Figure 5.
Mevalonate Pathway-Derived Metabolites Are Necessary for Adipocytes
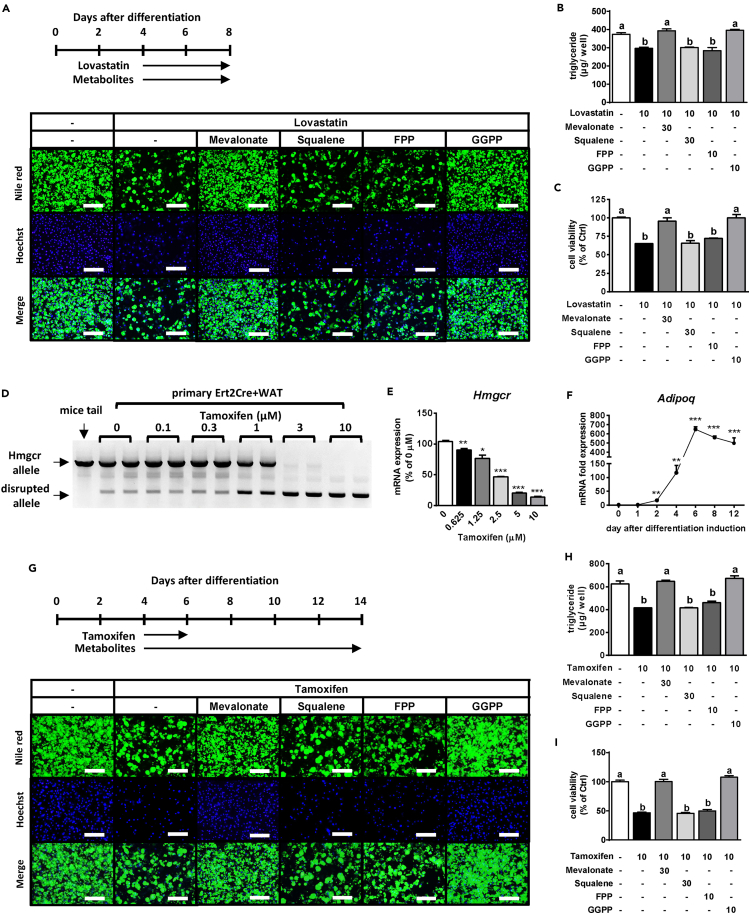
(A–C) After the primary WAT cells were differentiated and kept until D4, they were treated with 10 μM Lova in combination with or without MVA metabolites for an additional 4 days (A). The lipids and nuclei in primary WAT cells were stained with Nile red (top panel) and Hoechst 33342 (middle panel), respectively (A). TG accumulation levels (B) and cell viability (C) are also shown. Merge (bottom), the combined images of Nile red and Hoechst. Scale bars, 200 μm.
(D and E) After the primary Ert2+ Cre WAT cells were differentiated and kept until D4 to D6, they were treated with Tamo for an additional 48 hr and used for analysis of genomic alleles (D) or Hmgcr mRNA levels (E).
(F) Adipoq mRNA expression during adipocyte differentiation.
(G–I) Primary Ert2+ Cre WAT cells were differentiated and kept until D4. Subsequently, the cells were treated with 10 μM Tamo for an additional 48 hr and co-treated with the MVA metabolites until D14 (G). The lipids and nuclei in primary WAT cells were stained with Nile red (top panel) and Hoechst33342 (middle panel), respectively (G). TG accumulation levels (H) and cell viability (I) are also shown. Merge (bottom), the combined images of Nile red and Hoechst. Scale bars, 200 μm. MVA metabolites included 30 μM MVA, 30 μM squalene, 10 μM FPP, and 10 μM GGPP.
Bars represent mean ± SE. (n = 6). Bars with different letters represent significant differences (p < 0.05) by one-way ANOVA with a post-hoc Tukey HSD test.