Abstract
The Pattern Sequence Forecasting (PSF) algorithm is a previously described algorithm that identifies patterns in time series data and forecasts values using periodic characteristics of the observations. A new method for univariate time series is introduced that modifies the PSF algorithm to simultaneously forecast and backcast missing values for imputation. The imputePSF method extends PSF by characterizing repeating patterns of existing observations to provide a more precise estimate of missing values compared to more conventional methods, such as replacement with means or last observation carried forward. The imputation accuracy of imputePSF was evaluated by simulating varying amounts of missing observations with three univariate datasets. Comparisons of imputePSF with well-established methods using the same simulations demonstrated an overall reduction in error estimates. The imputePSF algorithm can produce more precise imputations on appropriate datasets, particularly those with periodic and repeating patterns.
1. Introduction
Time series data analysis is applicable across several disciplines, including traffic engineering (Ran et al., 2015), eco-nomics (Taylor, 2008), environmental studies (Mendola, 2005), ecology (Nakagawa and Freckleton, 2011), energy research (Billinton et al., 1996), and signal processing (Gemmeke et al., 2010), among others. Time series data can be processed with various methods that characterize emergent data patterns and predict future behaviour to inform the implementation of policies or use of control measures. Extraction of information from time series data is dependent on data reliability and completeness of information. While measuring, collecting or generating data, missing observations can commonly occur as a result of numerous processes, such as communication errors, failure of data-generating sensors, or power outages. Missing data in analytical studies may lead to defective and undesirable outcomes, including inaccurate predictions or poor decisions in policy making. Hence, methods to replace missing data with appropriate alternative values are needed.
Missing data in time series can be estimated with various methods that are broadly classified as imputation-based or model-based techniques. Model-based methods differ from direct imputation by solving likelihood equations applied to models of the missing data (McLachlan and Krishnan, 2007; Little and Rubin, 2014). Conversely, imputation-based methods estimate missing values either by complete removal or estimating with suitable values using a more generic approach. The majority of methods for filling missing observations in univariate time series are imputation-based, as compared to model-based methods for multivariate time series. Moritz et al. (2015) reviewed different imputation methods for univariate time series and categorized each approach as one of three types; univariate algorithms, univariate time series algorithms, and multivariate algorithms on lagged data. Moritz et al. (2015) also noted several limitations of imputation methods for univariate time series as demonstrated by other studies (Engels and Diehr, 2003; Spratt et al., 2010; Twisk and de Vente, 2002), most of which did not explicitly consider the statistical properties of the time series data. Commonly-used methods for univariate time series are relatively simple and include the arithmetic mean, interpolation, and last observation carried forward (locf). Overall, there is a need for more robust imputation methods for univariate time series, particularly those that can better leverage statistical characteristics of the observations.
This paper presents a univariate time series method that modifies an existing forecasting algorithm for imputation to evaluate relative positions of missing values. The imputePSF method is a modification of the pattern sequence based forecasting (PSF) method (Martínez-Álvarez et al., 2011b,a) to incorporate control structures that evaluate repeating characteristics of the time series as a statistical basis for imputation. We describe the methodology and rationale of the imputePSF algorithm and compare imputation precision to other methods using three different data sets. The implications of this research and practical applications of imputePSF are also discussed.
2. Methodology
2.1. Pattern Sequence-based Forecasting (PSF) algorithm
The PSF algorithm was first proposed in Martínez-Álvarez et al. (2008) and compared with other methods in Martínez-Álvarez et al. (2011b). The algorithm depends primarily on sequence patterns in the dataset and is broadly divided into two steps. The first step labels the time series data using a clustering technique and the second step processes the sequence-based predictions. The novelty of this algorithm is the use of labels for different pattern sequences rather than actual time series data. In the PSF algorithm, labelling is based on k-means clustering to partition the time series with different cluster centres. The number of clusters is identified using combined information from the Silhouette index (Kaufman and Rousseeuw, 2008), Dunn index (Dunn, 1974) and Davies-Bouldin index (Davies and Bouldin, 1979). The optimum number is based on concordance between two of the three indices as a minimim criteria for choosing clusters.
The second step in PSF is sequence prediction in three steps: selecting the optimum window size, matching pattern sequences, and estimation. First, the label sequence generated with clustering is used as input data and the last W sized sequence is used for searching. This sequence of labels of size W is collected from a backward position of labelled time series data and is searched within the input dataset. If this window pattern is found to be repeated in the labelled dataset, then the very next value of each window replica is considered and stored in a vector. The mean of this vector is considered as a label for the next predicted value. The window size is reduced by one unit if no replicates are found for a sequence. The process is repeated until the pattern in a chosen window is found at least once in the input dataset. This ensures at least one replicate is found in a labelled sequence, having a minimium W of 1. When the process is complete, de-normalization is performed such that the predicted labels are transformed into their alternative values in the actual dataset. In this way, PSF predicts one future sample (which can be of an arbitrary size) at a time. If more than one value is to be predicted, the predicted value in an earlier step is appended to the original dataset. This process is continued until the desired number of future values is obtained. Fig. 1 shows the general scheme for the PSF algorithm.
Fig. 1.
Pseudocode for the PSF algorithm. See Bokde et al. (2017) for details.
The R package PSF (Bokde et al., 2017) automates all processes and includes a simple user interface. In this package, the number of optimal clusters and window sizes are estimated automatically. Further, pattern searching and prediction is performed with the psf() function.
2.2. The imputePSF algorithm
The imputePSF algorithm builds on principles of the PSF algorithm for univariate time series. The algorithm begins by decomposing the input time series with missing values into a matrix as a preliminary step to quantify the characteristics of missing observations. This matrix is defined as the ‘missing profile matrix’ and includes relevant information for the imputePSF algorithm, i.e., patch sizes of missing data and position in the time series. The missing profile matrix includes three parameters for each patch, where a patch is defined as a continuous block of missing observations with a potential minimim size of one. The first parameter is the starting index of each missing patch in the time series, the second parameter is the ending index, and the third parameter is the overall patch size. The parameters in the matrix are used as reference points for fast indexing of the missing patches.
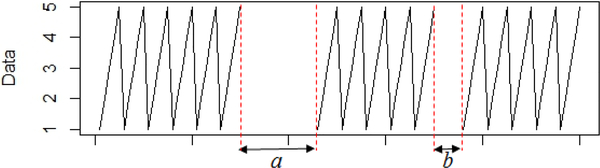
Each patch is evaluated in sequence with imputePSF. First, rows of the missing profile matrix are arranged in decreasing order of patch size described by the third parameter. The largest missing value patch is positioned at the top of the matrix. Processing larger patches first improves the precision of imputed values for the shorter patches and further ensures that shorter patches are given less importance earlier in the imputation process. For example, if patch a is considered the first and largest patch to impute, then the smaller patch b is imputed with greater accuracy (Fig. 2). Imputation of patch a will not be as dependent on missing values of patch b, as compared to first imputating the smaller patch b where more values are missing in a.
Fig. 2.
Sample waveform with two missing patches a and b.
As such, the imputePSF algorithm first evaluates the longest missing patch in the time series and removes all other missing patches from the original dataset (D). The dataset is then separated in two parts; the decomposed dataset (D1) consists of a single missing patch and the remainder of the complete observations from D. The data in D1 is processed differently depending on location before or after the missing patch. Data prior to the missing patch is processed with the PSF algorithm to predict future values (DF1). The length of these predictions is the same length as those in the missing patch. Similarly, observations following the missing patch are processed in reverse with the PSF algorithm. The reverse prediction is for the same values (DR1) as those in the missing patch. Consider a time series of length 100 with a missing patch from indices 60 to 65. The values for DF1 are those that are forecasted using the preceding 59 observations using the PSF method. Conversely, DR1 is estimated by reversing the values from index 66 to 100 and hindcasting using PSF. Averages of the vectors DF1 and reverse vector DR1 are used to create the vector Dimp for the imputed values corresponding to the missing patch in the first row of the missing profile matrix.
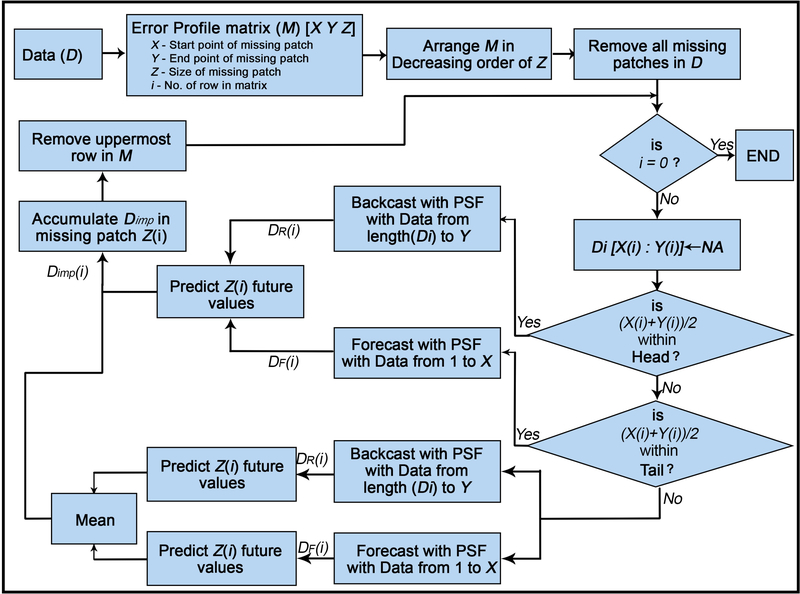
The imputePSF algorithm is implemented with control structures to ensure results are accurate within the characteristics of the data. First, if the missing patch is present in the first 20% of the dataset D1, only reverse prediction with PSF is performed. If a missing patch is within the last 20% of the time series D1, only forward prediction with PSF is performed. The initial 20% of the time series is referred to as the ‘head section’, the middle 60% as the ‘body’, and the last 20% as the ‘tail’. Lengths of each section were selected using cross-validation as described below. The imputed values (Dimp) are then substituted in the corresponding patch in the original dataset (D). The process is repeated until all missing patches in the time series are imputed. The block diagram and general scheme for the imputePSF algorithm are shown in Fig. 3 and Fig. 4.
Fig. 3.
Block diagram demonstrating the methods of imputePSF. Details are provided in section 2.2 and psueudocode is provided in Fig. 4. D: dataset with missing observations, M: error profile matrix, X: starting point of missing patch, Y: end point of missing patch, Z: size of missing patch, i: row number in matrix, DF: forward forecast of patch, DR: backwards hindcast of patch, D>imp: average of DF and DR
Fig. 4.
Pseudocode for the imputePSF algorithm.
2.3. Optimum head and tail size selection
As discussed above, a preliminary step of imputePSF is the segmentation of the observed time series into three sections (head, body, and tail). The PSF method predicts future values by evaluating patterns in the time series and it is necessary to maintain reference patterns prior to the location where imputation is performed. The imputePSF algorithm builds on this approach by averaging the forecast and backcast values estimated with PSF. Missing observations in the head and tail sections are problematic because of insufficient reference patterns for prediction with PSF. Appropriate sizes of the head and tail sections were chosen using cross-validation of imputation results with the nottem dataset (described in detail below, Anderson, 1977). Missing values in the dataset were randomly created as 10 to 50% of the total sample sizes. The cross-validation analyses evaluated head and tail sections of equal size, ranging from 10, 20, 30, 40, 45 and 50% of the data for each section. Table 1 show the imputation errors corresponding to missing values for different head and tail sizes. The results demonstrate that the imputation errors are minimized for head and tail sections as 20% of the total size of each time series. Hence, the head and tail sizes are fixed at these values for imputePSF.
Table 1.
Comparison of RMSE values for 10 to 50 % of missing values for different sizes of head/tail sections in the nottem dataset
| Nottem dataset | Percentages of Missing Values (%) |
|||||
|---|---|---|---|---|---|---|
| 10 | 20 | 30 | 40 | 50 | ||
| Head/Tail Section Size (%) | 10 | 1.05 | 1.80 | 2.78 | 4.01 | 5.50 |
| 20 | 0.95 | 1.76 | 2.37 | 3.72 | 4.72 | |
| 30 | 1.01 | 1.84 | 2.63 | 4.55 | 5.91 | |
| 40 | 1.65 | 2.46 | 3.39 | 4.36 | 5.64 | |
| 45 | 1.65 | 2.51 | 3.43 | 4.43 | 5.64 | |
| 50 | 1.15 | 1.77 | 2.72 | 3.78 | 5.39 | |
3. Experimental results
Results using the imputePSF algorithm were compared with those from conventional algorithms to demonstrate the potential of the proposed method to improve imputation. Three time series obtained from field-based observations and simulation experiments were evaluated. These datasets were chosen specifically as representative examples that were expected to exhibit characteristics that are favorable for use with imputePSF. Specifically, the datasets are generally characterized as having periodic or cyclical components with different dominant frequencies and degree of temporal correlation between observations. The precision of imputed values in each dataset were compared between methods using the root mean square error (RMSE):
| (1) |
where Xi is the original data, is the corresponding predicted data, and N is the number of samples in X. All calculations were performed on a personal computer with an Intel Core i3 Processor and 8GB of RAM. All analyses were conducted with the statistical programming language R (v3.2.5) using the imputeTestbench package (Beck et al., 2018). This package serves as a test bench environment for method comparison by generating profiles of prediction errors from each imputation method under consideration. All datasets under consideration were complete and missing values were simulated using imputeTestbench by varying the amount of missing observations from 10 to 60% of the total size of the complete dataset at increasing intervals of 10%. For each interval, all methods were tested with 1000 repetitions of missing at random (MAR) simulations, where missing data were taken as continuous blocks varying in size from 25 to 100% of the total missing data. The average RMSE of simulated repetitions at each amount of missing values was used to compare imputation methods. In addition to imputePSF, the compared methods included historic means and linear interpolation using the imputeTS package (Moritz, 2017), ARIMA with Kalman filter imputation using the forecast package (Hyndman and Khandakar, 2008), the random forest method using the imputeMissing package (Matthijs Meire, 2016), and the locf method using the zoo package (Zeileis and Grothendieck, 2005).
3.1. Traffic speed data
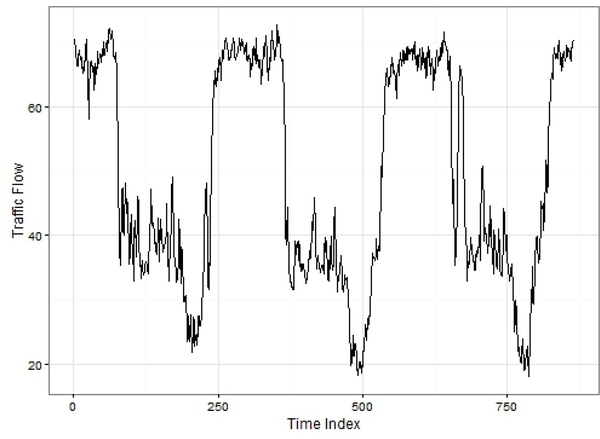
The first comparison evaluated imputation of traffic speed data (http://pems.dot.ca.gov/). This dataset is a univariate time series of traffic speeds from a loop detector in District 7 of Los Angeles County, USA. Data from 10 consecutive Wednesdays at a five minute time step were evaluated following a similar analysis in Ran et al. (2015). The time series is of interest given periodic variation and seasonal patterns that are common in time-variant processes. Data for the first three Wednesdays is plotted in Fig. 5.
Fig. 5.
Patterns of the traffic speed time series for data collected on Wednes-days.
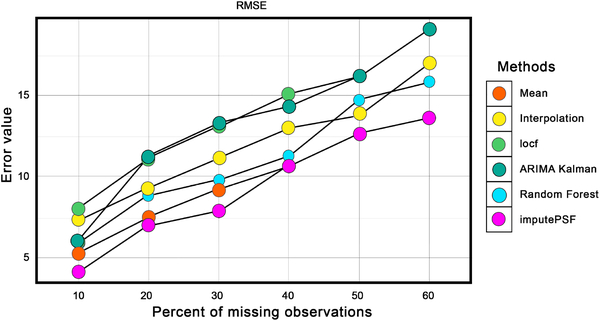
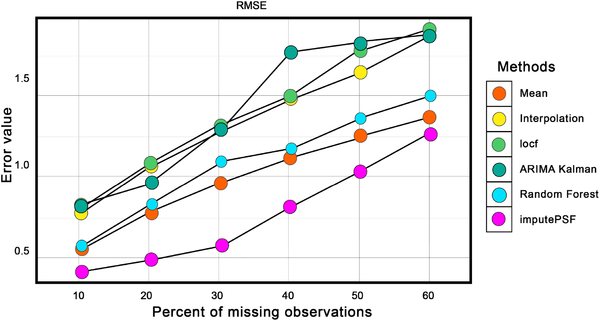
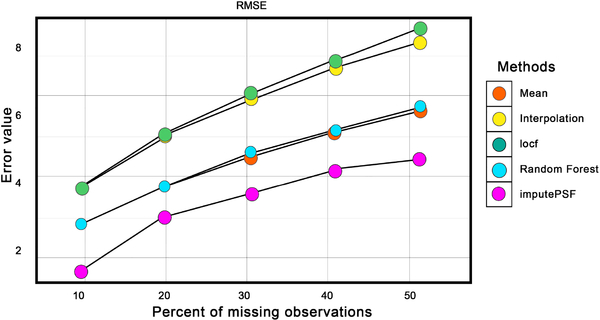
Fig. 6 compares the RMSE of imputed values from each method for varying amounts of missing observations in the dataset. Imputed values using the ARIMA with Kalman filter and locf methods had the highest RMSE values, whereas historic means, linear interpolation and random forest had slightly improved results. For all percentages of missing observations, imputePSF outperformed all other methods.
Fig. 6.
Comparison of imputePSF with mean, interpolation, locf, Arima with Kalman filter, and random forest methods for missing values in the traffic speed data set. RMSE is used as the error performance metric.
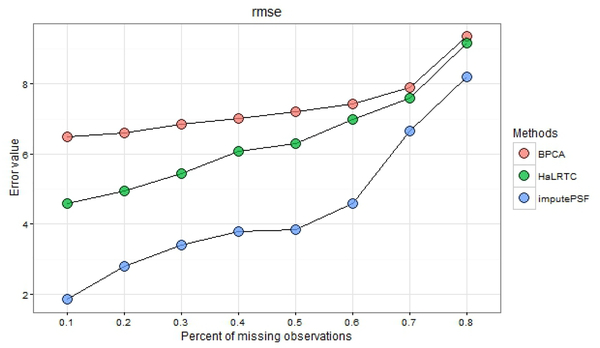
For the same traffic speed dataset, Ran et al. (2015) proposed an imputation method based on Tensor completion (HaL-RTC). The HalRTC method was compared with imputePSF and Bayesian Principal Component Analysis (BPCA), where the latter was used as a benchmark for comparison with HaLRTC (Ran et al., 2015). Output from the imputeTestBench package showed that imputation using HaLRTC produced lower RMSE than BPCA, whereas results from imputePSF had lower RMSE than both BPCA and HaLRTC (Fig. 7). Although the results demonstrated the potential improvement in accuracy using imputePSF, direct comparisons were done following procedures in Ran et al. (2015). Methods comparison using a simulation design with MAR observations produced similar performance between methods. As such, characteristics of the missing data have an effect on imputation accuracy such that that multiple methods should be considered. Regardless, imputePSF produced imputed values no worse, and potentially more precise, than those provided by alternative approaches.
Fig. 7.
RMSE comparison for traffic speed data with methods studied in Ran et al. (2015).
3.2. Water flow rate data
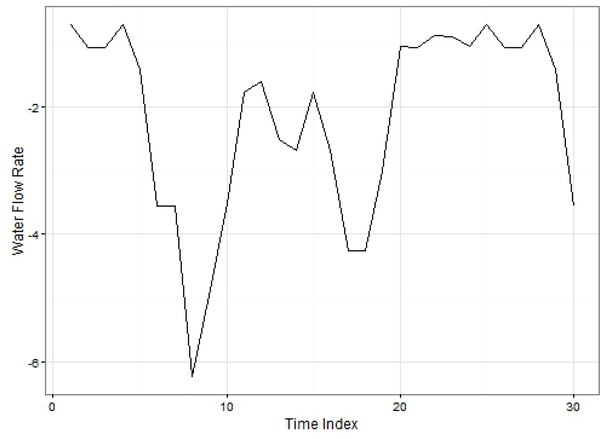
Smart water systems are engineering-based approaches to measure and efficiently control water usage (Gupta et al., 2016). This comparison used a univariate time series of water flow rates generated from hydraulic simulations with EPANET (Rossman et al., 2000). EPANET models water movement and distribution within simulated piping systems. For our analysis, one tank is simulated to provide water to 1240 people following a base demand of 145 litres per day. The dataset describes water consumption in the tank as a function of water flow (litres per second) on an hourly time step for 2 years (n = 17520). The simulation data further included parametric effects of day and night, seasonal variation effects on characteristics of the time series, and considerable random variation to evaluate effects of stochasticity on imputation. A sample pattern for the dataset is shown in Fig. 8. RMSE values for each imputation method at increasing amounts of missing data are shown in Fig. 9. The imputePSF method produced the highest accuracy of the imputed values among methods for all levels of missing data.
Fig. 8.
Patterns of Water flow rate time series data.
Fig. 9.
Comparison of imputePSF with mean, interpolation, locf, Arima with Kalman filter, and random forest methods for the water flow rate dataset. RMSE is used as the error performance metric.
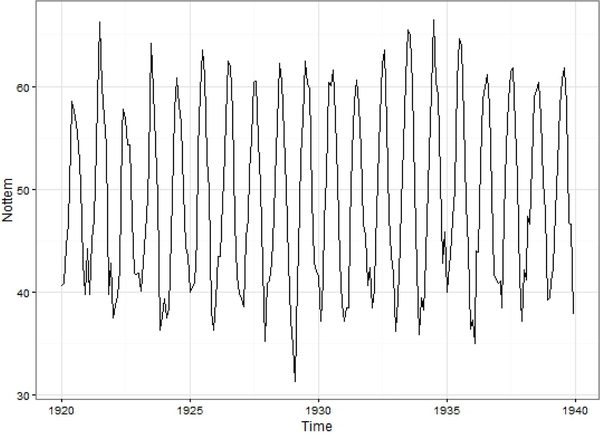
3.3. Nottem Dataset
The nottem dataset is a twenty year time series of the monthly average air temperature (◦F) at Nottingham Castle, England (Anderson, 1977) (Fig. 10). Similar to the above analyses, imputePSF is compared with alternative imputation methods using the imputeTestbench package. Comparisons between the methods are shown in Fig. 11, excluding the results for the ARIMA with Kalman filter method, which had very high error values. As before, these results demonstrate how imputePSF can outperform alternative imputation methods.
Fig. 10.
Patterns of the Nottem data set.
Fig. 11.
Comparison of imputePSF with mean, interpolation, locf, and random forest methods for the nottem dataset. RMSE is used as the error performancef metric.
3.4. Evaluation of imputePSF limitations
The imputePSF algorithm provides superior imputations of missing values for time series with prominent internal correlation structures (e.g., sections 3.1 – 3.3). However, the algorithm may not be appropriate for time series that are dominated by noisy trends or non-cyclical patterns. In such cases, more conventional algorithms may be more appropriate given less computational effort and less bias that may be introduced during imputation. A final analysis demonstrates when the imputePSF algorithm provides superiod, equal, or inferior predictions relative to a simpler and unbiased imputation algorithm that replaces missing values with the mean of all observations.
Several time series were simulated that represented differing contributions of random noise and periodicity. Twenty-five time series were created to simulate a gradient of increasing random noise from none to a chosen maximum and twenty-five time series were created to simulate a gradient of increasing periodicity from none to a chosen maximum. The first set of time series were simulated as a random variable with a normal distribution, where the standard deviation was varied from zero to one to creat the twenty-five time series. The second set of time series were simulated using the nottem dataset by varying the amplitude of the repeating pattern in the observed data from zero (centered at the average of observations) to the maximum observed in the data. All combinations of increasing random noise and increasing periodicity were then simulated as the cross-product of the two gradients yielding 625 total time series (examples in Fig. 12). The cross-product represents time series with characteristics bounded by 1) no random noise and no periodicity, 2) no random noise and maximum periodicity, 3) maximum random noise and no periodicity, and 4) maximum noise and maximum periodicity. Thirty percent of the observations were randomly removed from each of the time series in a continuous chunk and then imputed using imputePSF and mean replacement. The imputations were compared to evaluate the time series characteristics that contributed to differing predictive accuracy of each method.
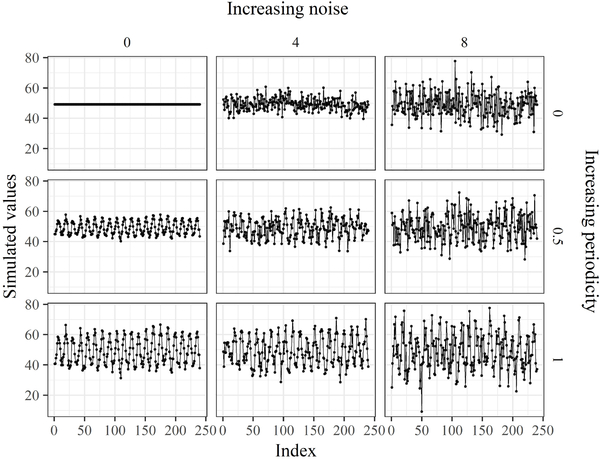
Fig. 12.
A subset of the simulated time series used to evaluate limitations of the imputePSF algorithm. Time series with increasing amounts and differing contributions of random noise (standard deviation from 0 to 8) and increasing periodicity (amplitude from zero to maximum observed in the nottem dataset) were evaluated. A portion of data was removed for imputation from each time series.
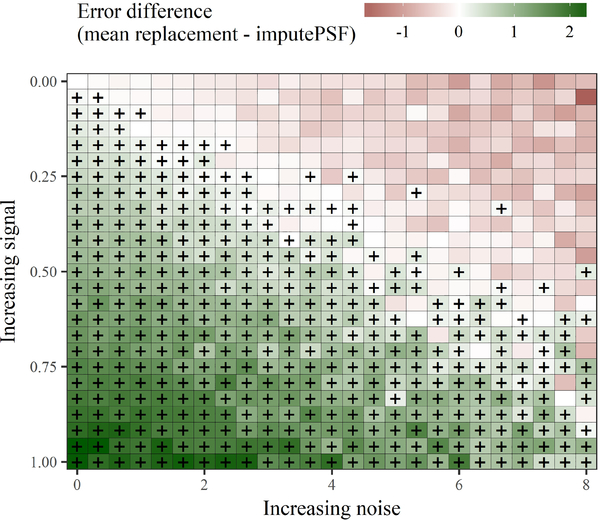
Fig. 13 shows the differences in imputation accuracy between the two methods for each of the simulated time series. The differences were based on the mean absolute percent error from mean replacement minus the error estimate from imputePSF. Thus, positive values represent superior performance for imputePSF (lower error than mean replacement) and negative values represent inferior performance (higher error than mean replacement). As expected, the largest differences in imputation accuracy between the two methods were observed in the lower left and upper right corners of Fig. 13. These locations show the extreme differences betwen the time series cross-products, where the lower left is a purely periodic signal (observed nottem dataset) and the upper right is a purely noisy signal. ImputePSF performs best with the periodic signal, whereas replacement by means produces an improved and unbiased imputation for the purely noise signal. The breaking point where imputePSF no longer had improved imputation accuracy occured along the diagonal of Fig. 13, such that mean replacement had improved accuracy for the time series in the upper triangle of the plot. Time series in the upper triangle had a larger contribution of noise relative to periodicity. General conclusions are that imputePSF 1) works well for time series with periodic components, and 2) provides more accurate imputations if the contribution of the periodic component is equal to or greater than that from random noise. These relationships likely apply to similar datasets where repeating periodicity and significant autocorrelation between observations are observed.
Fig. 13.
Differing imputation accuracy of the imputePSF algorithm compared to imputation by mean replacement using 625 simulated time series. The colors indicate the pairwise differenced in imputation errors (as mean absolute percent error) between the algorithms, where green cells are positive differences indicating imputePSF had higher accuracy and red cells are negative differences indicating mean replacement had higher accuracy. All cells marked with a plus sign indicate improved accuracy with imputePSF. Fig. 12 shows representative examples of all the evaluated time series.
4. Conclusions
This paper introduced the imputePSF method for estimating missing observations in univariate time series. The imputePSF method modifies the PSF algorithm to forecast or backcast missing values based on position and characteristics of missing data patches. In particular, the new algorithm leverages its forecasting ability to automatically impute values for an arbitrary length of missing values. The proposed method was evaluated with simulations of missing observations on four time series and the results showed that imputePSF generally had improved performance in estimates of missing values relative to alternative imputation methods. To date, imputePSF has been applied to univariate time series and future work could focus on application to multivariate time series.
As with forecasting, choosing an appropriate imputation method is dependent on characteristics of the dataset to evaluate. Our above analyses demonstrated that imputePSF works well for datasets with periodic components and when observations are missing in continuous blocks. Based on these characteristics, application of imputePSF to time series that are more random (e.g., white noise or random walk) would not be appropriate, nor would application to datasets with values missing completely at random (i.e., not in chunks). Simpler algorithms, such as interpolation with mean values or last observation carried forward, could work equally well or better for datasets with non-periodic characteristics. ImputePSF is also computationally intensive for larger datasets such that the improvement in precision compared to simpler methods should be considered when choosing an imputation algorithm. Regardless, the above results suggest that imputePSF can provide more accurate imputations and its potential advantages should be considered relative to the dataset and alternative algorithms.
Research Highlights (Required).
An imputation method to impute missing values in univariate time series is described
The imputePSF method imputes missing values using repeating patterns of observed data
The performance of imputePSF is tested on three datasets with distinct patterns
Acknowledgments
The authors would like to thank the Spanish Ministry of Economy and Competitiveness, Junta de Andalucía for the support under projects TIN2014–55894-C2-R and P12-TIC-1728, respectively and the R Community for feedback on the method and analysis.
The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency.
Footnotes
Supplementary information
The imputePSF method is developed for use with the R Statistical Programming Language and is available for download at https://github.com/neerajdhanraj/imputePSF. Analysis code to reproduce results in section 3.4 is available here.
References
- Anderson OD, 1977. Time series analysis and forecasting: another look at the Box-Jenkins approach. The Statistician 26, 285–303. [Google Scholar]
- Beck MW, Bokde N, Asencio-Cortés G, Kulat K, 2018. R Package im-puteTestbench to Compare Imputation Methods for Univariate Time Series. The R Journal 10 URL: https://journal.r-project.org/archive/2018/RJ-2018-024/index.html. [PMC free article] [PubMed] [Google Scholar]
- Billinton R, Chen H, Ghajar R, 1996. Time-series models for reliability evaluation of power systems including wind energy. Microelectronics Reliability 36, 1253–1261. [Google Scholar]
- Bokde N, Asencio-Cortés G, Martínez-Álvarez F, Kulat K, 2017. PSF: Introduction to R package for pattern sequence based forecasting algorithm. The R Journal 9, 324–333. [Google Scholar]
- Davies DL, Bouldin DW, 1979. A cluster separation measure. IEEE Transactions on Pattern Analysis and Machine Intelligence PAMI-1, 224–227. [PubMed] [Google Scholar]
- Dunn JC, 1974. Well-separated clusters and optimal fuzzy partitions. Journal of Cybernetics 4, 95–104. [Google Scholar]
- Engels JM, Diehr P, 2003. Imputation of missing longitudinal data: a comparison of methods. Journal of Clinical Epidemiology 56, 968–976. [DOI] [PubMed] [Google Scholar]
- Gemmeke JF, Van Hamme H, Cranen B, Boves L, 2010. Compressive sensing for missing data imputation in noise robust speech recognition. IEEE Journal of Selected Topics in Signal Processing 4, 272–287. [Google Scholar]
- Gupta A, Mishra S, Bokde N, Kulat K, 2016. Need of smart water systems in India. International Journal of Applied Engineering Research 11, 2216–2223. [Google Scholar]
- Hyndman RJ, Khandakar Y, 2008. Automatic time series forecasting: the forecast package for R. Journal of Statistical Software 26, 1–22.19777145 [Google Scholar]
- Kaufman L, Rousseeuw JP, 2008. Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons. [Google Scholar]
- Little RJ, Rubin DB, 2014. Statistical analysis with missing data. John Wiley & Sons. [Google Scholar]
- Martínez-Álvarez F, Troncoso A, Riquelme JC, Aguilar-Ruiz JS, 2008. LBF: A labeled-based forecasting algorithm and its application to electricity price time series, in: Data Mining, 2008. ICDM’08. Eighth IEEE International Conference on, IEEE. pp. 453–461. [Google Scholar]
- Martínez-Álvarez F, Troncoso A, Riquelme JC, Aguilar-Ruiz JS, 2011a. Discovery of motifs to forecast outlier occurrence in time series. Pattern Recognition Letters 32, 1652–1665. [Google Scholar]
- Martínez-Álvarez F, Troncoso A, Riquelme JC, Aguilar-Ruiz JS, 2011b. Energy time series forecasting based on pattern sequence similarity. IEEE Transactions on Knowledge and Data Engineering 23, 1230–1243. [Google Scholar]
- Meire Matthijs, Ballings Michel, D.V., 2016. imputemissings: Impute missing values in a predictive context. R package version 0.0.3, https://CRAN.Rproject.org/package=imputeMissings. [Google Scholar]
- McLachlan G, Krishnan T, 2007. The EM algorithm and extensions. volume 382 John Wiley & Sons. [Google Scholar]
- Mendola D, 2005. Imputation strategies for missing data in environmental time series for an unlucky situation, in: Innovations in Classification, Data Science, and Information Systems. Springer, pp. 275–282. [Google Scholar]
- Moritz S, 2017. imputets: Time series missing value imputation. R package version 2.5, https://CRAN.R-project.org/package=imputeTS. [Google Scholar]
- Moritz S, Sardá A, Bartz-Beielstein T, Zaefferer M, Stork J, 2015. Comparison of different methods for univariate time series imputation in r. arXiv preprint arXiv:1510.03924.
- Nakagawa S, Freckleton RP, 2011. Model averaging, missing data and multiple imputation: a case study for behavioural ecology. Behavioral Ecology and Sociobiology 65, 103–116. [Google Scholar]
- Ran B, Tan H, Feng J, Liu Y, Wang W, 2015. Traffic speed data imputation method based on tensor completion. Computational intelligence and neuroscience 2015, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman LA, et al. , 2000. EPANET 2: Users Manual. US Environmental Protection Agency. Office of Research and Development. National Risk Management Research Laboratory. [Google Scholar]
- Spratt M, Carpenter J, Sterne JA, Carlin JB, Heron J, Henderson J, Tilling K, 2010. Strategies for multiple imputation in longitudinal studies. American Journal of Epidemiology 172, 478–487. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, 2008. Modelling Financial Time Series. 2nd ed., World Scientific. [Google Scholar]
- Twisk J, de Vente W, 2002. Attrition in longitudinal studies: how to deal with missing data. Journal of Clinical Epidemiology 55, 329–337. [DOI] [PubMed] [Google Scholar]
- Zeileis A, Grothendieck G, 2005. zoo: S3 infrastructure for regular and irregular time series. Journal of Statistical Software 14, 1–27. doi: 10.18637/jss.v014.i06. [DOI] [Google Scholar]