numerous ways.14 Because human mtDNA has a mutation rate that is 10 to 20 times higher than that of nuclear DNA,15, 16 it is likely that most of the mtDNA variation that affects function is rare in frequency and detectable only by direct sequencing. We have previously shown that common and rare mtDNA variants are important contributors to numerous diseases and aging‐related traits.17, 18, 19, 20, 21, 22, 23 Given the tremendous prevalence of hypertension among elderly patients and prior reports of mtDNA contributions to hypertension, we aimed to evaluate these associations in 2 large, well‐characterized cohorts of older adults. In the current study, we present results from the first comprehensive examination, to our knowledge, of mitochondrial genomic variation and blood pressure among older adults. These novel data are likely to provide important biological information that could help advance the understanding interindividual variation in blood pressure and ultimately improve hypertension treatment paradigms.
Materials and Methods
Population
This study used data from 2 samples of older adults from large, diverse studies of aging: LIFE (Lifestyle Interventions and Independence for Elders) and Health ABC (Health, Aging, and Body Composition). Baseline data from the LIFE and Health ABC cohorts were used in this study. Design, recruitment, baseline characteristics, and main outcomes of the LIFE and Health ABC studies are published and described elsewhere.24, 25, 26 Participants in the LIFE study originated from 2 samples—380 from the LIFE‐Pilot and 1375 from the main LIFE study. These studies were combined into 1 sample because both were multicenter, single‐blinded, parallel, randomized trials and the eligibility criteria were identical. The LIFE study enrolled participants in 8 field centers across the United States between February 2010 and December 2013. The LIFE‐Pilot study enrolled participants across 4 field centers between April 2004 and February 2005. The Health ABC cohort study enrolled participants for observational data collection from 2 field centers between 1997 and 1998.
The eligibility criteria for LIFE consisted of men and women aged 70 to 89 years (1) who were inactive (reporting <20 min/week in the past month performing regular physical activity and <125 min/week of moderate physical activity); (2) who were at high risk of mobility disability based on lower extremity functional limitations measured by a Short Physical Performance Battery (SPPB) score ≤9 of 12; (3) who could walk 400 m in ≤15 minutes without sitting, leaning, or needing the help of another person or walker; (4) who had no major cognitive impairment; and (5) who could safely participate in the intervention as determined by medical history, physical examination, and resting ECG. Given their similarity, the LIFE and LIFE‐Pilot studies were pooled and referred to as LIFE for simplicity thereafter. For Health ABC, the eligibility criteria included age >70 to 79 years at baseline, self‐reporting no difficulty in walking a distance of 0.4 km or climbing at least 10 stairs, independently performing activities of daily living, planning to live in the area for the next 3 years, and presenting no evidence of life‐threatening illnesses. The sample was approximately balanced for sex (53% women), and 50% of participants were black.
The institutional review board at all field sites approved each study protocol, and written informed consent was obtained from all study participants. Both LIFE studies were additionally monitored by a data and safety monitoring board appointed by the National Institute on Aging. LIFE and LIFE‐Pilot are registered at http://ClinicalsTrials.gov with the identifiers NCT01072500 and NCT00116194, respectively. The study data used in this analysis can be downloaded from the Health ABC and LIFE study websites.27 Users must register for an account, submit an analysis plan for review by study sponsors, and accept the terms of a data use agreement before downloading or exploring study data.
Blood Pressure
For each study, blood pressure was measured at 1 time point with standardized methods used for numerous blood pressure trials (eg, SHEP (Systolic Hypertension in the Elderly Program) trial). The measurement was done with the participant in a seated position after ≈5 minutes of rest, in a quiet room with feet on the floor and legs uncrossed. The method involved determining the pulse obliteration pressure level by adding 30 mm Hg to the obtained maximal inflation level. Two measurements were taken with a 30‐second rest in between each measurement. The 2 measurements were averaged for the analysis. The right arm was used unless contraindicated. SBP and DBP as well as MAP were used as an estimate of overall blood pressure calculated as MAP=[DBP+0.33 (SBP−DBP)]. Importantly, observed pressures were adjusted for antihypertensive medication usage, as done elsewhere for other cohorts included in hypertension genetic association studies.8 Given the direct role of the medications in altering the phenotype, this adjustment is critical for proper interpretation of the genetic associations because the medication can mask familial associations with blood pressure (eg, someone taking ≥2 antihypertensive medications but with “normal” observed blood pressure would inappropriately reflect genetic associations if left unadjusted). As such, pressures were adjusted according to the methods of Cui et al,28 whereby adjusted SBP is calculated by increasing the recorded measure by 8, 14, and 20 mm Hg for 1, 2, and ≥3 medication classes taken, respectively. Similarly adjusted DBP is calculated by increasing the recorded measure by 4, 10, and 16 mm Hg for 1, 2, and ≥3 medication classes taken, respectively. Adjusted MAP was calculated using the adjusted blood pressure measures in the equation above.
Across cohorts, 28% of participants were taking no antihypertensive medications, 27% took 1 medication, 23% took 2 medications, and 22% took ≥3 medications. As expected based on study cohort recruitment criteria, LIFE participants displayed more antihypertensive medication usage (29%, 26%, and 25% on 1, 2, and ≥3 medications, respectively) compared with Health ABC (24%, 17%, and 14%). The mean adjustments in SBP, DBP, and MAP, respectively, were 10.9, 7.7, and 8.8 mm Hg in LIFE and 7.1, 4.9, and 5.6 mm Hg in Health ABC. Importantly, correlations between adjusted and unadjusted blood pressures (for those using medications and thus adjusted) were high across both studies. Correlation coefficients between adjusted and unadjusted blood pressures in LIFE were 0.97, 0.89, and 0.91 for SBP, DBP, and MAP, respectively; these coefficients were 0.97, 0.94, and 0.94, respectively, in Health ABC.
Demographic and Health Factors
Participants self‐designated their sex, race, and ethnicity from a fixed set of options that were used to categorize individuals as male, female, white, black, and other (Hispanic, Asian, Native Hawaiian/Pacific Islander, Native American/Alaskan Native, and other). Age was calculated from birthdate. Medical and hospitalization history was obtained by interview. Weight and height were measured by trained staff using calibrated equipment, and body mass index was calculated as weight in kilograms divided by height in meters squared.
MtDNA Sequencing
DNA was extracted from buffy coat using whole blood collected from Health ABC study participants at baseline and from LIFE study participants collected at the baseline or 12‐month visit. Briefly, the entire mitochondrial genome is first amplified in 2 long‐range polymerase chain reactions (PCRs; 9.1 and 11.2 kb) spanning the entire human mtDNA genome using the LA PCR Kit (Takara Bio). This kit was chosen because it combines the Takara LA Taq enzyme with an optimized buffer system, resulting in longer and more accurate PCR products compared with conventional PCR kits. The size distribution of each PCR amplicon is then verified using an Agilent Technologies 2100 Bioanalyzer. Using the Nextera XT DNA sample preparation kit designed specifically for human mtDNA, the PCR amplicons were simultaneously fragmented and tagged with adapter sequences. Before sequencing, each sample is amplified using a limited‐cycle PCR program that adds unique indexes to each sample for a total of 384 indexes per sequencing run. In preparation for cluster generation and sequencing, equal volumes of bead‐based normalized library are combined, diluted in hybridization buffer, and heat denatured before MiSeq sequencing. After sequencing and alignment to the Cambridge reference sequence (NC_12920), sequence processing is accomplished with the MiSeq Reporter and the mtDNA Analysis Tool for variant calling. Multiplex sequencing of 384 bar‐coded samples yielded an average depth of coverage >250× and a minimum of 100× per individual.
Statistical Analyses
Genetic association analyses followed established norms for performing analyses via replication and meta‐analysis.29, 30, 31, 32 Associations were first conducted in the LIFE study cohort and then replicated in the Health ABC cohort. Meta‐analysis was then performed using METAL33 with a fixed‐effects model. Meta‐analysis provides an opportunity to evaluate the heterogeneity of genetic effect across studies and reduces the probability of false‐negative results and the “winner's curse” phenomenon.34, 35 Blood pressure was compared between the major and minor alleles for individual mtDNA variants in both LIFE and Health ABC studies using the generalized linear model. Allele thresholds were set to 5% minor allele frequency (MAF) for white, black, and other participants. All analyses were stratified by race (white, black, and other) and adjusted for age and sex using custom R scripts and SAS version 9.4 (SAS Institute).
We also leveraged the Sequence Kernel Association Test (SKAT)36 method that collapses and tests the collective effects of common and rare variants. The SKAT method is designed for resequencing data in which rare mutations are observed directly and generally has higher power than alternative methods for genetic burden analyses.36 This test was developed based on random‐effects models, which assume a common distribution for the genetic effects of variants at different sites and test for the null hypothesis that the distribution has no variation. We applied the SKAT approach in association tests for the continuous SBP, DBP, and MAP measures and computed statistical significance for each test using 10 000 independent simulations. For the SKAT testing we considered a “hierarchy” of annotations within 7 mtDNA regions that will allow us to refine the influence of collections of rare and common variants, including each of the 4 individual oxidative phosphorylation complexes (I, III, IV, and V), 2 rRNAs, all tRNAs combined, and combined HV regions.
For the current analysis, associations were examined for all variants in both the white and black participants from the LIFE and Health ABC studies. Given limited sample size in the other race category, we used this group for additional validation of significant associations identified in black and white participants from both studies. To avoid false‐positive results caused by population stratification, all analyses were adjusted for 6 eigenvectors of mitochondrial genetic ancestry derived from principal component analysis. In our previous mtDNA sequencing work, the first 6 eigenvectors accounted for 71% of the variance in the mtDNA sequence data set.37 We used the in silico PhyloP38 method to examine mtDNA nucleotide conservation for all associated variants.
Results
Participant Characteristics
MtDNA sequence variants were evaluated in 1755 and 788 participants in the LIFE and Health ABC studies, respectively (Table 1). Participants in the LIFE studies were older, had higher body mass index, and generally had higher rates of chronic disease. SBP in LIFE participants (adjusted mm Hg: mean: 139.2; SD: 19.6) was slightly lower than in Health ABC participants (adjusted mm Hg: mean: 143.5; SD: 23.5). DBP in LIFE participants (adjusted mm Hg: mean: 76.1; SD: 11.1) was similar to that in Health ABC participants (adjusted mm Hg: mean: 76.7; SD: 13.8). MAP was also similar in participants from LIFE (adjusted mm Hg: mean: 97.2; SD: 12.4) and Health ABC (adjusted mm Hg: mean: 98.9; SD: 15.3) participants. Additional details regarding relevant blood pressure characteristics of the cohorts are provided in Table S1. Per study designs, participants in the Health ABC study were more likely to be black and sex balanced than participants in the LIFE study.
Table 1.
Baseline Characteristics of LIFE and Health ABC Cohorts
| LIFE (n=1755) | Health ABC (n=788) | |
|---|---|---|
| Age, y, mean (SD) | 78.4 (5.1) | 74.0 (2.9) |
| Race, n (%) | ||
| White | 1344 (76.6) | 395 (50.1) |
| Black | 290 (16.5) | 393 (49.9) |
| Other | 121 (6.9) | … |
| Women, n (%) | 1166 (66.4) | 418 (53.0) |
| BMI, kg/m2, mean (SD) | 30.2 (5.9) | 27.6 (4.9) |
| SBP, mean (SD)a | ||
| White | 138.2 (19.6) | 139.3 (22.8) |
| Black | 144.2 (19.3) | 147.7 (23.5) |
| Other | 138.6 (18.7) | … |
| DBP, mean (SD)a | ||
| White | 75.9 (11.0) | 73.4 (12.2) |
| Black | 78.0 (11.7) | 80.0 (14.5) |
| Other | 74.5 (10.7) | … |
| MAP, mean (SD)a | ||
| White | 96.7 (12.4) | 95.3 (14.0) |
| Black | 100.1 (12.5) | 102.6 (15.8) |
| Other | 95.9 (12.1) | … |
| Prevalent health conditions, n (%) | ||
| Hypertension | 1167 (66.5) | 358 (45.4) |
| Diabetes mellitus | 342 (19.5) | 135 (17.1) |
| Cardiovascular diseaseb | 240 (13.7) | 95 (12.1) |
| Cancer | 367 (20.9) | 107 (13.6) |
| Chronic pulmonary disease | 253 (14.4) | 52 (6.6) |
| Osteoarthritis with pain | 278 (15.8) | 25 (3.2) |
| Depressive symptoms, CESD, mean (SD) | 8.2 (7.5) | 4.7 (5.1) |
| Morbid obesity (BMI ≥35) | 346 (19.7) | 60 (7.6) |
BMI indicates body mass index; CESD, Center for Epidemiologic Studies Depression Scale; DBP, diastolic blood pressure; Health ABC, Health, Aging, and Body Composition study; LIFE, Lifestyle Interventions and Independence for Elders studies; MAP, mean arterial pressure; SBP, systolic blood pressure.
Values adjusted for antihypertensive medication use.8
Includes myocardial infarction, stroke, or heart attack.
Sequencing of ≈16.5 kb of mtDNA from LIFE and Health ABC participants yielded a cumulative total of 3364 variants including 89 common variants (MAF ≥5%), 459 low‐frequency variants (MAF 1% to <5%), and 2816 rare variants (MAF <1%). Among white and black participants, a total of 89 mtDNA variants with MAF ≥5% were examined for associations with blood pressure variation resulting in a Bonferroni adjusted P=6E−4 for significance that accounts for multiple comparisons.
Associations of mtDNA variants with SBP (Table 2), DBP (Table 3), and MAP (Table 4) are reported. Race‐specific meta‐analysis results of the LIFE and Health ABC studies are based on self‐report of white and black ancestry. We identified a total of 11 common mtDNA variants associated with SBP, DBP, and MAP among white and black participants from both the LIFE and Health ABC studies (Tables 2, 3 through 4). Of these, 6 variants yielded statistically significant associations after adjustment for multiple testing (Tables 2, 3 through 4).
Table 2.
Meta‐analytic Variant Associations With SBP Adjusted for Antihypertensive Medication Use8
| Race and Variant | LIFE (n=1755) | Health ABC (n=788) | Meta‐analysis P Value | ||||
|---|---|---|---|---|---|---|---|
| β (SE) | P Value | MAF | β (SE) | P Value | MAF | ||
| White | |||||||
| m.1811A>G, 16S rRNAa | −3.6 (1.56) | 0.021 | 0.14 | −4.8 (1.25) | 0.006 | 0.30 | 0.0013 |
| m.3197T>C, 16S rRNAa | 3.91 (1.96) | 0.0460 | 0.08 | 4.9 (1.68) | 0.0010 | 0.13 | 0.0005 |
| m.14182T>C, ND6a | 4.26 (2.24) | 0.058 | 0.06 | 4.58 (2.03) | 0.008 | 0.09 | 0.004 |
| Black | |||||||
| m.93A>G, HVIIa | 9.79 (3.5) | 0.006 | 0.12 | 7.98 (1.66) | 0.002 | 0.15 | 0.0001 |
| m.16183A>C, HVIb | 10.92 (3.84) | 0.005 | 0.10 | 10.47 (1.87) | 0.002 | 0.11 | 0.0001 |
| m.16189T>C, HVIb | 4.78 (2.31) | 0.039 | 0.50 | 4.3 (1.19) | 0.037 | 0.58 | 0.011 |
Health ABC indicates Health, Aging, and Body Composition study; LIFE, Lifestyle Interventions and Independence for Elders studies; MAF, minor allele frequency; rRNA, ribosomal RNA; SBP, systolic blood pressure.
Variant not present in other race.
Variant present at MAF <5% but not significant in other race.
Table 3.
Meta‐analytic Variant Associations With DBP Adjusted for Antihypertensive Medication Use8
| Race and Variant | LIFE Studies (n=1755) | Health ABC (n=788) | Meta‐analysis P Value | ||||
|---|---|---|---|---|---|---|---|
| β (SE) | P Value | MAF | β (SE) | P Value | MAF | ||
| White | |||||||
| m.15924A>G, tRNA‐thra | 2.85 (1.15) | 0.013 | 0.07 | 2.42 (1.51) | 0.007 | 0.21 | 0.0009 |
| Black | |||||||
| m.16172T>C, HVIb | 5.61 (2.01) | 0.006 | 0.13 | 5.95 (1.83) | 0.005 | 0.20 | 0.0003 |
| m.16189T>C, HVIb | 3.86 (1.38) | 0.005 | 0.50 | 5.73 (1.49) | 0.003 | 0.58 | 0.0002 |
DBP indicates diastolic blood pressure; Health ABC, Health, Aging, and Body Composition study; LIFE, Lifestyle Interventions and Independence for Elders studies; MAF, minor allele frequency; tRNA‐thr, threonine transfer RNA.
Variant not present in other race.
Variant present at MAF <5% but not significant in other race.
Table 4.
Meta‐analytic Variant Associations With MAP Adjusted for Antihypertensive Medication Use8
| Race and Variant | LIFE Studies (n=1755) | Health ABC (n=788) | Meta‐analysis P Value | ||||
|---|---|---|---|---|---|---|---|
| β (SE) | P Value | MAF | β (SE) | P Value | MAF | ||
| White | |||||||
| m.15924A>G, tRNA‐thra | 3.64 (1.3) | 0.005 | 0.07 | 3.92 (1.74) | 0.008 | 0.21 | 0.0004 |
| m.16069C>T, HVIa | −2.17 (1.07) | 0.043 | 0.11 | −2.72 (2.2) | 0.005 | 0.12 | 0.002 |
| m.3197T>C, 16S rRNAa | 2.33 (1.23) | 0.060 | 0.08 | 4.39 (2.09) | 0.007 | 0.13 | 0.004 |
| Black | |||||||
| m.10586G>A, ND4La | −5.41 (2.61) | 0.039 | 0.09 | −5.22 (2.26) | 0.064 | 0.15 | 0.017 |
| m.16172T>C, HVIb | 6.38 (2.16) | 0.003 | 0.13 | 8.11 (1.99) | 0.007 | 0.20 | 0.0002 |
| m.16183A>C, HVIb | 7.63 (2.48) | 0.002 | 0.10 | 11.9 (2.53) | 0.003 | 0.11 | 0.00008 |
| m.16189T>C, HVIb | 4.17 (1.48) | 0.005 | 0.50 | 6.68 (1.62) | 0.003 | 0.58 | 0.00018 |
| m.93A>G, HV2a | 5.92 (2.27) | 0.010 | 0.12 | 6.92 (2.25) | 0.054 | 0.15 | 0.0046 |
Health ABC indicates Health, Aging, and Body Composition study; LIFE, Lifestyle Interventions and Independence for Elders studies; MAF, minor allele frequency; MAP, mean arterial pressure; rRNA, ribosomal RNA; tRNA‐thr, threonine transfer RNA.
Variant not present in other race.
Variant present at MAF <5% but not significant in other race.
Each significant associated variant exhibited associations with >1 blood pressure trait either nominally or significantly after adjustment for multiple testing as follows. Among white participants, we identified 2 common mtDNA variants significantly associated with SBP and MAP (Tables 2, 3 through 4, Figures 1 and 2). These results include the m.3197T>C, 16S rRNA variant, which was associated with significantly higher SBP (P=5E−4) and was nominally associated with higher MAP (P=4E−3). In addition, the m.15924A>G, threonine tRNA (tRNA‐thr) variant was associated with significantly higher MAP (P=4E−4) and was nominally associated with higher DBP (P=9E−4). Among black participants, we identified 5 common mtDNA variants that were significantly associated with SBP, DBP, and MAP (Tables 2, 3 through 4, Figures 1 and 2). These results include the m.16183A>C, HVI variant, which was significantly associated with both higher SBP (P=1E−4) and higher MAP (P=8E−5). The m.16189T>C, HVI variant was significantly associated with both higher DBP (P=2E−4) and MAP (P=1.8E−4) while being nominally associated with SBP (P=1.1E−2). In addition, the m.16172T>C, HVI variant was associated with significantly higher MAP (P=2E−4) and was nominally associated with higher DBP (P=3E−4). The m.12705C>T variant was also associated with significantly higher MAP (P=4E−4). Finally, the m.93A>G, HVII variant was associated with significantly higher SBP (P=1E−4) and was nominally associated with higher MAP (P=4.6E−3).
Figure 1.
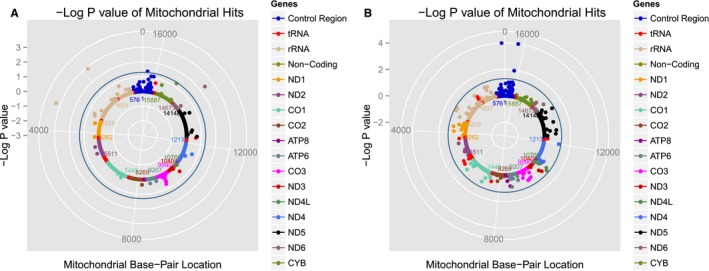
Solar plots representing mitochondrial DNA (mtDNA) variant associations with systolic blood pressure (SBP) among white (A) and black (B) participants from the LIFE (Lifestyle Interventions and Independence for Elders) and Health ABC (Health, Aging, and Body Composition) studies. Each dot represents an mtDNA variant association with SBP color‐coded by mitochondrial gene. The blue circle represents P=0.05. rRNA indicates ribosomal RNA; tRNA, transfer RNA.
Figure 2.
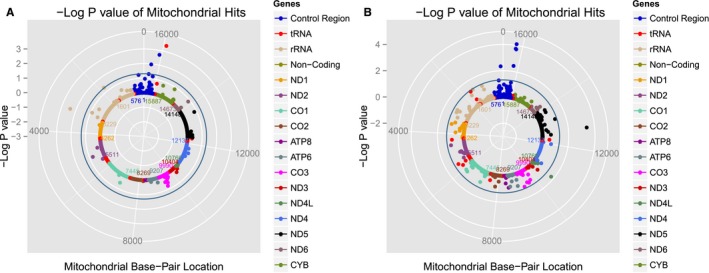
Solar plots representing mitochondrial DNA (mtDNA) variant associations with mean arterial pressure (MAP) among white (A) and black (B) participants from the LIFE (Lifestyle Interventions and Independence for Elders) and Health ABC (Health, Aging, and Body Composition) studies. Each dot represents an mtDNA variant association with MAP color‐coded by mitochondrial gene. The blue circle represents P=0.05. rRNA indicates ribosomal RNA; tRNA, transfer RNA.
Figure 3 illustrates SBP values for each allele at the m.3197T>C, m.93A>G, and m.16183A>C sites. Figure 4 illustrates MAP values for each allele at the m.15924A>G, m.16172T>C, m.16183A>C, HV, and m.16189T>C, HVI sites. The majority of the associated variants were unique to each racial group, making meta‐analysis impossible for these loci. None of the variants present in all races (white, black, and other) were significantly associated with blood pressure across races.
Figure 3.
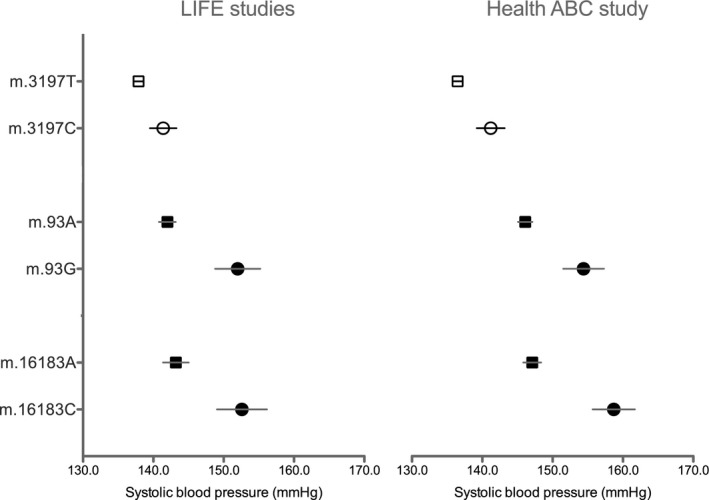
Average systolic blood pressure values for the m.3197T>C position in participants with European ancestry and the m.93A>G and m.16183A>C positions in participant with African ancestry. Values are expressed as mean (SEM) in the LIFE (Lifestyle Interventions and Independence for Elders) and Health ABC (Health, Aging, and Body Composition) studies.
Figure 4.
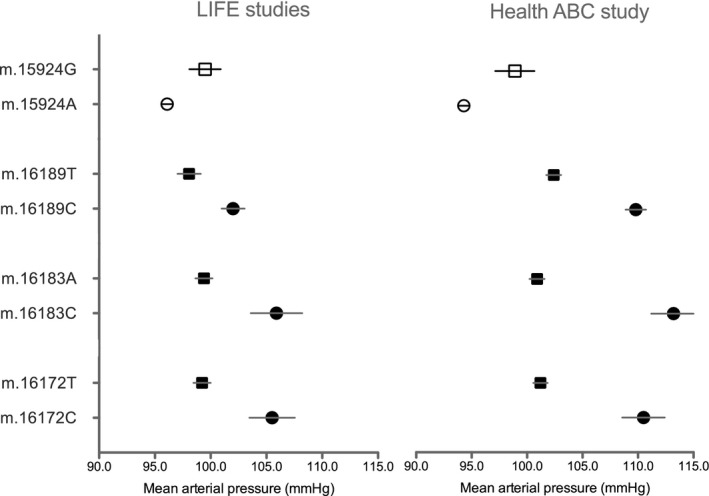
Average mean arterial pressure values for the m.15924A>G position in participants with European ancestry and the m.16172T>C, m.16183A>C, and m.16189T>C positions in participants with African ancestry. Values are expressed as mean (SEM) in the LIFE (Lifestyle Interventions and Independence for Elders) and Health ABC (Health, Aging, and Body Composition) studies.
Significant pooled effects (P=0.007 due to multiple test correction for 7 mtDNA regions) on SBP were observed across all tRNA regions (P=0.0056) in white participants from the LIFE and Health ABC studies. Nominally significant pooled effects across all tRNA regions (P=0.014) were observed among white participants for MAP.
We focused in silico prediction methods on the assessment of nucleotide conservation generated via phyloP38 because none of the associated variants conferred nonsynonymous substitutions. In brief, phyloP performs multiple alignments of 100 vertebrate species and calculates metrics of evolutionary conservation for each variant under a null hypothesis of neutral evolution. The absolute values of phyloP scores represent −log P values, with positive scores reflecting evolutionary conservation across taxa (ie, slower rates than expected) and negative scores reflecting accelerated evolution (ie, faster than expected). The 6 associated variants exhibited negative phyloP scores, implying that these variants are not highly conserved among multiple vertebrate taxa: m.93A>G=−0.79, m.3197T>C=−5.13, m.15924A>G=−0.63, m.16172T>C=−1.79, m.16183A>C=−1.09, and m.16189T >C=−12.39.
Discussion
Cardiovascular function is a complex trait attributable to multiple genetic and nongenetic factors.5, 6, 7, 8, 9, 10 To date, few studies have evaluated the role of the mitochondrial genome on cardiovascular functions such as blood pressure. Moreover, many questions remain regarding the role of mtDNA in age‐related changes in health.39 As such, no large, multiethnic, comprehensive examination exists regarding the role of mtDNA variation in regulating blood pressure among older adults. In the current study, we present results from the first comprehensive examination of mitochondrial genomic variation in late‐life blood pressure. These data provide evidence that specific mtDNA variants are associated with elevated SBP, DBP, and MAP in meta‐analyses involving participants sampled from 2 well‐characterized older cohorts (LIFE and Health ABC). Each of the variants identified among white and black study participants was associated with >1 measure of blood pressure variation; however, none of the variants was associated across racial groups. The mtDNA associations with blood pressure variation reported herein suggest that mitochondrial variants may have an important role regarding their effects on biological pathways that underlie blood pressure variation. Moreover, it appears that relevant variants may be unique to specific racial groups, creating yet further need for follow‐up studies in this area.
The interpretation of mtDNA associations with measured traits is often complicated by the observation that recurrent mutations at the same mtDNA site arise independently across populations of divergent ancestries. By examining participants of African and European ancestry, we leveraged the extreme diversity of mitochondrial genetics among these groups to identify unique and shared biological mechanisms underlying cardiovascular traits among genetically unique populations. The single‐copy and maternally inherited nature of the mtDNA ensures that specific variants unique to a single ancestral group (eg African, European, Asian) will persist only in that group unless the mutation either arises de novo (independently) in a new population or is introduced to a new population through admixture among populations harboring ancestral groups. Previous observations of the same mtDNA mutations on different ancestral backgrounds has been cited as evidence of convergent adaptive evolution of particular mtDNA mutations.40 The lack of consistent associations among participants of African and European ancestries in this and other studies, however, illustrates the need to report results across these diverse human populations. For example, of the 5 variants associated with cardiovascular traits in participants of European ancestry in this study, none were present in participants with African ancestry. Of the 5 variants associated with cardiovascular traits in participants of African ancestry, 2 were not present in participants of European ancestry, whereas the remaining 3 variants were detected at extremely low frequencies in participants of European ancestry (1–4%), thus limiting statistical power to detect associations among European ancestry participants. By presenting statistically significant results from 2 independent studies made up of participants with African and European ancestries, we are contributing a more comprehensive and biologically relevant study of cardiovascular function across diverse populations.
Among white participants, variants in tRNA‐thr (m.15924A>G) and 16S rRNA (m.3197T>C) were significantly or nominally associated with higher SBP, DBP, and/or MAP. More than half of pathogenic mtDNA mutations are located in genes controlling tRNA expression.41 Such mutations lead to reduced tRNA steady‐state levels, decreased mitochondrial protein synthesis, and destabilized tRNA secondary or tertiary structure42, 43 and impaired oxidative phosphorylation and oxygen consumption.43 Elsewhere, tRNA‐thr m.15294T>C and other tRNA‐thr variants have been associated with dilated cardiomyopathy.44, 45 Several additional tRNA‐thr mutations located in close proximity to the m.15924A>G identified in the present study have also been associated with diseases including encephalomyopathy (m.15915G>A) and mitochondrial myopathy (m.15923A>G and m.15940T>delT).46 Moreover, several lysidine tRNAs and methionine tRNAs have been associated with the presence of essential hypertension.13 In addition, several 16S rRNA mutations have been associated with a variety of mitochondrial diseases.46 The m.3197 T>C variant identified in the current study is located in domain IV of the 16S rRNA and is adjacent to the m.3196G>A site, which has been associated with Alzheimer and Parkinson diseases.46 With regard to mitochondrial haplogroup associations, the m.15924A>G variant is found in multiple lineages from superhaplogroups L, M, and N (including sublineages of European haplogroups U, K, H, and J). The m.3197T>C, 16S rRNA variant is found in multiple lineages of superhaplogroup N including specific sublineages of European haplogroups H and U. Though speculative, it is possible that these mutations contribute to mitochondrial dysfunction, which impairs nutrient metabolism—ultimately, increasing blood pressure.47
Among black participants, variants in the HVI (m.16172T>C, m.16183A>C, and m.16189T>C) and HVII (m.93A>G) regions were nominally or significantly associated with higher SBP, DBP, and/or MAP. Prior studies suggest that HV sequence variation may be involved in regulating mtDNA copy number48, 49 and may directly affect mitochondrial biogenesis (increase in mitochondrial number and/or mass). The m.16172T>C and m.93A>G variants are found in multiple lineages from superhaplogroups L, M, and N (including African haplogroups L0, L1, L2, L3, and L4). The m.16183A>C is highly variable and provides little phylogenetic value50 but has been associated with melanoma risk.46 The m.16189T>C variant encodes the ancestral including split between African haplogroups L0/L1/L5 and L2/L3/L4/L6 and has been associated with risk of diabetes mellitus, cardiomyopathy, metabolic syndrome, melanoma, and endometrial cancer.46 A polycytosine variant at m.16189 has been associated with mtDNA copy number in human peripheral blood cells.48
Collections of variants within genes or genomic regions influence phenotypes in diverse ways,14 and examining the combined effect of rare variants may also reveal the role of specific genes in disease etiology. Significant pooled effects on SBP were observed across all tRNA regions in white participants from both LIFE and Health ABC studies (nominally significant pooled effects across all tRNA regions were observed for MAP). As described previously, individual and collective variation in the tRNA and region may affect mitochondrial function by affecting the rate or efficiency of mitochondrial biogenesis that may decline with age.51 Impaired ability to turn over may allow for defective mitochondria to accumulate, especially in older, postmitotic cells, affecting blood pressure variation.52
As the first data of their kind, the ultimate physiological influence of these data remains unclear. It is possible that specific mtDNA variants contribute to issues in the heart or vascular endothelium that manifest most dramatically as the tissues age. It is also possible that these variants contribute to altered kidney function in late life. Mitochondria are particularly important in the kidney tubules, where oxygen consumption is utilized primarily for active solute transport.53, 54 Proximal tubule cells, in particular, are densely packed with mitochondria, given their high energy requirements and limited capacity for anaerobic metabolism,55 and inherited mitochondrial disorders are often characterized by severe proximal tubular dysfunction. Defective mitochondria are deleterious at the intracellular level both because they are unable to produce adequate ATP, and they leak reactive oxygen species, which can trigger inflammation and cell death.56 Although speculative, it is possible that these variants contribute to altered kidney function, which contributes to hypertension development. Future studies are needed to confirm this hypothesis.
This study had several strengths, including complete mtDNA sequencing, which allows for an unbiased assessment of mitochondrial genomic variation; 2 well‐characterized cohorts that included participants from multiple ancestries; and a large range in blood pressure variation. Some limitations are also acknowledged, including limited power to detect an effect of individual rare variants. It is also possible that the mtDNA variants identified in this study may not be causally related to blood pressure variation, thus additional replication cohorts beyond LIFE and Health ABC are needed.
It should be noted that the results of the present study should not be overinterpreted. As the first study to identify these mtDNA variant associations with blood pressure, it is important that the winner's curse phenomenon—whereby identified variants may be assigned overly optimistic roles in a physiological or disease process—be considered. For instance, given prior effect sizes for blood pressure observed in the literature, it seems unlikely that the true effect size for m.16183A>C is >10 mm Hg. This finding, among those others found significant, highlights the need to be cautious with interpretation of these data as well as for the need for replication in future studies. It is also critical to confine interpretations of these data to older adults because the variants may not have the same associations with phenotypic outcomes in younger cohorts. Still, these data from 2 relatively large studies in the field of aging are the first to indicate associations between mtDNA variants and BP among older people. Consequently, these novel data provide an important contribution to the literature and a foundation for further follow‐up study regarding the role of mtDNA on blood pressure levels in late life.
In conclusion, these data reveal identify mtDNA variants associated with interindividual variation in blood pressure among older adults. These novel data may eventually be used to identify older people at risk of developing hypertension and associated complications. Despite being the primary source of cellular energy production within most cell types—including the heart, kidney, and vascular endothelium—relatively few studies have focused on the mitochondrial genome in blood pressure regulation. In particular, data are lacking from older adults—a group predominantly affected by hypertension. These data may also provide the foundation for generation and/or testing of novel hypotheses surrounding the involvement of mitochondria in the regulation of blood pressure during advanced age.
Sources of Funding
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Institute on Aging, contracts N01‐AG‐6‐2101, N01‐AG‐6‐2103, and N01‐AG‐6‐2106; NIH grants R01‐AG028050, R03‐AG032498, R01‐NR012459, Z01A6000932, and R01‐HL121023; and grants from the Research and Education Leadership Committee of the CPMC Foundation and the L. K. Whittier Foundation. The LIFE (Lifestyle Interventions and Independence for Elders) study was also supported by cooperative agreements U01 AG22376, 3U01AG022376‐05A2S as well by the Claude D. Pepper Older Americans Independence Centers at the University of Florida (1 P30 AG028740), Tufts University (1P30AG031679), University of Pittsburgh (P30 AG024827), Wake Forest University (P30AG021332), and Yale University (P30AG021342), the NIH/National Center for Research Resources Clinical and Translational Science Award at Stanford University (UL1 RR025744), and the Boston Rehabilitation Outcomes Center (1R24HD065688‐01A1) at Tufts University. Dr Fielding's work on this project was partially supported by the US Department of Agriculture (USDA), under agreement no. 58‐1950‐4‐003, and the Boston Claude D. Pepper Center Older American Independence Centers (1P30AG031679). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the USDA.
Disclosures
None.
Supporting information
Appendix S1. Research investigators for the LIFE (Lifestyle Interventions and Independence for Elders) study.
Table S1. Blood Pressure Characteristics of Study Cohorts
Acknowledgments
A full list of LIFE investigators is provided in Appendix S1.
(J Am Heart Assoc. 2018;7:e010009 DOI: 10.1161/JAHA.118.010009.)
Contributor Information
Thomas W. Buford, Email: twbuford@uabmc.edu.
Gregory J. Tranah, Email: gtranah@psg.ucsf.edu.
References
- 1. Roberts L. 9 billion? Science. 2011;333:540–543. [DOI] [PubMed] [Google Scholar]
- 2. Hodgson TA, Cohen AJ. Medical care expenditures for selected circulatory diseases: opportunities for reducing national health expenditures. Med Care. 1999;37:994–1012. [DOI] [PubMed] [Google Scholar]
- 3. Bramlage P, Hasford J. Blood pressure reduction, persistence and costs in the evaluation of antihypertensive drug treatment—a review. Cardiovasc Diabetol. 2009;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P; American Heart Association Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics—2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hottenga JJ, Whitfield JB, de Geus EJ, Boomsma DI, Martin NG. Heritability and stability of resting blood pressure in Australian twins. Twin Res Hum Genet. 2006;9:205–209. [DOI] [PubMed] [Google Scholar]
- 6. Finkel D, Pedersen NL, Reynolds CA, Berg S, de Faire U, Svartengren M. Genetic and environmental influences on decline in biobehavioral markers of aging. Behav Genet. 2003;33:107–123. [DOI] [PubMed] [Google Scholar]
- 7. Steves CJ, Spector TD, Jackson SH. Ageing, genes, environment and epigenetics: what twin studies tell us now, and in the future. Age Ageing. 2012;41:581–586. [DOI] [PubMed] [Google Scholar]
- 8. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace‐Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O'Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome‐wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. International Consortium for Blood Pressure Genome‐Wide Association S , Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg‐Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw‐Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland‐Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND; Consortium CA, Consortium CK, KidneyGen C, EchoGen c, Consortium C‐H , Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace‐Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala‐Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska‐Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton‐Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Liu K, Morrison AC, Ganesh S, Kutlar A, Ramachandran VS, Polak JF, Fabsitz RR, Dries DL, Farlow DN, Redline S, Adeyemo A, Hirschorn JN, Sun YV, Wyatt SB, Penman AD, Palmas W, Rotter JI, Townsend RR, Doumatey AP, Tayo BO, Mosley TH Jr, Lyon HN, Kang SJ, Rotimi CN, Cooper RS, Franceschini N, Curb JD, Martin LW, Eaton CB, Kardia SL, Taylor HA, Caulfield MJ, Ehret GB, Johnson T; International Consortium for Blood Pressure Genome‐wide Association Studies (ICBP‐GWAS) , Chakravarti A, Zhu X, Levy D. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lemieux H, Hoppel CL. Mitochondria in the human heart. J Bioenerg Biomembr. 2009;41:99–106. [DOI] [PubMed] [Google Scholar]
- 12. Davidson SM, Duchen MR. Endothelial mitochondria: contributing to vascular function and disease. Circ Res. 2007;100:1128–1141. [DOI] [PubMed] [Google Scholar]
- 13. Ding Y, Xia B, Yu J, Leng J, Huang J. Mitochondrial DNA mutations and essential hypertension (review). Int J Mol Med. 2013;32:768–774. [DOI] [PubMed] [Google Scholar]
- 14. Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wallace DC, Stugard C, Murdock D, Schurr T, Brown MD. Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc Natl Acad Sci USA. 1997;94:14900–14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merriwether DA, Clark AG, Ballinger SW, Schurr TG, Soodyall H, Jenkins T, Sherry ST, Wallace DC. The structure of human mitochondrial DNA variation. J Mol Evol. 1991;33:543–555. [DOI] [PubMed] [Google Scholar]
- 17. Tranah GJ, Lam ET, Katzman SM, Nalls MA, Zhao Y, Evans DS, Yokoyama JS, Pawlikowska L, Kwok PY, Mooney S, Kritchevsky S, Goodpaster BH, Newman AB, Harris TB, Manini TM, Cummings SR; Health, Aging and Body Composition Study . Mitochondrial DNA sequence variation is associated with free‐living activity energy expenditure in the elderly. Biochem Biophys Acta. 2012;1817:1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tranah GJ. Mitochondrial‐nuclear epistasis: implications for human aging and longevity. Ageing Res Rev. 2011;10:238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katzman SM, Strotmeyer ES, Nalls MA, Zhao Y, Mooney S, Schork N, Newman AB, Harris TB, Yaffe K, Cummings SR, Liu Y, Tranah GJ; for the Health, Aging and Body Composition Study . Mitochondrial DNA sequence variation associated with peripheral nerve function in the elderly. J Gerontol A Biol Sci Med Sci. 2015;70:1400–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lam ET, Bracci PM, Holly EA, Chu C, Poon A, Wan E, White K, Kwok PY, Pawlikowska L, Tranah GJ. Mitochondrial DNA sequence variation and risk of pancreatic cancer. Cancer Res. 2012;72:686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tranah GJ, Yokoyama JS, Katzman SM, Nalls MA, Newman AB, Harris TB, Cesari M, Manini TM, Schork NJ, Cummings SR, Liu Y, Yaffe K; Health, Aging and Body Composition Study . Mitochondrial DNA sequence associations with dementia and amyloid‐beta in elderly African Americans. Neurobiol Aging. 2014;35:442.e441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tranah GJ, Yaffe K, Katzman SM, Lam ET, Pawlikowska L, Kwok PY, Schork NJ, Manini TM, Kritchevsky S, Thomas F, Newman AB, Harris TB, Coleman AL, Gorin MB, Helzner EP, Rowbotham MC, Browner WS, Cummings SR; Health, Aging and Body Composition Study . Mitochondrial DNA heteroplasmy associations with neurosensory and mobility function in the elderly. J Gerontol A Biol Sci Med Sci. 2015;70:1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tranah GJ, Santaniello A, Caillier SJ, D'Alfonso S, Boneschi FM, Hauser SL, Oksenberg JR. Mitochondrial DNA sequence variation in multiple sclerosis. Neurology. 2015;85:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fielding RA, Rejeski WJ, Blair S, Church T, Espeland MA, Gill TM, Guralnik JM, Hsu FC, Katula J, King AC, Kritchevsky SB, McDermott MM, Miller ME, Nayfield S, Newman AB, Williamson JD, Bonds D, Romashkan S, Hadley E, Pahor M. The lifestyle interventions and independence for elders study: design and methods. J Gerontol A Biol Sci Med Sci. 2011;66:1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rejeski WJ, Fielding RA, Blair SN, Guralnik JM, Gill TM, Hadley EC, King AC, Kritchevsky SB, Miller ME, Newman AB, Pahor M. The lifestyle interventions and independence for elders (LIFE) pilot study: design and methods. Contemp Clin Trials. 2005;26:141–154. [DOI] [PubMed] [Google Scholar]
- 26. Visser M, Kritchevsky SB, Goodpaster BH, Newman AB, Nevitt M, Stamm E, Harris TB. Leg muscle mass and composition in relation to lower extremity performance in men and women aged 70 to 79: the Health, Aging and Body Composition Study. J Am Geriatr Soc. 2002;50:897–904. [DOI] [PubMed] [Google Scholar]
- 27. Health ABC . Available at: https://healthabc.nia.nih.gov/. LIFE . Available at: https://www.thelifestudy.org/public/index.cfm. Accessed August 1, 2018.
- 28. Cui JS, Hopper JL, Harrap SB. Antihypertensive treatments obscure familial contributions to blood pressure variation. Hypertension. 2003;41:207–210. [DOI] [PubMed] [Google Scholar]
- 29. Kraft P, Zeggini E, Ioannidis JP. Replication in genome‐wide association studies. Stat Sci. 2009;24:561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeggini E, Ioannidis JP. Meta‐analysis in genome‐wide association studies. Pharmacogenomics. 2009;10:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skol AD, Scott LJ, Abecasis GR, Boehnke M. Optimal designs for two‐stage genome‐wide association studies. Genet Epidemiol. 2007;31:776–788. [DOI] [PubMed] [Google Scholar]
- 32. Nguyen TT, Pahl R, Schafer H. Optimal robust two‐stage designs for genome‐wide association studies. Ann Hum Genet. 2009;73:638–651. [DOI] [PubMed] [Google Scholar]
- 33. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta‐analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta‐analyses of genome‐wide association investigations. PLoS One. 2007;2:e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta‐analyses. BMJ. 2003;327:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare‐variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2010;89:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tranah GJ, Manini TM, Lohman KK, Nalls MA, Kritchevsky S, Newman AB, Harris TB, Miljkovic I, Biffi A, Cummings SR, Liu Y. Mitochondrial DNA variation in human metabolic rate and energy expenditure. Mitochondrion. 2011;11:855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shokolenko IN, Wilson GL, Alexeyev MF. Aging: a mitochondrial DNA perspective, critical analysis and an update. World J Exp Med. 2014;4:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wallace DC. Colloquium paper: bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci USA. 2010;107(suppl 2):8947–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zifa E, Giannouli S, Theotokis P, Stamatis C, Mamuris Z, Stathopoulos C. Mitochondrial tRNA mutations: clinical and functional perturbations. RNA Biol. 2007;4:38–66. [DOI] [PubMed] [Google Scholar]
- 42. Hao H, Moraes CT. A disease‐associated G5703A mutation in human mitochondrial DNA causes a conformational change and a marked decrease in steady‐state levels of mitochondrial tRNA(Asn). Mol Cell Biol. 1997;17:6831–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guan MX, Enriquez JA, Fischel‐Ghodsian N, Puranam RS, Lin CP, Maw MA, Attardi G. The deafness‐associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long‐range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998;18:5868–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arbustini E, Diegoli M, Fasani R, Grasso M, Morbini P, Banchieri N, Bellini O, Dal Bello B, Pilotto A, Magrini G, Campana C, Fortina P, Gavazzi A, Narula J, Vigano M. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am J Pathol. 1998;153:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruppert V, Nolte D, Aschenbrenner T, Pankuweit S, Funck R, Maisch B. Novel point mutations in the mitochondrial DNA detected in patients with dilated cardiomyopathy by screening the whole mitochondrial genome. Biochem Biophys Res Commun. 2004;318:535–543. [DOI] [PubMed] [Google Scholar]
- 46. Mitomap: a human mitochondrial genome database. Available at: http://www.Mitomap.Org. Accessed March 15, 2018.
- 47. Bernal‐Mizrachi C, Gates AC, Weng S, Imamura T, Knutsen RH, DeSantis P, Coleman T, Townsend RR, Muglia LJ, Semenkovich CF. Vascular respiratory uncoupling increases blood pressure and atherosclerosis. Nature. 2005;435:502–506. [DOI] [PubMed] [Google Scholar]
- 48. Liou CW, Lin TK, Chen JB, Tiao MM, Weng SW, Chen SD, Chuang YC, Chuang JH, Wang PW. Association between a common mitochondrial DNA D‐loop polycytosine variant and alteration of mitochondrial copy number in human peripheral blood cells. J Med Genet. 2010;47:723–728. [DOI] [PubMed] [Google Scholar]
- 49. Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. [DOI] [PubMed] [Google Scholar]
- 51. Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E. Age‐related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J Gerontol A Biol Sci Med Sci. 2001;56:B288–B293. [DOI] [PubMed] [Google Scholar]
- 52. de Grey AD. A proposed refinement of the mitochondrial free radical theory of aging. Bioessays. 1997;19:161–166. [DOI] [PubMed] [Google Scholar]
- 53. Weinberg JM, Venkatachalam MA, Roeser NF, Nissim I. Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc Natl Acad Sci USA. 2000;97:2826–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hansell P, Welch WJ, Blantz RC, Palm F. Determinants of kidney oxygen consumption and their relationship to tissue oxygen tension in diabetes and hypertension. Clin Exp Pharmacol Physiol. 2013;40:123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hall AM, Unwin RJ. The not so ‘mighty chondrion’: emergence of renal diseases due to mitochondrial dysfunction. Nephron Physiol. 2007;105:p1–p10. [DOI] [PubMed] [Google Scholar]
- 56. Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial damps cause inflammatory responses to injury. Nature. 2010;464:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Research investigators for the LIFE (Lifestyle Interventions and Independence for Elders) study.
Table S1. Blood Pressure Characteristics of Study Cohorts


