Abstract
Objective:
Technological advances in radiation therapy are evolving with the use of hadrons, such as protons, indicated for tumors where conventional radiotherapy does not give significant advantages or for tumors located in sensitive regions, which need the maximum of dose-saving of the surrounding healthy tissues. The genomic response to conventional and non-conventional linear energy transfer exposure is a poor investigated topic and became an issue of radiobiological interest. The aim of this work was to analyze and compare molecular responses in term of gene expression profiles, induced by electron and proton irradiation in breast cancer cell lines.
Methods:
We studied the gene expression profiling differences by cDNA microarray activated in response to electron and proton irradiation with different linear energy transfer values, among three breast cell lines (the tumorigenic MCF7 and MDA-MB-231 and the non-tumorigenic MCF10A), exposed to the same sublethal dose of 9 Gy.
Results:
Gene expression profiling pathway analyses showed the activation of different signaling and molecular networks in a cell line and radiation type-dependent manner. MCF10A and MDA-MB-231 cell lines were found to induce factors and pathways involved in the immunological process control.
Conclusion:
Here, we describe in a detailed way the gene expression profiling and pathways activated after electron and proton irradiation in breast cancer cells. Summarizing, although specific pathways are activated in a radiation type-dependent manner, each cell line activates overall similar molecular networks in response to both these two types of ionizing radiation.
Advances in knowledge:
In the era of personalized medicine and breast cancer target-directed intervention, we trust that this study could drive radiation therapy towards personalized treatments, evaluating possible combined treatments, based on the molecular characterization.
Introduction
Over the last decade, the technological development of radiation therapy (RT) has led to more efficient processing techniques which can deliver, with high precision, increasing doses saving the organ at risk, and high dose values even on small-sized tumor targets. Approximately, 50% of all cancer patients will receive some form of RT (such as, external beam or internal RT), either alone or in combination with other treatment modalities such as surgery or chemotherapy.1 Indeed, a great amount of scientific evidence reports that RT remains a vital component of the curative multimodality therapy for many types of cancer including breast cancer (BC). BC is a heterogeneous disease, at both clinical and molecular levels, presenting distinct subtypes associated with different clinical outcomes.2 In this sense, a clinical point of interest is to better define the most successful treatment plan, including the choice of the best RT modality and schedule, according also to specific BC molecular characterization. Thus, many efforts from research teams are needed to help clinicians in understanding the molecular portrait of a specific cancer type, in order to propose successful combinatorial anticancer therapies in clinical practice. These multimodalities approaches are based on the principle that standalone, chemo- or radiotherapeutic regimens are generally unable to control neoplastic lesions, whereas combining therapeutic agents with dissimilar action mechanisms potentially results in synergistic anti neoplastic effects, as recently described by several authors.3, 4
Nowadays, technological advances in RT are evolving with the use of hadrons, such as protons, carbon ions and other ions, like helium. Proton radiation therapy (PRT) offers a number of potential advantages over conventional (photon/electron based) RT for cancer, due to a more localized delivery of the radiation dose thanks to their characteristic Bragg Peak.5–7 Precisely, the heavy particle enters tissue and deposits a minimal radiation dose on its track to the tumor. The radiation dose increases very gradually with greater depth and lower energy, suddenly rising to the Bragg Peak when the proton is ultimately stopped.7, 8 Although PRT may offer clinical advantages compared with conventional RT, it is expensive and therefore, important to evaluate whether the relative medical benefits are large enough to motivate the higher costs. On the other hand, it is important to study breast PRT in order to minimize the exposure of healthy tissue and, in this way, to reduce risk of coronary and heart disease.9
A number of challenges arise, such as to compare unconventional (proton) and conventional (photon/electron) treatment plans when both modalities are available. Such comparison, should firstly evaluate the potential clinical benefit for the individual patient based on the dose distribution, as well as more information regarding cell networks activated after conventional and unconventional RT exposure.10 A critical point of interest is the assumed biological effectiveness of protons. It is known that the dose deposition is fundamentally different between photon and proton RT, thus, the biological equivalent dose should be compared and not the physical dose. This is handled today by using a constant relative biological effectiveness (RBE) of 1.1 for protons, reflecting the assumption that the physical proton dose has a biological effect equivalent to 10% higher than photon dose.5, 9 The use of this generic spatially invariant RBE within tumors and normal tissues disregards the evidence that proton RBE varies with linear energy transfer (LET). The LET values showed by particles, range from a few hundreds of electronvolts to many hundreds of keV. Electrons LET values go from about 0.25 keV µm–1 to about 2.30 keV µm–1 and, as a consequence, electrons are considered low LET radiations. On the other hand, the LET values range of heavy ions goes from about 15–170 keV µm–1 and they are considered high LET radiations. Protons show halfway values and they range between 1 and 30 keV µm–1.11
The proton RBE increases with increasing LET, which grow when the beam energy decreases.12 In addition, regarding LET parameter, a large fraction of tumors show resistance to low LET ionizing radiation (IR; such as those delivered during photon or electron irradiation) by mechanisms that are only partly elucidated.12–17 Precisely, conventional low LET IR is known to cause relatively well separated ionizations, resulting almost in DNA single strand breaks.12 In contrast, high LET ions (such as carbon ions), release energy densely along their track through the cell nucleus, creating several double strand breaks (DSB) in a narrow region, resulting in more complex and less repairable DNA damage. Understanding the molecular response to high and low LET irradiation RT, also in term of gene expression changes, became a critical point of interest. According to us, this issue has not been entirely understood and investigated, in order to select the most successful treatment plan or combinatorial treatment regimens.
In turn, the principal aim of this work was to analyze and compare molecular responses in term of gene expression profiles (GEPs), induced by electron and proton irradiation in BC cell lines.
In addition, we performed a Monte Carlo simulation using the Geant4 toolkit in order to assess LET distribution inside the target, thus providing crucial physical information about the particle beam and highlighting how the different kind of interaction with the matter could have a role in gene expression regulation.
In the era of personalized medicine and BC target-directed intervention, we trust that this study could drive RT towards personalised treatments, evaluating possible combined treatments, based also on the molecular characterization in BC.
Methods and Materials
Cell cultures
The human non-tumorigenic breast epithelial MCF10A cell line and human breast adenocarcinoma MCF7 and MDA-MB-231 cell lines, were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured according to manufacturer’s guidelines. Cells were maintained using standard conditions in an exponentially growing culture.
Radiation treatment
2 days before irradiation cells were seeded and following irradiated at subconfluence (70–80%). In order to analyze and compare molecular responses induced by conventional and non-conventional radiation treatment modalities, cells were exposed to the same dose of IR (9 Gy). The dose value was chosen according to two assumptions: (i) a sublethal proton radiation dose of 8 Gy was recently described by Gameiro and colleagues able to mediate tumor changes in combination with immunotherapy8 and thus, we decide to investigate the molecular changes PRT-induced with those activated by conventional RT; (ii) we analyzed the GEPs induced by 9 Gy of IR dose because of its use during electron intraoperative RT (boost scheme) in BC care, about which a cell and molecular characterization was recently described by our group.18, 19
Electron irradiation configuration
In order to study the biological response in term of gene GEP profiles after low LET irradiation in BC cells, we used electrons beam configurations. The NOVAC7 (Sordina IORT Technologies, Vicenza, Italy) IORT system was used to perform the cell irradiations as previously described. The beam collimation was performed through a polymethylmethacrylate cylindrical tubes with a diameter of 10 cm and face angle of 0˚. The electron accelerator system was calibrated under reference conditions.20 Cell irradiations were conducted placing the cell at the build-up of the depth dose distribution with 9 Gy of IR dose (100% isodose) and at a dose rate of 3.2 cGy/pulse.
Proton irradiation configuration
The proton beam irradiation was performed at INFN-LNS (Catania) within the CATANA experimental room. When reaching the CATANA treatment room, proton beam of a maximum energy of 62 MeV A went out in the air and flew for 3 m before hitting the target. Through its path, the beam was intercepted by various elements such as scattering foils to spread the beam laterally, collimators to define beam profile in accordance to the tumour shape and monitor chambers to measure the dose delivered.21 Moreover, the beam was also modified in term of energy and shape to be used for applications. More precisely, by using a superconductive cyclotron, the accelerated beam was converted in a uniform clinical beam able to cover the entire target region passing through different passive elements. In addition, to cover entirely the flask surface, we used different beam shots and an automatic flask positioning and collimation system. We performed a dosimetric check before each beam time irradiation: the lateral beam profile was verified using a semi-conductor diode while the depth dose profiles and dosimetric calibrations were performed using a motorized Markus chamber within a water tank. The dosimetric system was calibrated under reference conditions.20 Cell irradiations were conducted placing the cell at the middle spread-out Bragg peak with dose value of 9 Gy and a dose rate of 15 Gy min-1.
Dose and LET distribution using Geant4 toolkit
As above described, we used the Monte Carlo code Geant4 (GEometry ANd Tracking),22–24 toolkit widely used to support technical and clinical issues in RT, in order to evaluate the depth absorbed dose and LET distributions of electron and proton beams. For the electron irradiations, the setup and the dose distribution were studied by modeling electron propagation with Monte Carlo methods using “iort-therapy” application, Advanced Example of GEANT4, developed by G. Russo et al.25 This application simulates the geometry of a real clinical intraoperative radiation therapy treatment (delivering a single high dose of IR immediately after surgical tumor removal in order to destroy the residual cancer cells) and each device component. The application, through the simulation of the electron accelerator, the beam collimation system and electron applicator, allowed us to calculate the real dose distribution released inside the cell irradiation flask.
The application used for the proton beam evaluation is called “HadronTherapy” and it is an Advanced Example of GEANT4. The code is developed by Cirrone et al. and Romano et al., and is able to simulate two typical proton and carbon transport beam lines utilized in clinical practice and cellular irradiation studies.11, 26 In this work, we modified the above mentioned open source application, in order to provide information (such as the absorbed dose and LET values) also for electron beam lines. In this way, we assessed the primary and total dose averaged LET reproducing LET distributions in a realistic manner. In addition, we included in the calculation the secondary particle contributions due to nuclear interactions and their spatial distribution in voxelized or sliced geometries.
Whole-genome cDNA microarray expression analysis
We analyzed GEPs of MCF10A, MCF7 and MDA-MB-231 cells induced by both electron and proton beam irradiations, using 9 Gy of IR dose.
Total RNA extraction and its qualitative and quantitative analyses were conducted as previously described.18, 19 Microarray analysis was performed using the Whole Human Genome 4 × 44 K cDNA kit according to the Agilent Two-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, Santa Clara, CA). Precisely, six replicates for each configuration assayed were performed. Statistical data analysis, background correction, normalization and summary of expression measures were conducted with GeneSpring GX 10.0.2 software (Agilent Technologies). Finally, genes were identified as being differentially expressed if they showed a fold change of at least 1.5 with a p-value < 0.05 compared to untreated cells of the same cell line analyzed, that was used as reference sample. The data discussed in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO)27 and are accessible through the following GEO Series accession number: GSE103472. Microarray data are available in compliance with minimum information about a microarray experiment standards.
Ingenuity pathway analysis
Differentially expressed gene lists obtained by GEP analysis were studied using Ingenuity pathway analysis (IPA- Ingenuity System, Montain View, CA). IPA is a software application that enables to identify the biological mechanisms, pathways and functions matching a particular selected data set of genes or proteins using a build-in scientific literature based database (according to web site https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/, Qiagen, Germany). IPA is based on a database obtained by abstracting and interconnecting a large fraction of the biomedical literature according to a very strict algorithm. The networks, displayed graphically as nodes, representing individual proteins and edges representing the biological relation between nodes and are ordered by score and optimized including as many differentially expressed proteins as possible. A p-score [i.e._log(p-value)] for each possible network is computed. Therefore, networks with scores of 2 or higher have at least 99% confidence of not being generated by random chance alone.
oPOSSUM transcription factor binding sites analysis
The oPOSSUM analysis system identifies over represented transcription factor binding sites (TFBS) in sets of co-expressed genes (target gene set) by comparing the target gene set against a pre-compiled background set of genes.28, 29 Two measures of statistical significance, the Z-score and the Fisher score, are then calculated. In general, a Z-score of >10 and a Fisher score of <0.01 identify significantly over represented transcription factor (TF) binding sites. We employed this system to identify TFs of interest in our studies. Microarray down- and upregulated genes lists of MCF10A, MCF7 and MDA-MB-231 cell lines treated with electron or proton irradiation with 9 Gy of IR dose, were analyzed for enrichment of TFBS using the oPOSSUM program.30 The conserved non-coding regions of the promoters were searched for matches to all TFBS profiles in the JASPAR database.28–30 For each transcript, the top conserved regions in the 2000 bp upstream/downstream sequences between mouse and humans with minimum conservation cutoff of 0.40 and matrix match threshold of 80% was scanned for TFBS using a position weight matrices algorithm.
PubMatrix
All genes assayed in this work were analyzed using the PubMatrix tool as previously described order to confirm our assumptions and to study bibliographic relationships between proteins and some selected queries such as IR, radiation, cancer, BC, electron, proton, DNA DSBs.18, 19,31
Results
LET evaluation in electron and proton simulations
In order to assess LET distribution inside the target, we performed a Monte Carlo simulation using the Geant4 toolkit. We used two advanced examples provided within the Geant4 official release: iort-therapy and hadrontherapy.32 Moreover, a hadrontherapy tool was used to simulate the clinical proton facility at LNS-INFN and to assess the dose and LET distribution in the targets.
As shown in Figure 1, LET values for electron beams (displayed in the black curve) were quite constant along the penetration depth in the matter ranging from about 0.19 to 0.22 KeV µm–1. During cellular irradiations, the samples were positioned at the build-up of the depth-dose curve where the LET value measured was of 0.19 KeV µm–1 at about 25 mm depth of water equivalent material.
Figure 1.
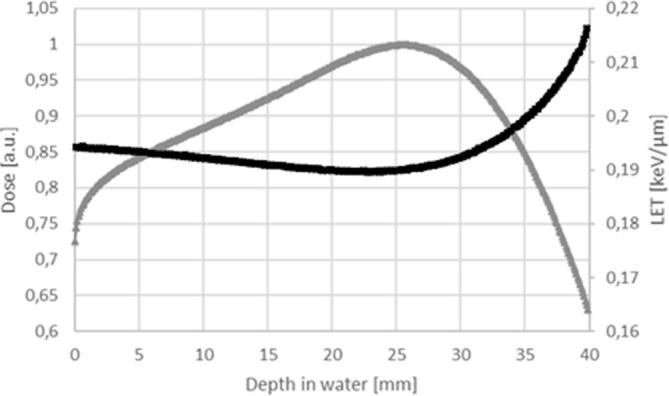
LET evaluation in electron simulation. The gray curve represents the depth dose curve and the black curve the LET values at different depth into the material. LET, linear energy transfer.
Regarding proton irradiation, LET values of a spread-out Bragg Peak (SOBP) generally ranging from 1 to 2 KeV µm–1 to 10 KeV µm–1 before the distal part of the modulated peak were obtained, as widely discussed in literature.26 During cell irradiation, a SOBP configuration was used. Precisely, the cell layer was positioned at the middle-SOBP, where LET value was of 5.4 KeV/µm, as shown in the black curve of Figure 2, and approximately, 29 times higher than LET values evaluated for the electron beam.
Figure 2.
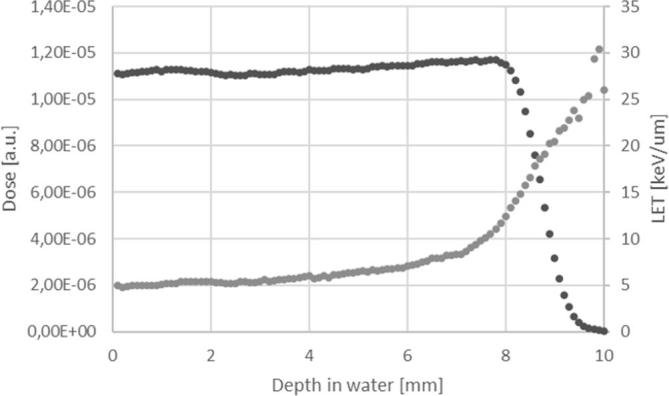
LET evaluation in electron simulation. The black dots represents the depth dose curve and the gray dots the LET values at different depth into the material. LET,linear energy transfer.
Overview of cDNA microarray gene expression induced by electron and proton beam irradiation
In this work, a Two-Color Microarray-Based Gene Expression Analyses were conducted on MCF10A, MCF7 and MDA-MB-231 cells treated with 9 Gy of IR dose delivered using electron
(e–) and proton (p+) beam irradiations. In details, MCF10A, MCF7 and MDA-MB-231 irradiated cells were named as follows: MCF10A/e– and MCF10A/p+; MCF7/e– and MCF7/p+; MDA-MB-231/e- and MDA-MB-231/p+. Respective MCF10A, MCF7 and MDA-MB-231 untreated cell lines were used as reference samples. Comparative differential gene expression analysis revealed that a conspicuous number of genes had significantly altered expression levels by 1.5-fold or greater, compared to the untreated reference group as shown in Table 1. Up and down regulated transcripts were available at GEO database (GEO ID: GSE103472).
Table 1.
Microarray differentially expressed gene lists
| Cell line | Cell details | Irradiation modality (particle type) | Configuration name | Genes differential expressed (>1.5 fold) | ||
| Number of genes | Down | Up | ||||
| MCF10A | Non-tumorigenic mammary epithelial cells | e– | MCF10A/e– | 558 | 179 | 379 |
| p+ | MCF10A/p+ | 1564 | 943 | 621 | ||
| MCF7A | Tumorigenic breast adenocarcinoma cell line | e– | MCF7/e– | 2554 | 1272 | 1282 |
| p+ | MCF7/p+ | 364 | 68 | 296 | ||
| MDA-MB-231 | Metastatic breast invasive ductal carcinoma | e– | MDA-MB-231/e– | 355 | 190 | 165 |
| p+ | MDA-MB-231/p+ | 1109 | 237 | 872 | ||
Deregulated transcripts for each configuration analyzed in this study and above mentioned, were selected according to their involvement in specific biological pathways using IPA. Precisely, the top enriched canonical pathways were analyzed and listed in Tables 2–4.
Table 2.
MCF10A GEPs IPA analysis
| IPA analysis of GEP IR induced in MCF7 breast cancer cell line | |||||
| Type of radiation treatment (particle type) | Top canonical pathway | enes involved | p-value | Overlap | % |
| e- | Cell cycle control of chromosomal replication | CDC6; CDC45; CDK6; MCM2;MCM4; MCM6; MCM7; ORC1;ORC6; POLA2; POLE; PRIM1; PRIM2; RPA4 |
2.63E-05 | 14/38 | 36.8 |
| p53 signaling | APAF1; ATM;BAX; BBC3;BRCA1; CDKN1A; CHEK1; DRAM1; FAS;GADD45A1; GADD45B; IRS1; MDM2; PIK3C2B; PIK3R3; PIK3R5; PML; PRKDC; SERPINB5; SNAI2; TIGAR; TLR9; TNFRSF10B; TP53I3; TP53INP1; TRIM29 |
1.35E-04 | 26/111 | 23.4 | |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | ATM; CALML5; CCL5; CEBPB; CEBPD; DAAM1; DKKL1; FN1; FOS; FZD2; FZD6; PZD7; GNAO1; ICAM; IL6; IL15; IL1RN; RAK4; IRS1; LEF1; LRP1; LTB; MAPK9; NFAT5; NFATC2; NFATC4NFKBIA; NGFR; PDGFA; PIK3R3; PIK3R5; PLCB3; PLCD4; PLCG2; PPP3R1; PRKCZ; PRKD3; RYK; SOCS1; SOCS3; TCF7L1; TCF7L2; TLR2; TLR9; TNF; TNFRSF11B; WNT4; WNT6; WNT5B |
1.28E-03 | 50/296 | 16.9 | |
| Renin-angiotensin signaling | ADCY4; ADCY5; AGTR1; ATM; CCL5; FOS; IRS1; ITPR1; MAP3K1; MAPK9; PMAPK13PIK3C2B; PIK3R3; PIK3R5; PLCG2; PRKAR2A; PRKAR2B; PRKD3; PTGER2; SHC2; SHC3; TLR9; TNF; |
2.27E-03 | 24/119 | 20.2 | |
| Netrin signaling | ABLIM3; ENAH; NFAT5; NFATC2; NFATC4; NTN1; PPP3R1; PRKAR2A; PRKAR2B; RAC2; RYR1 |
2.45E-03 | 11/39 | 28.2 | |
| p+ | p53 signaling | BBC3; CDKN1A; FAS; GADD45A; MDM2; PIK3CD; PIK3R5; SNAI2; TIGAR; TLR9; TNFRSF10B; TP53INP1 |
4.80E-07 | 10.8 | 12/111 |
| MSP-RON signaling pathway | ACTA1; ACTA2; ITGAM; PIK3CD; PIK3R5; TLR9 |
5.25E-04 | 10.2 | 6/59 | |
| PEDF signaling | DOCK3; FAS; PIK3CD; PIK3R5; SERPINF1; SRF; TLR9 |
6.18E-04 | 8.3 | 7/84 | |
| Paxilin signaling | ACTA1; ACTA2; ITGAM; ITGAX; PIK3CD; PIK3R5; SOS1; TLR9 |
6.40E-04 | 7.3 | 8/110 | |
| Fak signaling | ACTA1; ACTA2; CAPN8; PIK3CD; PIK3R5; SOS1;TLR9 |
1.54E-03 | 7.1 | 7/98 | |
| e-/p+ (common genes) | p53 signaling | BBC3; CDKN1A; FAS; GADD45A; MDM2; PIK3R5; SNAI2;TIGAR; TLR9;TP53INP1 |
5.56E-09 | 9 | 10/111 |
| Melanoma signaling | CDKN1A; MDM2;PIK3R5; TLR9 | 5.93E-04 | 7.3 | 4/55 | |
| PEDF signaling | FAS; PIK3R5; SERPINF1; TLR9; | 2.87E-03 | 4.8 | 4/84 | |
| Hereditary breast cancer signaling | CDKN1A; GADD45A; PIK3R5; SMARCA2;TLR9 |
2.96E-03 | 3.6 | 5/139 | |
| Role of p14/P19arf in tumor suppression | MDM2; PIK3R5;TLR9 | 3.16E-03 | 7.1 | 3/42 | |
GEP, gene expression profile; IPA, Ingenuity pathway analysis; IR, ionizing radiation.
Table 3.
MCF7 GEPs IPA analysis
| IPA analysis of GEP IR induced in MCF7 breast cancer cell line | |||||
| Type of radiation treatment (particle type) | Top canonical pathway | Genes involved | p-value | Overlap | % |
| e- | Cell cycle control of chromosomal replication | CDC6; CDC45; CDK6; MCM2;MCM4; MCM6; MCM7; ORC1; ORC6; POLA2; POLE; PRIM1; PRIM2; RPA4 |
2.63E-05 | 14/38 | 36.8 |
| p53 signaling | APAF1; ATM; BAX; BBC3; BRCA1; CDKN1A; CHEK1; DRAM1; FAS; GADD45A1; GADD45B; IRS1; MDM2; PIK3C2B; PIK3R3; PIK3R5; PML; PRKDC; SERPINB5; SNAI2; TIGAR; TLR9; TNFRSF10B; TP53I3; TP53INP1; TRIM29 |
1.35E-04 | 26/111 | 23.4 | |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | ATM; CALML5; CCL5; CEBPB; CEBPD; DAAM1; DKKL1; FN1; FOS; FZD2; FZD6; PZD7; GNAO1; ICAM; IL6; IL15; IL1RN; IRAK4; IRS1; LEF1; LRP1; LTB; MAPK9; NFAT5; NFATC2; NFATC4NFKBIA; NGFR; PDGFA; PIK3R3; PIK3R5; PLCB3; PLCD4; PLCG2; PPP3R1; PRKCZ; PRKD3; RYK; SOCS1; SOCS3; TCF7L1; TCF7L2; TLR2; TLR9; TNF; TNFRSF11B; WNT4; WNT6; WNT5B |
1.28E-03 | 50/296 | 16.9 | |
| Renin-angiotensin signaling | ADCY4; ADCY5; AGTR1; ATM; CCL5; FOS; IRS1; ITPR1; MAP3K1; MAPK9; PMAPK13PIK3C2B; PIK3R3; PIK3R5; PLCG2; PRKAR2A; PRKAR2B; PRKD3; PTGER2; SHC2; SHC3; TLR9; TNF; |
2.27E-03 | 24/119 | 20.2 | |
| Netrin signaling | ABLIM3; ENAH; NFAT5; NFATC2; NFATC4; NTN1; PPP3R1; PRKAR2A; PRKAR2B; RAC2; RYR1 |
2.45E-03 | 11/39 | 28.2 | |
| p+ | p53 signaling | BBC3; CDKN1A; FAS; GADD45A; MDM2; PIK3CD; PIK3R5; SNAI2; TIGAR; TLR9; TNFRSF10B; TP53INP1 |
4.80E-07 | 10.8 | 12/111 |
| MSP-RON signaling pathway | ACTA1; ACTA2; ITGAM; PIK3CD; PIK3R5; TLR9 |
5.25E-04 | 10.2 | 6/59 | |
| PEDF signaling | DOCK3; FAS; PIK3CD; PIK3R5; SERPINF1; SRF; TLR9 |
6.18E-04 | 8.3 | 7/84 | |
| Paxilin signaling | ACTA1; ACTA2; ITGAM; ITGAX; PIK3CD; PIK3R5; SOS1; TLR9 |
6.40E-04 | 7.3 | 8/110 | |
| Fak signaling | ACTA1; ACTA2; CAPN8; PIK3CD; PIK3R5; SOS1; TLR9 |
1.54E-03 | 7.1 | 7/98 | |
| e-/p+ (common genes) | p53 signaling | BBC3; CDKN1A; FAS; GADD45A; MDM2; PIK3R5; SNAI2; TIGAR; TLR9; TP53INP1 |
5.56E-09 | 9 | 10/111 |
| Melanoma signaling | CDKN1A; MDM2; PIK3R5; TLR9 | 5.93E-04 | 7.3 | 4/55 | |
| PEDF signaling | FAS; PIK3R5; SERPINF1; TLR9; | 2.87E-03 | 4.8 | 4/84 | |
| Hereditary breast cancer signaling | CDKN1A; GADD45A; PIK3R5; SMARCA2; TLR9 |
2.96E-03 | 3.6 | 5/139 | |
| Role of p14/P19arf in tumor suppression | MDM2; PIK3R5; TLR9 | 3.16E-03 | 7.1 | 3/42 | |
Table 4.
MDA-MB-231 GEPs IPA analysis
| IPA analysis of GEP IR induced in MDA-MB-231 breast cancer cell line | |||||
| Type of radiation treatment (particle type) | Top canonical pathway | Genes involved | p value | Overlap | % |
| e- | Tec kinase signaling | BLK; GNAO1; GNAZ; GNG13; PIK3R3; PTPN11:; STAT3; TNFRSF2 | 6.77E-03 | 4,8/162 | 4.9 |
| Histidine degradation III | FTCD; MTHFD2L | 7.61E-03 | 2/8 | 25 | |
| Acute phase response signaling | CD; IL-; IL36G; PIK3R3:PTPN11; RBP5; SERPIND; STAT3 | 7.79E-03 | 8/166 | 4,8 | |
| Role of Jak family kinase in IL-6-type cytokine signaling | IL-6; PTPN11; STAT3 | 8.59E-03 | 3/25 | 12 | |
| Regulation of the epithelial–mesenchymal transition pathway | EGR1; FZD4; ID2; PIK3R3PTPN11; STAT3; ZEB1; ZEB2 | 1.35E-02 | 8/183 | 4,4 | |
| p+ | IL-17A signaling | CXCL1; CXCL3; CXCL8; IL-17; IL17RA | 3.18E-04 | 5/13 | 38.5 |
| IL-17A signaling in gastric CELLS | CXCL1; CXCL8; FOS; IL17A; IL17RA; TNF | 1.36E-03 | 6/25 | 24 | |
| Altered T-cell and B-cell signaling in rheumatoid arthritis | HLA-DOA; HLA-DQB1; IL37; IL17A; IL36G; LTA; SLAMF1; TNF; TNFRSF17; TNFRSF13C;TRAF3 | 2.01E-03 | 11/78 | 14.1 | |
| CD40 signaling | FOS; ICAM1; JAK3; LTA;MAP2K6; PTGS1; PTGS2; STAT3; TNFAIP3; TRAF3 | 5.78E-03 | 10/77 | 13 | |
| Airway pathology in chronic obstructive pulmonary disease | CXCL3; CXCL8; TNF | 6.18E-03 | 3/8 | 37.5 | |
| e-/p+ (common genes) | TR/RXR activation | ATP2A1; RAB3B; SLC16A2 | 2.27E-02 | 3/98 | 3.1 |
| Transcriptional regulatory network in mbryonic stem cells | PAX6; STAT3 | 2.52E-02 | 2/40 | 5 | |
| Mouse embryonic stem cell pluripotency | FZD4; ID2; STAT3 | 2.78E-02 | 3/106 | 2,8 | |
| Agranulocyte adhesion and diapedesis | CD34; IL36G; MMP15; MSN | 2.96E-02 | 4/189 | 2,1 | |
| Regulation of the epithelial-mesenchymal transition pathway | FZD4;ID2;STAT3;ZEB2 | 2.96E-02 | 4/189 | 2,1 | |
The result of this mapping revealed the involvement of factors controlling specific pathways in each cell line tested, also different between the two irradiation modalities, as following reported. Moreover, candidate genes were studied using the PubMatrix tool31 in order to test their involvement in selected queries radiation related and to draw assumptions described in “Discussion” section.
MCF10A non-tumorigenic cell line
GEPs analysis by IPA conducted on MCF10A cells reveals the activation of different intracellular signaling in a cell line and radiation type-dependent manner, as displayed in Table 2. Precisely, after e- irradiation, MCF10A cells activated the following top five statistically relevant pathways: Ephrin A, LPS/IL-1, APR, JAK2 and mineralocorticoid signaling. Some of them, were often activated in cancer cells, where these signaling drive the inflammation process and the cancer survival/death balance. On the other hand, even after p+ irradiation, the inflammation process was achieved and sustained by HIPPO, oncostatin, IL-17 and chemokine signaling, representing four out of the five statistically relevant highlighted pathways. In addition, after p+ exposure, mitochondrial dysfunction were induced in MCF10A cell line, sustained by genes listed in Table 2 and principally involved in the mitochondrial respiratory chain. Finally, in Table 2, we provided the top five canonical pathways selected using the list of common deregulated genes after e- and p + exposure in MCF10A cell line. In particular, after both irradiation modalities, JAK, dolichol and mineralocorticoid signaling were modulated, and driven by the common deregulate genes listed in the final part of Table 2. Their role in radiation cell response need to be further explored.
MCF7 BC cell line
As shown in Table 3, MCF7 BC cell line, after e- irradiation, was able to activate specific set of genes (listed in Table 3), controlling cell cycle block and DNA damage IR-induced repair. So, among the top statistically relevant pathway modulated after this kind of radiation, we selected cell cycle control and TP53 signaling, processes known to be modulated by IR. In addition, MCF7 e- treated cells, were found to activate macrophages, renin-angiotensin and netrin signaling, involved in the regulation of the immune responses and known to be able to modify tumor and its microenvironment also after radiation exposure.
Even after p+ exposure, TP53 signaling was selected as the top statistically relevant pathway, underlying once again, its driving role in survival/death balance after irradiation. Moreover, MSP-RON, PEDF, Paxilin and FAK pathways were also selected, and described as able to modulate invasiveness in cancer cells.
Finally in Table 3, we provided the top five canonical pathways selected using the list of the common deregulated genes after e- and p+ exposure in MCF7 BC cell line. Specifically, after both irradiation modalities, TP53, melanoma, PEDF, BC and p14/p19arf signaling were modulated, and driven by the common deregulated genes listed in the final part of Table 3.
Summarizing, as shown in Table 3, only MCF7 treated cells activated TP53 intracellular signaling after both e- and p+ irradiation by using multiple key genes.
MDA-MB-231 BC cell line
As above described for the MCF10A and MCF7 cell lines, we performed IPA analysis of the MDA-MB-231 DEG lists, e- and p+ induced. As shown in Table 4, these BC cells were able to activate different intracellular signaling in a cell line and radiation type-dependent manner. In particular, after e- irradiation, MDA-MB-231 cell line activated the following top five statistically relevant pathways: Tec Kinase, histidine, APR, JAK and epithelial to mesenchymal transition pathways, known to play a crucial role in cell fate also after stress stimuli such as those induced by IR exposure. Conversely, a major induction of inflammation signaling is activated after p+ exposure and sustained by IL-17, T/B cell and CDC40 signaling, as shown in Table 4.
Finally, as above performed for the other cell lines used in this work, we provided the top five canonical pathways selected using the list of the common deregulated genes after e- and p+ exposure in MDA-MB-231 BC cells. Therefore, after both irradiation modalities, TR/RXR, stem cells, adhesion and epithelial to mesenchymal transition signaling were modulated, and driven by the common deregulated genes listed in the final last part of Table 4. Their role in radiation cell response need to be further clarified. Interestingly, as shown in Table 4, after both the irradiation conditions, STAT3 seems to play a key role in the inflammation network and in response to IR, as also recently described by our group,3 representing an interesting biomarker of IR exposure.
Gene expression signatures of proton and electron irradiations
Finally, in order to evaluate the unique and common deregulated gene lists after e- and p+ irradiation in MCF10A, MCF7 and MDA-MB-231 cell lines, we performed Venn diagrams as shown in Figure 3.
Figure 3. .
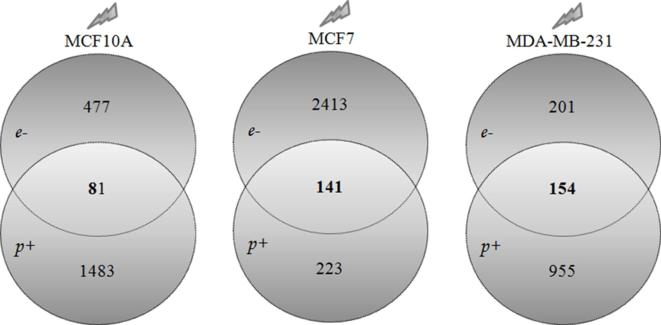
Venn diagrams of the number of unique and shared differentially expressed genes in breast cell lines, after proton and electron irradiations. (A) MCF10A cell line deregulate 477 and 1483 genes after electron and proton irradiations, respectively. Moreover, a 81-gene signature of shared deregulate genes after both irradiation modalities, was also selected. (B) MCF7 BC cell line deregulate 2413 and 223 genes after electron and proton irradiations, respectively. Moreover, a 141-gene signature of shared deregulate genes after both irradiation modalities, was also selected. (C) MDA-MB-231 BC cell line deregulate 201 and 995 genes after electron and proton irradiations, respectively. Moreover, a 154-gene signature of shared deregulate genes after both irradiation modalities, was also selected.
In particular, MCF10A cell line deregulates 477 and 1483 different genes after e- and p+ irradiations, respectively. Moreover, a 81-gene signature of shared deregulated genes after both irradiation modalities, was also selected.
On the other hand, MCF7 BC cell line deregulates 2413 and 223 different genes after e- and p+ irradiations, respectively. In addition, a 141-gene signature of shared deregulated genes after both irradiation modalities, was also selected.
Finally, MDA-MB-231 BC cell line deregulate 201 and 995 different genes after e- and p+ irradiations, respectively. Moreover, a 154-gene signature of shared deregulated genes after both irradiation modalities, was also selected.
IPA analyses of the 81-, 141-gene and 154-signatures, are shown in Tables 2–4 as described above. Moreover, all the mentioned GEP lists (with the up- and downregulated transcripts), are available in a Supplementary Material 1.
We trust that all collected data and in particular those included in the supplementary material 1, could represent a useful tool in order to select specific biomarkers of e- and p+ cell response for future clinical applications.
Identification of over-represented transcription factor binding sites in GEPs
Microarray gene data sets were loaded into oPOSSUM tool in order to identify the top-3 most statistically relevant TFs able to regulate gene expression changes induced by radiation treatments. Table 5 displays the top-3 TFs selected for each experimental configurations, by oPOSSUM analysis.
Table 5.
Top-5 statistically relevant pathways of the selected gene signatures
| Pathway name | Genes found in GEP list | Entities (total) | p-value | ||
| MCF10A 81-gene signature | 1 | Mineralocorticoid biosynthesis | 2 | 20 | 7.89E-3 |
| 2 | Glucocorticoid biosynthesis | 2 | 23 | 1.03E-2 | |
| 3 | Androgen biosynthesis | 2 | 27 | 1.4E-2 | |
| 4 | Regulation of IFNA signaling | 2 | 28 | 1.49E-2 | |
| 5 | Interleukin-7 signaling | 2 | 31 | 1.81E-2 | |
| MCF7 141-gene signature | 1 | Transcriptional regulation by TP53 | 22 | 486 | 4.11E-8 |
| 2 | TP53 regulates transcription of cell death genes | 8 | 83 | 5.88E-6 | |
| 3 | Transcriptional activation of p53 responsive genes | 3 | 6 | 0.00005 | |
| 4 | Transcriptional activation of cell cycle inhibitor p21 | 3 | 6 | 0.00005 | |
| 5 | TP53 regulates transcription of death receptors and ligands | 4 | 18 | 0.00006 | |
| MDA-MB-231 154-gene signature | 1 | Histone acetyltrasferases (HATs) acetylate histones | 6 | 110 | 1.68E-3 |
| 2 | RUNX1 regulates transcription of genes involved in B cell Receptor (BCR) signaling | 2 | 7 | 2.95E-3 | |
| 3 | Arginine methyltransferase (RMTs) methylate histone arginines | 4 | 53 | 3.24E-3 | |
| 4 | Metalloprotease Deubiquitinating enzymes (DUBs) | 3 | 32 | 5.93E-3 | |
| 5 | Histone deacetylase (HDACs) deacetylate histones | 4 | 63 | 5.94E-3 |
GEP, gene expression profile.
We analyzed whether the three different breast cell lines used in this work (MCF10A, MCF7 and MDA-MB-231), irradiated with e- and p+ particle beams, shared common gene expression regulators. Thus, TFBS analysis was performed on down- and upexpressed gene lists taking together (unique list with up and down genes) and alone (up or down gene lists), as shown in Table 5. Overall, not always the same TFs were selected by oPOSSUM tool among different irradiation modalities, thus, no simple generalization could be proposed.
Generalizing, we speculate that TFs of the forkhead box (FOX) family (in particular FoxD3, FoxI1 and FoxA1) and ATrich interaction domain 3 (ARID3) could be proposed as general regulators of transcriptome changes IR-induced (for both e- and p+), in almost all the cell lines used in this work. FOX TFs were described as regulators of growth, invasion, metastasis processes, and also in radiation gene expression response.33, 34 In particular, here we identify FoxD3, FoxI1 and FoxA1 as specific TFs IR-induced in breast cell lines, up to today poorly described in this topic. On the other hand, ARID family members were described to have a role in cell cycle control, transcriptional regulation and in chromatin structure modification. In particular, our data are in line with those described by Ma and colleagues regarding ARID3 protein, also known as DRIL1, proposed as a gene expression key regulator, activated by TP53 following radiation induced DNA damages.35 In summary, we think that TFs regulators of gene expression changes induced by e- and p+ irradiation are multiple for all breast cell lines tested, although ARID and FOX TFs members seem to be have a driving role in GEPs changes IR-induced. No simple generalized conclusions can be proposed, and other specific TFs regulators could be involved in other analysed cell lines .
Discussion
In the era of personalized medicine, prognostic and therapy-predictive molecular markers are required to guide cancer therapeutic decisions between different RT modalities and schedules.36 As above described, PRT may offer clinical advantages compared with conventional RT with X-rays (photons) or electrons for many cancer patients, mainly as a result of a more favorable distribution of the radiation dose and increased relative biological effectiveness.6, 37,38 The use of PRT in treating tumors is, therefore, a topic of great interest and early reports of clinical outcomes after these kind of treatments have been encouraging. However, few data are available regarding the PRT-induced molecular changes and a comparison between those induced by conventional RT. So, this topic needs to be more elucidated.6, 37,38
Thanks to the high-throughput technologies such as cDNA microarray analysis, at the molecular level numerous genes have been shown to be responsive to radiation exposure and specific gene signatures have been used to predict radiosensitivity in many cancer types including glioblastoma, cervical, breast, colorectal, head and neck cancer cells.18, 19,39 The idea that different radiation qualities induce differential expression of genes related to intracellular signaling (cell cycle control, apoptosis pathways, DNA damage responses etc), has been for many years one of the recognized explanations for the increased effectiveness of charged particles compared to photons/electrons. However, recent evidence supports the concept that cell response to radiation is influenced at different levels of complexity, including the molecular one.5, 40 In this sense, many efforts by research teams are needed to help clinicians in understanding the molecular portrait of a specific cancer type, in order to propose successful combinatorial anticancer therapies in clinical practice.
As known, IR causes direct or indirect damage to principal biological molecules according to its LET value. When the radiation has a high LET, cell damages are mainly induced by direct ionization of macromolecules including DNA (causing DSBs), RNA, lipids, and proteins. On the other hand, low LET radiations mainly cause DNA single strand breaks and indirect damage to macromolecules, due to the generation of reactive oxygen species and reactive nitric oxide species, which can both oxidate macromolecules and activate several intracellular signaling pathways, leading to stress responses and inflammation.12, 18,19,41,42
Here, we analyzed and compared molecular responses induced by conventional and non-conventional radiation treatment modalities (using electron and proton beams respectively), delivering the same dose of IR (9 Gy), in tumorigenic and non-tumorigenic breast cell lines. In particular, we considered to use a sublethal proton radiation dose (9 Gy), which is able to mediate tumor changes in combination with immunotherapy, as recently described by Gameiro et al.8 We compared PRT-induced molecular changes with those activated by conventional RT. In addition, we analyzed the GEPs induced by a high IR dose such as 9 Gy because of its use during electron intraoperative RT (boost scheme) in BC care, a cell and molecular characterization which has been recently described by our group.18, 19
In this study, we used the Monte Carlo code Geant4, in order to evaluate the depth dose and LET distributions of electron and proton beams. As known, Monte Carlo methods are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. They are based on the idea to use randomness to solve problems that might be deterministic in principle and are mainly used for optimization, numerical integration, and generating draws from a probability distribution.43 Our results showed that LET values calculated for electron beams were quite constant along the penetration depth in the matter and during cellular irradiations. Otherwise, LET values calculated during proton irradiation at the middle-SOBP, were approximately 29 times higher than those evaluated during electron irradiations (Figures 1 and 2).
Comparative differential gene expression analysis performed in the MCF10A non-tumorigenic breast cells and in tumorigenic MCF7, MDA-MB-231 BC cell lines after electron and proton irradiations, revealed that the GEPs were significantly altered compared to those of the untreated reference groups as shown in Table 1. In addition, top enriched canonical pathway analysis highlighted the involvement of a set of factors controlling specific pathways for each cell line tested and overall different between the irradiation treatment modalities (Tables 2–4).
Summarizing, as shown in Table 2, MCF10A non-tumorigenic cell line, exposed to both electron or proton irradiation, deregulated multiple factors involved in inflammatory and oxidative cell signaling.
On the other hand, MCF7 BC cells exposed to electron and proton irradiation deregulated several genes involved in cell cycle regulation and cell death process, as reported in Table 3. Particularly, in this work we highlighted that TP53 signaling plays a key role in MCF7 cells after both irradiation conditions. TP53 intracellular signaling was selected among the top-five statistically relevant pathways in MCF7 BC cells, after proton or electron irradiations, but probably was activated by different genes. This result represents, in our opinion ,one of the main findings of this work. As known, TP53 is often described as “the genome guardian”, because it exerts a crucial role in cell fate decision following IR-induced DNA damage. Indeed, the influence of TP53 status represents a relevant characteristic for cell survival because after IR exposure this factor is activated by the ATM-ATR pathway and is known to be able to regulate strand breaks repair (survival) or cell death induction (i.e. apoptosis).39 In particular, in MCF7 cells after both electron and proton irradiation, an interesting number of genes belonging to the TP53 signaling, were deregulated, some of which are involved in cell cycle arrest, DNA repair and cell death processes (such as BRCA1, BAX, CDKN1A, GADD45A, FAS etc.). Moreover, these genes were often described as deregulated after IR exposure and could be further investigated as a target of therapeutic combinatorial BC interventions.
In addition, more similarly to MCF10 cells, the IR-induced GEP profiles (by electron and proton irradiation), in MDA-MB-231 BC cell line highlighted the deregulation of several genes involved in the immunological and inflammatory signaling (Table 4). These data are in line with those recently published by our group regarding the cytokine signatures released in MCF10A, MCF7 and MDA-MB-231 BC cell lines after single high radiation dose. In particular, our results revealed that MCF10A and MDA-MB-231 cell lines were characterized by a secretion of almost all the cytokines assayed revealing the activation of several inflammatory signaling.44
In this work, we highlighted the activation of the JAK signaling after electron exposure in both MCF10A and MDA-MB-231 cells. JAK-STAT is another pathway that plays a key role in regulating the immune response to IR. Indeed, the STAT proteins are considered to be important for cell viability in response to different stimuli such as IR.3 In BC cells it has been described that the IL-6 JAK/STAT3 pathway could promote BC progression, metastasis, resistance to treatment45 and, at the same time, IL-6 through STAT3 can then activate IL-6/STAT3 signaling in neighboring cells. Today, this pathway is well studied because of its key role in several cancer types and also as a molecular driver and a potential therapeutic target of inflammatory and invasive ductal BC disease, after neoadjuvant chemotherapy.46, 47
In addition, as shown in Tables 2 and 4, MCF10A and MDA-MB-231 proton irradiated shared a common deregulated IL-17 signaling. IL-17 protein is a proinflammatory cytokine produced by activated T-cells. This cytokine regulates the activities of NF-kappaB (NFκB) and mitogen-activated protein kinases.48 NFκB is a well-defined radiation-responsive TF that regulates more than 200 target genes able to influence cell cycle regulation after irradiation, to suppress apoptosis and to induce cellular transformation, proliferation, metastasis and inflammation in a wide variety of tumors.35 NFκB is also able to induce radioresistance and thus, it has recently become an important target in the therapy of several chemoresistant/radioresistant types of cancer.49–51
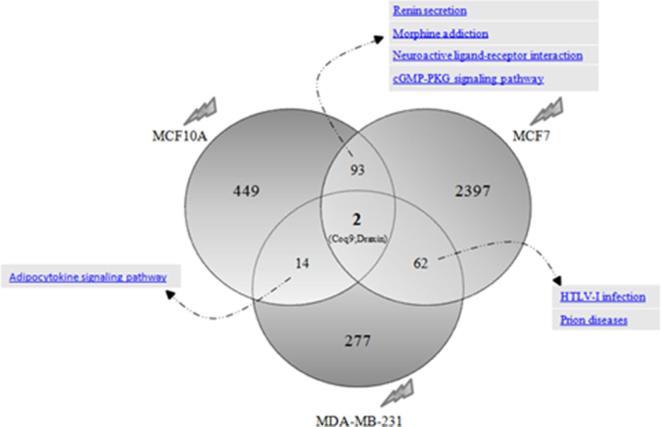
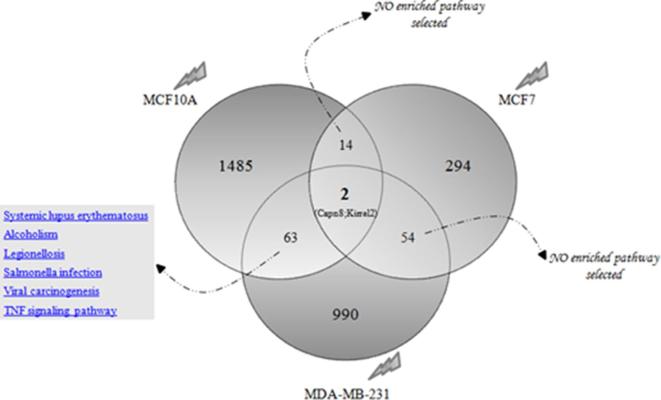
Interestingly, as shown in Figures 4 and 5, a limited number of differentially expressed genes were shared among the three cell lines studied after both electron and proton irradiations. Regarding these genes, no relevant enriched pathways were selected and thus, no general conclusion could be proposed. More precisely, only Coq9 and Draxin genes were deregulated after electron exposure among all the breast cell lines used in this work. Coq9-encoded protein is likely necessary for biosynthesis of coenzyme Q10, a requisite component of the mitochondrial oxidative phosphorylation machinery that produces more than 90% of cellular energy in term of ATP production.52 On the other hand, Draxin was recently described as a novel neural-specific secreted antagonist to canonical Wnt signaling that needs further investigation.53
Figure 4. .
Venn diagram of the number of unique and shared differentially expressed genes after electron irradiation in breast cell lines. Specifically, 449, 2397 and 277 unique genes were deregulated after electron irradiation in MCF10A, MCF7 and MDA-MB-231 cells, respectively. Moreover, 93 deregulate genes were shared between MCF10A and MCF7; 14 genes between MCF10A and MDA-MB-231 and 62 between MCF7 and MDA-MB-231 BC cell lines. Finally, only 2 genes were modulated after electron exposure, in all the three cell lines.
Figure 5. .
Venn diagram of the number of unique and shared differentially expressed genes after proton irradiation in breast cell lines. Specifically, 1485, 294 and 990 unique genes were deregulated after proton irradiation in MCF10A, MCF7 and MDA-MB-231 cells, respectively. Moreover, 14 deregulate genes were shared between MCF10A and MCF7; 63 genes between MCF10A and MDA-MB-231 and 54 between MCF7 and MDA-MB-231 BC cell lines. Finally, only two genes were modulated after proton exposure, in all the three cell lines.
In addition, as shown in Figure 5 Capn8 and Kirrel2 genes were deregulated after proton exposure among breast cell lines used. Capn8 was recently described as a member of the calpain protein family,54 while Kirrel2 as a member of the nephrin-like protein family.55 Up to today, no information regarding their relationship with radiation exposure is available in literature.
Finally, in order to evaluate the unique and common deregulated gene lists after electron and proton irradiation in MCF10A, MCF7 and MDA-MB-231 cell lines, we performed Venn diagrams as reported in Figure 3 and in the supplementary material 1 (with gene lists and fold change values). Moreover, we selected three cell-dependent gene signatures of the two irradiation modalities. Particularly, 81, 141 and 154-gene signature of shared deregulate genes after electron and proton exposures, in MFC10A, MCF7 and MDA-MB-231 cell lines respectively, were found.
In addition, we performed oPOSSUM analysis in order to test whether cells exposed to the two different irradiation modalities, using electron and proton beams, shared a common TF that controls gene expression changes. Probably, as shown in Table 6, the IR-induced GEPs changes were driven by ARID and FOX TFs members, but no simple generalized conclusions could be proposed.
Table 6.
oPOSSUM transcription factor binding sites analysis
| oPOSSUM transcription factor binding sites analysis | |||||||
| Particle | Deregulated genes (Down-Up) | TF | Downregulated genes | TF | Upregulated genes | TF | |
| MCF10A/e- | e- | 558 | Sry; FoxD3; Sox5 | 179 | HoxA5; Sox5; Gfi | 379 | Sry;FoxD3;Sox5 |
| p+ | 1564 | Egr1; Sp1; Klf4 | 943 | Egr1; Sp1; Klf4 | 621 | Sry;FoxI1;Arid3A | |
| MCF7/e- | e- | 2554 | FoxI1; Arid3A; NFkB | 1272 | Arid3A; FoxD3; Sry | 1282 | Sp1;Mzf1;Klf4 |
| p+ | 364 | FoxA1; Sry; FoxI1 | 68 | FoxA1; Mzf1; SpIB | 296 | FoxI1;FoxA1;Ap1 | |
| MDA-MB-231/e- | e- | 355 | Arid3A; FoxD3; FoxI1 | 190 | Arid3A; FoxD3; FoxI1 | 165 | Sp1;Mzf1;Klf4 |
| p+ | 1109 | ESR2; Pax4; Irf1 | 237 | Arid3A; TBP; Nkv2-5 | 872 | ESR2;Pax4;Myf | |
TF, transcription factor.
In summary, we analyzed for the first time to our knowledge, the GEP profiles induced by irradiation with particles showing different LET values, highlighting consistent differences in gene transcription among different breast cell lines used (tumorigenic MCF7 and MDA-MB-231 and non-tumorigenic MCF10A cell lines) as well as in molecular networks activated. We hope that the identification of target genes IR-induced, could be useful in the case of cancer cells resistant to multiple therapies, and could support proton irradiation in combination with targeted therapy schedules.
Conclusion
Many efforts by research teams are needed to help clinicians in understanding the molecular portrait of a specific cancer type, in order to propose successful combinatorial anticancer therapies in clinical practice.18,19,56–58 In addition, understanding the molecular response to multiple irradiation modalities, also in term of gene expression changes, is a critical point of interest not entirely understood and investigated, in order to select the most successful treatment plan.58
In this work, we compared and described in detailed way for the first time for our knowledge, the GEP and pathways activated after electron and proton irradiation, in BC cells. GEP pathway analyses showed the activation of different signaling in a cell line and radiation type-, dependent manner. MCF10A and MDA-MB-231 cell lines were found to induce many factors and pathways involved in immunological process control. However, although specific pathway are activated in a radiation type-dependent manner, each cell line activates overall similar molecular networks in response to both these two types of IR.
Footnotes
Funding: This study was funded by FIRB/MERIT project (RBNE089KHH) and INFN-funded.
Ethical approval: This study does not require ethical approval and informed consent, as the research project uses commercial immortalized cell lines samples from the American Type Culture Collection (ATCC, Manassas, VA, USA).
Contributor Information
Valentina Bravatà, Email: valentina.bravata@ibfm.cnr.it.
Luigi Minafra, Email: luigi.minafra@ibfm.cnr.it.
Francesco Paolo Cammarata, Email: francesco.cammarata@ibfm.cnr.it.
Pietro Pisciotta, Email: pietro.pisciotta@ibfm.cnr.it.
Debora Lamia, Email: debora.lamia@ibfm.cnr.it.
Valentina Marchese, Email: marchesev@lns.infn.it.
Lorenzo Manti, Email: manti@na.infn.it.
Giuseppe AP Cirrone, Email: pablo.cirrone@lns.infn.it.
Maria Carla Gilardi, Email: mariacarla.gilardi@ibfm.cnr.it.
Giacomo Cuttone, Email: cuttone@lns.infn.it.
Giusi Irma Forte, Email: giusi.forte@ibfm.cnr.it.
Giorgio Russo, Email: giorgio.russo@ibfm.cnr.it.
REFERENCES
- 1.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer 2011; 11: 239–53. doi: 10.1038/nrc3007 [DOI] [PubMed] [Google Scholar]
- 2.Cuaron JJ, MacDonald SM, Cahlon O. Novel applications of proton therapy in breast carcinoma. Chin Clin Oncol 2016; 5: 52. doi: https://doi.org/10.21037/cco.2016.06.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Maggio FM, Minafra L, Forte GI, Cammarata FP, Lio D, Messa C, et al. Portrait of inflammatory response to ionizing radiation treatment. J Inflamm 2015; 12: 14. doi: 10.1186/s12950-015-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Formenti SC, Demaria S. Combining radiotherapy and cancer immunotherapy: a paradigm shift. J Natl Cancer Inst 2013; 105: 256–65. doi: 10.1093/jnci/djs629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tommasino F, Durante M. Proton radiobiology. Cancers 2015; 7: 353–81. doi: 10.3390/cancers7010353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundkvist J, Ekman M, Ericsson SR, Isacsson U, Jönsson B, Glimelius B. Economic evaluation of proton radiation therapy in the treatment of breast cancer. Radiother Oncol 2005; 75: 179–85. doi: 10.1016/j.radonc.2005.03.006 [DOI] [PubMed] [Google Scholar]
- 7.Grant SR, Grosshans DR, Bilton SD, Garcia JA, Amin M, Chambers MS, et al. Proton versus conventional radiotherapy for pediatric salivary gland tumors: acute toxicity and dosimetric characteristics. Radiother Oncol 2015; 116: 309–15. doi: 10.1016/j.radonc.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gameiro SR, Malamas AS, Bernstein MB, Tsang KY, Vassantachart A, Sahoo N, et al. Tumor cells surviving exposure to proton or photon radiation share a common immunogenic modulation signature, rendering them more sensitive to T cell-mediated killing. Int J Radiat Oncol Biol Phys 2016; 95: 120–30. doi: 10.1016/j.ijrobp.2016.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz-Hector S, Trott KR. Radiation-induced cardiovascular diseases: is the epidemiologic evidence compatible with the radiobiologic data? Int J Radiat Oncol Biol Phys 2007; 67: 10–18. doi: 10.1016/j.ijrobp.2006.08.071 [DOI] [PubMed] [Google Scholar]
- 10.Ödén J, Eriksson K, Toma-Dasu I. Inclusion of a variable RBE into proton and photon plan comparison for various fractionation schedules in prostate radiation therapy. Med Phys 2017; 44: 810–22. doi: 10.1002/mp.12117 [DOI] [PubMed] [Google Scholar]
- 11.Romano F, Cirrone GA, Cuttone G, Rosa FD, Mazzaglia SE, Petrovic I, et al. A Monte Carlo study for the calculation of the average linear energy transfer (LET) distributions for a clinical proton beam line and a radiobiological carbon ion beam line. Phys Med Biol 2014; 59: 2863–82. doi: 10.1088/0031-9155/59/12/2863 [DOI] [PubMed] [Google Scholar]
- 12.Ståhl S, Fung E, Adams C, Lengqvist J, Mörk B, Stenerlöw B, et al. Proteomics and pathway analysis identifies JNK signaling as critical for high linear energy transfer radiation-induced apoptosis in non-small lung cancer cells. Mol Cell Proteomics 2009; 8: 1117–29. doi: 10.1074/mcp.M800274-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph B, Ekedahl J, Lewensohn R, Marchetti P, Formstecher P, Zhivotovsky B. Defective caspase-3 relocalization in non-small cell lung carcinoma. Oncogene 2001; 20: 2877–88. doi: 10.1038/sj.onc.1204402 [DOI] [PubMed] [Google Scholar]
- 14.Joseph B, Marchetti P, Formstecher P, Kroemer G, Lewensohn R, Zhivotovsky B. Mitochondrial dysfunction is an essential step for killing of non-small cell lung carcinomas resistant to conventional treatment. Oncogene 2002; 21: 65–77. doi: 10.1038/sj.onc.1205018 [DOI] [PubMed] [Google Scholar]
- 15.Sirzén F, Zhivotovsky B, Nilsson A, Bergh J, Lewensohn R. Spontaneous and radiation-induced apoptosis in lung carcinoma cells with different intrinsic radiosensitivities. Anticancer Res 1998; 18: 695–9. [PubMed] [Google Scholar]
- 16.Viktorsson K, Ekedahl J, Lindebro MC, Lewensohn R, Zhivotovsky B, Linder S, et al. Defective stress kinase and Bak activation in response to ionizing radiation but not cisplatin in a non-small cell lung carcinoma cell line. Exp Cell Res 2003; 289: 256–64. doi: 10.1016/S0014-4827(03)00264-7 [DOI] [PubMed] [Google Scholar]
- 17.Viktorsson K, Lewensohn R, Zhivotovsky B. Apoptotic pathways and therapy resistance in human malignancies. Adv Cancer Res 2005; 94: 143–96. doi: 10.1016/S0065-230X(05)94004-9 [DOI] [PubMed] [Google Scholar]
- 18.Minafra L, Bravatà V, Russo G, Forte GI, Cammarata FP, Ripamonti M, et al. Gene expression profiling of MCF10A breast epithelial cells exposed to IOERT. Anticancer Res 2015; 35: 3223–34. [PubMed] [Google Scholar]
- 19.Bravatà V, Minafra L, Russo G, Forte GI, Cammarata FP, Ripamonti M, et al. High-dose ionizing radiation regulates gene expression changes in MCF7 breast cancer cell Line. Anticancer Res 2015; 35: 5. [PubMed] [Google Scholar]
- 20. International Atomic Energy Agency (IAEA). Absorbed dose determination in external beam radiotherapy an international code of practice for dosimetry based on standards of absorbed dose to water technical reports series no. 398. International Atomic Energy Agency Vienna 2000;ISBN:92-0-102200-X. [Google Scholar]
- 21.Russo G, Pisciotta P, Cirrone GAP, Romano F, Cammarata F, Marchese V, et al. Preliminary study for small animal preclinical hadrontherapy facility. Nucl Instrum Methods Phys Res A 2017; 846: 126–34. doi: 10.1016/j.nima.2016.10.021 [DOI] [Google Scholar]
- 22.Agostinelli S, Allison J, Amako K, Apostolakis J, Araujo H, Arce P, et al. Geant4—a simulation toolkit. Nucl Instrum Methods Phys Res A 2003; 506: 250–303. doi: 10.1016/S0168-9002(03)01368-8 [DOI] [Google Scholar]
- 23.Allison J, Amako K, Apostolakis J, Araujo H, Arce Dubois P, Asai M, et al. Geant4 developments and applications. IEEE Trans Nucl Sci 2006; 53: 270–8. doi: 10.1109/TNS.2006.869826 [DOI] [Google Scholar]
- 24.Allison J, Amako K, Apostolakis J, Arce P, Asai M, Aso T, et al. Recent developments in G eant 4. Nucl Instrum Methods Phys Res A 2016; 835: 186–225. doi: 10.1016/j.nima.2016.06.125 [DOI] [Google Scholar]
- 25.Russo G, Casarino C, Arnetta G, Candiano G, Stefano A, Alongi F, et al. Dose distribution changes with shielding disc misalignments and wrong orientations in breast IOERT: a Monte Carlo - GEANT4 and experimental study. J Appl Clin Med Phys 2012; 13: 74–92. doi: 10.1120/jacmp.v13i5.3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirrone GAP, Cuttone G, Di Rosa F, Raffaele L, Russo G, Guatelli S, et al. The GEANT4 toolkit capability in the hadron therapy field: simulation of a transport beam line. Nucl Phys B Proc Suppl 2006; 150: 54–7. doi: 10.1016/j.nuclphysbps.2005.04.061 [DOI] [Google Scholar]
- 27.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 2013; 41(Database issue): D991–D995. doi: 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, et al. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res 2005; 33: 3154–64. doi: 10.1093/nar/gki624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S, Fulton D, Arenillas DJ, Perco P, Ho Sui SJ, al MJR. Identification of over-represented combinations of transcription factor binding sites in sets of co-expressed genes. Advances in Bioinformatics and Computational Biology London: Imperial College Press 2006; 3: 247–56. [Google Scholar]
- 30.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res 2007; 35(Web Server issue): W245–W252. doi: 10.1093/nar/gkm427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker KG, Hosack DA, Dennis G, Lempicki RA, Bright TJ, Cheadle C, et al. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics 2003; 4: 61. doi: 10.1186/1471-2105-4-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cirrone GAP, Cuttone G, Mazzaglia SE, Romano F, Sardina D, Agodi C, et al. Hadrontherapy: a Geant4-based tool for proton/iontherapy studies. Prog Nucl Sci 2011; 2: 207–12. doi: https://doi.org/10.15669/pnst.2.207 [Google Scholar]
- 33.Li X, Wang W, Wang J, Malovannaya A, Xi Y, Li W, et al. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol Syst Biol 2015; 11: 775. doi: https://doi.org/10.15252/msb.20145504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heckman MG, Robinson JL, Tzou KS, Parker AS, Wu KJ, Hilton TW, et al. An examination of the association between FOXA1 staining level and biochemical recurrence following salvage radiation therapy for recurrent prostate cancer. PLoS One 2016; 11: e0151785. doi: 10.1371/journal.pone.0151785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma K, Araki K, Ichwan SJ, Suganuma T, Tamamori-Adachi M, Ikeda MA. E2FBP1/DRIL1, an AT-rich interaction domain-family transcription factor, is regulated by p53. Mol Cancer Res 2003; 1: 438–44. [PubMed] [Google Scholar]
- 36.Roberson JD, Burnett OL, Robin N. Radiogenomics: towards a personalized radiation oncology. Curr Opin Pediatr 2016; 28: 713–7. doi: 10.1097/MOP.0000000000000408 [DOI] [PubMed] [Google Scholar]
- 37.Lundkvist J, Ekman M, Ericsson SR, Jönsson B, Glimelius B. Proton therapy of cancer: potential clinical advantages and cost-effectiveness. Acta Oncol 2005; 44: 850–61. doi: 10.1080/02841860500341157 [DOI] [PubMed] [Google Scholar]
- 38.Bradley JA, Dagan R, Ho MW, Rutenberg M, Morris CG, Li Z, et al. Initial report of a prospective dosimetric and clinical feasibility trial demonstrates the potential of protons to increase the therapeutic ratio in breast cancer compared with photons. Int J Radiat Oncol Biol Phys 2016; 95: 411–21. doi: 10.1016/j.ijrobp.2015.09.018 [DOI] [PubMed] [Google Scholar]
- 39.Minafra L, Bravat V. Cell and molecular response to IORT treatment. Transl Cancer Res 2014; 3: 32–47. [Google Scholar]
- 40.Kim B, Bae H, Lee H, Lee S, Park JC, Kim KR, et al. Proton beams inhibit proliferation of breast cancer cells by altering DNA methylation status. J Cancer 2016. 7; 7: 344–52. doi: 10.7150/jca.13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keta OD, Todorović DV, Bulat TM, Cirrone PGA, Romano F, Cuttone G, et al. Comparison of human lung cancer cell radiosensitivity after irradiations with therapeutic protons and carbon ions. Exp Biol Med 2017; 242: 1015–24. doi: 10.1177/1535370216669611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finnberg N, Wambi C, Ware JH, Kennedy AR, El-Deiry WS. Gamma-radiation (GR) triggers a unique gene expression profile associated with cell death compared to proton radiation (PR) in mice in vivo. Cancer Biol Ther 2008; 7: 2023–33. doi: 10.4161/cbt.7.12.7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroese DP, Brereton T, Taimre T, Botev ZI. Why the Monte Carlo method is so important today. Wiley Interdiscip Rev Comput Stat 2014; 6: 386–92. doi: 10.1002/wics.1314 [DOI] [Google Scholar]
- 44.Bravatà V, Minafra L, Forte GI, Cammarata FP, Russo G, Di Maggio FM, et al. Cytokine profile of breast cell lines after different radiation doses. Int J Radiat Biol 2017; 93: 1217–26. doi: 10.1080/09553002.2017.1362504 [DOI] [PubMed] [Google Scholar]
- 45.Chang Q, Bournazou E, Sansone P, Berishaj M, Gao SP, Daly L, et al. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia 2013; 15: 848–IN45. doi: 10.1593/neo.13706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jhaveri K, Teplinsky E, Silvera D, Valeta-Magara A, Arju R, Giashuddin S, et al. Hyperactivated mTOR and JAK2/STAT3 pathways: molecular drivers and potential therapeutic targets of inflammatory and invasive ductal breast cancers after neoadjuvant chemotherapy. Clin Breast Cancer 2016; 16: 113–22. doi: 10.1016/j.clbc.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du Y, Peyser ND, Grandis JR. Integration of molecular targeted therapy with radiation in head and neck cancer. Pharmacol Ther 2014; 142: 88–98. doi: 10.1016/j.pharmthera.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 48.Liao W, Hei TK, Cheng SK. Radiation-induced dermatitis is mediated by IL17-expressing γδ T cells. Radiat Res 2017; 187: 464–74. doi: 10.1667/RR007CC.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Shen B, Xia L, Khaletzkiy A, Chu D, Wong JY, et al. Activation of nuclear factor κB in radioresistance of TTP53-inactive human keratinocytes. Cancer Res 2002; 62: 1213–21. [PubMed] [Google Scholar]
- 50.Starenki D, Namba H, Saenko V, Ohtsuru A, Yamashita S. Inhibition of nuclear factor-κB cascade potentiates the effect of a combination treatment of anaplastic thyroid cancer cells. J Clin Endocrinol Metab 2004; 89: 410–8. doi: 10.1210/jc.2003-031216 [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J Clin Invest 2001; 107: 135–42. doi: 10.1172/JCI11914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci USA 2014; 111: E4697–E4705. doi: 10.1073/pnas.1413128111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyake A, Takahashi Y, Miwa H, Shimada A, Konishi M, Itoh N. Neucrin is a novel neural-specific secreted antagonist to canonical Wnt signaling. Biochem Biophys Res Commun 2009; 390: 1051–5. doi: 10.1016/j.bbrc.2009.10.113 [DOI] [PubMed] [Google Scholar]
- 54.Huang Y, Wang KKW. The calpain family and human disease. Trends Mol Med 2001; 7: 355–62. doi: 10.1016/S1471-4914(01)02049-4 [DOI] [PubMed] [Google Scholar]
- 55.Sun C, Kilburn D, Lukashin A, Crowell T, Gardner H, Brundiers R, et al. Kirrel2, a novel immunoglobulin superfamily gene expressed primarily in β cells of the pancreatic islets☆. Genomics 2003; 82: 130–42. doi: 10.1016/S0888-7543(03)00110-1 [DOI] [PubMed] [Google Scholar]
- 56.Pucci M, Bravat V, Forte GI, Cammarata FP, Messa C, Gilardi MC, et al. Caveolin-1, breast cancer and ionizing radiation. Cancer Genomics Proteomics 2015; 12: 143–52. [PubMed] [Google Scholar]
- 57.Minafra L, Bravat V, Cammarata FP, Russo G, Gilardi MC, Forte GI. Radiation gene-expression signatures in primary breast cancer cells. Anticancer Res 2018; 38: 2707–15. [DOI] [PubMed] [Google Scholar]
- 58.Paganetti H. Relative biological effectiveness (RBE) values for proton beam therapy. Variations as a function of biological endpoint, dose, and linear energy transfer. Phys Med Biol 2014; 59: R419–R472. doi: 10.1088/0031-9155/59/22/R419 [DOI] [PubMed] [Google Scholar]