Abstract
Background
Resistance of Plasmodium falciparum to anti-malarial drugs has hampered efforts to eradicate malaria. Recent reports of a decline in the prevalence of chloroquine-resistant P. falciparum in several countries, including Malawi and Zambia, is raising the hope of reintroducing chloroquine in the near future, ideally in combination with another anti-malarial drug for the treatment of uncomplicated malaria. In Côte d’Ivoire, the decrease in the clinical efficacy of chloroquine, in addition to a high proportion of clinical isolates carrying the Thr-76 mutant allele of the pfcrt gene, had led to the discontinuation of the use of chloroquine in 2004. Previous studies have indicated the persistence of a high prevalence of the Thr-76 mutant allele despite the withdrawal of chloroquine as first-line anti-malarial drug. This present study is conducted to determine the prevalence of the Thr-76T mutant allele of the Pfcrt gene after a decade of the ban on the sale and use of chloroquine in Côte d’Ivoire.
Results
Analysis of the 64 sequences from all three study sites indicated a prevalence of 15% (10/64) of the Thr-76 mutant allele against 62% (40/64) of the Lys-76 wild-type allele. No mutation of the allele Thr-76 was observed at Anonkoua Kouté while this mutant allele was in 31% (5/16) and 25% (5/20) of isolate sequences from Port-Bouët and Ayamé respectively.
Conclusion
More than a decade after the discontinuation of the use of chloroquine in Côte d’Ivoire, the proportion of parasites sensitive to this anti-malarial seems to increase in Anonkoua-kouté, Port-bouët and Ayamé.
Electronic supplementary material
The online version of this article (10.1186/s12936-018-2551-7) contains supplementary material, which is available to authorized users.
Keywords: Pfcrt, Thr-76, Chloroquine sensitivity, Côte d’Ivoire, Antimalarial drug resistance
Background
Malaria remains a major public health problem in the world. According to the World Health Organization (WHO), 212 million cases of malaria were recorded in 2015, of which 429,000 led to death, of which 92% occurred in Africa with 70% of children under 5 years of age [1]. Up to 1990, chloroquine (CQ) was the main malaria treatment therapy thanks to its efficacy, safety, low cost and antipyretic properties. In the late 1950s, resistance to CQ emerged in different parts of the world, first in South-East Asia and South America (Colombia and Venezuela) [2–5]. The resistance to CQ spread rapidly and was detected in West Africa in the 80s and 90s [6, 7].
In Côte d’Ivoire, the prevalence of CQ-resistant parasites that used to be very low in 1987 with only three confirmed cases of chloroquine resistance (CQR) appeared to have increased [8, 9]. Indeed, high prevalence of the K76T mutation, a key mutation associated with Plasmodium falciparum resistance to CQ, have been reported in Yopougon in Abidjan (65%), Bonoua (100%), Samo (95%) and Adzopé (62%) [10]. Recent reports of a decline in the prevalence of chloroquine-resistant P. falciparum in several countries, including Malawi and Zambia, raise the hope of reintroducing chloroquine in the near future ideally in combination with another anti-malarial drug for the treatment of uncomplicated malaria [11, 12]. It could possibly be given to non-vulnerable groups, but it requires close monitoring of possible reemergence of CQ resistance development. Indeed, it has been suggested that effective and sustained withdrawal of CQ could lead to the reappearance of CQ-sensitive P. falciparum. It is the case in East Africa, particularly in Malawi and Kenya where a re-emergence of this sensitivity to CQ was reported after the discontinuation of its uses in 1993 and 1999, respectively [11, 12]. Some studies in Cameroon (Central Africa) and in Senegal (West Africa) [13–15], have reported the same trend for susceptibility to CQ after withdrawal as first-line malaria treatment.
In Côte d’Ivoire, the decreasing clinical efficacy of chloroquine, in addition to a high proportion of resistant isolates, led the Ivorian health authorities to call for withdrawal and discontinuation of the use of chloroquine in 2004 in favor of artemisinin-based combination therapy (ACT) as a first-line treatment for uncomplicated malaria. This study aimed to investigate the prevalence of the K76T mutation a little more than a decade after the official withdrawal of CQ in uncomplicated malaria treatment management in Côte d’Ivoire.
Methods
Study site
This was a prospective study that took place in three different health centers, Anonkoua Kouté, Port-Bouët general hospital and Ayamé from February to August 2015. All these sites are located in the southern region of Côte d’Ivoire where the climate is equatorial with annual rainfall exceeding 1700 mm of rain and the temperature varies between 27 and 33 °C. Malaria is seasonal, more frequent during the rainy season from June to September with peaks prevalence rate and incidence in October–November. Plasmodium falciparum is the dominant species with more than 90% of malaria parasites identified. The main vectors of malaria in this study area (the southern forest zone of Côte d’Ivoire) are the members of the complexes Anopheles gambiae sensu lato (s.l.) and Anopheles funestus s.l. [16].
Anonkoua-kouté Health Centre and Ayamé General Hospital were selected based on the high annual incidences of malaria cases records. In addition, these health centers are chosen for several years as the main sites for performing multicenter clinical efficacy tests by the Malaria Unit of Institut Pasteur of Côte d’Ivoire. Port Bouët general hospital was selected for this study not only because of the constantly high annual incidences of malaria cases, but also and especially because of its swampy environment often used for market garden produces.
Study population and sample collection
All patients clinically suspected to have malaria at Anonkoua Kouté Health Center, Port-Bouët general hospitals and Ayamé during the study period were eligible. However, after informed consent, blood samples were collected from patients who are over 2 years of age with an axillary temperature greater than 37.5 °C and suffering from uncomplicated P. falciparum malaria confirmed by microscopic examination.
Blood sample
In each patient who have been confirmed of having malaria by microscopic examination, approximately 2–5 mL of venous blood was collected in an EDTA tube. Approximately 50 μL of whole blood was dropped on Whatman 3 MM filter paper discs [dried blood spots (DBS)] using a micropipette with filter cones. The papers containing the blood spots were dried for about 60–120 min at room temperature away from dust. Unused blood contained in the EDTA tube was stored in microtubes at − 20 °C for possible subsequent utilization.
Extraction of Plasmodium falciparum genomic DNA
Plasmodial DNA was extracted with methanol from DBS cut into small pieces were immersed in 1 mL of washing buffer (950 μL of 1× PBS plus 50 μL of 10% saponin) and incubated overnight at 4 °C [17]. The wash buffer was removed and 150 μL of methanol were added. After a 20 min incubation, the methanol was gently removed and the samples were dried at room temperature for 2 h before adding 300 μL of sterilized water. The samples were then heated at 99 °C in a thermo-mixer for 30 min to elute the DNA. After removing the confetti debris, the DNA extracts were aliquoted into a 1.5 mL Eppendorf tube and stored at − 20 °C.
Amplification of the pfcrt gene
The pfcrt gene was amplified by nested PCR using a specific pair of primers and a commercial DNA polymerase kit called 5× FIREPol Blend Master Mix with mM MgCl2. This kit is a pre-mix (for the reaction mixture) ready to use composed of DNA polymerase (FIREPol® DNA polymerase), buffer (5× Blend Master Mix Buffer), MgCl2 (7.5 mM MgCl2) and dNTPs (2 mM dNTPs of each). For primary PCR, the primer pairs used for amplification of the pfcrt gene were 72_97EF (5′GACCTTAACAGGTGGCTCAC)/72_97ER (5′TTTATTGGTAGGTGGAATAG). The primary PCR of this gene was carried out in a reaction volume of 25 μL containing: 0.625 μL of each primer, 3 μL of plasmodial DNA, 5 μL of Taq DNA polymerase and 15.75 μL of milliQ water. The mixture was then put into a PTC-100TM thermocycler (Eppendorf Mastercycler, PTC-100 Peltier Thermal Cycler), programmed as follows: Initial denaturation at 95 °C for 15 min followed by 30 denaturation cycles at 95 °C for 30 s, hybridization at 58 °C for 2 min and extension at 72 °C for 2 min. Finally, a terminal extension at 72 °C for 10 min.
The second PCR was carried out on the amplification products of the primary PCR in a reaction volume of 50 μL containing: 1.25 μL of each primer, 5 μL of amplification product (amplicon) of the first PCR, of 5 μL of Taq DNA polymerase and 37.5 μL of milliQ water. The primer pairs used for the secondary PCR were SecIF (5′ GGTAAATGTGCTCATGTGTTTAAACTTATT)/SecIR (5′ TTACTTTTGAATTTCCCTTTTTATTTCCA). The secondary PCR was performed with the same thermocycler used for the primary PCR with the following program: Initial denaturation at 95 °C for 15 min followed by 30 denaturation cycles at 95 °C for 30 s, hybridization at 60 °C for one minute and extension at 72 °C for 1 min. Finally, a terminal extension at 72 °C for 10 min.
Detection and analysis of PCR products
The amplification products were migrated on a 1.5% agarose gel containing ethidium bromide (EtBr). After migration, the gel was recovered and then observed under a UV lamp using the UV transilluminator (Gel DocTM EZ Imager). The presence or absence of bands made it possible to judge the effectiveness of the PCR.
Sequencing amplification
The amplified DNA fragments (pfcrt gene) of P. falciparum were sequenced according to the Sanger method by the company Eurofins MWG operon (Cochin sequencing platform). Samples were dropped to the platform in a microplate (Greiner Bio-one-652270B) along with a deposit slip that was sent to the platform’s email address. A reaction medium was prepared for the PCR-nested sense primer (sequencing primer) from the amplification products. In each well of the microplate, a volume of 13 μL of amplification product was added to 2 μL of sequencing primer at 10 μM. Wells containing the sequencing reaction medium were sealed with cap strips (4titude-044737) before covering the entire surface of the microplate with an adhesive film (AmpliSeal, Greiner Bio-one-676040). This microplate containing the samples was sent to the platform for sequencing.
After the sequencing reaction, the received DNA sequences were recovered as fasta. In this study, it is the sequences corresponding to the pfcrt gene of the isolates collected. The use of the software BioEdit made it possible to analyse the sequences to search for possible mutations. Indeed, the loci of interest, namely the codons at position 74, 75, 76 of the PfCRT polypeptide or the nucleotides at position 222, 225, 228 of the pfcrt gene were identified and analysed after a parallel alignment of two or more DNA sequences, including the reference sequence pfcrt gene, by maximizing the number of identical nucleotides or residues, while minimizing the number of mismatches and voids.
Statistical analysis of data
Data were collected on a standard questionnaire tested and validated. They were then entered and analysed on the statistical software R; version 3.2.2 [18]. The χ2 comparison test of three averages was used to compare the prevalence of the molecular marker of CQ resistance (pfcrt K76T). The χ2 test was used to determine whether the molecular marker prevalence can be considered to be all equal (null hypothesis H0) or if two or more prevalence are different (alternative hypothesis Ha). A difference and/or statistical association was considered significant if p of the test χ2 < 0.05.
Results
A total of 64 persons infected with P. falciparum were selected for this study, including 41 (64%) women and 23 (36%) men. The average age of the patients was 17 years (age ranging from 2 to 62 years) (Table 1). In addition, the parasite densities varied from 1200 to 200,000 parasites/μL with average parasite densities of 22,900; 9193 and 42,327 parasites/μL at Anonkoua Kouté, Port-Bouët and Ayamé, respectively (Table 1 and Fig. 1). A significant difference (p = 0.002) was observed between the parasite densities at Port-Bouët and Ayamé. The mean parasite density in all three sites was 24,806 parasites/μL.
Table 1.
Samples used for molecular analysis of chloroquine chemoresistance
| Sites | Period of collection in 2015 | Age group (years) | DBS collected | Average Parasite density (µL/mm3) |
|---|---|---|---|---|
| Anonkoua-kouté | February–March | 2–53 | 28 | 22,900 |
| Port-Bouët | April–May–July | 2–62 | 16 | 9193 |
| Ayamé | June–July–August | 2–55 | 20 | 42,327 |
| Total | 64 | 24,806 |
Fig. 1.
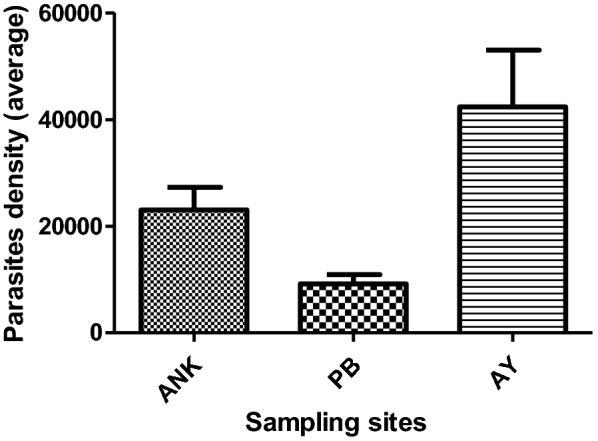
Mean parasite density versus sampling site. ANK Anonkoua-Kouté, PB Port-Bouët, AY Ayamé
Prevalence of the individual alleles of the pfcrt gene and molecular analysis of the corresponding genotypes
Across all the three study sites, the results indicated that the prevalence of wild-type isolates Met-74 (73%), Asn-75 (75%), Lys-76 (62%) is higher than those isolates carrying mutations of the pfcrt gene Ile-74 (7%), Glu-75 (7%) and Thr-76 (15%) (Table 2). Molecular analysis of the genotypes corresponding to the pfcrt gene shows that the MNK genotype (wild type) was predominant with a prevalence of 62% (Table 3). In contrast, single genotypes mutant, double mutant and triple mutant were observed with respective prevalence of 12%, 6% and 18%.
Table 2.
Prevalence of the individual alleles of the pfcrt gene in the study sites
| Codons | Alleles | Sample size (N = 64) | |
|---|---|---|---|
| n = 59 | % | ||
| Crt_74 | Wild | ||
| Met-74 | 47 | 73 | |
| Ile-74 | 5 | 7 | |
| Mutants | |||
| Lys-74 | 1 | 1 | |
| Leu-74 | 5 | 7 | |
| Trp-74 | 1 | 1 | |
| Crt_75 | n = 64 | % | |
| Wild | |||
| Asn-75 | 48 | 75 | |
| Glu-75 | 5 | 7 | |
| Mutants | |||
| Lys-75 | 6 | 9 | |
| Tyr-75 | 5 | 7 | |
| Crt_76 | n = 64 | % | |
| Wild | |||
| Lys-76 | 40 | 62 | |
| Thr-76 | 10 | 15 | |
| Mutants | |||
| Gly-76 | 1 | 1 | |
| Ile-76 | 1 | 1 | |
| Gln-76 | 16 | 25 | |
“N” represents the total number of isolates sequenced at the three sites. “n” represents the number of isolates sequenced successfully by codon
Table 3.
Prevalence of genotypes corresponding to pfcrt
| pfcrt key codons | Genotypes | Sample size (N = 64) | ||
|---|---|---|---|---|
| M74I | N75E | K76T | n (%) | |
| Wild type | M | N | K | 40 (62) |
| Single mutations | 8 (12) | |||
| M | N | T | 6 (9) | |
| Others | 2 (3) | |||
| Double mutations | M | Y | Q | 4 (6) |
| Triple mutations | 12 (18) | |||
| L | K | Q | 6 (9) | |
| I | E | T | 4 (6) | |
| K | E | G | 1 (1) | |
| I | Y | I | 1 (1) | |
A capital letter in the “genotype” column represents the one-letter code of amino acids. The amino acids resulting from the mutation of pfcrt are underlined and in italic. The determined prevalences correspond to the number of observations on the number of successes per gene
Among the triple mutant genotypes, there was a predominance of isolates carrying IET and LKQ with respective prevalence of 6% and 9%. The analysis also found single mutant MNT (9%) and double mutant MYQ (6%) genotypes (Table 3).
Prevalence of wild-type Lys-76 and Thr-76 mutants of the pfcrt gene at Anonkoua-Kouté, Port-Bouët and Ayamé
No Thr-76 mutation was observed in Anonkoua-Kouté while this mutant allele was found in 31% and 25% of the sequences of the isolates from Port-Bouët and Ayamé, respectively, as indicated in Table 4. The highest prevalence of the Thr-76 allele were found in Port-Bouët (31%) and Ayamé (25%) (Table 4). Moreover, in all the study sites no significant difference (P = 0.955) was observed in the prevalence of the wild-type Lys-76 allele, Anonkoua-Kouté (60%), Port-Bouët (62%) and Ayamé (65%).
Table 4.
Prevalence of the wild-type Lys-76 and the mutant Thr-76 of the pfcrt gene in Anonkoua-Kouté, Port-Bouët and Ayamé
| Codon | Alleles | Anonkoua-Kouté: N = 28 | Port-Bouët: N = 16 | Ayamé: N = 20 | p-value (χ2 test) |
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | |||
| Lys-76-Thr | Lys-76 | 17 (60) | 10 (62) | 13 (65) | 0.955 |
| Thr-76 | 0 (0) | 5 (31) | 5 (25) | – |
The mutated amino acids are italic. “N” represents the total number of isolates sequenced successfully per study site. “n” represents the number of isolates sequenced successfully at the codon Crt_76
The list of other mutants is in Table 2
The χ2 test could not be performed for the mutants because of the value less than 5 in a cell
Discussion
Previous studies carried out in West Africa and particularly in Côte d’Ivoire showed a strong correlation between the Thr-76 mutation of the pfcrt gene and the therapeutic failures on one hand, and between the Thr-76 mutation of the pfcrt gene and in vitro chemoresistance of P. falciparum isolates to chloroquine [19–22] on the other hand.
Results indicate that in all three study sites, the mutant allele Thr-76 (15%) was associated with mutant Ile-74 (7%) and Glu-75 (7%) in isolates at very low proportion compared to wild-type alleles Lys-76 (62%), Met-74 (73%) and Asn-75 (75%). In addition, the wild allele Lys-76 was observed in Anonkoua-Kouté, Port-Bouët and Ayamé 60%, 62% and 65%, respectively.
These results contrasted those obtained (for the Thr-76 mutation) in 2006 (65%) in Yopougon, Abidjan [8], in 2005 (100% and 95%) in Bonoua and Samo respectively [10] and in 2010 (62%) in Adzopé [9]. However, the data are in accordance with Gharbi et al. [23] who, after modelling chloroquine resistance in Côte d’Ivoire from the Thr-76 allele with samples from travelers returning to Côte d’Ivoire reported a decrease in prevalence from 63 to 37% of Thr-76 allele.
Normally, the prevalence of the Thr-76 mutation should be reduced because parasites that carry the Lys-76 wild-type allele have a survival advantage in the absence of drug pressure [24]. Indeed, when drug pressure is low, drug resistance is accompanied by a reduction in the genetic performance of resistant parasites compared to susceptible parasites [25, 26]. Thus, when the drug pressure decreases, the proportion of sensitive parasites increases and that of the resistant parasites decreases [11].
The low prevalence observed for Thr-76 isolates may be due to a number of factors, the main one being the effective withdrawal of CQ in Côte d’Ivoire. Indeed, CQ has always been prescribed and/or delivered in Côte d’Ivoire for the treatment of uncomplicated malaria [27] until 2007, when it was withdrawn in favor of amodiaquine (AQ) [28]. However, the substitution of amodiaquine would have delayed the decline since AQ also selects for the Thr-76 allele of pfcrt [29] in contrary to the countries where the decline has been most precipitous thanks to an intensive deployment of lumefantrine as part of Coartem. The data generated are in line with these observations, since artemether-lumefantrine (AL) was officially used as first-line treatment in Côte d’Ivoire by year 2013. It was, therefore, necessary to wait a few years to observe a possible significant decrease in chloroquine-resistant P. falciparum isolates. Thus, the public authorities have had to intensify awareness campaigns to inform the population and the medical staff, to take action to fight against the illegal sale of anti-malarial drug on parallel markets (street drugs). All these actions eventually made effective the removal of the CQ and that would be the basis of the increase in the prevalence of chloroquine-sensitive parasites. In addition, government control over pharmaceutical distribution channels and drug supply chains in the public and private sectors has reduced the use of non-recommended drugs such as CQ [30, 31]. For example, in Malawi, the successful implementation of national information campaigns and the effective control of drug delivery patterns has led, 10 years later, to the re-emergence of sensitivity to CQ [32]. The context in Côte d’Ivoire is also different from what was reported in French Guyana where, despite the fixation of the IET genotype, the return to sensitivity was observed thanks to the acquisition of the C350R mutation in parasites carrying the Lys-76 allele [33]. No mutation of the Thr-76 allele was observed in Anonkoua Kouté while this mutant allele was carried by 31% and 25% of isolates from Port-Bouët and Ayamé, respectively.
These prevalence of the Thr-76 mutation in Port-Bouët and Ayamé could be related to the effects of migratory movements of the populations towards these areas. Indeed, these two communities are characterized by a strong agroeconomic activity (livestock, farming, and fishing) with many rivers, large farms that attract many indigenes and non-indigenes from the sub-region. These populations mainly occupy the villages (Ayamé) and the many precarious neighbourhoods (Port-Bouët) where most households often do not have access to national information thus depriving them of awareness campaigns for the withdrawal and abandon of the CQ. Thus, these households opt for anti-malarial treatments without consultation (self-medication), thus maintaining drug pressure [34]. The populations migrating from neighbouring West African countries to these localities may also be potential carriers of resistant parasites, which could explain the prevalence of the Thr-76 mutation observed in these areas.
Therefore, the withdrawal of chloroquine and the introduction of ACT seem to promote the re-emergence of CQ-sensitive isolates. It would be desirable to carry out another study that could be extended to several localities with a larger number of samples to confirm this decrease of CQ-resistant parasites in Côte d’Ivoire. Thus, if the proportion of chloroquine-resistant parasites decreases at the national level to an undetectable level of pfcrt mutants, a reintroduction of chloroquine in combination with other anti-malarial drug for malaria treatment and prophylaxis may be considered, as Malawi has done [11].
Conclusion
The present study showed that the CQ resistance has decreased in Côte d’Ivoire in Anonkoua-Kouté, Port-Bouët and Ayamé communities since its withdrawal in 2004. This decrease in CQ resistance seems to be related to the efficiency and the success of the policy of abandoning the use of CQ in Côte d’Ivoire. Therefore, the withdrawal of chloroquine (CQ) and the introduction of ACT for the treatment of uncomplicated malaria in Côte d’Ivoire appear to favour the re-emergence of isolates sensitive to CQ. However, even if the proportion of chloroquine-sensitive parasites seems to increase in Anonkoua-Kouté, Port-Bouët and Ayamé, a reintroduction of chloroquine in malaria treatment cannot be recommended currently in Côte d’Ivoire.
Additional files
Additional file 1: Data on sampling sites and on subjects included in the study.
Authors’ contributions
DO: blood collection, designed and realized the technical manipulations, CND directed technical manipulations, AAB and DDN realized the statistical treatment, OL contributed to the drafting of the manuscript, TOA followed the realization of the project, DAJ conceived the project. Each author participated sufficiently in the work to take public responsibility for appropriate portions of the content. All authors read and approved the final manuscript.
Acknowledgements
The authors express their profound gratitude to the Pasteur Institute of Côte d’Ivoire who allowed us to use the facilities of the molecular biology platform of the Institut Pasteur of Côte d’Ivoire to perform the PCR tests.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data related to the current manuscript are available in Additional file 1.
The consent for publication
This consent was also given for publication of the results obtained.
Ethics approval and consent to participate
The study was conducted in accordance with the Helsinki Declaration and approval was received from the National Committee on Ethics and Research (NCER) of the Ivorian Ministry of Health and AIDS. After the appropriate information and explanations, adult participants, parents or legal guardians of all children wishing to participate in the study gave their written consent prior to sampling.
Funding
The study was carried out on own funds.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ACT
artemisinin-based combination therapy
- AQ
amodiaquine
- CQ
chloroquine
- CQR
chloroquine resistance
- DNA
deoxyribonucleic acid
- EDTA
ethylene diamine tetraacetic
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- NCER
National Committee on Ethics and Research
- WHO
World Health Organization
References
- 1.WHO . World malaria report. Geneva: World Health Organization; 2016. [Google Scholar]
- 2.Harinasuta T, Suntharasamai P, Viravan C, Ravel P, Ma L, Tichit M, et al. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965;286:657–660. doi: 10.1016/S0140-6736(65)90395-8. [DOI] [PubMed] [Google Scholar]
- 3.Young MD, Moore DV. Chloroquine resistance in Plasmodium falciparum. Am J Trop Med Hyg. 1961;10:317–320. doi: 10.4269/ajtmh.1961.10.317. [DOI] [PubMed] [Google Scholar]
- 4.Payne D. Did medicated salt hasten the spread of chloroquine resistance in Plasmodium falciparum? Parasitol Today. 1988;4:112–115. doi: 10.1016/0169-4758(88)90042-7. [DOI] [PubMed] [Google Scholar]
- 5.Peters W. Chemotherapy and drug resistance in malaria. New York: Academic Press Ltd; 1987. p. 542. [Google Scholar]
- 6.Hellgren U, Ardal OK, Lebbad M, Rombo L. Is chloroquine-resistant Plasmodium falciparum malaria emerging in Senegal or The Gambia? Trans R Soc Trop Med Hyg. 1987;81:728. doi: 10.1016/0035-9203(87)90008-3. [DOI] [PubMed] [Google Scholar]
- 7.Kyrönseppä H, Lumio J, Ukkonen R, Pettersson T. Chloroquine-resistant malaria from Angola. Lancet. 1984;1:1244. doi: 10.1016/S0140-6736(84)91731-8. [DOI] [PubMed] [Google Scholar]
- 8.Djaman J, Ahibo H, Yapi HF, Bla KB, Ouattara L, Yavo W, et al. Molecular monitoring of Plasmodium falciparum malaria isolates in Côte d’Ivoire: genetic markers (dhfr-ts, dhps, pfcrt, pfmdr-1) for antimalarial-drugs resistance. Eur J Sci Res. 2010;40:461–470. [Google Scholar]
- 9.Ouattara L, Bla KB, Assi SB, Yavo W, Djaman AJ. pfcrt and dhfr-ts sequences for monitoring drug resistance in Adzopé Area of Côte d’Ivoire after the withdrawal of chloroquine and pyrimethamine. Trop J Pharm Res. 2010;9:568. [Google Scholar]
- 10.Johansson M, Penali LK, Assanvo SP, Ako BA, Offianan AT, Johansson M, et al. Molecular analysis of markers associated with chloroquine and sulfadoxine/pyrimethamine resistance in Plasmodium falciparum malaria parasites from southeastern Côte-d’Ivoire by the time of artemisinin-based combination therapy adoption in 2005. Infect Drug Resist. 2012;5:113–120. doi: 10.2147/IDR.S31409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 12.Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ndam NT, Basco LK, Ngane VF, Ayouba A, Ngolle EM, Deloron P, et al. Reemergence of chloroquine-sensitive pfcrt K76 Plasmodium falciparum genotype in southeastern Cameroon. Malar J. 2017;16:130. doi: 10.1186/s12936-017-1783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieng Y, Gaye O, Faye B, Tine R, Ndiaye M, Ndiaye JL, et al. Assessment of the molecular marker of Plasmodium falciparum chloroquine resistance (Pfcrt) in Senegal after several years of chloroquine withdrawal. Am J Trop Med Hyg. 2012;87:640–645. doi: 10.4269/ajtmh.2012.11-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurtz N, Fall B, Pascual A, Diawara S, Sow K, Baret E, et al. Prevalence of molecular markers of Plasmodium falciparum drug resistance in Dakar, Senegal. Malar J. 2012;11:197. doi: 10.1186/1475-2875-11-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adja AM, N’goran EK, Koudou BG, Dia I, Kengne P, Fontenille D, et al. Contribution of Anopheles funestus, An. gambiae and An. nili (Diptera: Culicidae) to the perennial malaria transmission in the southern and western forest areas of Côte d’Ivoire. Ann Trop Med Parasitol. 2011;105:13–24. doi: 10.1179/136485910X12851868780388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miguel RB, Coura JR, Samudio F, Suárez-Mutis MC. Evaluation of three different DNA extraction methods from blood samples collected in dried filter paper in Plasmodium subpatent infections from the Amazon region in Brazil. Rev Inst Med Trop Sao Paulo. 2013;55:205–208. doi: 10.1590/S0036-46652013000300012. [DOI] [PubMed] [Google Scholar]
- 18.Team RC. R: A language and environment for statistical computing. R Found Stat Comput 2008.
- 19.Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 20.Djaman JA, Bla BK, Yavo W, Yapi HF, Mazabraud A, Basco LK. Polymorphism of PFCRT and PFMDR-1 genes of Plasmodium falciparum and chloroquine susceptibility in Côte d’Ivoire. Acta Protozool. 2007;46:361–365. [Google Scholar]
- 21.Figueiredo P, Benchimol C, Lopes D, Bernardino L, do Rosario VE, Varandas L, et al. Prevalence of pfmdr1, pfcrt, pfdhfr and pfdhps mutations associated with drug resistance, in Luanda, Angola. Malar J. 2008;7:236. doi: 10.1186/1475-2875-7-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bridges DJ, Molyneux M, Nkhoma S. Low level genotypic chloroquine resistance near Malawi’s northern border with Tanzania. Trop Med Int Health. 2009;14:1093–1096. doi: 10.1111/j.1365-3156.2009.02340.x. [DOI] [PubMed] [Google Scholar]
- 23.Gharbi M, Flegg J, Pradines B, Berenger A, Ndiaye M, Djimde AA, et al. Surveillance of travellers: an additional tool for tracking antimalarial drug resistance in endemic countries. PLoS One. 2013;8:e77775. doi: 10.1371/journal.pone.0077775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laufer MK, Plowe CV. Withdrawing antimalarial drugs: impact on parasite resistance and implications for malaria treatment policies. Drug Resist Updat. 2004;7:279–288. doi: 10.1016/j.drup.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Gadalla NB, Elzaki SE, Mukhtar E, Warhurst DC, El-Sayed B, Sutherland CJ. Dynamics of pfcrt alleles CVMNK and CVIET in chloroquine-treated Sudanese patients infected with Plasmodium falciparum. Malar J. 2010;9:74. doi: 10.1186/1475-2875-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ord R, Alexander N, Dunyo S, Hallett R, Jawara M, Targett G, et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J Infect Dis. 2007;196:1613–1619. doi: 10.1086/522154. [DOI] [PubMed] [Google Scholar]
- 27.Kiki-Barro CP, Konan FN, Yavo W, Kassi R, Menan EIH, Djohan V, et al. [Antimalaria drug delivery in pharmacies in non-severe malaria treatment. A survey on the quality of the treatment: the case of Bouake (Côte d’Ivoire)](in French). Sante. 2004;14:75–9. [PubMed]
- 28.MSHP: Arrêté No 24/CAB/MSHP Du 12 Janvier 2007 Portant Institution d’un Schéma Thérapeutique pour Traitement du Paludisme en Côte d’Ivoire. Abidjan; 2007.
- 29.Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether–lumefantrine in Africa. J Infect Dis. 2009;199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talisuna AO, Adibaku S, Amojah CN, Amofah GK, Aubyn V, Dodoo A, et al. The affordable medicines facility-malaria—a success in peril. Malar J. 2012;11:370. doi: 10.1186/1475-2875-11-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tougher S, Ye Y, Amuasi JH, Kourgueni IA, Thomson R, Goodman C, et al. Effect of the Affordable Medicines Facility-Malaria (AMFm) on the availability, price, and market share of quality-assured artemisinin-based combination therapies in seven countries: a before-and-after analysis of outlet survey data. Lancet. 2012;380:1916–1926. doi: 10.1016/S0140-6736(12)61732-2. [DOI] [PubMed] [Google Scholar]
- 32.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 33.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouadio AS, Ciss‚ G, Obrist B, Wyss K, Zinsstag J, Yao YJ, et al. Fardeau économique du paludisme sur les ménages démunis des quartiers défavorisés d’Abidjan, Cote d’Ivoire. VertigO. 2006, Hors Serie 3.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Data on sampling sites and on subjects included in the study.
Data Availability Statement
All data related to the current manuscript are available in Additional file 1.


