Abstract
Deep learning has demonstrated tremendous revolutionary changes in the computing industry and its effects in radiology and imaging sciences have begun to dramatically change screening paradigms. Specifically, these advances have influenced the development of computer-aided detection and diagnosis (CAD) systems. These technologies have long been thought of as “second-opinion” tools for radiologists and clinicians. However, with significant improvements in deep neural networks, the diagnostic capabilities of learning algorithms are approaching levels of human expertise (radiologists, clinicians etc.), shifting the CAD paradigm from a “second opinion” tool to a more collaborative utility. This paper reviews recently developed CAD systems based on deep learning technologies for breast cancer diagnosis, explains their superiorities with respect to previously established systems, defines the methodologies behind the improved achievements including algorithmic developments, and describes remaining challenges in breast cancer screening and diagnosis. We also discuss possible future directions for new CAD models that continue to change as artificial intelligence algorithms evolve.
INTRODUCTION
Computer-aided detection and diagnosis (CAD) systems have been employed for over three decades and have often been considered as “second-opinion” tools. CAD systems work by utilizing radiographic images with known diagnostic features to train highly specialized software solutions that are equipped with machine learning and pattern recognition algorithms. These systems can then recognize the imaging patterns they were trained with, on test images (i.e. unseen or not used in training), allowing them to participate in detection and diagnosis of various diseases. These systems have the potential to be very useful in the field of oncology, aiding with improved detection and diagnosis of a wide variety of tumor types.1, 2
There have been numerous studies in the literature investigating the use of CAD systems for breast cancer detection and diagnosis. These studies have used various imaging modalities and machine learning algorithms, some of which have even gone through clinical workflow for feasibility tests.3, 4 However, the success of these studies has been limited due to high phenotypic variations in tumors, large number of false positives, and poor diagnosis rates.5 For these reasons, many studies were dedicated to improving these systems. Lately, research in this field is moving towards a more favorable direction due to exciting new advances in machine learning, specifically “deep learning”.6, 7
Deep learning, i.e. as deep neural networks, has been a rapidly growing subfield of machine learning. The main reasons behind this breakthrough over the past few years are increased availability of more advanced computer algorithms that are inspired by human intelligence, updates on contemporary hardware technology for processing and storing large data sets, and an increased availability of massive amounts of labeled data to train these algorithms with better precision. This revolutionary and cutting-edge approach to computer vision has had a broad spectrum of applications including graphics, genetics, medicine, the automotive industry, the Internet, and ultimately, radiology and imaging sciences.8–12
In this review, we aim to evaluate the impact of deep learning based diagnostic systems that can help clinicians with screening and diagnosing breast cancer. Not only do we summarize the details of recently developed deep learning based CAD systems, but we also explain various deep neural network designs for image analysis, explore the benefits and limitations of recently developed decision support systems, and elucidate future perspectives that radiologists and clinicians can benefit from in their routine diagnostic tasks. To address the concerns associated with conventional imaging techniques, CAD and decision support systems have made considerable advancements allowing for precise characterization of various pathologies by recognizing imaging features that are not easily visible to human eyes. These enhanced diagnostic capabilities allow for a reduced number of missed tumor cases and, ultimately, assistance in the diagnostic decision-making process.
UNDERSTANDING BREAST CANCER DIAGNOSIS IN THE DEEP LEARNING ERA
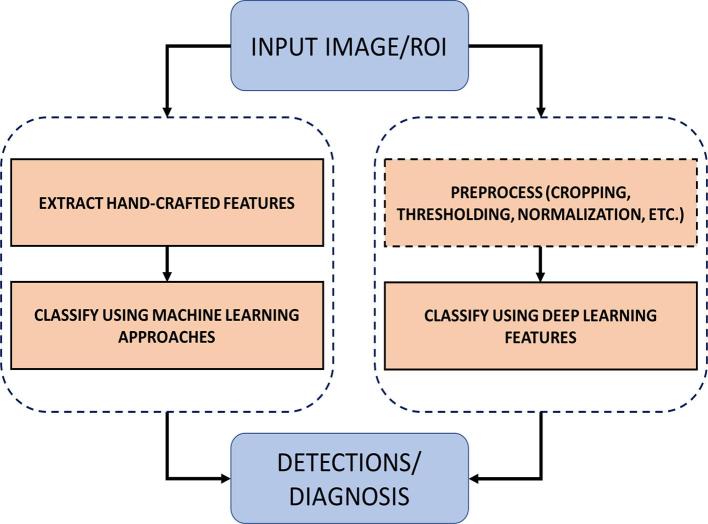
Machine learning algorithms such as Naive Bayes, Genetic Algorithms, Fuzzy Logic, Clustering, Neural Networks, Support Vector Machines, Decision Trees and Random Forests etc. have been used for more than two decades for detection, diagnosis, classification, and risk assessment of breast cancer. Figure 1 shows a representative comparison of conventional machine learning CAD systems and deep learning based CAD systems, both of which utilize radiographic images for breast cancer diagnosis. The conventional machine learning approach for image classification is trained using carefully designed hand-engineered features (e.g. visual descriptions such as elongation, sphericity, or low gradients in borders) that are learned from radiologists and can be coded into algorithms. In contrast, deep learning employs high-level imaging features from large sets of images for training purposes. The literature pertaining to these machine learning methodologies, prior to the deep learning era, is vast. Interested readers may refer to the literature13–26 for further description of conventional machine learning methods in breast cancer, which include a large number of methods that are beyond the scope of this review.
Figure 1.
Comparison of conventional machine learning approach vs deep learning based approaches. ROI, Region of interest.
Literature review and search strategy
For the literature survey, we used Pubmed™, IEEEXplore™, Google Scholar™, and ScienceDirect™ to search for publications relating to deep learning applications towards breast cancer detection and diagnosis. Keywords searched included “deep learning”, “breast cancer”, “breast tissue”, “convolutional neural network”, “machine learning”, “diagnosis”, and “detection”. Only papers that were published in peer-reviewed conferences and journals were selected for review. Our search yielded 28 research articles.
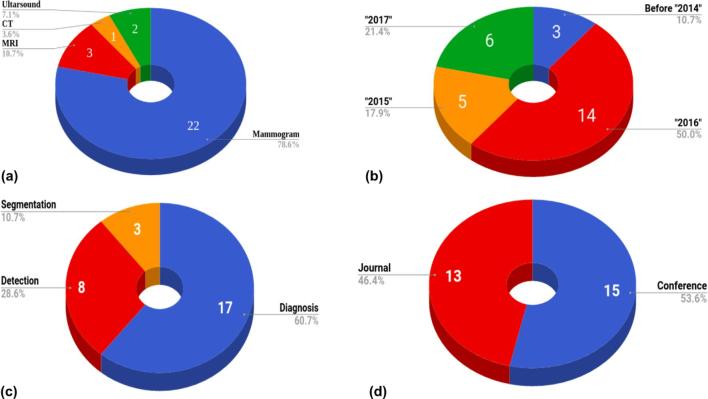
Figure 2a shows the distribution of imaging modality used for breast cancer detection and diagnosis, among which mammography was the most commonly used modality. The search was conducted to include all previous publications up to and including December 2017.
Figure 2.
Statistical distribution of 28 deep learning papers selected for this review. The distribution regarding imaging modalities (a), year of publication (b), deep learning applications (c), and type of publication (d) are shown.
The distribution of the deep learning papers published over time can be seen in Figure 2b, where half of the studies found were published in the year 2016. Figure 2c shows the distribution of the studies based on the application of deep learning with respect to diagnosis, detection, or segmentation of breast cancer, diagnosis being the most commonly studied field of investigation. Furthermore, Figure 2d shows the distribution of these studies between peer-reviewed journals and conferences.
Five authors worked on the literature survey. The topics were divided into subsections of “detection” and “diagnosis”. Two authors worked on detection and three on diagnosis. Papers corresponding to each subsection were found from the above mentioned sources by members of each group independently. The senior authors of the study (i.e. the first and last) verified and reviewed the pool of papers from both subsections as well as resolved any disagreements.
Deep learning in simplified details
The 28 publications, we selected for review utilized various approaches and applications of deep learning towards breast cancer detection and diagnosis. In this section, we briefly outline the inner workings of deep learning.
Machine learning enables computers with the capacity to tackle real-world problems. These systems are trained with representative data, and when new input information is received, they utilize computer algorithms to identify regularities and make outcome decisions/predictions.27 The basis of these algorithms and learned relationships, called features, is widely varied; they can be as simple as detecting differences in intensity values of individual pixels, or as complex as recognizing advanced relationships between position, texture, and shape of the tumors.
Although machine learning is useful in effectively extracting features for certain tasks, the remaining challenge is deciding which specific features should be extracted to feed into the algorithms for accurate diagnosis. Deep learning, in this regard, provides a basis for developing new and improved algorithms that are better equipped to generalize data, by enabling the computer to build complex concepts out of simple ideas.27 In the following section, we describe basic definitions that will help radiologists and clinicians understand the basic principles of deep learning as applied to radiology image analysis and CAD development.
Neural networks, with respect to artificial intelligence, are inspired by the biological basis of neural networks, in which neurons can sense their environment and communicate information to surrounding neurons. In artificial intelligence, neural networks are typically represented by layers. These layers are, essentially, computational functions that process input information, as it compares to training data, to predict an outcome (i.e. f(x) =y, where x is the input information, and y is the outcome prediction). Input neurons can sense new data and pass information onto neurons within different layers, processing this information. Connections between neurons are called “synaptic weights”, which are coefficients used to amplify or dampen the input signal by multiplication, assigning significance to the input to obtain the corresponding output.28 The computational power of these networks relies on the extent of training data that is available, allowing these neural networks to update weights of the connections. Simple network structures with only a few layers are known as “shallow” learning neural networks, whereas network structures which employ numerous and large layers are referred to as “deep” learning neural networks.
Deep neural networks are distinct from ancestral neural networks in that they have much improved universal approximation properties (i.e. the ability to represent any non-linear function/associations) by comparison.27 This is largely due to the use of large numbers of layers, providing flexibility of approximations of different function classes. Convolutional Neural Networks (CNNs) are a subclass of deep neural networks that employ a specialized mathematical function, called “convolution”, in their layers instead of direct multiplication operations.29 CNNs are biologically-inspired variants of neural networks and the convolution operation in their layers provides invariance (i.e. output remains unchanged regardless of changes in input measurement), sparsity of learned features (i.e. most of the entries of the image features/signals have a value of zero; hence, the computer needs to store only non-zero entries), and sharing of parameters, allowing the entire system to work more accurately and efficiently.
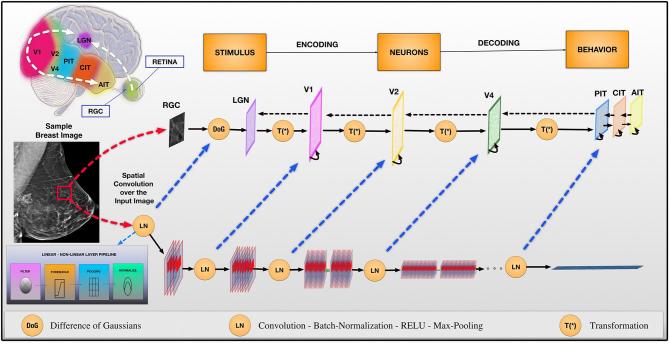
Figure 3 illustrates the relationship between CNNs and the human vision recognition pathway. With CNNs, a given image (e.g. a sample breast image as shown in Figure 3) is represented by edges, curves, or lines (i.e. local features) in the initial layers. Further layers combine these features to detect geometric shapes, or other midlevel feature representations; this continues until the system ends up detecting entire sections of the input image (i.e. global features). CNNs have been shown to be very successful in numerous practical applications.30
Figure 3.
The connection between convolutional networks and the recognition pathway of the human visual cortex is illustrated. The recognition pathway in the visual cortex has multiple relays of information processing: retina, LGN, V1, V2, V4, PIT, AIT. Similarly, these relays of processing are represented in convolutional neural network layers. DoG, difference of Gaussians; LGN, lateral geniculate nucleus; LN, convolution—batch-normalization—RELU—max-pooling; RGC, retinal ganglion cell; T(*), transformation.
End-to-End learning (or training) often refers to the joint training of all parameters in a network such as the approach taken in Jia et al31, Mortazi et al32 and Sukhbaatar et al33. In neural networks, the input is accepted from one end, and the network produces an output at the other end. Training of parameters between these two ends (input to output) is called End-to-End training or learning.
A pre-trained network, as the name implies, uses a network that has been previously trained with images and has optimized parameters for the task it will be performing. If a pre-trained network is used, then the parameters can be used for testing without the need for training the entire system, which can otherwise be a costly endeavor in terms of computation. Pre-training network will tend to work if the target task is similar to the base task (i.e. base task is the one that the network is trained and features are learned from). When the target data set is significantly smaller than the base data set, and the tasks are considerably different from each other (e.g. base network is trained to classify natural images while the target network is aimed to classify tumor images from mammography data), then the phenomena called transfer learning can be used to the transfer the knowledge from base task (i.e. features) into the target task.
Data augmentation, in the context of neural networks, refers to increasing the number of data points. With images, particularly radiologic images, data augmentation works in the following ways: (i) rotating the images in several different angles and saving those images as new training samples, (ii) adding different levels of noise, (iii) cropping the images, and (iv) geometrically transforming the images (e.g. rotation, translation, and scaling) without distorting its structural integrity. Data augmentation can increase the data size by more than 1000-fold by concatenating operations defined in (i–iv). It is effective when using insufficient quantities of data to train a generalizable model. This is often the case for radiology images and applications where data augmentation acts as a regularization.34–36
Literature review: Deep Learning for Breast Cancer Detection and diagnosis
Tables 1 and 2 summarize the deep learning methods for detection and diagnosis of breast cancer, respectively. Evaluation of detection and diagnosis in those studies were performed using the standard metrics such as sensitivity (true-positive rate), specificity (true-negative rate), false-positive rate, false-negative rate, receiver operating characteristics (ROC) and free-response receiver operating characteristics curves. With respect to tissue segmentation evaluation, the Dice Similarity Coefficient metric is used (Tables 1 and 2: fifth and sixth columns).
Table 1.
A summary of different detection methods with data sets and results
| Paper | Approach | Data set | # Images used | AUC | ACC. | Pros (+)/Cons (−) |
| Dhungel et al3 | m-DBN and R-CNN + SVM | DDSM-BCRP/INbreast | 79/115 cases | 0.75 TPR at 4.8 FP/scan for DDSM-BCRP and 0.96 TPR at 1.2 FP/scan on INbreast | +Public data set, transparent results −Complex system (four modules) −Performance gain of cascade of two R-CNNs is not justified |
|
| Akselrod-Ballin et al37 | Faster R-CNN | In-house | 850 | 0.72 | 77.0% | +Includes finer layer information +Classification based on BI-RADS −Patch-based (suboptimal global information, slow testing time) −Impact of preprocessing performance not evaluated |
| Kooi et al38 | CNN + RF | In-house | 62,272 images | 0.941 | +Hand crafted, deep and combined feature performances compared −Suboptimal candidate detector −Patch-based (suboptimal global information, slow testing time) |
|
| Gallego-Posada et al39 | CNN (AlexNet and VGG)+SVM | mini-MIAS | 200 | 60.01 and 64.52% | +Simple system +Presented performance gain of data-augmentation and balanced data set −SVM is not justified |
|
| Jadoon et al7 | CNN-DW and CNN-CT | IRMA | 2796 patches | 81.83 and 83.74%, respectively | +SVM and soft-max compared −Patch-based (suboptimal global information, slow testing time) −Hand-crafted features used (not-deep, and suboptimal) |
|
| Samala et al40 | Various CNN comparisons | In-house | 127 | 0.89 and 0.93 | +216 CNN architectures compared −Exhaustive search of the architecture |
|
| Samala et al41 | CNN (four convolutional layers and three fully connected layers) | In-house | 2282 digitized film and digital mammograms and 324 DBT | 0.90 | +Transfer learning from mammogram to DBT - DBT is acquired with a different geometry prototype |
|
| Fotin et al42 | CNN AlexNet | In-house | 2607 | ROI Sensitivity of 93.0% | +Conventional CNN approach −Patch-based (suboptimal global information, slow testing time) −Testing time five times slower than other systems |
|
| Ertosun et al43 | Two back-to-back CNNs | DDSM | 2420 images | 85% | +Comparisons among different architectures −Back-to-back CNNs −Complex system |
ANN, artificial neural network; BCRP, breast cancer research program (DoD); CNN, convolutional neural network; DBT, digital breast tomosynthesis; DDSM, digital dataset for screening mammography; IRMA, image retrieval in medical applications; RF, random forest; SVM, support vector machine.TPR, true positive rate.
Table 2.
A summary of published diagnostic methods with data sets and results
| Paper | Approach | Data set | # Images used | AUC | Acc (%) | Pros (+)/Cons (−) |
| Dheeba et al44 | WNN | In-house | 216 | – | 85.4 | +Use of texture features and WNN −No empirical/theoretical evidence of preferring WNN to CNN. |
| Dhungel et al4 | CNN + RF | INBreast | 116 | – | 95 | +Incorporates low and high level features −Requires both training from scratch and fine-tuning |
| Jiao et al45 | 18 layer CNN + SVM | DDSM | 600 | – | 96.7 | +Deeper network −Remains subjective as requires gray-level intensities |
| Arevalo et al46 | Two conv layers and FC layers | BCDR | 736 | 0.82 | – | +Pre-processing and extensive data-augmentation −No regularizer is used, so optimality of the method is unknown. |
| Becker et al47 | ANN | In-house | 286 | 0.82 | – | +Relationship between breast density and accuracy was studied. −Model details missing |
| Sahiner et al48 | Texture feature imagesthree + layer CNN | In-house | 168 | – | 86.0 | +Comparison among different texture feature for classification −Using shallow network |
| Huynh et al49 | CNN (AlexNet) | In-house | 607 | 0.86 | – | +Increasing training data by using augmented ROI −Insufficient network depth, use extra features for classification |
| Peterson et al50 | Unsupervised sparse autoencoder | Dutch-breast cancer screening data set/MMHS/Dutch-breast cancer screening data set | 493/668/1576 | 0.65 | – | +Using whole image for train network with autoencoder features −Using shallow network for supervisor learning |
| Qiu et al51 | Three conv-max pooling layers and two FC layers | In-house | 270 | – | 71.4 | +Using bilateral mammographic tissue density as features −Using shallow network |
| Carneiro et al6 | CNN-F | INBreast/DDSM | 115/172 | 0.91/ 0.97 |
+Using pre-trained model and multiview −No comparison with other methods |
|
| Li et al52 | 3D-CNN | In-house | 143 | 0.801 | 78.1 | +3D model - Small real training data set for 3D |
| Zhou et al53 | CNN (AlexNet) | In-house | 463 | – | 76 | +Faster classification −Non-3D approach |
| Zhang et al54 | PGBM | In-house | 227 | – | 93.4 | +Multistage architecture −No empirical advantage of PGBM over CNN shown |
| Cheng et al55 | SDAE | 10,133 slices | – | 82.4 | +Slice selection strategy to account for depth −Needs pre-training and training |
|
| Kooi et al56 | CNN + gradient boosted tree classifier | In-house | 1804 | 0.8 | – | +Large data set for training −Requires both training from scratch and fine-tuning |
3D, three-dimensional; AUC, area under the curve; CNN,Convolutional Neural Network; PGBM, PointwiseGated Boltzmann Machine; RF, random forest; ROI, region of interest; SDAE, Stacked Denoising Autoencoders; WNN, Wavelet Neural Network.
Breast cancer detection and diagnosis using a pre-trained network
Mammography
Literature on deep learning-based breast cancer detection from mammography shares very similar network designs: pre-trained network, data augmentation (or transfer learning), and extracting features to be used with classifiers such as support vector machine, random forest, or others. In other words, deep networks were used only for extracting discriminative features (i.e. imaging features that are unique to tumor type). Advantages and disadvantages of detection methods are summarized in Table 1.
The use of pre-trained networks for breast cancer diagnosis started in 2015 (Table 2). Most pre-trained networks are trained on the ImageNet data set owing to a large number of images (>1 million) and thousands of classes.57 Several studies showed that pre-trained models could be used to boost classification results for mammograms.6 However, there is no consensus on what features should be used for classification.58–60 The only study, to our knowledge, that puts some sort of feature interpretations into the diagnostic task is by Becker et al.44 Uniquely, the authors studied the relationship of breast density to classification accuracy and found that low density was easier to classify. Despite the reported effectiveness of transfer learning, studies (see also Table 2) share also a concern: the differences in source task and target task in transfer learning should be solved to have an effective diagnosis system. Till now, the only viable way to approach this problem is to use data augmentation.
Ultrasound
Another modality used for classification of breast tissues is ultrasound. To perform automatic classification using shear-wave elastography, Zhang et al proposed Pointwise Gated Boltzmann Machines-based approach, where local and global features were combined to identify tissue types.54 In another work, Cheng et al used Stacked Denoising Autoencoders.55 These methods are able to extract higher-level features but they are still using not so deep networks compared to conventional CNN-based methods. Thus, their accuracies were limited by the discriminative power of the extract features affected by the network choices.
MRI
Recently, the application of deep learning was explored for tissue classification using MRI. Li et al proposed a three-dimensional (3D) CNN-based architecture for breast tumor classification in dynamic contrast enhanced MRI.52 A 3D-CNN consisting of 10 layers was employed for feature learning and classification. Experimental evaluations demonstrated that the 3D-CNN-based approach outperformed the two-dimensional-CNN based approach by around 8% in terms of AUC, showing a promising future for deep learning with MRI in breast cancer diagnosis.
Breast cancer detection and diagnosis using end-to-end training
Mammography
Another trend in deep learning based cancer detection is to use end-to-end training instead of using pre-trained models. Briefly, in end-to-end learning/training, post-processing steps are added to have one system learn its parameters jointly instead of connecting multiple individual parts. Studies3, 7,38,40 showed that classification accuracies with end-to-end training-based networks were higher than a single network implementation. Other than that, network architecture, the use of alternative classifiers, and combination of handcrafted features with deep features were conceptually identical to each other. These studies used the three publicly available data sets, Digital Dataset for Screening Mammography-Breast Cancer Research Program [DoD (Department of Defense)],Image Retrieval in Medical Applications, and INbreast. Although promising results have been shown in end-to-end training approaches (Table 1), each module in the end-to-end training should be made sure contributing to the whole learning process because interactions of such modules with each other can slow down the learning significantly if learning dynamics of each module is different. Hence, this can lead into difficulties in converging into an optimal training model.
Within the breast cancer classification studies (Table 2), end-to-end training has been used for the first time by Sahiner et al in 1996, where gray-level difference statistics of mammogram images and spatial gray-level dependence images were used to train a three layer CNN.48 Similarly, since then, various studies have investigated different handcrafted features to train CNNs.4, 46,47,61,62 These methods conceptually work in a similar way: combining multiple modules and jointly training all parameters to demonstrate the power of end-to-end learning as a single architecture.
Digital breast tomosynthesis
An in-depth comparison of different CNN architectures was performed by Samala et al40 where authors automatically differentiated micro-calcification in digital breast tomosynthesis scans from other tissues. A total of 216 unique deep learning CNN architectures were trained by varying the number of filters, filter kernel sizes and partial sums. Although this study is one of the few that evaluated the impact of architecture on a deep learning application, it presents an inefficient solution for finding the best architecture. Instead, an automated search algorithm such as genetic algorithms or simulated annealing would be more efficient.
Another comparison study was conducted by Fotin et al.42 where authors found that the deep learning approaches outperform conventional approaches in terms of sensitivity and specificity (Table 1).
Breast tumor segmentation
Mammography
Deep learning-based mammogram segmentation approaches are not significantly different than patch-based CNN-based detection approaches.39, 63 In Dhungel et al61, authors extracted the most discriminative imaging features to classify each voxel either as normal or abnormal. Although this increases the specificity rates, false-positive findings increase as well. The authors had to utilize the last layer as a combination of random field and structural support vector machine. Similarly in Chuquicusma et al63, a CNN with overlapping patches was used for segmentation to increase segmentation accuracy. However, a drawback to this approach was the complicated model, which included two different network structures, causing instability when training. Furthermore, the patch-based classification was not able to incorporate spatial constraints and a post-processing step was required, which puts additional computational cost into the framework.
MRI
Earlier segmentation studies used patch-based systems (i.e. CNNs were trained using patches) to delineate tumors.64 Although the accuracy of such studies was lower than clinically acceptable, the algorithm design is promising for future improvements. Recently, a U-net architecture has been used for image segmentation applications on MRI.62 The U-net is an architecture based on CNN and takes its name because of the “U” shape of the network. This network is specifically tailored for successful biomedical image segmentation applications. Although promising results were obtained for segmentation (as in Dalmis et al43 where breast and fibroglandular tissues were delineated), it should be noted that success of the U-net strongly depends on data augmentation procedure, and precise labeling of the tissues. It should be also noted that U-net has a slow convergence rate, raising questions about training efficiency.
DISCUSSION AND CONCLUDING REMARKS
With the help of deep neural networks, the diagnostic capabilities of learning algorithms are approaching levels of human expertise, shifting the CAD paradigm from a “second-opinion” tool to a more collaborative utility. In this study, we have systematically analyzed and summarized the latest status of the deep learning-based CAD approaches for breast cancer detection and diagnosis. Examined studies showed significant improvement with respect to conventional machine learning approaches and the state of the art results in autodetection and diagnosis of breast cancer from medical imagery. Deep learning has achieved enormous successes in several different fields; however, its true potential in medical imaging has yet to be achieved. Breast cancer detection and diagnosis using deep learning methods have unique challenges that must be addressed prior to clinical adoption.
Lack of imaging data in big data era
One of the major problems in developing deep learning based CAD systems for breast cancer is the lack of the sufficient data for training models with millions of parameters. Some approaches that address this issue include: (1) building and training a model with a very shallow network (only a few thousand parameters), (2) data augmentation.
Each of these approaches has their own drawbacks. In the first approach, limiting the number of parameters will lead to potentially significant inaccuracies. On the other hand, data augmentation will either add noise to the images or require sampling of overlapping image patches. The augmented samples, however, can be highly correlated with each other resulting in overfitting. Overfitting is a well-known machine learning and statistical modeling problem, which occurs when a learning model memorizes the training data and is not able to generalize to the new data. Moreover, local patches cannot incorporate the global and spatial context of the image, which can lead to inaccuracies.
Technical challenges of deep learning
There are challenges associated with the use of transfer learning in deep learning including architecture selection, number of examples sufficient to fine-tune, as well as the numbers of layers used on top of the pre-trained model. Moreover, the effectiveness of transfer learning decreases when the target task (mammogram diagnosis) is different from the source task (pre-trained network’s task).65
The success of deep learning methods currently relies on high capacity models requiring several iterative updates across many labeled examples. However, obtaining millions of labeled examples is not an easy task, especially in the medical imaging field. Nonetheless, once these systems have been perfected, deep learning can be used in prevention and treatment programs for optimal results, radically transforming clinical practice and public health. In the following, we envision the potential of deep learning to transform other imaging modalities in the context of breast cancer detection and diagnosis.
Potential role of PET/MRI
The increasing availability of PET/MRI will most certainly lead to continued improvements in the accuracy of diagnosing breast cancer. PET/CT is limited by relatively high radiation dose and low spatial resolution resulting in inadequate sensitivity for detection of cancers ≤ 2 cm.66 In contrast, Pinker et al showed that fused multiparametric MRI and PET imaging had significant improvements in accuracy with an AUC of 0.935 (0.835–1), when compared with delayed contrast enhanced MRI or PET alone.67 This technique has the potential to significantly decrease the number of unnecessary biopsies. There are also advantages in combining the sensitivity of MRI, to determine the extent of disease, and the sensitivity of PET, to detect axillary and chest nodal disease.68 No published studies to date have evaluated the use of machine learning and PET/MR for breast cancer.
What would it take for radiologists to accept deep learning tools for daily use?
The use of this advanced technology has the potential to update breast imaging techniques that have changed very little over the past 40–50 years. Many of our current practices in breast screening and diagnosis suffer from limited specificity, requiring an image-guided biopsy to reach a definitive diagnosis. Computer assisted diagnosis using machine learning, including deep CNNs, has the ability to efficiently make accurate diagnoses of breast pathology, potentially without the need of biopsy. With the help of user-interface development and commercialization, these artificial intelligence algorithms will certainly be part of an exciting future in breast imaging.
However, there are more steps that must be taken before radiologists accept these decision-support systems in their daily routine workflows. First, a global real-life application should account for widespread geographic, ethnic, and genetic variations.69 Common cases in certain regions of the world may be quite rare in other areas. However, from the current hardware point of view, it may not be possible to train the deep-learning systems with a large amount of worldwide data. Sampling from this large database requires the criteria of “best representative” positive and negative samples of an illness. Thus, with the current hardware limitations, it may be beneficial to use locally trained versions of the same application, and to integrate and adapt their outputs as needed for the worldwide stage. The commercialized real-life applications should also make clear how to interpret the “rare cases”. In general, these applications are going to miss the “rare cases” for which they are not trained. For the time being, a critical step will be to have radiologists provide a final verification of the outcomes of the real-life applications. In addition, real-life applications should allow radiologists to upload new data into the training system. However, it is not clear yet how training can be repeated, from scratch or as a pre-trained network, by each piece of uploaded data, and how training duration and overfitting can be handled by frequent training with slightly modified data sets. Finally, moving these systems to a virtual cloud environment and having them accessible at any time would be a very useful feature.
How deep learning tools will impact the practice of radiology in breast cancer diagnosis and evaluation?
Success in the development of accurate deep learning algorithms for breast cancer diagnosis has the potential to significantly impact the practice of radiology. These algorithms can aid radiologists in detecting and discriminating normal and suspicious tissues. Deep learning tools can provide important cues, which may be left out in a single screening session by the radiologist. Moreover, these tools can be applied to reduce the interpretation time required radiologists, thus improving clinical efficiency.
Recently, deep learning is being applied to generate artificial but realistic images across different applications of computer vision. As it is both cumbersome and time-consuming to manually annotate radiological images, the use of Generative Adversarial Networks can help generate better synthetic images to train deeper architectures.63, 70 Moreover, these images can also be used to train radiology residents and interns.
FDA approval process, current status, and future steps
One may wonder if the development in deep learning-based CAD system will be available shortly in clinical routine. One big step is review and approval from the Food and Drug Administration (FDA) and other regulatory bodies.71 Although studies on CAD systems date back to 1960, Image CheckerTM was the first CAD system that got FDA approval in 1998 by R2® technology for screening mammography (R2 Technology Inc, Sunnyvale, CA). This began an era of shift from research phase to commercialized industry practice for CAD systems. To date there are several FDA-approved CAD systems in the market. Specific to breast cancer, some examples include iCAD, Inc. that got an FDA approval for the SecondLook® system in 2001 (iCAD Inc, Nashua, NH). VuCOMP, Inc. got FDA approval for their M-Vu CAD and M-Vu Breast Density systems, which use computer vision techniques to identify areas of a mammogram which are consistent with breast cancer in 2013 and 2014. This company was later bought by iCAD. QView Medical Inc. is another company which announced the FDA approval of their CAD system, QVCAD, in early November 2016. QVCAD is a deep learning-based CAD for 3D automated breast ultrasound analysis (QView Medical Inc, Los Altos, CA). The number of FDA-approved CAD systems are increasing and many companies have pending FDA approvals for their breast cancer CAD systems. For a CAD system to be approved for clinical use, the FDA recommends a generalizability test to establish the system’s performance on different imaging devices. Furthermore, a standalone and clinical performance assessment is necessary to test the performance and safety of the device or tool. For clinical assessment, performance of clinicians using the newly designed system (CAD) is to be compared against their performance on an existing tool, if applicable. With the increased use of deep learning algorithms, hardware support, and compelling properties of the deep learning algorithms such as robustness, generalizability, and expert-level accuracies, we expect to see more and more CAD system approved by the FDA for breast cancer detection and diagnosis.
Contributor Information
Jeremy R Burt, Email: Jeremy.Burt.MD@flhosp.org.
Neslisah Torosdagli, Email: neslimsaht@gmail.com.
Naji Khosravan, Email: naji.khosravan@gmail.com.
Harish RaviPrakash, Email: harishr@knights.ucf.edu.
Aliasghar Mortazi, Email: aamortazi@gmail.com.
Fiona Tissavirasingham, Email: fiona.tissavirasingham@gmail.com.
Sarfaraz Hussein, Email: sarfarazhusein@gmail.com.
Ulas Bagci, Email: ulasbagci@gmail.com.
Funding
The study is partially funded by CRCV, UCF.
REFERENCES
- 1.El-Baz A, Beache GM, Gimel'farb G, Suzuki K, Okada K, Elnakib A, et al. Computer-aided diagnosis systems for lung cancer: challenges and methodologies. Int J Biomed Imaging 2013; 2013: 1–46. doi: 10.1155/2013/942353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee H, Chen Y-PP. Image based computer aided diagnosis system for cancer detection. Expert Systems with Applications 2015; 42: 5356–65. doi: 10.1016/j.eswa.2015.02.005 [DOI] [Google Scholar]
- 3.Dhungel N, Carneiro G, Bradley AP. Automated mass detection in mammograms using cascaded deep learning and random forests. in digital image computing: techniques and applications (DICTA), 2015 international conference on: The British Institute of Radiology.; 2015. [Google Scholar]
- 4.Dhungel N, Carneiro G, Bradley AP. The automated learning of deep features for breast mass classification from mammograms. In MICCAI. 2; 2016. [Google Scholar]
- 5.Fenton JJ, Taplin SH, Carney PA, Abraham L, Sickles EA, D’Orsi C, et al. Influence of computer-aided detection on performance of screening mammography. N Engl J Med 2007; 356: 1399–409. doi: 10.1056/NEJMoa066099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carneiro G, Nascimento JC, Bradley AP. Unregistered multiview mammogram analysis with pre-trained deep learning models. In MICCAI. 3; 2015. [Google Scholar]
- 7.Jadoon MM, Zhang Q, Haq IU, Butt S, Jadoon A. Three-class mammogram classification based on descriptive CNN features. Biomed Res Int 2017; 2017: 1–11. doi: 10.1155/2017/3640901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussein S, Cao K, Song Q, Bagci U. Risk stratification of lung nodules using 3D CNN-based multi-task learning In: International conference on information processing in medical imaging; 2017. pp 249–60. [Google Scholar]
- 9.Lane ND, Bhattacharya S, Georgiev P, Forlivesi C, Kawsar F. An early resource characterization of deep learning on wearables, smartphones and internet-of-things devices : Proceedings of the 2015 international workshop on internet of things towards applications: The British Institute of Radiology.; 2015. [Google Scholar]
- 10.Luckow A. Deep learning in the automotive industry: applications and tools : Big data (Big Data), 2016 IEEE international conference on: The British Institute of Radiology.; 2016. [Google Scholar]
- 11.Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform 2016; 31: bbw068. doi: 10.1093/bib/bbw068 [DOI] [PubMed] [Google Scholar]
- 12.Nalbach O, Arabadzhiyska E, Mehta D, Seidel H-P, Ritschel T. Deep shading: convolutional neural networks for screen space shading : Computer graphics forum. 36 The British Institute of Radiology.; 2017. 65–78. doi: 10.1111/cgf.13225 [DOI] [Google Scholar]
- 13.Abreu PH, Santos MS, Abreu MH, Andrade B, Silva DC. Predicting breast cancer recurrence using machine learning techniques. ACM Computing Surveys 2016; 49: 1–40. doi: 10.1145/2988544 [DOI] [Google Scholar]
- 14.Akay MF. Support vector machines combined with feature selection for breast cancer diagnosis. Expert Syst Appl 2009; 36: 3240–7. doi: 10.1016/j.eswa.2008.01.009 [DOI] [Google Scholar]
- 15.Al-Ghaib H, Wang Y, Adhami R. A new machine learning algorithm for breast and pectoral muscle segmentation. Eur J Adv Eng Technol 2015; 2: 21–9. [Google Scholar]
- 16.Begum S, Bera SP, Chakraborty D, Sarkar R. Breast cancer detection using feature selection and active learning : Computer, communication and electrical technology: proceedings of the international conference on advancement of computer communication and electrical technology (ACCET 2016), West Bengal, India, 21-22 October 2016: The British Institute of Radiology.; 2017. 43–8. [Google Scholar]
- 17.Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform 2006; 2: 59. doi: 10.1177/117693510600200030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durai SG, Ganesh SH, Christy AJ. Prediction of breast cancer through classification algorithms: a survey. 9: The British Institute of Radiology.; 2016. 359–65. [Google Scholar]
- 19.Huang MW, Chen CW, Lin WC, Ke SW, Tsai CF. SVM and SVM ensembles in breast cancer prediction. PLoS One 2017; 12: e0161501. doi: 10.1371/journal.pone.0161501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kharel N, Alsadoon A, Prasad PWC, Elchouemi A. Early diagnosis of breast cancer using contrast limited adaptive histogram equalization (CLAHE) and morphology methods : Information and communication systems (ICICS), 2017 8th international conference on: The British Institute of Radiology.; 2017. [Google Scholar]
- 21.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 2015; 13: 8–17. doi: 10.1016/j.csbj.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polat K, Güneş S. Breast cancer diagnosis using least square support vector machine In: digital signal processing. 17; 2007. pp 694–701. doi: 10.1016/j.dsp.2006.10.008 [DOI] [Google Scholar]
- 23.Qayyum A, Basit A. Automatic breast segmentation and cancer detection via SVM in mammograms : Emerging technologies (ICET), 2016 international conference on: The British Institute of Radiology.; 2016. [Google Scholar]
- 24.Shajahaan SS, Shanthi S, ManoChitra V. Application of data mining techniques to model breast cancer data. IJETAE 2013; 3: 362–9. [Google Scholar]
- 25.Xi X, Shi H, Han L, Wang T, Ding HY, Zhang G, et al. Breast tumor segmentation with prior knowledge learning. Neurocomputing 2017; 237: 145–57. doi: 10.1016/j.neucom.2016.09.067 [DOI] [Google Scholar]
- 26.Xie W, Li Y, Ma Y. Breast mass classification in digital mammography based on extreme learning machine. Neurocomputing 2016; 173: 930–41. doi: 10.1016/j.neucom.2015.08.048 [DOI] [Google Scholar]
- 27.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–44. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 28.Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators. Neural Networks 1989; 2: 359–66. doi: 10.1016/0893-6080(89)90020-8 [DOI] [Google Scholar]
- 29.Girshick R. Fast r-cnn In: Proceedings of the IEEE international conference on computer vision; 2015. [Google Scholar]
- 30.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw 2015; 61: 85–117. doi: 10.1016/j.neunet.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 31.Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, et al. Caffe: convolutional architecture for fast feature embedding : Proceedings of the 22nd ACM international conference on multimedia: The British Institute of Radiology.; 2014. [Google Scholar]
- 32.Mortazi A, Karim R, Rhode K, Burt J, Bagci U. CardiacNET: segmentation of left atrium and proximal pulmonary veins from MRI using multi-view CNN. arXiv preprint arXiv 2017; 1705: 06333. [Google Scholar]
- 33.Sukhbaatar S, Weston J, Fergus R. End-to-end memory networks In: Advances in neural information processing systems; 2015. [Google Scholar]
- 34.Srivastava N. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014; 15: 1929–58. [Google Scholar]
- 35.Wan L, Zeiler M, Zhang S, LeCun Y, Fergus R. Regularization of neural networks using dropconnect In: Proceedings of the 30th international conference on machine learning (ICML-13); 2013. [Google Scholar]
- 36.Zeiler MD, Fergus R. Stochastic pooling for regularization of deep convolutional neural networks. arXiv preprint arXiv 2013; 1301: 3557. [Google Scholar]
- 37.Akselrod-Ballin A, Karlinsky L, Alpert S, Hasoul S, Ben-Ari R, Barkan E. A region based convolutional network for tumor detection and classification in breast mammography : International workshop on large-scale annotation of biomedical data and expert label synthesis: The British Institute of Radiology.; 2016. 197–205. [Google Scholar]
- 38.Kooi T, Litjens G, van Ginneken B, Gubern-Mérida A, Sánchez CI, Mann R, et al. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 2017; 35: 303–12. doi: 10.1016/j.media.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 39.Gallego-Posada J, Montoya-Zapata D, Quintero-Montoya O. Detection and diagnosis of breast tumors using deep convolutional neural networks; 2016. [Google Scholar]
- 40.Samala RK, Chan H-P, Hadjiiski LM, Cha K, Helvie MA. Deep-learning convolution neural network for computer-aided detection of microcalcifications in digital breast tomosynthesis : SPIE medical imaging. 97850Y 9785: The British Institute of Radiology.; 2016. [Google Scholar]
- 41.Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K. Mass detection in digital breast tomosynthesis: deep convolutional neural network with transfer learning from mammography. Med Phys 2016; 43: 6654–66. doi: 10.1118/1.4967345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fotin SV, Yin Y, Haldankar H, Hoffmeister JW, Periaswamy S. Detection of soft tissue densities from digital breast tomosynthesis: comparison of conventional and deep learning approaches : Proc: The British Institute of Radiology.; 2016. [Google Scholar]
- 43.Ertosun MG, Rubin DL. Probabilistic visual search for masses within mammography images using deep learning : Bioinformatics and biomedicine (BIBM), 2015 IEEE International Conference on: The British Institute of Radiology.; 2015. [Google Scholar]
- 44.Dheeba V, Albert Singh N, Pratap Singh JA. Breast cancer diagnosis: an intelligent detection system using wavelet neural network In: Proceedings of the international conference on frontiers of intelligent computing: theory and applications (FICTA) 2013; 2014. pp 111–8. [Google Scholar]
- 45.Jiao Z, Gao X, Wang Y, Li J. A deep feature based framework for breast masses classification. Neurocomputing 2016; 197: 221–31. doi: 10.1016/j.neucom.2016.02.060 [DOI] [Google Scholar]
- 46.Arevalo J, González FA, Ramos-Pollán R, Oliveira JL, Guevara Lopez MA. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed 2016; 127: 248–57. doi: 10.1016/j.cmpb.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 47.Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol 2017; 52: 434–40. doi: 10.1097/RLI.0000000000000358 [DOI] [PubMed] [Google Scholar]
- 48.Sahiner B, Chan HP, Petrick N, Wei D, Helvie MA, Adler DD, et al. Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE Trans Med Imaging 1996; 15: 598–610. doi: 10.1109/42.538937 [DOI] [PubMed] [Google Scholar]
- 49.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging 2016; 3: 034501. doi: 10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen K, Nielsen M, Diao P, Karssemeijer N, Lillholm M. Breast tissue segmentation and mammographic risk scoring using deep learning : International workshop on digital mammography: The British Institute of Radiology.; 2014. [Google Scholar]
- 51.Qiu Y. An initial investigation on developing a new method to predict short-term breast cancer risk based on deep learning technology : SPIE medical imaging: The British Institute of Radiology.; 2016. [Google Scholar]
- 52.Li J, Fan M, Zhang J, Li L. Discriminating between benign and malignant breast tumors using 3D convolutional neural network in dynamic contrast enhanced-MR images : SPIE medical imaging: The British Institute of Radiology.; 2017. [Google Scholar]
- 53.Zhou X, Kano T, Koyasu H, Li S, Zhou X. Automated assessment of breast tissue density in non-contrast 3D CT images without image segmentation based on a deep CNN. In proc. Vol. 10134: The British Institute of Radiology.; 2017. [Google Scholar]
- 54.Zhang Q, Xiao Y, Dai W, Suo J, Wang C, Shi J, et al. Deep learning based classification of breast tumors with shear-wave elastography. Ultrasonics 2016; 72: 150–7. doi: 10.1016/j.ultras.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 55.Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep 2016; 6: 24454. doi: 10.1038/srep24454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kooi T, van Ginneken B, Karssemeijer N, den Heeten A. Discriminating solitary cysts from soft tissue lesions in mammography using a pretrained deep convolutional neural network. Med Phys 2017; 44: 1017–27. doi: 10.1002/mp.12110 [DOI] [PubMed] [Google Scholar]
- 57.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks In: Advances in neural information processing systems; 2012. [Google Scholar]
- 58.Chen X, Udupa JK, Bagci U, Zhuge Y, Yao J. Medical image segmentation by combining graph cuts and oriented active appearance models. IEEE Trans Image Process 2012; 21: 2035–46. doi: 10.1109/TIP.2012.2186306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis J, Goadrich M. The relationship between precision-recall and ROC curves : Proceedings of the 23rd international conference on machine learning: The British Institute of Radiology.; 2006. [Google Scholar]
- 60.Gur D, Bandos AI, Rockette HE, Zuley ML, Sumkin JH, Chough DM, et al. Localized detection and classification of abnormalities on FFDM and tomosynthesis examinations rated under an FROC paradigm. AJR Am J Roentgenol 2011; 196: 737–41. doi: 10.2214/AJR.10.4760 [DOI] [PubMed] [Google Scholar]
- 61.Dhungel N, Carneiro G, Bradley AP. Deep learning and structured prediction for the segmentation of mass in mammograms In: MICCAI. 1; 2015. [Google Scholar]
- 62.Dalmış MU, Litjens G, Holland K, Setio A, Mann R, Karssemeijer N, et al. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys 2017; 44: 533–46. doi: 10.1002/mp.12079 [DOI] [PubMed] [Google Scholar]
- 63.Chuquicusma MJ, Hussein S, Burt J, Bagci U. How to fool radiologists with generative adversarial networks? A visual turing test for lung cancer diagnosis. arXiv preprint arXiv 2017; 1710: 09762. [Google Scholar]
- 64.Moeskops P, Wolterink JM, Velden BHMvander, Gilhuijs KGA, Leiner T, Viergever MA, et al. Deep learning for multi-task medical image segmentation in multiple, modalities In: Medical image computing and computer-assisted intervention – MICCAI. 2016; 2016. pp 478–86. [Google Scholar]
- 65.Yosinski J. How transferable are features in deep neural networks? In: Advances in neural information processing systems; 2014. [Google Scholar]
- 66.Avril N, Rosé CA, Schelling M, Dose J, Kuhn W, Bense S, et al. Breast imaging with positron emission tomography and fluorine-18 fluorodeoxyglucose: use and limitations. J Clin Oncol 2000; 18: 3495–502. doi: 10.1200/JCO.2000.18.20.3495 [DOI] [PubMed] [Google Scholar]
- 67.Pinker K, Bogner W, Baltzer P, Karanikas G, Magometschnigg H, Brader P, et al. Improved differentiation of benign and malignant breast tumors with multiparametric 18fluorodeoxyglucose positron emission tomography magnetic resonance imaging: a feasibility study. Clin Cancer Res 2014; 20: 3540–9. doi: 10.1158/1078-0432.CCR-13-2810 [DOI] [PubMed] [Google Scholar]
- 68.Tabouret-Viaud C, Botsikas D, Delattre BMA, Mainta I, Amzalag G, Rager O et al. PET/MR in breast cancer : Seminars in nuclear medicine. 45: The British Institute of Radiology.; 2015. 304–21. doi: 10.1053/j.semnuclmed.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 69.Hedlund M, Tollstadius A. Artificial intelligence — the future is now. Diagnostic Imaging Europe 2017: 20–3. [Google Scholar]
- 70.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets In: Advances in neural information processing systems; 2014. [Google Scholar]
- 71. Food and Drug Administration. Computer-assisted detection devices applied to radiology images and radiology device data - premarket notification [510(k)] submissions; 2012. [Google Scholar]