Abstract
A low-cost and easy-to-fabricate microchip remains a key challenge for the development of true point-of-care (POC) diagnostics. Cellulose paper and plastic are thin, light, flexible, and abundant raw materials, which make them excellent substrates for mass production of POC devices. Herein, a hybrid paper–plastic microchip (PPMC) is developed, which can be used for both single and multiplexed detection of different targets, providing flexibility in the design and fabrication of the microchip. The developed PPMC with printed electronics is evaluated for sensitive and reliable detection of a broad range of targets, such as liver and colon cancer protein biomarkers, intact Zika virus, and human papillomavirus nucleic acid amplicons. The presented approach allows a highly specific detection of the tested targets with detection limits as low as 102 ng mL−1 for protein biomarkers, 103 particle per milliliter for virus particles, and 102 copies per microliter for a target nucleic acid. This approach can potentially be considered for the development of inexpensive and stable POC microchip diagnostics and is suitable for the detection of a wide range of microbial infections and cancer biomarkers.
Keywords: electrical sensing, flexible electronics, paper microfluidics, plastic microfluidics, point-of-care diagnostics
1. Introduction
Advances in point-of-care (POC) diagnostics can potentially set the pace of modern health-care settings and global health.[1–3] Microchips fabricated in a variety of substrates, including silicon, glass, quartz, paper and plastics, have shown great promise in the development of POC diagnostics.[4,5] Flexible substrates such as paper and plastics are particularly interesting because they offer greater potential for making devices on a cost-effective basis.[6–11] Several studies have described the use of paper and plastic substrates for producing different platforms for the detection of various diseases and analytes using multiple colorimetric, fluorescent, electrical, electrochemical, photoelectrochemical, chemiluminescence, and electrochemilu-minescence modalities.[9–16] The fabrication process of such platforms relied on different techniques, including photolithography, wax printing, inkjet printing, screen printing, spraying, stamping, etching, plotting, and laser cutting.[13,17]
Electrical sensing modality is simple and sensitive and does not require bulky components that are usually used in optical and fluorescence-based assays, which makes it as one of the most common sensing modalities used in the development of POC devices.[8,18] The recent advances in developing highly conductive electrode nanomaterials, such as carbon nanotubes, graphene, and metal nanoparticles, have greatly enhanced the performance of electrical sensing-based POC diagnostics.[19–24] Among these nanomaterials, graphene is unique in structure with a single atom layer thick of carbon and has been widely described to have extraordinary electrical double layer capacitance, high mechanical strength, high carrier electron mobility, high surface-to-volume ratio, and low signal-to-noise ratio.[21,25,26] Previous experiments involving deposition of metal structures, such as silver, platinum, and gold on exfoliated graphene, have shown that metal–graphene nanocompos-ites can act as robust materials for the development of electrical sensing-based diagnostics with enhanced electrical conductivity and flexibility.[21,27,28] Using paper and plastic materials for electrical sensing-based POC testing is of high interest and can be clinically relevant and economically sustainable. However, individually both plastic and paper substrates have drawbacks that limit their wide use in microchip fabrication and POC testing. Difficult surface modification, hydrophobicity of most plastic materials, and nonspecific adsorption are some of the limitations of using plastic substrates when complex biological samples are used, which can lead to poor device performance.[9,29,30] Compared to plastic, paper has limited mechanical stability and flexibility, making the fabrication of durable chips difficult. In addition, the most common method for paper-based microchip fabrication is wax printing, which can be time-consuming and is sensitive to temperature changes, thus increasing the manufacturing cost and complexity.[13,31] Paper–plastic composites have been developed with enhanced flexibility while keeping the properties of both substrates. However, they require complex and expensive manufacturing processes.[32–34] Thus, hybrid materials that combine the advantages of both paper and plastic without extensive processing or expensive modifications can allow the development of high-performance, high-throughput, low-cost, and simple POC diagnostics. Here, we developed a microchip with an upper surface made of a cellulose paper and a lower back layer of plastic. We adopted this new design for electrical sensing of different targets, including liver and colon cancer biomarkers, Zika virus (ZIKV), and human papilloma-virus (HPV).
2. Results and Discussion
2.1. Paper–Plastic Microchip Electrodes Design and Fabrication
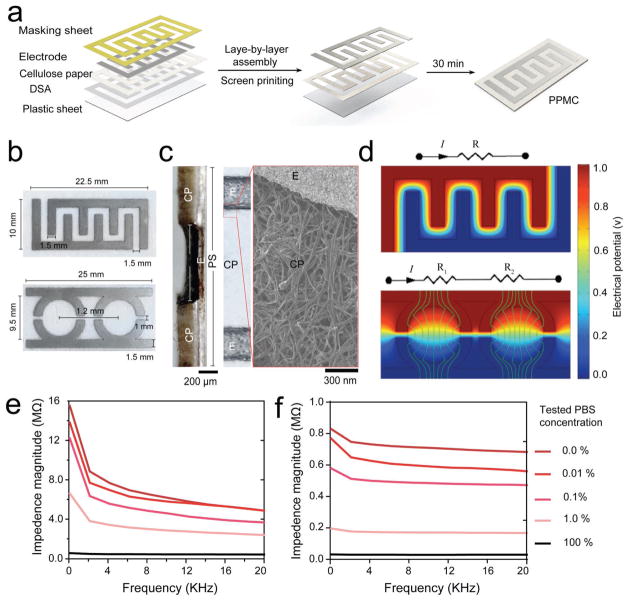
The developed paper–plastic microchip (PPMC) comprises three-layer hybrid substrates prepared of a cellulose paper substrate assembled together with a transparent plastic sheet by double-sided adhesive (DSA). The fabrication process of PPMC is simple and leverages the advantages of the well-known layer-by-layer assembly and screen-printing protocols. The entire process can be completed in <1 h. Figure 1a shows the fabrication process and the layers of materials used in this process. Figure S1 (Supporting Information) shows the actual image of a fabricated PPMC. The upper layer of cellulose paper was sandwiched with the masking sheet and DSA, patterned by laser cutting technique and then sealed with a thin sheet of plastic (0.1 mm in thickness). The electrodes are prepared by the normal screen-printing protocol using preoptimized gra-phene-modified silver nanocomposite ink. In our study, PPMC electrodes were developed with two main designs in order to allow both single and multiplexed detection: 1) PPMC with four-finger interdigitated electrodes for single target detection and 2) PPMC with two-semi-circular parallel electrodes for multitarget detection (Figure 1b and Figure S2, Supporting Information). The detailed structure of both designs is shown in Figure 1b. The use of laser cutting allowed accurate patterning and screen-printing of electrodes on the surface of the paper substrate as confirmed by optical and electron microscopy. Transverse and surface sections of freshly prepared chips showed the layer structure of the prepared electrodes (Figure 1c). Specifically, the formed electrode layers had an increased contact area with the cellulose paper layer, which can significantly enhance the performance of the developed systems. The distribution of electrical field on the surface paper layer of electrodes was simulated using COMSOL software to confirm the design and potential of the developed chips for electrical impedimetric sensing (Supplementary Methods in the Supporting Information and Figure 1d). In addition, we evaluated the performance of the prepared PPMC electrodes by impedance spectroscopy using different dilutions from phosphate buffer saline (1x PBS, pH 7.2). The impedance spectra of 100%, 1%, and 0.01% PBS samples diluted in deionized (DI) water at frequencies between 1 and 20 000 Hz and 1 V are shown in Figure 1e,f. The results showed that the developed microchip can be used to differentiate between different concentrations of PBS samples using impendence spectroscopy. Furthermore, we compared the performance of the microchips prepared using (i) our presented approach in screen-printing electrodes within the cellulose paper and (ii) the common protocol of screen-printing electrodes on the surface of the chip. Figure S3 (Supporting Information) presents the impedance spectra (at frequencies between 1 and 20 000 Hz and 1 V) and the impedance magnitudes at 10 000 Hz and 1 V of for 100%, 1%, and 0.01% PBS samples tested on PPMC with semicircular electrodes prepared by the two different methods (i.e., screen printing on the surface vs within the cellulose paper). The results indicated that PPMC with electrodes printed within the cellulose paper has better signal resolution when tested under different PBS concentrations. In addition, optical microscopy analysis of different sections of microchips fabricated by the two different methods showed that the electrodes printed within paper using our approach were more uniform and had an actual thickness of 0.171 mm ± 0.0143 mm, which was four times more than the electrodes formed by the surface screen-printing approach (Figure S4, Supporting Information).
Figure 1.
PPMC fabrication and characterization. a) Fabrication of PPMC system using the screen-printing protocol coupled with layer-by-layer assembly. b) Digital images of PPMC developed with interdigitated four-finger electrodes for singleplex detection and two-semi-circular electrodes for multiplex detection. c) Optical and electron microscopy images of the transverse section and the top surface of a fabricated PPMC device, showing the detailed layer structure and organization of electrodes. CP: cellulose paper; E: electrode; and PS: plastic sheet. d) Electrical field simulation of PPMC designed with finger (top) and semi-circular (down) electrodes. Impedance spectroscopy of different dilutions of phosphate buffer saline (1x PBS, pH 7.2) tested over a range of frequencies between 1 Hz and 20 KHz using PPMC with e) finger and f) semi-circular electrodes.
2.2. Virus Particles Detection
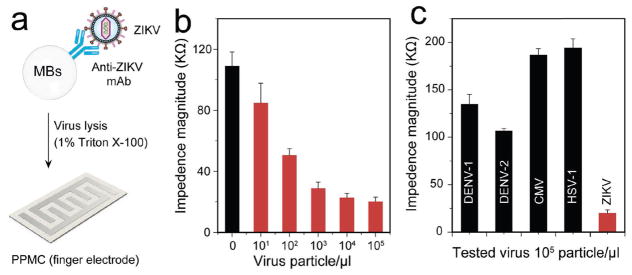
Virus particle detection is of high importance in early infectious disease detection and treatment monitoring. Here, we used PPMC with finger electrodes to detect ZIKV through electrical sensing of viral lysate. ZIKV is a newly emerging flavivirus that has been of major international public health concern following large outbreaks in the Americas.[35,36] The increasing number of studies that confirm the linkage between ZIKV and birth defects calls for the urgent need for the development of low-cost, simple, and rapid POC tests. ZIKV particles were captured using magnetic beads modified with anti-ZIKV envelope monoclonal antibody (anti-ZIKV mAb) and the virus lysate was prepared using 1% Triton X-100 solution (Figure 2a). The modification of magnetic beads with anti-ZIKV mAb and the virus capture was confirmed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Figure S5, Supporting Information). The results indicated the presence of proteins bands at 25 and 51 kDa that is characteristic for IgG antibody molecules, while an additional intense band appeared at 87 kDa for samples containing captured virus.[37,38] Viral lysates of different ZIKV concentrations (100 particle per microliter to 106 particle per microliter spiked in 1x PBS, pH 7.2) were loaded on four-finger PPMC electrodes and tested using an LCR meter for impedance measurement. The results showed that with the increase in the tested virus concentrations, the impedance value decreased, which could be attributed to the increase in the presence of charged molecules (i.e., viral nucleic acid and proteins) released during the lysis step, correlating with the increase in virus concentrations tested (Figure 2b). Following this protocol, we achieved a detection limit down to 102 particle per microliter of ZIKV in PBS, considering signal-to-noise ratio (S/N) = 2 compared to the control (no virus was added). To confirm the specificity of this protocol, we measured the impedance values of samples of the target ZIKV and non-target viruses, such as dengue virus (DENV) type 1 and type 2, herpes simplex virus-1 (HSV-1), and cytomegalovirus (CMV) at the same concentration of 105 particle per microliter. The impedance magnitude generated from the target ZIKV was at least four times lower than the impendence magnitudes generated from all the nontarget viruses tested, including DENV-1, DENV-2, HSV-1, and CMV (n = 3, P < 0.001) (Figure 2c).
Figure 2.
ZIKV particle detection using PPMC with finger electrodes. a) Schematic presentation of the developed PPMC-based assay for ZIKV particle detection through electrical sensing of viral lysate. Magnetic beads (MBs) modified with anti-ZIKV monoclonal antibody (anti-ZIKV mAb) were used to capture virus particles. The captured viruses on MBs were washed with a low electrically conductive solution, lysed using 1% Triton X-100, and loaded onto the paper microchip for impedance measurements. b) Impedance magnitude of viral lysate for samples with different virus concentrations at 8000 Hz and 1 V. c) Impedance magnitude of viral lysate for samples spiked with ZIKV, dengue virus (DENV) type 1 and type 2, cytomegalovirus (CMV), and herpes simplex virus-1 (HSV-1) at 8000 Hz and 1 V. Error bars represent standard error of the mean calculated of at least three independent trials.
2.3. Nucleic Acid Biomarkers Detection
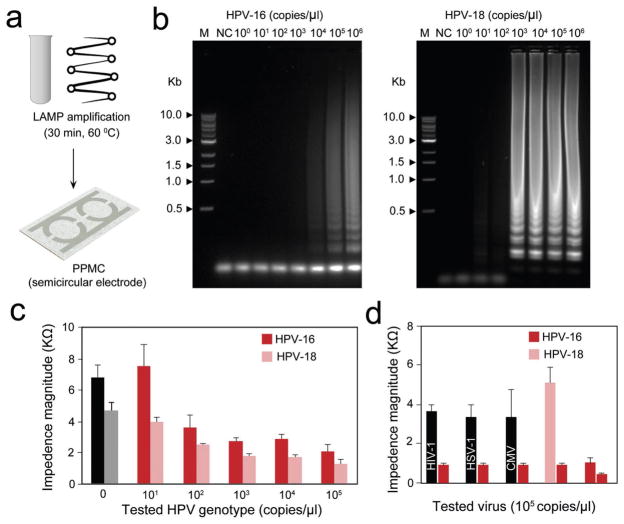
Nucleic acid detection is routinely used to test for diseases and monitoring medical treatments. Coupled with loop-mediated isothermal amplification (LAMP), we tested the developed PPMC electrodes for nucleic acid testing and genotyping of HPV (Figure 3a). HPV is one of the most common sexually transmitted diseases and widely described to be the main cause of cervical cancer in women.[39–41] Two sets of primers, each comprising of four specific primers, which are specific for two different genotypes of HPV-16 and HPV-18, were used for LAMP amplification (Tables S1 and S2, Supporting Information).[42] Two independent amplification reactions were performed using the specified set of primers for 30 min at 60 °C for generating DNA amplicons. The successful LAMP reaction and the formation of DNA amplicons in each reaction was confirmed using agarose gel electrophoresis. The formed LAMP amplicons in each reaction were simultaneously tested on PPMC designed with two detection zones (one was speci-fied for HPV-16 and other for HPV-18). Figure 3b shows the results of agarose gel electrophoresis analysis of LAMP amplification products. There was no amplification observed for control samples (no HPV plasmids were added) while ladder-like amplicons that are characteristic to LAMP were observed for both HPV-16 and HPV-18, confirming the spe-cific amplification of the target HPV genotype (Figure 3b). The loading of LAMP amplicons to the surface of PPMC resulted in a significant decrease in the impedance magnitudes measured at 8000 Hz and 1 V. The change in the impedance magnitude was inversely proportional to the tested concentration of the target HPV plasmid used in the samples of both HPV genotypes tested (Figure 3c). Using this approach, the proposed PPMC can detect concentrations as low as 102 copies per microliter and 103 copies per microliter of HPV-18 and HPV-16, respectively. To confirm the efficiency of this approach to spe-cifically detect HPV, LAMP reactions were performed using nucleic acids of different viruses, including human immuno-deficiency virus-1 (HIV-1), HSV-1, and CMV. Gel electrophoresis indicated that there were no visible amplification products formed with any of the nontargeted nucleic acids (Figure S6, Supporting Information). The final LAMP reaction products of nontarget viruses (i.e., HIV-1, HSV-1, and CMV) were loaded on the PPMC electrodes along with the amplicons generated from the target genotype HPV-16. The results showed that the change in the impedance of PPMC electrodes caused by the addition of HPV-16 was significant when compared to the addition of nontarget viruses (Figure 3d).
Figure 3.
HPV nucleic acid detection and genotyping using PPMC with semi-circular electrodes. a) Schematic presentation of the developed PMMC-based nucleic acid assay for HPV DNA detection. Loop mediated isothermal amplification (LAMP) technique was used to amplify the target HPV DNA using a set of four specific primers (see Table S2 in the Supporting Information) for each tested genotype by independent reactions and the formed amplicons were loaded on PPMC for impedance measurements. b) Gel electrophoresis of LAMP amplification products generated from different concentrations of HPV DNA template. LAMP reaction was performed using tenfold serial dilutions of HPV DNA template (1 × 100 copies per microliter to 1 × 106 copies per microliter). M: 1-kb DNA ladder marker; NC: negative control (without target DNA template). c) Impedance magnitude of LAMP amplicons prepared from different concentrations of target HPV templates at 8000 Hz and 1 V. For each concentration, the impedance magnitude was initially measured for LAMP amplicons of HPV-16 loaded on one of the testing zones and then for LAMP amplicons of HPV-18 loaded on the other testing zone. d) Impedance magnitude of LAMP amplicons prepared from the target HPV-16 and nontarget viruses and genotype of human immunodeficiency virus-1 (HIV-1), herpes simplex virus-1 (HSV-1), and cytomegalovirus (CMV). Error bars represent standard error of the mean calculated of at least three independent trials.
2.4. Cancer Biomarker Detection
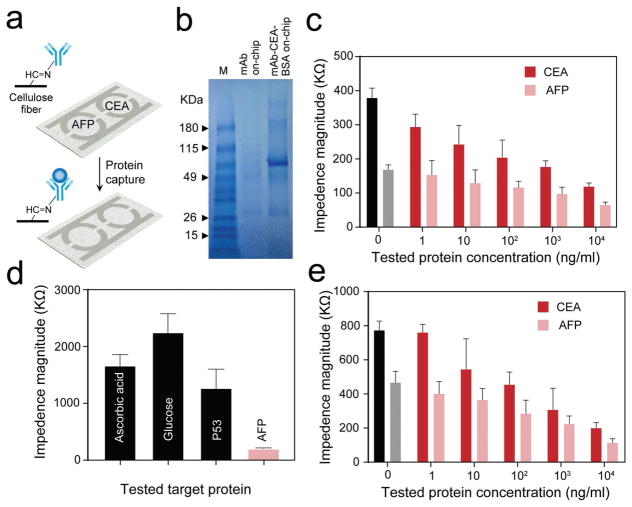
The developed PPMC was tested for the detection of alpha-fetoprotein (AFP) and carcinoembryonic antigen (CEA) that have been identified as biomarkers of hepatocellular carcinoma (HCC) and colorectal cancer (CRC), respectively. HCC and CRC are among the most common types of cancers in humans and represent around 8.0% of total cancer cases reported in 2017 with five year survival rates, thus its early detection is critical.[43–45] PPMC designed with two semi-circular electrodes and two detection zones modified with biomarker specific mono-clonal antibodies was used for multiplex detection of AFP and CEA (Figure 4a). The modification with antibodies was followed with a blocking step using bovine serum albumin (BSA), which allowed for specific capture of the target protein biomarkers and its specific detection by measuring the impedance magnitude. The testing protocol starts by direct capturing of the target proteins from the tested samples followed by impedance measurement using an LCR meter (see the Experimental Section). The surface modification with antibody and capture of the target protein were confirmed using Fourier-transform infrared spectroscopy (FT-IR) and SDS-PAGE. Figure S7 (Supporting Information) illustrates the generated FT-IR spectra of unmodified and oxidized cellulose paper used in the preparation of PPMC electrodes. The peak at 1683 cm−1 corresponds to stretching vibration of C=O group in oxidized paper, which confirms the oxidation of cellulose paper and the formation of aldehyde groups on the surface of papers ready for covalent conjugation of the antibodies.[46] We used CEA as a model to test the efficiency of the developed PPMC to capture the target antigen. Figure 4b shows the SDS-PAGE analysis of CEA captured on PPMC modified with anti-CEA monoclonal antibody. The results indicated the presence of bands around 25 and 50 kDa in lane 2 corresponds to the covalently immobilized CEA antibody on the surface of PPMC, and in lane 3 the band at 180 kDa corresponds to captured CEA and at 90 and 190 kDa corresponds to BSA on the surface of the chip.[47] The image of full gel with the bands of pure CEA and anti-CEA proteins tested is shown in Figure S8 (Supporting Information). To test the sensitivity and specificity of the developed protocol for testing protein biomarkers using PPMC, we used serial dilutions of the target CEA and AFP antigens (1 ng mL−1 to 1 mg mL−1 in PBS) and nontarget common blood biomarkers, such as glucose, ascorbic acid, and P53. The results indicated that the increase in AFP and CEA concentrations resulted in significant decrease in impedance magnitude at 10 000 Hz and 1 V through the designated testing zone for each target (Figure 4c). The impedance measurements of AFP and CEA on the PPMC exhibited a detection limit of 102 ng mL−1 for both targets using the multiplex detection format. However, this detection limit can go down to 10 ng mL−1 and 1 ng mL−1 for CEA and AFP, respectively, when the PPMC was used to capture single antigen in both detection zones (Figure S9, Supporting Information). On the other hand, the decrease in the impedance caused by nontarget analytes (i.e., glucose, ascorbic acid, and P53) was insignificant compared to the decrease in impedance magnitude measured in the presence of the target AFP antigen (Figure 4d). Figure 4e shows the impedance magnitudes measured for different concentrations of CEA and AFP spiked in plasma samples using the proposed protocol. The results indicated a significant decrease in the impedance magnitude with the increase in the concentrations of both CEA and AFP. Following S/N = 2, this protocol allowed the multiplex detection of the target antigens with concentrations down to 102 ng mL−1. To further confirm the potential of the developed PPMC for quantitative detection of targets in biological samples, plasma samples (n = 7) spiked with different concentrations of CEA and AFP were tested and the concentration of target antigen was calculated using the results of previous plasma testing with known antigen concentrations as presented in Figure 4e. The results in Table 1 indicate a good correlation between the expected (from Figure 4e) and measured values with a maximum error <7% of the expected values. These results confirm the potential of the developed protocol using PPMC for multiplex quantitative impedance based detection of biomarkers.
Figure 4.
Protein detection using PPMC with semi-circular electrodes. a) Schematic presentation of the PMMC-based protein assay for the detection of carcinoembryonic antigen (CEA) and α-fetoprotein (AFP) cancer biomarkers. The cellulose paper surface of the chip was modified with antibody to allow specific capturing of target antigen CEA or AFP on the corresponding detection zone. After capturing, the impedance magnitude was measured sequentially for each detection zone to determine the presence or absence of each target. b) SDS gel electrophoresis for PPMC modified with anti-CEA monoclonal antibody and captured CEA on-chip. M: protein marker; mAb: monoclonal antibody; BSA: bovine serum albumin. c) Impedance magnitude of different concentrations of target antigens at 10000 Hz and 1 V. For each concentration, the impedance magnitude was initially measured for CEA captured on one of the testing zones and then for AFP captured on the other testing zone. d) Impedance magnitude of the target AFP antigens and nontarget biomarkers, including ascorbic acid, glucose, and tumor protein P53. e) Impedance magnitude of plasma samples spiked with different concentrations of the target AFP and CEA antigens. Error bars represent standard error of the mean calculated of at least three independent trials.
Table 1.
Detection of AFP and CEA spiked in human plasma samples.
| Impedance (Ω)
|
||||
|---|---|---|---|---|
| Antigen | Concentration [ng mL−1] | Expected | Measured | Error [%] |
| AFP | 10 000 | 71 778 | 77 161 | 6.97 |
| 1000 | 91 711 | 97 849 | 6.27 | |
| 100 | 111 644 | 120 336 | 7.22 | |
| 10 | 131 577 | 129 288 | 1.74 | |
| 1 | 151 510 | 159 630 | 5.09 | |
| CEA | 10 000 | 115 190 | 118 849 | 3.08 |
| 1000 | 163 464 | 158 129 | 3.26 | |
3. Conclusions
Integrated micro/nanosystems with electronic and fluidic elements have broad applications in medicine and biology including POC diagnostics for disease detection and treatment monitoring. Paper/plastic-based microdevices with printed electronics offer mass producibility of such systems with less complexity in design and manufacturing compared to traditional micro total analysis systems. Currently developed paper-based systems with screen-printed electronics benefit from the advantages of either cellulose fibers or plastic substrates. Here, we presented a new category of hybrid paper-based systems with printed electronics by seamlessly integrating cellulose and plastic substrates to overcome some of the major limitations in traditional paper-based systems such as poor contact adhesion between the electrode and cellulose paper, reduced fluid flow due to hydrophobicity of silver or carbon electrodes, and limited surface area between the printed electrode and the sample on cellulose substrates. Such a hybrid microchip fabrication method offers high-resolution screen printing of the electrodes with a relatively simple manufacturing process and higher surface contact area between the electrodes and the sample compared to the traditional paper-based manufacturing. This work demonstrates a major progress toward the development of low-cost, simple paper-based micro-chips with electrical sensing modality by uniquely integrating the hydrophobicity, flexibility, and durability of plastic substrates, wicking property of cellulose substrates, and durable screen printing of conductive inks on plastic substrates.
This platform garners the benefit of the unique wicking properties of paper to greatly help sensitive and efficient sample electrical testing.[8] In addition, the cellulose fibers with known chemical structure allow feasible and cost-effective surface modification with specific antibodies with the flexibility and stability of plastics.[9,15] The presence of plastic tightly bound to the paper allowed efficient and high-resolution screen printing of the electrodes completely through the paper rather than just over the surface of paper, increasing the contact between the tested samples and electrodes, which directly enhances the detection sensitivity and analytical performance of the developed microchip. In addition, the stability of the hybrid substrates allowed better control over the electrode design and geometry during the fabrication process. Using the PPMC platform with two different electrode designs, we were able to develop paper/plastic-based devices with printed electrodes for the detection of multiple bio-targets, including liver and colorectal cancer biomarkers, ZIKV, and LAMP-based HPV amplicons. The platform developed in our study is simple, easy to fabricate, and cost effective and can be adopted to develop more complex systems for rapid and sensitive multiplex detection of biomarkers using portable electric devices.
4. Experimental Section
PPMC Electrodes Fabrication and Characterization
The fabrication of PPMC combines layer-by-layer assembly and screen-printing protocols. Each electrode is prepared of a surface layer of cellulose paper substrate (Whatman 3MM Chromatography Paper, Fisher Scientific) and a lower layer of thin transparent plastic sheet (0.1 mm thickness, CG5000-Dual-Purpose Transparency Film) assembled together by a DSA that is 80 μm thick. The paper with the DSA and mask was cut using a laser cutter (Laser Cutting, VLS2.30 from Universal Laser System). The power, scan speed, and pulse per inch rate were set at 11 W, 5 mm s−1, and 500 pulses per inch, respectively, with the laser at a height of 3.175 mm. After removing the protective layer from the other side of the DSA, the paper was attached to the transparent plastic sheet. Then a silver/ graphene nanocomposite ink prepared by mixing graphene conductive dispersion (Graphene Supermarket, UHC-NPD-100ML) and silver ink (Engineered Conductive Materials, CI-1001) in the ratio of 4:1 was used for screen printing the electrodes over the laser machined mask layer followed by a drying step for 45 min at 60 ºC. PPMCs were designed with two electrode geometries to allow single and multiplex target detection. The first type was designed with four-finger integrated electrodes for singleplex detection of virus particles using a cellulose paper substrate that is 0.34 mm thick. The second type was designed with semicircular parallel electrodes with two detection zones for multiplex detection for nucleic acid and cancer biomarkers using cellulose paper substrate that is 0.18 mm thick. The prepared PPMC electrodes were characterized using optical and scanning electron microscopy and impedance spectroscopy using different concentrations of 1x PBS. In addition, the electrical filed distribution on the surface of electrodes with both geometries was simulated using COMSOL 5.3 software (see the Supporting Information).
Detection of Virus Particles on PPMC Electrodes
The testing protocol starts by virus particles capture using magnetic beads modified with anti-ZIKV monoclonal antibodies from EastCoast Bio, Inc. North Berwick, ME, USA (see the Supporting Information). The magnetic beads in 50 μL of antibody-conjugated magnetic beads solution were concentrated using MagnaGrIP magnetic stand (Millipore) and then mixed with 50 μL of ZIK sample for virus capture. This reaction mixture was incubated on a shaker at 25 rpm for 30 min at room temperature followed by four washing steps using 10% v/v glycerol to remove any electrically conductive solution present in the sample. After washing, glycerol solution was completely removed and 50 μL of 1% v/v Triton X-100 for virus particle lysis. The lysis reaction was kept for 5 min at room temperature and then the magnetic beads were isolated and the virus lysate was collected for impedance measurements. An amount of 8 μL of the lysate was loaded on four-finger PPMC electrodes and impedance magnitudes were recorded using an LCR meter (LCR8110G, GW Instek, CA) at 8000 Hz and 1 V.
Detection of Nucleic Acid on PPMC Electrodes
DNA plasmids with HPV genome (i.e., HPV-16 and HPV-18) were used to confirm the potential of PPMC electrodes for multiplex detection of nucleic acid and genotyping. The target plasmid was first amplified with independent LAMP reactions using two different sets of primers (each comprises four specific primers for each genotype). Serial dilutions of each plasmid (100, 101, 102, 103, 104, 105, 106 copies per microliter) were prepared and amplified using the specific set of LAMP primers (see the Supporting Information). The generated LAMP amplicons were detected on PPMC with semi-circular electrodes in which each electrode contains two detection zones marked by HPV-16 and HPV-18. An amount of 4 μL of the amplicons formed for each genotype was added to the specified testing zone. PPMC electrodes loaded with LAMP amplicons were washed with deionized water to remove any excess electrolytes from LAMP reaction (Figure S10 in the Supporting Information shows the presence of DNA on the chip using fluorescence microscopy). Then the impedance was measured for each testing zone at 8000 Hz and 1 V by adding 4 μL of deionized water to the center of the testing zone. The detection specificity of this protocol was tested using the nucleic acid of different nontarget viruses including HIV-1, HSV-1, and CMV.
Detection of Protein Markers on PPMC Electrodes
Cancer biomarkers of AFP and CEA antigens were used to test PPMC electrodes for multiplex protein detection. For this assay, PPMCs with semi-circular electrodes that are modified with monoclonal antibodies against CEA and AFP were used (see the Supporting Information). The target antigens were captured on PPMC with semi-circular electrodes by adding 4 μL of the target antigen to the specific detection zone and incubated for 30 min at room temperature. The chip was washed with deionized water and then the impedance of each detection zone was measured at 10 000 Hz and 1 V. In all the measurements, 4 μL of deionized water was loaded on the first well of the chip and impedance values were measured after 1 min. After the initial measurement of impedance, another 4 μL of deionized water was loaded again on the second detection zone of the tested PPMC electrode. The detection specificity was tested by performing a series of experiments using nonspecific targets, including glucose, ascorbic, and P53 in clinical range (10–700 ng mL−1). In addition, different concentrations of the target CEA and AFP (10 000, 1000, 100, 10, and 1 ng mL−1) spiked 1 × PBS were tested to evaluate the detection sensitivity of the developed assay.
Testing of PPMC Electrodes for Quantitative Detection of Biomarkers in Plasma
Fresh whole blood sample was purchased from Research Blood Components (Boston, MA). Plasma was separated from the blood by centrifuging the whole blood for 15 min at 2500 rpm using Eppendorf centrifuge 5417R. The obtained plasma was pipetted into a clean Eppendorf tube. Different concentrations of AFP and CEA were spiked in the plasma and then the proteins were captured on the surface of modified chips. After 30 min incubation, the chips were washed with deionized water and kept to dry. The aforementioned procedure for protein detection was repeated accordingly.
Supplementary Material
Acknowledgments
The authors would like to thank Donald Coen and Seamus McCarron Xu Yu, Behzad Etemad, Stephane Hua, Francoise Giguel, Mohammadali Safavieh, and Shantanu Kallakuri. Research reported in this publication was partially supported by the National Institute of Health under award numbers R01AI118502, R21HD092828, and P30ES000002; King Abdulaziz University, Saudi Arabia through Scientific WAQF Fund under grant number 17/1436; Harvard T.H. Chan School of Public Health, Harvard Center for Environmental Health through Harvard NIEHS Grant; American Board of Obstetrics and Gynecology, American College of Obstetricians and Gynecologists, American Society for Reproductive Medicine, Society for Reproductive Endocrinology and Infertility through ASRM Award; and Harvard University Center for AIDS Research (CFAR) under award number 5P30AI060354-14.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Mohamed Shehata Draz, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA, Department of Medicine, Harvard Medical School, Boston, MA 02115, USA, Faculty of Science, Tanta University, Tanta 31527, Egypt
Maryam Moazeni, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Manasa Venkataramani, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Harini Lakshminarayanan, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Ecem Saygili, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Nivethitha Kota Lakshminaraasimulu, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Kamyar Mehrabi Kochehbyoki, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Manoj Kumar Kanakasabapathy, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Shirin Shabahang, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Anish Vasan, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Mohamad Ali Bijarchi, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA.
Prof. Adnan Memic, Center for Nanotechnology, King Abdulaziz University, Jeddah 21589, Saudi Arabia
Prof. Hadi Shafiee, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s, Hospital, Harvard Medical School, Boston, MA 02115, USA, Department of Medicine, Harvard Medical School, Boston, MA 02115, USA
References
- 1.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 2.Daar AS, Thorsteinsdóttir H, Martin DK, Smith AC, Nast S, Singer PA. Nat Genet. 2002;32:229. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 3.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 4.McDonald JC, Whitesides GM. Acc Chem Res. 2002;35:491. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 5.Gervais L, De Rooij N, Delamarche E. Adv Mater. 2011:23. doi: 10.1002/adma.201100464. [DOI] [PubMed] [Google Scholar]
- 6.Tobjörk D, Österbacka R. Adv Mater. 2011;23:1935. doi: 10.1002/adma.201004692. [DOI] [PubMed] [Google Scholar]
- 7.Klemm D, Heublein B, Fink HP, Bohn A. Angew Chem, Int Ed. 2005;44:3358. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- 8.Connelly JT, Rolland JP, Whitesides GM. Anal Chem. 2015;87:7595. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- 9.Becker H, Locascio LE. Talanta. 2002;56:267. doi: 10.1016/s0039-9140(01)00594-x. [DOI] [PubMed] [Google Scholar]
- 10.Printed flexible plastic microchip for viral load measurement through quantitative detection of viruses in plasma and saliva. Scientific Reports. 2015;5:e9919. doi: 10.1038/srep09919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Scientific Reports. 2015;5:8719. doi: 10.1038/srep08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin CD, Chin SY, Laksanasopin T, Sia SK. In: Point-of-Care Diagnostics on a Chip. Issadore D, Westervelt RM, editors. Springer, Springer-Verlag; Berlin, Heidelberg: 2013. p. 3. [Google Scholar]
- 13.Sher M, Zhuang R, Demirci U, Asghar W. Expert Rev Mol Diagn. 2017;17:351. doi: 10.1080/14737159.2017.1285228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liana DD, Raguse B, Gooding JJ, Chow E. Sensors. 2012;12:11505. doi: 10.3390/s120911505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yetisen AK, Akram MS, Lowe CR. Lab Chip. 2013;13:2210. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 16.Lebiga E, Edwin Fernandez R, Beskok A. Analyst. 2015;140:5006. doi: 10.1039/c5an00720h. [DOI] [PubMed] [Google Scholar]
- 17.Xia Y, Si J, Li Z. Biosens Bioelectron. 2016;77:774. doi: 10.1016/j.bios.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 18.Lan WJ, Maxwell EJ, Parolo C, Bwambok DK, Subramaniam AB, Whitesides GM. Lab Chip. 2013;13:4103. doi: 10.1039/c3lc50771h. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed MU, Hossain MM, Safavieh M, Wong YL, Rahman IA, Zourob M, Tamiya E. Crit Rev Biotechnol. 2016;36:495. doi: 10.3109/07388551.2014.992387. [DOI] [PubMed] [Google Scholar]
- 20.Cui Z, Poblete FR, Cheng G, Yao S, Jiang X, Zhu Y. J Mater Res. 2015;30:79. [Google Scholar]
- 21.Syedmoradi L, Daneshpour M, Alvandipour M, Gomez FA, Hajghassem H, Omidfar K. Biosens Bioelectron. 2017;87:373. doi: 10.1016/j.bios.2016.08.084. [DOI] [PubMed] [Google Scholar]
- 22.da Silva ET, Miserere S, Kubota LT, Merkoci A. Anal Chem. 2014;86:10531. doi: 10.1021/ac503029q. [DOI] [PubMed] [Google Scholar]
- 23.Safavieh M, Kaul V, Khetani S, Singh A, Dhingra K, Kanakasabapathy MK, Draz MS, Memic A, Kuritzkes DR, Shafiee H. Nanoscale. 2017;9:1852. doi: 10.1039/c6nr06417e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Draz MS, Shafiee H. Theranostics. 2018;8:1985. doi: 10.7150/thno.23856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JR, Outlaw R, Holloway B. Science. 2010;329:1637. doi: 10.1126/science.1194372. [DOI] [PubMed] [Google Scholar]
- 26.Pumera M. Mater Today. 2011;14:308. [Google Scholar]
- 27.Kim J, Lee MS, Jeon S, Kim M, Kim S, Kim K, Bien F, Hong SY, Park JU. Adv Mater. 2015;27:3292. doi: 10.1002/adma.201500710. [DOI] [PubMed] [Google Scholar]
- 28.Pasricha R, Gupta S, Srivastava AK. Small. 2009;5:2253. doi: 10.1002/smll.200900726. [DOI] [PubMed] [Google Scholar]
- 29.Belder D, Ludwig M. Electrophoresis. 2003;24:3595. doi: 10.1002/elps.200305648. [DOI] [PubMed] [Google Scholar]
- 30.Guo J, Wang Z, Sun P, Tang Q, Li H, Wang X, Guo G, Pu Q. Sens Actuators, B. 2017;250:250. [Google Scholar]
- 31.Li X, Tian J, Shen W. Cellulose. 2010;17:649. [Google Scholar]
- 32.Yao Y, Tao J, Zou J, Zhang B, Li T, Dai J, Zhu M, Wang S, Fu KK, Henderson D, Hitz E, Peng J, Hu L. Energy Environ Sci. 2016;9:2278. [Google Scholar]
- 33.Zhu H, Fang Z, Wang Z, Dai J, Yao Y, Shen F, Preston C, Wu W, Peng P, Jang N, Yu Q, Yu Z, Hu L. ACS Nano. 2016;10:1369. doi: 10.1021/acsnano.5b06781. [DOI] [PubMed] [Google Scholar]
- 34.Jung YH, Chang TH, Zhang H, Yao C, Zheng Q, Yang VW, Mi H, Kim M, Cho SJ, Park DW, Jiang H, Lee J, Qiu Y, Zhou W, Cai Z, Gong S, Ma Z. Nat Commun. 2015;6:7170. doi: 10.1038/ncomms8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckham JD, Pastula DM, Massey A, Tyler KL. JAMA Neurol. 2016;73:875. doi: 10.1001/jamaneurol.2016.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Driggers RW, Ho C-Y, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G. N Engl J Med. 2016;374:2142. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 37.Brecher M, Li Z, Liu B, Zhang J, Koetzner CA, Alifarag A, Jones SA, Lin Q, Kramer LD, Li H. PLoS Pathog. 2017;13:e1006411. doi: 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong SS, Poon RW, Wong SC. J Formosan Med Assoc. 2016;115:226. doi: 10.1016/j.jfma.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Low N, Broutet N, Adu-Sarkodie Y, Barton P, Hossain M, Hawkes S. Lancet. 2006;368:2001. doi: 10.1016/S0140-6736(06)69482-8. [DOI] [PubMed] [Google Scholar]
- 40.Gillison ML, Chaturvedi AK, Lowy DR. Cancer. 2008;113:3036. doi: 10.1002/cncr.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burd EM. Clin Microbiol Rev. 2003;16:1. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saetiew C, Limpaiboon T, Jearanaikoon P, Daduang S, Pientong C, Kerdsin A, Daduang J. J Virol Methods. 2011;178:22. doi: 10.1016/j.jviromet.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Lu J, Ren H, Chen T, Gao L, Di L, Song Z, Zhang Y, Yang T, Thakur A. Mol Clin Oncol. 2013;1:153. doi: 10.3892/mco.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. C. Society. American Cancer Society; Atlanta, GA: 2017. [Google Scholar]
- 45.Yang Z, Liu S, Zhu M, Zhang H, Wang J, Xu Q, Lin K, Zhou X, Tao M, Li C, Zhu H. Sci Rep. 2016;6:22090. doi: 10.1038/srep22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silverstein RM, Webster FX, Kiemle DJ, Bryce DL. Spectrometric Identification of Organic Compounds. 3. John Wiley & Sons; New York: 1974. [Google Scholar]
- 47.Das A, Barik S, Bose A, Roy S, Biswas J, Baral R, Pal S. Immunol Lett. 2014;162:132. doi: 10.1016/j.imlet.2014.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.