Abstract
MRI is becoming more frequently used in studies involving measurements of adipose tissue and volume and composition of skeletal muscles. The large amount of data generated by MRI calls for automated analysis methods. This review article presents a summary of automated and semi-automated techniques published between 2013 and 2017. Technical aspects and clinical applications for MRI-based adipose tissue and muscle composition analysis are discussed based on recently published studies. The conclusion is that very few clinical studies have used highly automated analysis methods, despite the rapidly increasing use of MRI for body composition analysis. Possible reasons for this are that the availability of highly automated methods has been limited for non-imaging experts, and also that there is a limited number of studies investigating the reproducibility of automated methods for MRI-based body composition analysis.
Introduction
Tomographic techniques, i.e. CT and MRI, are recognized as state-of-the-art methods for body composition analysis, in particular for quantification of compartmental volumes of adipose tissue (AT)1 and muscles.2 High soft-tissue contrast, in combination with increasing availability and absence of ionizing radiation, makes MRI more frequently the preferred method of choice. In several large population studies, such as the UK Biobank,3 the German National Cohort,4 the KORA-MRI study,5 the Netherlands Epidemiology of Obesity Study6 and the Dallas Heart Study,7 MR images are acquired to enable advanced body composition analysis. For example, in the UK Biobank Imaging Study, MR image volumes from 100,000 subjects are being collected, each volume containing 332 axial slices covering 1.1 m of the abdomen and upper legs. Although the overall aim of these studies is to collect data for future research, and the analysis methods have not been prescribed in the study design, it is quite obvious that automated methods are required in order to analyse such large amounts of imaging data.
The direct volumetric measurements of single muscles or fat compartments obtained by MRI enable much higher accuracy compared to indirect measurements such as anthropometric measures. For example, visceral AT (VAT) can vary significantly between people with identical body mass index or waist circumference.8, 9 It is also well-known that the metabolic risk related to fat accumulation is strongly dependent on its distribution.10, 11 In particular, large amounts of VAT are related to increased risk for cardiac disease,10, 12,13 type-2 diabetes (T2D),14, 15 liver disease16 and cancer.17, 18 Furthermore, increased muscle fat infiltration has been associated with reduced mobility,19 increased risk for T2D20 and higher mortality in patients with liver cirrhosis.21
While two-dimensional (2D) projections of the body using dual-energy X-ray absorptiometry (DXA) have high agreement with volumetric measurements using MRI for whole-body fat and lean tissue quantification, compartmental measurements such as VAT have a relatively low agreement.22, 23 Also, while CT technically has the same capability as MRI to acquire complete three-dimensional (3D) image volumes, body composition analysis with CT is commonly restricted to one or a limited number of slices in order to reduce the radiation exposure. But the use of 2D area measures from one or a limited set of slices as a proxy for volume reduces the precision compared to volumetric measurements when measuring abdominal AT distribution.24, 25 The challenge, of course, with 3D volumetric body composition analysis is the huge amount of data needed to be analysed. Completely manual analysis of full tomographic image volumes covering a large part of the body is extremely time-consuming and hence, generally not feasible except in very small studies. This has generated a demand for automated methods for quantifying AT and muscle composition in 3D MR images. The feasibility of using a highly automated analysis method in a large population study was, e.g. demonstrated by West et al26 quantifying VAT, abdominal subcutaneous adipose tissue (ASAT) and thigh muscles on 3000 subjects from the UK Biobank Imaging Study.
Another advantage with automated analysis methods is their potentially higher precision because of reduced or eliminated dependency on human operator variability. For example, in a study by Newman et al27 significantly higher test–retest repeatability of VAT quantification was achieved with a highly automated method compared to using manual analysis (1.8 vs 6.3% coefficient of variation).
The aim of this review was to give an overview of automated and semi-automated methods for MRI-based body composition analysis by presenting a summary and discussion of methods published between 2013 and 2017. Recent clinical applications of automated methods for MRI-based AT quantification are also discussed based on clinical studies on the subject published during 2017.
Methods for MRI-based body composition analysis
Any method for body composition analysis using MRI can be divided into three main steps: image acquisition, image segmentation, and tissue quantification. All three steps affect the results of a body composition analysis, and although this review focuses on automation of image post-processing, and not image acquisition, relevant image acquisition methods are also discussed here, since the choice of acquisition method has an impact on the subsequent analysis.
Image acquisition
MRI is often not used in a quantitative way, i.e. the image intensity values do not directly reflect a physical property of the imaged object (as opposed to CT, which is calibrated against the Hounsfield scale). Imperfections in the MRI system, as well as interactions between the imaged object and the electromagnetic field, cause the sensitivity and hence, the image intensity scale to vary over the image. In such cases, the interpretation of the images has to rely solely on the visual appearance of the contrast between tissues in the image. This limits the possibilities of accurate quantification of the image data. Still, both T1- and T2 weighted images have good contrast between water and fat. This means that lean tissue (LT) and AT can be segmented from each other (as long as the image resolution is high enough), enabling quantification of geometric properties such as area or volume of different tissue compartments using many standard T1- or T2 weighted imaging protocols.
In the context of AT and muscle analysis, the fat–water (FW) separated imaging (aka “Dixon imaging”) techniques28 are particularly useful. FW-separated imaging is based on gradient recalled echo imaging,29 which uses the chemical shift between the resonance frequencies of protons bound in water and in fat. This is the same effect which is used in magnetic resonance spectroscopy. FW separated imaging can be seen as a special case of magnetic resonance spectroscopy, where the spectral resolution has been sacrificed in favour for spatial resolution. In FW-MRI, two or more echoes are acquired after each excitation pulse, where the fat and water signals' relative phase differ due to their chemical shifts. Besides the separation of the fat and water components of the MR signal, measurements of multiple echoes enable estimation of several unknown confounding factors in the signal equations, such as T1 and T2*, which can then be corrected for.30 A well-known example of FW-MRI is the IDEAL reconstruction method.31
Besides the excellent FW contrast enabled by FW-MRI, it also enables quantitative fat imaging. One common method of achieving quantitative fat images is to compute the fat fraction (FF), which is the fat signal divided by the sum of the fat and water signals. A well-known version of FF is proton density FF (PDFF),32 which can be obtained by using proton-density weighted FW separated imaging. Another example is fat-referenced MRI.33, 34 The difference between FF and fat-referenced MRI lies in the reference used for calibrating the fat image. In FF (including PDFF), the sum of fat and water is used as reference, while in fat-referenced MRI, the reference is a fat signal interpolated from pure adipose tissue. Hence, PDFF measures the proportion of the total MR signal that originates from fat-bound protons, and fat-referenced MRI measures the fat signal in a given voxel in relation to the fat signal in pure adipose tissue. An example of a whole-body fat image that has been calibrated using fat-referenced MRI is shown in Figure 1.
Figure 1.
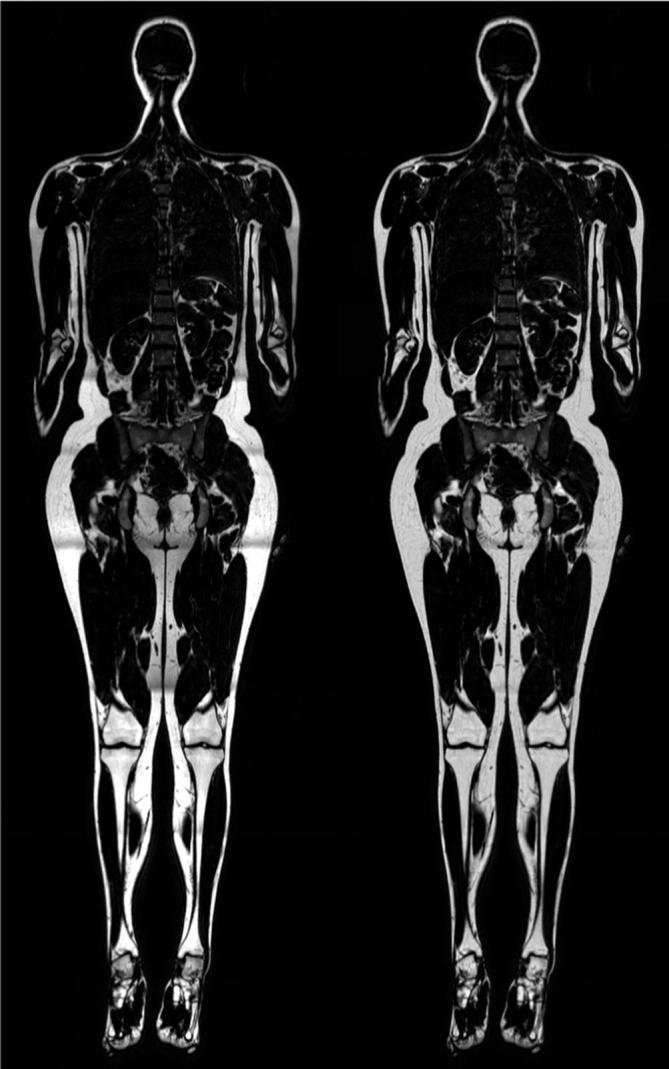
Example of quantitative imaging. To the left is an original fat image acquired using 2-point Dixon fat-water separated imaging. To the right is the corresponding quantitative fat image after calibration using the fat-referenced method. Inhomogeneities in the intensity of adipose tissue can be observed in the original image (left) that are almost completely remove in the calibrated image (right).
Also, brown adipose tissue (BAT) imaging using MRI requires quantitative imaging. BAT has most commonly been imaged using positron emission tomography but, more recently, dual energy CT and MRI have also been used to detect and quantify BAT.35 Quantitative FW separated MRI can be used for identification and characterization of BAT by its lower fat content and higher water content compared to white AT.36–38 A challenge when using FW-MRI to detect BAT is that it requires a high resolution in order to separate the BAT from partial volume effects in the interfaces between white AT and LT, while maintaining sufficiently high signal-to-noise ratio.38 Another contrast mechanism which can be used to characterize BAT is T2* relaxation.39 T2* mapping can be combined with FW separation using multiecho chemical-shift imaging.40, 41 In addition to detecting the presence of BAT, the activation of BAT can be detected by changes in T2* due to an increase in blood deoxyhemoglobin levels, which is caused by increased oxygen consumption in active BAT.42
Image segmentation
There is often not a clear distinction made between tissue classification and compartmental segmentation, but usually the tissue classification is done before the segmentation into compartments. Classification, in this context, refers to the labelling of each voxel into a tissue class, e.g. LT, AT or background. Segmentation, on the other hand, refers to distinguishing between different compartments containing the same type of tissue (e.g. VAT from ASAT or one muscle from another) by labelling or delineating relevant anatomical regions. If the image is quantitative, i.e. calibrated in relation to a defined grey scale, tissue classification is rather straight forward as it can be based on thresholding the image intensity; in particular in FW separated images.43
In non-quantitative imaging, the tissue classification can be facilitated by reducing the image inhomogeneities present in non-quantitative MR images. Such intensity inhomogeneities are caused by imperfections in the image acquisition system, such as inhomogeneities in the magnetic field and variations in the coil sensitivities. This causes variations in the intensity scale over the image. A number of methods have been proposed for correcting MR images for such inhomogeneities, and a comprehensive review and classification of such methods is given by Vovk et al.44 They divided the methods generally into prospective and retrospective methods, where the prospective methods try to solve the problem during the image acquisition while retrospective methods use image analysis post-processing to estimate and correct for the inhomogeneities. An important advantage with retrospective methods is that they can also correct for inhomogeneities induced by the body in the scanner, since they make very few assumptions about the physical sources of the inhomogeneities.44 The most common assumption being made is that the inhomogeneities can be modelled as a spatially slowly varying bias field and the different methods use different image processing methods to estimate and remove this bias field. There have also been other methods published after the review by Vovk et al, e.g. Consistent Intensity Inhomogeneity Correction45 which is based on multiscale normalized averaging.46
A common way to classify voxels into different tissues when the image is non-quantitative and the grey scale is undefined, is k-means clustering47 or fuzzy c-means clustering.48 In both methods, a pre-defined number k (or c) of clusters is used to categorize the data. In the case of body composition analysis, k or c are usually set to 3, indicating the classes AT, LT and background. The difference between the two methods is that k-means uses crisp (mutually exclusive) class memberships, while fuzzy c-means uses continuous (“fuzzy”) class membership values. Note that classification is actually not required when quantitative images are used, since each pixel or voxel then implicitly contains a class-membership value.
Proposed segmentation methods have been more or less automated, ranging from computer-aided manual drawing tools to fully automated segmentation algorithms.49–63 Less automated methods are often more generally applicable to different segmentation tasks, but at the cost of more time-demanding (and therefore more expensive) manual work. In 2D images, a common segmentation method is active contours (aka “snakes”),64 where a deformable contour is fitted to a structure in the image. This class of methods works by minimizing an energy function with one term depending on the image (e.g. the image gradient) and a regularization term depending on the curvature of the contour. While there are 3D extensions of active contours (“balloons"),65 a more common class of 3D segmentation methods in this context is multiatlas segmentation,66 where several anatomical atlases with pre-defined compartments are fitted to the image and whose anatomical definitions are combined using a voting scheme or expectation maximization.67 While atlas-based segmentation has mainly been used for brain image segmentation,68 they have recently also been applied to abdominal and whole-body image analysis.33, 53,61 Beside active contours and multiatlas segmentation, there are a multitude of different segmentation methods in the literature, and even more ways of combining such methods. These two segmentation methods were, however, found to be the most commonly used in the studies covered in this review. Atlas-based methods are, in principle, easier to generalize to other compartments compared to methods based on rule-based morphological operations that rely on specific anatomical assumptions.
Few fully automated segmentation methods have been extensively evaluated on different data sets with different properties (e.g. from different scanner models and different field strengths). Notable exceptions are the work by Addeman et al52 where a fully automated algorithm for segmentation of abdominal AT into VAT and ASAT was evaluated on data from scanners of three different brands and two different field strengths, and the work by Karlsson et al61 where fully automated segmentation of muscle groups from whole-body MRI was evaluated on data from two scanners with different field strengths.
A fully automated segmentation tool does not necessarily allow the user to modify the result, which obviously limits the range of images that can successfully be segmented. Such methods may also face a challenge from a regulatory perspective different to methods where the result is controlled by a human operator. One class of methods, which can be referred to as supervised segmentation (following the terminology by Hu et al69), combines the efficiency of fully automated methods with the flexibility and control enabled by manual interaction. The difference between a supervised automated tool and a semi-automated tool is that the latter can never do the complete segmentation without human interaction, while the supervised tool, at least in principle, can always autonomously perform a segmentation, albeit one that may need to be manually adjusted to give the desired result.
Segmentation methods can also be dived into 2D and 3D methods. 2D methods operate on one image slice at a time and are often used to segment one or a limited number of slices. Analysing only one or a few image slices with a manual or semi-automated tool is, of course, much less time consuming than analysing all slices in a complete volume, and many studies using MRI for body composition analysis have used one or a limited set of 2D slices, mostly due to the lack of efficient image analysis tools for handling 3D image segmentation. However, software tools for slice-wise semi-automated segmentation took on average more than 10 min per slice for a trained expert,70 which limits their use to small studies and to measurements of a limited set of slices. The rationale of using area as a proxy for volume is based on modelling the body as a cylinder. While such a model might be reasonable for the extremities, it is not well suited for the abdominal compartment, where single-slice analysis cannot accurately measure intra abdominal AT.24 When the tissue area in one or a few slices is used as proxy for VAT and ASAT volume, the location of the slices is critical. While such area measurements can have a good correlation with the absolute volume, single-slice imaging does not have the accuracy required to measure VAT and ASAT changes in an intervention study.25 For automated segmentation, 3D methods should (at least theoretically) be more powerful than 2D methods, since the connectivity of a non-convex 3D object may be lost when viewed as a 2D slice. Also, information from neighbouring slices (which is inherent in 3D methods) increases the redundancy and, hence can alleviate detection of significant structures and thereby support the segmentation in noisy images.
Tissue quantification
In non-quantitative imaging, the actual tissue quantification is usually performed by simply counting the number of voxels of each tissue class within each compartment and multiplying with the volume of a voxel. Such methods will be referred to as discrete methods. When using quantitative images, on the other hand, the amount of fat in a segmented compartment can be quantified by integrating the fat signal within that compartment, multiplied by the voxel volume. Such methods will be referred to as continuous methods. While continuous methods do not require a classification between AT and LT, they usually include a classification of soft tissue vs background (i.e. air and MR-invisible tissue) in order to remove noise contributions from the background.
The disadvantage with discrete methods is that partial volume effects, i.e. voxels with mixed content, will lead to a bias in the volume estimates;71, 72 a bias that will change with the resolution, hence limiting the reproducibility across different scanning protocols. This effect is illustrated in Figure 2 where an original quantitative fat image (top left) has been subsampled a number of steps (left column). A discretisation of the images on each resolution has been made using a threshold of 0.5 (second column). The diagram to the right shows, for different resolutions, the estimated AT area using a continuous method (solid line) and a discrete method with different thresholds (dashed lines). In the continuous method, all pixel values in the quantitative fat image are summed and then multiplied with the pixel area. The discrete method applies a threshold on the fat image and then multiplies the number of pixels above the threshold by the pixel area. A brief explanation of this resolution-dependent bias is as follows: for large objects, partial volume effects will only occur at the tissue–background interface. Peripheral voxels covering the object to a fraction higher than the threshold will be classified as object and, hence, overestimate the volume, while voxels covering the object less than the threshold will be considered as background, hence under estimating the volume. For a threshold of 0.5, these errors will, on average, cancel each other out, hence giving an unbiased estimate71 (assuming a symmetric noise distribution with zero mean). But objects (or parts of objects) that are smaller than half the voxel size in any dimension (e.g. a thin sheet of tissue) will all be classified as background and lost. Hence, a discrete classification of voxels will under estimate the object volume, and this bias will increase with decreasing image resolution. The more irregularly shaped and thin the tissues are, the more prominent this effect will be, and in the case of muscle fat infiltration, the effect can be quite significant.72 This will make discrete methods difficult to use in multicentre studies, where the scanner model and scanning protocol may differ.
Figure 2.
Illustration of continuous and discrete quantification. Continuous methods (left column and black solid line in diagram) are less dependent on the image resolution than discrete methods (second column and dashed lines in diagram). See text for details.
Continuous methods are less sensitive to partial volume effects, which makes them less prone to the resolution-dependant bias discussed above. But, perhaps more importantly, in contrast to discrete methods, continuous methods enable measurements of diffuse infiltration of thin AT structures and ectopic fat in non-adipose tissue, such as liver and muscles, since these methods can quantify weak fat signals that would not reach any reasonable threshold for classifying a voxel as AT.
Accuracy and precision
Few studies have investigated repeatability and reproducibility. Reproducibility measures how well the measurement can be reproduced with different scanners while repeatability measures how well the analysis of a subject scanned in the same scanner agrees with the analysis of a second scan, or between two or more analyses made by the same or different operators. Most studies have only evaluated accuracy of the segmentation against manual segmentation. But from a clinical perspective, repeatability and reproducibility are at least as important as accuracy. Evaluation of the segmentation alone does not address the quantitative properties of the complete imaging chain. For example, sensitivity to differences in scanning parameters such as image resolution is not reflected by comparison to manual segmentation, and different methods may be more or less sensitive to partial volume effects, which are directly related to image resolution.71, 72
Review of recent literature
Starting in the early 1990s,73–75 a wide range of papers on AT quantification using MRI have been published. An excellent review of methods for segmentation of AT was recently presented by Hu et al.69 The present review is constrained to the last 5 years (2013–2017), during which a range of papers on more or less automated methods for quantification of abdominal AT,27,49–55,57,76–83 BAT84, 85 and muscles58–63,86–93 have been published. A search was made on PubMed for studies using MRI with automated or semi-automated methods for quantifying fat or muscles. 34 publications are summarized in Supplementary Table 1 (Supplementary Material 1) with respect to measurements; image pre-processing (e.g. inhomogeneity correction); segmentation method; if the image analysis was performed slice-wise in 2D or if a full 3D processing method was used; if a complete volume measure was obtained or if an area-measure was used as a proxy; the level of automation; the type of validation reported, and the repeatability if reported.
Quantitative imaging and inhomogeneity correction
More than half of the 34 papers used either continuous methods or applied some kind of inhomogeneity correction to aid the segmentation. Seven papers used continuous quantification based on quantitative MRI, either PDFF52, 84,85 or fat-referenced MRI.27, 79,82,91 11 of the papers that used discrete quantification methods used inhomogeneity correction before AT quantification51, 52,54,57,76,78,81 or muscle analysis.62, 63,88,92
Segmentation methods
Several of the discrete methods used k-means50, 51,53,58,77,86 or fuzzy c-means clustering54, 62,78,81,88 for tissue classification. About half of the studies used 3D segmentation.27, 49,52,53,59,61,63,76,79,82,84,85,87,88,91 The remaining studies used 2D segmentation of each slice separately50, 51,54,55,57,77,83 or analysed only one or a limited number of slices62, 86,89,92,93 using 2D area measures as proxy for the volume. Active contours were used for segmentation in most of the 2-D methods,50, 51,54,62,81,86,90,92 while in the studies using 3D segmentation, multiatlas-based segmentation was the most common method, used in eight of the studies.59, 61,79,82,84,85,87,91
Level of automation
17 studies used fully automated methods for AT quantification49–57,84 and muscle measurements.55,58–63 One study76 used completely manual segmentation of baseline images and automated non-rigid registration for transferring the baseline segmentations to follow-up images. The remaining studies used semi-automated77, 78,80,81,83,88,89,93 or supervised27, 79,82,86,91,92 segmentation methods.
Validation
Most of the studies reported accuracy in terms of agreement with manual segmentation, but only 14 of them reported precision in terms of repeatability.27, 50,52,56,57,59,61,79,82,86,89,90,92,93 All of the studies that reported precision showed very good repeatability, with coefficients of variation of up to a few percent and the intraclass correlation was very close to one in most of the studies.
The only study that investigated the between-scanner reproducibility was presented in the paper by Karlsson et al61 who compared fully automated quantification of 10 different muscle groups in data acquired on a 1.5 T and a 3 T scanner with excellent agreement. Also, Addeman et al52 addressed the issue of multicentre studies, showing their method's ability to analyse data from three different cohorts using different scanners and field strengths. They did, however, not evaluate between-scanner reproducibility since the different scanners were used on different cohorts.
Availability
Most of the methods used in these studies are not readily accessible for other researchers and non-image analysis experts. Three exceptions are FATCALC55; AMRA® Researcher, which was used in six27, 59,61,79,82,91 of the studies listed above, and SliceOmatic used in one of the studies.83
FATCALC55 is an add-on to ImageJ,94 which is a free image processing software originally developed by NIH for analysis of microscopy images. FATCALC is a discrete 2D quantification method for VAT and ASAT. It works on images acquired using FW-MRI, using 2D analysis of individual slices and fuzzy c-means clustering to classify AT pixels in the fat image. This is followed by morphological operations (erosions and dilations) of the binary classification result in order to separate VAT from ASAT. Finally, fat pixels belonging to the arms, abdominal muscles and paravertebral fat are removed. The complete process is fully automated.
AMRA Researcher (AMRA Medical AB, Linköping, Sweden) is a cloud-based analysis service, in which the volumes of VAT, ASAT and different muscle groups, as well as muscle fat infiltration and liver fat are quantified. It uses a continuous quantification method based on quantitative fat-referenced MRI and 3D multiatlas segmentation to separate different muscle groups and AT compartments. It has efficiently been used for body composition analysis of thousands of subjects in the UK Biobank imaging study.26, 95 An example of segmentations of VAT, ASAT and different muscle groups using AMRA Researcher is shown in Figure 3.
Figure 3.

Example of segmentation of adipose tissue and muscle groups using AMRA Researcher. To the left is the fat image with ASAT (blue) and VAT (red), and to the right is the water image with 10 different muscle groups coloured. Reproduced with permission from AMRA Medical AB. ASAT, abdominal subcutaneous adipose tissue; VAT, visceral AT.
A less automated generally available software tool that can be used for AT and muscle quantification is SliceOmatic (Tomovision, Quebec, Canada). This is a general interactive image segmentation tool that operates on one image slice at a time using a set of general-purpose image analysis tools to aid the operator. Even though a segmentation can be propagated from one slice to another, the time required for experienced operators was approximately 40 min for segmenting the complete VAT volume27 and 30 min for segmenting the calf muscles.59 Other similar tools are Analyze (AnalyzeDirect Inc. Overland Park, KS), and Hippo Fat.96 Hippo Fat has been evaluated against SliceOmatic in the context of VAT segmentation.97 These three methods were also evaluated by Bonekamp et al.70
Clinical applications
Among the publications reviewed above, only seven had a specific clinical application in focus. Five were related to clinical aspects of abdominal AT distributions, one on intramuscular fat and one on both. Shen et al53 investigated changes in volumes of abdominal organs and AT compartments in obese females during weight loss. Radmard et al81 investigated the relationship between abdominal AT distribution and carotid atherosclerosis. Eichler et al80 investigated lipodystrophy in HIV patients during antiviral intervention. Karlsson et al78 investigated gender differences in abdominal AT distribution in pre-school children. Lareau-Trudel et al86 measured intramuscular fat in the legs in a study on facioscapulohumeral muscular dystrophy, and Orsso et al83 investigated abdominal AT and intramuscular AT composition in youth with Prader-Willi syndrome.
In order to find more clinical applications of MRI-based quantification of AT, a wider search was made on PubMed for papers published in 2017 (as e-pub or print date) with “adipose tissue” and “MRI” in the title or abstract. Among the 137 papers found, 43 described clinical studies including quantification of VAT and/or SAT. Out of these 43 clinical studies, 27 used some kind of automated analysis, but most of those used tools with a low degree of automation. Hence, as many as 20 of these 27 studies used area measures in one or a few slices instead of measuring complete volumes of VAT and ASAT, likely due to the laborious task of using a 2D tool with low automation for analysing complete volumetric data. Among these area-based studies, SliceOmatic was the most common tool, used in 13 of the 43 clinical studies. Only a few studies98–101 used volumetric analysis tools.
The most common clinical areas in the studies found in this search were obesity (12), T2D (9), liver disease (8) and cardiovascular disease (CVD) (5 studies). Six of the studies addressed BAT. The most common focus of the obesity studies concerned the effect of interventions (medical, surgical or life-style) on body composition. Most of the diabetes studies either investigated the effect of some intervention on abdominal AT distribution or the relationship between AT distribution and insulin resistance. The studies addressing liver fat either measured relations between body composition and liver fat or investigated intervention effects on liver fat and abdominal AT distribution. Most of the CVD-related studies investigated the relation between AT distribution and cardio-metabolic risk factors. The aims of the BAT studies were diverse. Two looked at hibernoma (a BAT tumour) and two looked at the effect of cold stimulation on BAT FF.
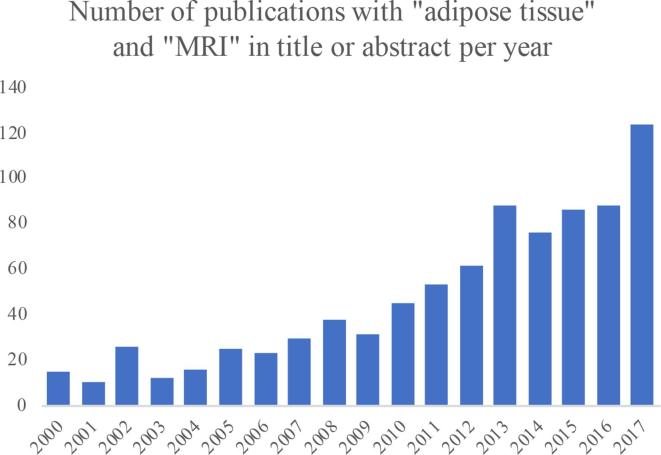
Extending the search on PubMed a number of years back in time shows an approximately exponential increase in the number of publications with “adipose tissue” and “MRI” in title or abstract (Figure 4). A search on NIH ClinicalTrials.gov on clinical trials with “adipose tissue” as outcome and “MRI” in other terms, shows a similar trend (Figure 5). Both these search results indicate a rapidly growing interest for MRI in clinical studies related to adipose tissue.
Figure 4.
Number of publications per year with “adipose tissue” and “MRI” in title or abstract found on PubMed.
Figure 5.
Number of clinical trials per year with “adipose tissue” as outcome and “MRI” in other terms found on NIH ClinicalTrials.gov.
One example of studies that would be extremely difficult, if indeed possible at all, to perform without highly automated analysis and quantitative MRI was recently published by Linge et al.95 MR images from 6021 subjects from the UK Biobank were analysed quantifying ASAT, VAT, thigh muscle volume, liver fat, and muscle fat infiltration and their multivariate associations to coronary heart disease and Type 2 diabetes were investigated. The study showed that different diseases were linked to different imbalances in fat accumulation, which could not be described by sex, age, lifestyle, generalized adiposity or by investigating a single fat compartment alone. The method used in that study was AMRA Researcher.
Conclusions
Apparently, up till now, rather few clinical studies have used highly automated methods for assessment of AT and muscle volume. Most of the studies used completely manual tools for segmentation. Some studies even used other imaging modalities (such as CT102 or DXA103 to measure VAT even though an MRI scan was included in the study, thereby increasing the study time for the patient and also, in the case of CT or DXA, unnecessarily exposing the subjects to ionizing radiation.
Quantitative imaging both simplifies tissue segmentation and fat quantification. The fact that most studies in this review did not use quantitative imaging could to some extent be explained by a need to analyse images that had been previously acquired for other purposes. However, the frequent application of an intermediate inhomogeneity correction step to alleviate some of the problems with non-quantitative MRI, shows the importance of using quantitative MRI (e.g. FW-MRI) in studies involving body composition analysis.
One requirement for a method to be widely used in clinical studies is that it has been carefully evaluated, in particular with respect to repeatability and reproducibility. Another is that the method is readily accessible for other researchers and non-image analysis experts. A number of software packages for abdominal AT quantification have earlier been evaluated by Bonekamp et al70 but all of them had a relatively low degree of automation. Still, the interest in using MRI in studies of adipose tissue is rapidly increasing, as can be seen in Figures 4 and 5, in particular in studies related to obesity, T2D, liver disease and CVD.
As shown by Linge et al95 simultaneous quantification of several body composition parameters reveals new relations between body fat distribution and different diseases. Such simultaneous volumetric analyses of multiple fat compartments and muscles would be a discouraging task, unless using automated analysis methods. The access to such methods enables efficient and detailed body composition assessment in patients with suspected metabolic disorders, particularly in cases when MRI is already an established examination, such as in liver diseases. In conclusion, there is a growing need for efficient and generally available methods for automated volumetric assessment of AT and muscle composition and, in particular, a need for reproducibility studies of such methods.
Footnotes
Conflicts of interest: The author is stock holder in AMRA Medical AB.
REFERENCES
- 1.Thomas EL, Fitzpatrick JA, Malik SJ, Taylor-Robinson SD, Bell JD. Whole body fat: content and distribution. Prog Nucl Magn Reson Spectrosc 2013; 73: 56–80. doi: 10.1016/j.pnmrs.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 2.Cruz-Jentoft AJ, Baeyens JP, Bauer JM. Sarcopenia: European consensus on definition and diagnosis: report of the European Working Group on Sarcopenia in Older People. United States, North America: The British Institute of Radiology.; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. . UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12: e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlett CL, Hendel T, Weckbach S, Reiser M, Kauczor HU, Nikolaou K, et al. . Population-based imaging and radiomics: rationale and perspective of the german national cohort MRI study. Rofo 2016; 188: 652–61. doi: 10.1055/s-0042-104510 [DOI] [PubMed] [Google Scholar]
- 5.Bamberg F, Hetterich H, Rospleszcz S, Lorbeer R, Auweter SD, Schlett CL, et al. . Subclinical disease burden as assessed by whole-body MRI in subjects with prediabetes, subjects with diabetes, and normal control subjects from the general population: The KORA-MRI study. Diabetes 2017; 66: 158–69. doi: 10.2337/db16-0630 [DOI] [PubMed] [Google Scholar]
- 6.Elffers TW, de Mutsert R, Lamb HJ, de Roos A, Willems van Dijk K, Rosendaal FR, et al. . Body fat distribution, in particular visceral fat, is associated with cardiometabolic risk factors in obese women. PLoS One 2017; 12: e0185403–10. doi: 10.1371/journal.pone.0185403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neeland IJ, Grundy SM, Li X, Adams-Huet B, Vega GL. Comparison of visceral fat mass measurement by dual-X-ray absorptiometry and magnetic resonance imaging in a multiethnic cohort: the Dallas Heart Study. Nutr Diabetes 2016; 6: e221: e221. doi: 10.1038/nutd.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas EL, Frost G, Taylor-Robinson SD, Bell JD. Excess body fat in obese and normal-weight subjects. Nutr Res Rev 2012; 25: 150–61. doi: 10.1017/S0954422412000054 [DOI] [PubMed] [Google Scholar]
- 9.Thomas EL, Parkinson JR, Frost GS, Goldstone AP, Doré CJ, McCarthy JP, et al. . The missing risk: MRI and MRS phenotyping of abdominal adiposity and ectopic fat. Obesity 2012; 20: 76–87. doi: 10.1038/oby.2011.142 [DOI] [PubMed] [Google Scholar]
- 10.Neeland IJ, Ayers CR, Rohatgi AK, Turer AT, Berry JD, Das SR, et al. . Associations of visceral and abdominal subcutaneous adipose tissue with markers of cardiac and metabolic risk in obese adults. Obesity 2013; 21: E439–E447. doi: 10.1002/oby.20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JJ, Pedley A, Hoffmann U, Massaro JM, Fox CS. Association of changes in abdominal fat quantity and quality with incident cardiovascular disease risk factors. J Am Coll Cardiol 2016; 68: 1509–21. doi: 10.1016/j.jacc.2016.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Fox CS, Hickson DA, May WD, Hairston KG, Carr JJ, et al. . Impact of abdominal visceral and subcutaneous adipose tissue on cardiometabolic risk factors: the Jackson Heart Study. J Clin Endocrinol Metab 2010; 95: 5419–26. doi: 10.1210/jc.2010-1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artham SM, Lavie CJ, Patel HM, Ventura HO. Impact of obesity on the risk of heart failure and its prognosis. J Cardiometab Syndr 2008; 3: 155–61. doi: 10.1111/j.1559-4572.2008.00001.x [DOI] [PubMed] [Google Scholar]
- 14.Iwasa M, Mifuji-Moroka R, Hara N, Ishidome M, Iwata K, Sugimoto R, et al. . Visceral fat volume predicts new-onset type 2 diabetes in patients with chronic hepatitis C. Diabetes Res Clin Pract 2011; 94: 468–70. doi: 10.1016/j.diabres.2011.09.016 [DOI] [PubMed] [Google Scholar]
- 15.Kurioka S, Murakami Y, Nishiki M, Sohmiya M, Koshimura K, Kato Y. Relationship between visceral fat accumulation and anti-lipolytic action of insulin in patients with type 2 diabetes mellitus. Endocr J 2002; 49: 459–64. doi: 10.1507/endocrj.49.459 [DOI] [PubMed] [Google Scholar]
- 16.van der Poorten D, Milner KL, Hui J, Hodge A, Trenell MI, Kench JG, et al. . Visceral fat: a key mediator of steatohepatitis in metabolic liver disease. Hepatology 2008; 48: 449–57. doi: 10.1002/hep.22350 [DOI] [PubMed] [Google Scholar]
- 17.Britton KA, Massaro JM, Murabito JM, et al. . Body fat distribution, incident cardiovascular disease, cancer, and all-cause mortality. J Am Coll Cardiol 2013; 62: 921–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle SL, Donohoe CL, Lysaght J, Reynolds JV. Visceral obesity, metabolic syndrome, insulin resistance and cancer. Proc Nutr Soc 2012; 71: 181–9. doi: 10.1017/S002966511100320X [DOI] [PubMed] [Google Scholar]
- 19.Marcus RL, Addison O, Dibble LE, Foreman KB, Morrell G, Lastayo P. Intramuscular adipose tissue, sarcopenia, and mobility function in older individuals. J Aging Res 2012; 2012: 629637–6. doi: 10.1155/2012/629637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodpaster BH, Thaete FL, Kelley DE. Thigh adipose tissue distribution is associated with insulin resistance in obesity and in type 2 diabetes mellitus. 885 United States: The British Institute of Radiology.; 2000. [DOI] [PubMed] [Google Scholar]
- 21.Praktiknjo M, Book M, Luetkens J, Pohlmann A, Meyer C, Thomas D, Michael P, Marius B, Julian L, et al. . Fat-free muscle mass in magnetic resonance imaging predicts acute-on-chronic liver failure and survival in decompensated cirrhosis. Hepatology 2018; 67: 1014–26. doi: 10.1002/hep.29602 [DOI] [PubMed] [Google Scholar]
- 22.Borga M, West J, Bell JD, Harvey NC, Romu T, Heymsfield SB, et al. . Advanced body composition assessment: from body mass index to body composition profiling. J Investig Med 2018; 66: 1.10–9. doi: 10.1136/jim-2018-000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamel EG, McNeill G, Van Wijk MC. Usefulness of anthropometry and DXA in predicting intra-abdominal fat in obese men and women. Obes Res 2000; 8: 36–42. doi: 10.1038/oby.2000.6 [DOI] [PubMed] [Google Scholar]
- 24.Thomas EL, Bell JD. Influence of undersampling on magnetic resonance imaging measurements of intra-abdominal adipose tissue. Int J Obes Relat Metab Disord 2003; 27: 211–218. doi: 10.1038/sj.ijo.802229 [DOI] [PubMed] [Google Scholar]
- 25.Shen W, Chen J, Gantz M, Velasquez G, Punyanitya M, Heymsfield SB. A single MRI slice does not accurately predict visceral and subcutaneous adipose tissue changes during weight loss. Obesity 2012; 20: 2458–63. doi: 10.1038/oby.2012.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West J, Dahlqvist Leinhard O, Romu T, Collins R, Garratt S, Bell JD, et al. . Feasibility of MR-based body composition analysis in large scale population studies. PLoS One 2016; 11: e0163332. doi: 10.1371/journal.pone.0163332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman D, Kelly-Morland C, Leinhard OD, Kasmai B, Greenwood R, Malcolm PN, et al. . Test-retest reliability of rapid whole body and compartmental fat volume quantification on a widebore 3T MR system in normal-weight, overweight, and obese subjects. J Magn Reson Imaging 2016; 44: 1464–73. doi: 10.1002/jmri.25326 [DOI] [PubMed] [Google Scholar]
- 28.Dixon WT. Simple proton spectroscopic imaging. Radiology 1984; 153: 189–94. doi: 10.1148/radiology.153.1.6089263 [DOI] [PubMed] [Google Scholar]
- 29.Elster AD. Gradient-echo MR imaging: techniques and acronyms. Radiology 1993; 186: 1–8. doi: 10.1148/radiology.186.1.8416546 [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB, Huanzhou Y, Ann S A. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med 2008; 60: 1122–34. doi: 10.1002/mrm.21737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeder SB, Pelc NJ, Alley MT, Gold GE. Rapid MR imaging of articular cartilage with steady-state free precession and multipoint fat-water separation. AJR Am J Roentgenol 2003; 180: 357–62. doi: 10.2214/ajr.180.2.1800357 [DOI] [PubMed] [Google Scholar]
- 32.Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. J Magn Reson Imaging 2012; 36: 1011–4. doi: 10.1002/jmri.23741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlqvist Leinhard O, Johansson A, Rydell J. Quantitative Abdominal Fat Estimation Using MRI. International Conference On Pattern Recognition 2008; 19: 2137–40. [Google Scholar]
- 34.Hu HH, Nayak KS. Quantification of absolute fat mass using an adipose tissue reference signal model. J Magn Reson Imaging 2008; 28: 1483–91. doi: 10.1002/jmri.21603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borga M, Virtanen KA, Romu T, Leinhard OD, Persson A, Nuutila P, et al. . Brown adipose tissue in humans: detection and functional analysis using PET (positron emission tomography), MRI (magnetic resonance imaging), and DECT (dual energy computed tomography). Methods Enzymol 2014; 537: 141–59. doi: 10.1016/B978-0-12-411619-1.00008-2 [DOI] [PubMed] [Google Scholar]
- 36.Lunati E, Marzola P, Nicolato E, Fedrigo M, Villa M, Sbarbati A. In vivo quantitative lipidic map of brown adipose tissue by chemical shift imaging at 4.7 Tesla. J Lipid Res 1999; 40: 1395–400. [PubMed] [Google Scholar]
- 37.Hu HH, Smith DL, Nayak KS, Goran MI, Nagy TR. Identification of brown adipose tissue in mice with fat-water IDEAL-MRI. J Magn Reson Imaging 2010; 31: 1195–202. doi: 10.1002/jmri.22162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romu T, Elander L, Leinhard OD, Lidell ME, Betz MJ, Persson A, et al. . Characterization of brown adipose tissue by water-fat separated magnetic resonance imaging. J Magn Reson Imaging 2015; 42: 1639–45. doi: 10.1002/jmri.24931 [DOI] [PubMed] [Google Scholar]
- 39.Hu HH, Yin L, Aggabao PC, Perkins TG, Chia JM, Gilsanz V. Comparison of brown and white adipose tissues in infants and children with chemical-shift-encoded water-fat MRI. J Magn Reson Imaging 2013; 38: 885–96. doi: 10.1002/jmri.24053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Regan DP, Callaghan MF, Wylezinska-Arridge M, Fitzpatrick J, Naoumova RP, Hajnal JV, et al. . Liver fat content and T2*: simultaneous measurement by using breath-hold multiecho MR imaging at 3.0 T--feasibility. Radiology 2008; 247: 550–7. doi: 10.1148/radiol.2472070880 [DOI] [PubMed] [Google Scholar]
- 41.Yu H, McKenzie CA, Shimakawa A, Vu AT, Brau AC, Beatty PJ, et al. . Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging 2007; 26: 1153–61. doi: 10.1002/jmri.21090 [DOI] [PubMed] [Google Scholar]
- 42.Khanna A, Branca RT. Detecting brown adipose tissue activity with BOLD MRI in mice. Magn Reson Med 2012; 68: 1285–90. doi: 10.1002/mrm.24118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kullberg J, Karlsson AK, Stokland E, Svensson PA, Dahlgren J. Adipose tissue distribution in children: automated quantification using water and fat MRI. J Magn Reson Imaging 2010; 32: 204–10. doi: 10.1002/jmri.22193 [DOI] [PubMed] [Google Scholar]
- 44.Vovk U, Pernus F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging 2007; 26: 405–21. doi: 10.1109/TMI.2006.891486 [DOI] [PubMed] [Google Scholar]
- 45.Andersson T, Romu T, Karlsson A, Norén B, Forsgren MF, Smedby Ö, et al. . Consistent intensity inhomogeneity correction in water-fat MRI. J Magn Reson Imaging 2015; 42: 468–76. doi: 10.1002/jmri.24778 [DOI] [PubMed] [Google Scholar]
- 46.Romu T, Borga M, Dahlqvist Leinhard O, Dahlqvist Leinhard O. MANA - Multi scale adaptive normalized averaging. 2011 IEEE International Symposium on Biomedical Imaging: from Nano to Macro. IEEE conference proceedings; 2011. pp 361–4. [Google Scholar]
- 47.MacQueen J. Some methods for classification and analysis of multivariate observations. In: Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics Berkeley, Calif., 1967. 1967; 281–97.
- 48.Dunn JC. A fuzzy relative of the ISODATA process and Its use in detecting compact well-separated clusters. J Cybern 1973; 3: 32–57. doi: 10.1080/01969727308546046 [DOI] [Google Scholar]
- 49.Silver HJ, Niswender KD, Kullberg J, Berglund J, Johansson L, Bruvold M, et al. . Comparison of gross body fat-water magnetic resonance imaging at 3 tesla to dual-energy X-ray absorptiometry in obese women. Obesity 2013; 21: 765–74. doi: 10.1002/oby.20287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thörmer G, Bertram HH, Garnov N, Peter V, Schütz T, Shang E, et al. . Software for automated MRI-based quantification of abdominal fat and preliminary evaluation in morbidly obese patients. J Magn Reson Imaging 2013; 37: 1144–50. doi: 10.1002/jmri.23890 [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Shi L, Chu WC, Hu M, Tomlinson B, Huang WH, Yeung DKW, et al. . Fully automatic and nonparametric quantification of adipose tissue in fat-water separation MR imaging. Med Biol Eng Comput 2015; 53: 1247–54. doi: 10.1007/s11517-015-1347-y [DOI] [PubMed] [Google Scholar]
- 52.Addeman BT, Kutty S, Perkins TG, Soliman AS, Wiens CN, McCurdy CM, McKenzie CA, et al. . Validation of volumetric and single-slice MRI adipose analysis using a novel fully automated segmentation method. J Magn Reson Imaging 2015; 41: 233–41. doi: 10.1002/jmri.24526 [DOI] [PubMed] [Google Scholar]
- 53.Shen J, Baum T, Cordes C, Ott B, Skurk T, Kooijman H, et al. . Automatic segmentation of abdominal organs and adipose tissue compartments in water-fat MRI: application to weight-loss in obesity. Eur J Radiol 2016; 85: 1613–21. doi: 10.1016/j.ejrad.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 54.Sun J, Xu B, Freeland-Graves J. Automated quantification of abdominal adiposity by magnetic resonance imaging. Am J Hum Biol 2016; 28: 757–66. doi: 10.1002/ajhb.22862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddalo M, Zorza I, Zubani S, Nocivelli G, Calandra G, Soldini P, et al. . Validation of a free software for unsupervised assessment of abdominal fat in MRI. Phys Med 2017; 37: 24–31. doi: 10.1016/j.ejmp.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 56.Waduud MA, Sharaf A, Roy I, Lopez-Gonzalez R, Hart A, McGill D, et al. . Validation of a semi-automated technique to accurately measure abdominal fat distribution using CT and MRI for clinical risk stratification. Br J Radiol 2017; 90: 20160662. doi: 10.1259/bjr.20160662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hui SCN, Zhang T, Shi L, Wang D, Ip CB, Chu WCW. Automated segmentation of abdominal subcutaneous adipose tissue and visceral adipose tissue in obese adolescent in MRI. Magn Reson Imaging 2018; 45: 97–104. doi: 10.1016/j.mri.2017.09.016 [DOI] [PubMed] [Google Scholar]
- 58.Valentinitsch A, Karampinos DC, Alizai H, Subburaj K, Kumar D, Link TM, et al. . Automated unsupervised multi-parametric classification of adipose tissue depots in skeletal muscle. J Magn Reson Imaging 2013; 37: 917–27. doi: 10.1002/jmri.23884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas MS, Newman D, Leinhard OD, Kasmai B, Greenwood R, Malcolm PN, et al. . Test-retest reliability of automated whole body and compartmental muscle volume measurements on a wide bore 3T MR system. Eur Radiol 2014; 24: 2279–91. doi: 10.1007/s00330-014-3226-6 [DOI] [PubMed] [Google Scholar]
- 60.Andrews S, Hamarneh G. The generalized log-ratio transformation: learning shape and adjacency priors for simultaneous thigh muscle segmentation. IEEE Trans Med Imaging 2015; 34: 1773–87. doi: 10.1109/TMI.2015.2403299 [DOI] [PubMed] [Google Scholar]
- 61.Karlsson A, Rosander J, Romu T, Tallberg J, Grönqvist A, Borga M, et al. . Automatic and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water-fat MRI. J Magn Reson Imaging 2015; 41: 1558–69. doi: 10.1002/jmri.24726 [DOI] [PubMed] [Google Scholar]
- 62.Orgiu S, Lafortuna CL, Rastelli F, Cadioli M, Falini A, Rizzo G. Automatic muscle and fat segmentation in the thigh from T1-Weighted MRI. J Magn Reson Imaging 2016; 43: 601–10. doi: 10.1002/jmri.25031 [DOI] [PubMed] [Google Scholar]
- 63.Yang YX, Chong MS, Tay L, Yew S, Yeo A, Tan CH. Automated assessment of thigh composition using machine learning for Dixon magnetic resonance images. MAGMA 2016; 29: 723–31. doi: 10.1007/s10334-016-0547-2 [DOI] [PubMed] [Google Scholar]
- 64.Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. International Journal of Computer Vision 1988; 1: 321–31. doi: 10.1007/BF00133570 [DOI] [Google Scholar]
- 65.Cohen LD. On active contour models and balloons. CVGIP: Image Understanding 1991; 53: 211–8. doi: 10.1016/1049-9660(91)90028-N [DOI] [Google Scholar]
- 66.Iglesias JE, Sabuncu MR. Multi-atlas segmentation of biomedical images: a survey. Med Image Anal 2015; 24: 205–19. doi: 10.1016/j.media.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 2004; 23: 903–21. doi: 10.1109/TMI.2004.828354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999; 9: 179–94. doi: 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- 69.Hu HH, Chen J, Shen W. Segmentation and quantification of adipose tissue by magnetic resonance imaging. MAGMA 2016; 29: 259–76. doi: 10.1007/s10334-015-0498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonekamp S, Ghosh P, Crawford S, Solga SF, Horska A, Brancati FL, et al. . Quantitative comparison and evaluation of software packages for assessment of abdominal adipose tissue distribution by magnetic resonance imaging. Int J Obes 2008; 32: 100–11. doi: 10.1038/sj.ijo.0803696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng Q, McColl RW, Ding Y, Wang J, Chia JM, Weatherall PT. Automated method for accurate abdominal fat quantification on water-saturated magnetic resonance images. J Magn Reson Imaging 2007; 26: 738–46. doi: 10.1002/jmri.21040 [DOI] [PubMed] [Google Scholar]
- 72.Romu T. Fat-Referenced MRI – Quantitative MRI for Tissue Characterization and Volume Measurement. PhD Thesis, Linköping University, Linköping. 2018.
- 73.Fowler PA, Fuller MF, Glasbey CA, Foster MA, Cameron GG, McNeill G, et al. . Total and subcutaneous adipose tissue in women: the measurement of distribution and accurate prediction of quantity by using magnetic resonance imaging. Am J Clin Nutr 1991; 54: 18–25. doi: 10.1093/ajcn/54.1.18 [DOI] [PubMed] [Google Scholar]
- 74.Sobol W, Rossner S, Hinson B, et al. . Evaluation of a new magnetic resonance imaging method for quantitating adipose tissue areas. Int J Obes 1991; 15: 589–99. [PubMed] [Google Scholar]
- 75.Lancaster JL, Ghiatas AA, Alyassin A, Kilcoyne RF, Bonora E, DeFronzo RA. Measurement of abdominal fat with T1-weighted MR images. J Magn Reson Imaging 1991; 1: 363–9. doi: 10.1002/jmri.1880010315 [DOI] [PubMed] [Google Scholar]
- 76.Joshi AA, HH H, Leahy RM, et al. . Automatic intra-subject registration-based segmentation of abdominal fat from 3D water-fat MRI. J Magn Reson Imaging 2013; 37: 423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poonawalla AH, Sjoberg BP, Rehm JL, Hernando D, Hines CD, Irarrazaval P, et al. . Adipose tissue MRI for quantitative measurement of central obesity. J Magn Reson Imaging 2013; 37: 707–16. doi: 10.1002/jmri.23846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karlsson AK, Kullberg J, Stokland E, Allvin K, Gronowitz E, Svensson PA, et al. . Measurements of total and regional body composition in preschool children: a comparison of MRI, DXA, and anthropometric data. Obesity 2013; 21: 1018–24. doi: 10.1002/oby.20205 [DOI] [PubMed] [Google Scholar]
- 79.Borga M, Thomas EL, Romu T, Rosander J, Fitzpatrick J, Dahlqvist Leinhard O, et al. . Validation of a fast method for quantification of intra-abdominal and subcutaneous adipose tissue for large-scale human studies. NMR Biomed 2015; 28: 1747–53. doi: 10.1002/nbm.3432 [DOI] [PubMed] [Google Scholar]
- 80.Eichler K, Bickel TM, Klauke S, Eisen J, Vogl TJ, Zangos S. Magnetic resonance imaging evaluation of lipodystrophy in HIV-positive patients receiving highly active antiretroviral therapy. Int J STD AIDS 2015; 26: 582–9. doi: 10.1177/0956462414546916 [DOI] [PubMed] [Google Scholar]
- 81.Radmard AR, Poustchi H, Ansari L, Khorasanizadeh F, Yoonessi A, Hashemi Taheri AP, et al. . Abdominal fat distribution and carotid atherosclerosis in a general population: a semi-automated method using magnetic resonance imaging. Jpn J Radiol 2016; 34: 414–22. doi: 10.1007/s11604-016-0540-8 [DOI] [PubMed] [Google Scholar]
- 82.Middleton MS, Haufe W, Hooker J, Borga M, Dahlqvist Leinhard O, Romu T, et al. . Quantifying abdominal adipose tissue and thigh muscle volume and hepatic proton density fat fraction: repeatability and accuracy of an MR imaging-based, semiautomated analysis method. Radiology 2017; 283: 438–49. doi: 10.1148/radiol.2017160606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orsso CE, Mackenzie M, Alberga AS, Sharma AM, Richer L, Rubin DA, et al. . The use of magnetic resonance imaging to characterize abnormal body composition phenotypes in youth with Prader-Willi syndrome. Metabolism 2017; 69: 67–75. doi: 10.1016/j.metabol.2017.01.020 [DOI] [PubMed] [Google Scholar]
- 84.Lundström E, Strand R, Forslund A, Bergsten P, Weghuber D, Ahlström H, et al. . Automated segmentation of human cervical-supraclavicular adipose tissue in magnetic resonance images. Sci Rep 2017; 7: 3064. doi: 10.1038/s41598-017-01586-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gifford A, Towse TF, Walker RC, Avison MJ, Welch EB. Characterizing active and inactive brown adipose tissue in adult humans using PET-CT and MR imaging. Am J Physiol Endocrinol Metab 2016; 311: E95–E104. doi: 10.1152/ajpendo.00482.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lareau-Trudel E, Le Troter A, Ghattas B, Pouget J, Attarian S, Bendahan D, et al. . Muscle quantitative MR imaging and clustering analysis in patients with facioscapulohumeral muscular dystrophy type 1. PLoS One 2015; 10: e0132717–16. doi: 10.1371/journal.pone.0132717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le Troter A, Fouré A, Guye M, Confort-Gouny S, Mattei JP, Gondin J, et al. . Volume measurements of individual muscles in human quadriceps femoris using atlas-based segmentation approaches. MAGMA 2016; 29: 245–57. doi: 10.1007/s10334-016-0535-6 [DOI] [PubMed] [Google Scholar]
- 88.Ugarte V, Sinha U, Malis V, Csapo R, Sinha S. 3D multimodal spatial fuzzy segmentation of intramuscular connective and adipose tissue from ultrashort TE MR images of calf muscle. Magn Reson Med 2017; 77: 870–83. doi: 10.1002/mrm.26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.ÁN M, Volken T, Elliott JM. Reliability of quantifying the spatial distribution of fatty infiltration in lumbar paravertebral muscles using a new segmentation method for T1-weighted MRI. BMC Musculoskelet Disord 2016; 17: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim S, Lee D, Park S, Oh KS, Chung SW, Kim Y. Automatic segmentation of supraspinatus from MRI by internal shape fitting and autocorrection. Comput Methods Programs Biomed 2017; 140: 165–74. doi: 10.1016/j.cmpb.2016.12.008 [DOI] [PubMed] [Google Scholar]
- 91.Ulbrich EJ, Nanz D, Leinhard OD, Marcon M, Fischer MA. Whole-body adipose tissue and lean muscle volumes and their distribution across gender and age: MR-derived normative values in a normal-weight Swiss population. Magn Reson Med 2018; 79: 449-458. doi: 10.1002/mrm.26676 [DOI] [PubMed] [Google Scholar]
- 92.Kemnitz J, Eckstein F, Culvenor AG, Ruhdorfer A, Dannhauer T, Ring-Dimitriou S, et al. . Validation of an active shape model-based semi-automated segmentation algorithm for the analysis of thigh muscle and adipose tissue cross-sectional areas. MAGMA 2017; 30: 489–503. doi: 10.1007/s10334-017-0622-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fortin M, Omidyeganeh M, Battié MC, Ahmad O, Rivaz H. Evaluation of an automated thresholding algorithm for the quantification of paraspinal muscle composition from MRI images. Biomed Eng Online 2017; 16: 61. doi: 10.1186/s12938-017-0350-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schneider CA, Rasband WS, Eliceiri KW. NIH image to imageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675. doi: 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Linge J, Borga M, West J, Tuthill T, Miller MR, Dumitriu A, et al. . Body Composition Profiling in the UK Biobank Imaging Study. Obesity 2018; 2. doi: 10.1002/oby.22210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Positano V, Gastaldelli A, Sironi AM, Santarelli MF, Lombardi M, Landini L. An accurate and robust method for unsupervised assessment of abdominal fat by MRI. J Magn Reson Imaging 2004; 20: 684–9. doi: 10.1002/jmri.20167 [DOI] [PubMed] [Google Scholar]
- 97.Demerath EW, Ritter KJ, Couch WA, Rogers NL, Moreno GM, Choh A, et al. . Validity of a new automated software program for visceral adipose tissue estimation. Int J Obes 2007; 31: 285–91. doi: 10.1038/sj.ijo.0803409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waniek S, di Giuseppe R, Plachta-Danielzik S, Ratjen I, Jacobs G, Koch M, et al. . Association of vitamin E levels with metabolic syndrome, and MRI-derived body fat volumes and liver fat content. Nutrients 2017; 9: 1143. doi: 10.3390/nu9101143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.di Giuseppe R, Koch M, Schlesinger S, Borggrefe J, Both M, Müller HP, et al. . Circulating selenoprotein P levels in relation to MRI-derived body fat volumes, liver fat content, and metabolic disorders. Obesity 2017; 25: 1128–35. doi: 10.1002/oby.21841 [DOI] [PubMed] [Google Scholar]
- 100.Fogel A, Goh AT, Fries LR, et al. . Faster eating rates are associated with higher energy intakes during an Ad libitum meal, higher BMI and greater adiposity among 4.5 year old children – Results from the GUSTO cohort. Br J Nutr 2017; 117: 1042–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lundkvist P, Sjöström CD, Amini S, Pereira MJ, Johnsson E, Eriksson JW, et al. . Dapagliflozin once-daily and exenatide once-weekly dual therapy: a 24-week randomized, placebo-controlled, phase II study examining effects on body weight and prediabetes in obese adults without diabetes. Diabetes, Obesity and Metabolism 2017; 19: 49–60. doi: 10.1111/dom.12779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lake JE, Popov M, Post WS, Palella FJ, Sacktor N, Miller EN, et al. . Visceral fat is associated with brain structure independent of human immunodeficiency virus infection status. J Neurovirol 2017; 23: 385–93. doi: 10.1007/s13365-016-0507-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pasha EP, Birdsill A, Parker P, Elmenshawy A, Tanaka H, Haley AP. Visceral adiposity predicts subclinical white matter hyperintensities in middle-aged adults. Obes Res Clin Pract 2017; 11: 177–87. doi: 10.1016/j.orcp.2016.04.003 [DOI] [PubMed] [Google Scholar]