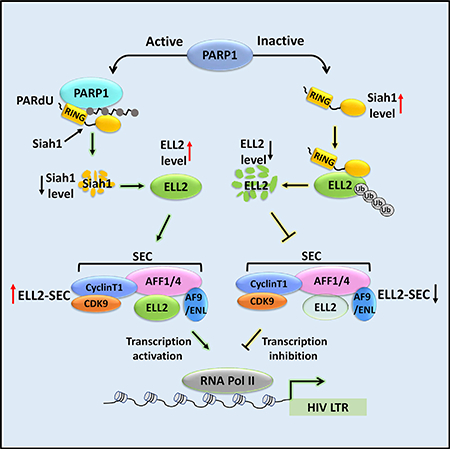
SUMMARY
Recentstudieshave revealeda keyrole ofPARP1that catalyzes the poly-ADP-ribosylation (PARylation) of substrates in regulating gene transcription. We show here that HIV-1 transcriptional activationalso requires PARP1 activity. Because efficient HIV-1 transactivation is knowntodependontheELL2-containing super elongation complex (SEC), we investigated the functional relationship between PARP1 and ELL2-SEC in HIV-1 transcriptional control. We show that PARP1 elevates ELL2 protein levels to form more ELL2-SEC in cells.This effect is caused by PARP1’ssuppression of expression of Siah1, an E3 ubiquitin ligase for ELL2, at both mRNA and protein levels. At the mRNA level, PARP1 coordinates with the co-repressor NCoR to suppress Siah1 transcription. At the protein level, PARP1 promotes Siah1 proteolysis, likely through inducing PARylation-dependent ubiquitination (PARdU) of Siah1. Thus, a PARP1-Siah1 axis activates HIV-1 transcription and controls the expression of ELL2 and other Siah1 substrates.
In Brief
Yu et al. reveal a critical role for a PARP1Siah1 axis in controlling HIV-1 viral transcription. The axis increases cellular levels of the transcription factor ELL2 and its associated SEC complex that is required for robust HIV-1 transcription.
Graphical Abstract
INTRODUCTION
The ELL1 protein was first reported in 1996 as a transcription factor that specifically promotes the elongation stage of RNA polymerase (Pol) II transcription (Shilatifard et al., 1996). A more recent study has provided further details about its mechanism of action in facilitating the pause site entry and release by Pol II (Byun et al., 2012). Like ELL1, ELL2, a member of the ELL family of transcription factors, has also been reported to stimulate Pol II elongation (Shilatifard et al., 1997). Studies performed by others and us show that ELL1 and ELL2 serve as alternative subunits of the super elongation complex (SEC) and can synergize with another SEC component, P-TEFb, to greatly promote Pol II elongation (He et al., 2010; Lin et al., 2010; Sobhian et al., 2010).
In HIV-1-infected cells, the viral-encoded Tat protein recruits the SEC to the viral gene promoter to activate transcriptional elongation (He et al., 2010; Sobhian et al., 2010). The human SEC is thus a specific host factor required for productive HIV-1 replication (Lu et al., 2013). Importantly, between ELL1 and ELL2, the ELL2-containing SEC (ELL2-SEC) is especially favored by Tat to transactivate HIV-1 (Li et al., 2016).
In cells, the expression of ELL2 but not ELL1 is often controlled at the protein stability level by an E3 ubiquitin ligase called Siah1 (Liu et al., 2012). In addition to ELL2, Siah1 also has other substrates and can thus control diverse biological and disease processes (House et al., 2009). Although Siah1 is an important regulatory protein, whose level is affected by various signals, the detailed mechanism that controls Siah1 gene expression is largely unknown.
PARP1 mainly functions as a post-translational modification enzyme that transfers the poly(ADP-ribose) (PAR) moiety to a variety of target proteins using NAD+ as a substrate. In addition to its well-known role in DNA damage repair, PARP1 also controls many other biological processes including transcription. The studies of the transcriptional role of PARP1 have largely focused on its effect on chromatin structure, serving as a transcriptional activator or repressor through direct binding to target genes, and acting as a coactivator or co-repressor for several other transcriptional regulators (Kraus, 2008).
In this study, we first determined if PARP1 plays a role in regulating HIV-1 transcription and found that its catalytic activity was indeed essential for robust Tat transactivation. This was due to PARP1’s promotion of ELL2 expression at the protein but not mRNA level, leading to an increased ELL2-SEC formation. Contributing to this positive effect on ELL2, PARP1 suppressed the expression of Siah1, the ubiquitin ligase for ELL2, at both the mRNA and protein levels. At the mRNA level, PARP1 coordinates with the co-repressor NCoR to suppress Siah1 transcription. At the protein level, PARP1 promotes Siah1 proteolysis, likely through inducing PARylation-dependent ubiquitination (PARdU) of Siah1. Together, these data reveal how PARP1 controls HIV transcription as well as the expression of ELL2 and other Siah1 substrates through a PARP1-Siah1 axis.
RESULTS
PARP1 Knockdown Inhibits Tat Activation of HIV-1 Transcription
To determine if PARP1 plays a role in HIV-1 transcription, we created two independent PARP1 knockdown (KD) clones (1–3 and 2–1; Figure 1A) that express short hairpin (sh) RNAs (shPARP1–1-3 and shPARP1–2-1; shPARP1–2-1 was used in all subsequent experiments and simply labeled shPARP1) targeting two separate regions of PARP1 mRNA in HeLa-based NH1 cells. This cell line contains an integrated HIV-1 LTR-luciferase reporter gene (Yang et al., 2001). The two KD clones were tested for luciferase expression in the absence or presence of increasing amounts of the Tat protein.
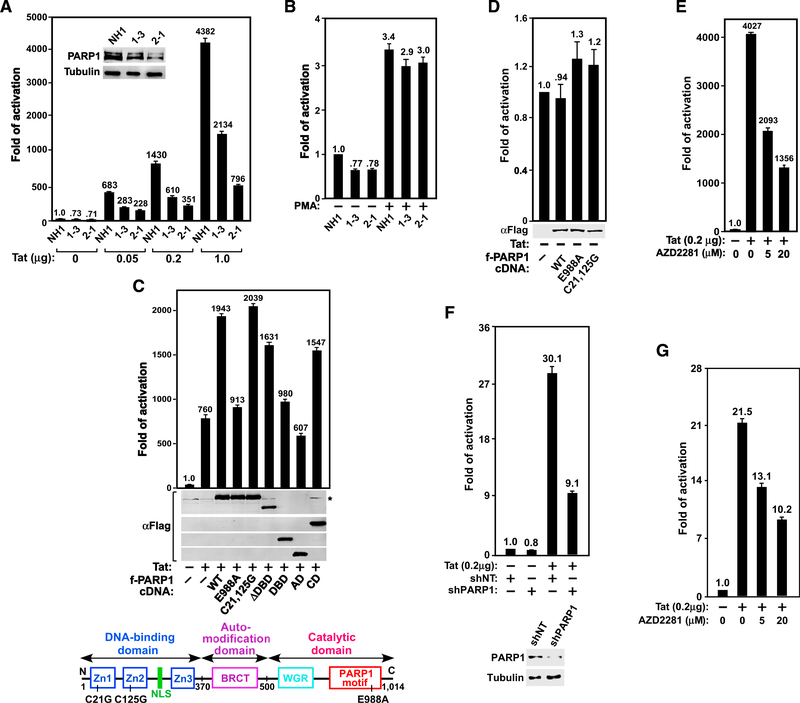
Figure 1. PARP1 and Its Catalytic Activity Are Required for Tat- but Not PMA-Induced Activation of HIV-1 Transcription.
(A and B) The HeLa-based NH1 cells with an integrated HIV-1 LTR-luciferase reporter construct and two NH1-derived PARP1 KD clones 1–3 and 2–1 were transfected with the indicated amounts of Tat cDNA (A) or treated with 200 nM PMA for 24 hr (B). Luciferase activities were measured in these cells and compared with that in Tat-free (A) or untreated NH1 cells (B), which was set to 1. The inset in (A) shows PARP1 levels in the three cell lines as revealed by western blotting (WB). (C and D) The indicated Flag-tagged PARP1 mutants were co-expressed with (C) or without (D) Tat in 2–1 cells and examined using anti-Flag WB. Luciferase activities were analyzed as in (A). The domain structure of PARP1 and point mutations are shown in (C).
(E and G) NH1 (E) or the Jurkat-based 1G5 cells containing an integrated HIV-1 LTR-luciferase reporter construct (G) were transfected with or without the Tat cDNA and then treated with the indicated concentrations of AZD2281. Luciferase activities were measured and analyzed as in (A).
(F) 1G5 cells were stably transfected with plasmids expressing the indicated shRNA (NT, non-target) and/or the Tat cDNA. Also shown are PARP1 levels in the KD cells. Luciferase activities were measured and analyzed as in (A).
Error bars in all graphs represent mean ± SD from three separate measurements. See also Figure S1
Compared with the parental NH1 cells, the basal level HIV-1 LTR activity was only mildly reduced (~30%) in the KD cells (Figure 1A). However, the Tat-dependent LTR activity was more severely affected (up to 5.5-fold reduction), and the degree of reduction correlated with the extent of PARP1 KD in the cells (Figure 1A). Notably, in addition to Tat, the mitogen phorbol myristate acetate (PMA) can also activate the HIV-1 LTR. Although Tat-transactivation requires the SEC, PMA acts largely through upregulating the activity of NF-kB that binds to the LTR to activate viral transcription (West et al., 2001). Although the PARP1 KD significantly decreased Tat transactivation, it produced only a mild effect on the PMA-induced LTR activation (Figure 1B). Thus, efficient HIV-1 transcription, especially the Tat/SECdependent process, requires PARP1.
Catalytic Activity of PARP1 Is Required for Optimal Tat Transactivation
To determine which region of PARP1 is required to support robust Tat transactivation, shPARP1-resistant wild-type (WT) or mutant PARP1 was introduced into the KD clone 2–1 to test their abilities to rescue the Tat-activated HIV LTR-luciferase expression. Although both WT PARP1 and the C21,125G mutant (Cys at positions 21 and 125 changed to Gly; Gradwohl et al., 1990) lacking the DNA-binding activity markedly increased Tat transactivation, the catalytically inactive mutant E988A (Rolli et al., 1997) and the deletion mutant that contains either the DNA-binding domain (DBD; aa 1–370) or the auto-modification domain (AD; aa 371–500) alone but lacks the catalytic domain (CD) were largely ineffective in this regard (Figure 1C). Moreover, introduction of the deletion mutant lacking the DBD (DDBD; aa 371–1014) or containing only the CD (aa 501–1014) rescued Tat transactivation to ~80% of the WT level (Figure 1C). In contrast to the Tat-activated process, the basal HIV transcription was not much affected by either WT or mutant PARP1 (Figure 1D). Thus, the catalytic but not DNA-binding or AD was essential for PARP1 to promote Tat transactivation. Consistently, AZD2281, a chemical inhibitor of the PARP1 catalytic activity, also suppressed Tat transactivation in a dose-dependent manner (Figure 1E).
Because AZD2281 also inhibits PARP2, we examined the impact of PARP2 KD on Tat transactivation in NH1 cells. In contrast to the decreased Tat transactivation observed in PARP1 KD cells (Figure 1A), the expression of two different PARP2-specific shRNAs promoted Tat transactivation (Figure S1). Given that AZD2281 inhibits both PARP1 and PARP2, its ability to decrease Tat transactivation overall (Figure 1E) implies that between the positive effect of PARP1 and negative effect of PARP2, the former plays a more predominant role in controlling Tat transactivation.
To test whether dependence on PARP1 for Tat transactivation can be generalized beyond HeLa cells, we inhibited PARP1 in the Jurkat T cell-based 1G5 cells, which contain an integrated HIV-1 LTR-luciferase reporter and are used widely to analyze HIV-1 Tat activity, inhibitors, and T cell activation effects (Aguilar-Cordova et al., 1994). Consistent with the results in HeLa cells (Figures 1A and 1E), both KD of PARP1 expression and inhibition of PARP1 activity by AZD2281 markedly decreased Tat transactivation in 1G5 cells (Figures 1F and 1G).
PARP1 Depletion/Inhibition Decreases Cellular Levels of ELL2 and ELL2-SEC
Because optimal Tat transactivation requires the SEC, especially the ELL2-SEC (Li et al., 2016), we examined by immunoblotting whether PARP1 KD could affect the expression of key SEC components as well as other P-TEFb-associated factors. Indeed, KD markedly reduced the level of ELL2 but not ELL1 or other SEC subunits in nuclear extract of clone 2–1 cells (Figure 2A). It also failed to alter the expression of key subunits within the 7SK snRNP (e.g., HEXIM1 and LARP7) and the Brd4-P-TEFb complex, which join the SEC to form a P-TEFb network for controlling HIV-1 and cellular gene transcription.
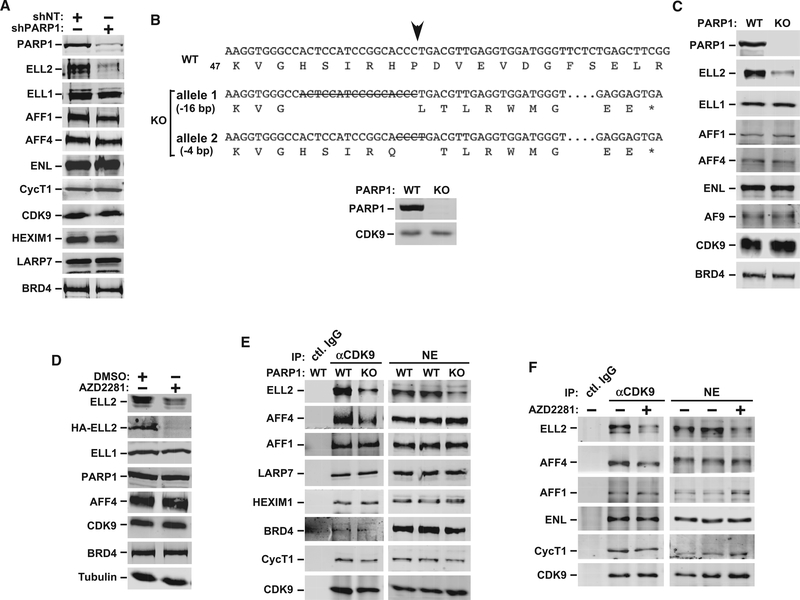
Figure 2. PARP1 Depletion or Inhibition Decreases Cellular Levels of ELL2 and ELL2-SEC.
(A) Levels of the indicated proteins in WCE of HeLa cells transfected with the plasmids expressing shPARP1 or shNT were analyzed using WB.
(B) Nucleotide and predicted amino acid sequences surrounding the intended Cas9 cleavage site (arrow) in WT PARP1 gene as well as the mutated alleles generated by CRISPR-Cas9 are shown. Deleted nucleotides are indicated by capital letters containing strikethroughs, the omitted nucleotides are marked by consecutive dots, and the premature stop codons due to frameshift mutations are indicated by an asterisk. The loss of PARP1 expression in the HeLa-based KO clone was confirmed using WB.
(C and D) Analysis using WB of the indicated proteins in extracts of parental HeLa (WT), the PARP1 KO cells (C), and WT HeLa cells transfected with HA-ELL2 and treated with DMSO or AZD2281 (D).
(E and F) Nuclear extracts (NEs) were prepared from WT HeLa or PARP1 KO cells (E) or from HeLa cells treated with or without AZD2281 (F) and then subjected to immunoprecipitation with the anti-CDK9 antibody or total rabbit IgG. The precipitates were examined using WB for the indicated proteins.
See also Figure S2.
In addition to the PARP1 KD cells, the ELL2 protein level also decreased in an engineered HeLa cell line, where the PARP1 gene was destroyed by CRISPR-Cas9 genome editing (Figures 2B and 2C). Finally, consistent with the above demonstration that the PARP1 catalytic activity was essential for optimal Tat transactivation, AZD2281 markedly decreased the protein levels of both endogenous ELL2 and the transiently overexpressed HAELL2 but not ELL1 in the treated cells (Figure 2D). Notably, the decrease was more pronounced for HA-ELL2 than for endogenous ELL2. This was probably caused by the fact that a major portion of the overexpressed HA-ELL2 was not incorporated into a SEC, thus making it more susceptible to proteolytic degradation. AZD2281 also reduced ELL2 level in 1G5 cells (Figure S2).
As expected, the decreased ELL2 level in the PARP1 knockout (KO) cells also reduced the amount of ELL2 bound to the immunoprecipitated CDK9, indicating a loss of the ELL2-SEC in these cells (Figure 2E). The AFF4 level in the immunoprecipitates also decreased for an unknown reason. A similar decrease in ELL2SEC was also caused by AZD2281 (Figure 2F). The reduced ELL2-SEC formation in cells in which the PARP1 gene was deleted or activity inhibited explains well the observed decrease in Tat transactivation under these conditions.
PARP1 Depletion/Inhibition Decreases Cellular ELL2 Protein but Not mRNA Level in a Process Dependent on ELL2 C-Terminal Region
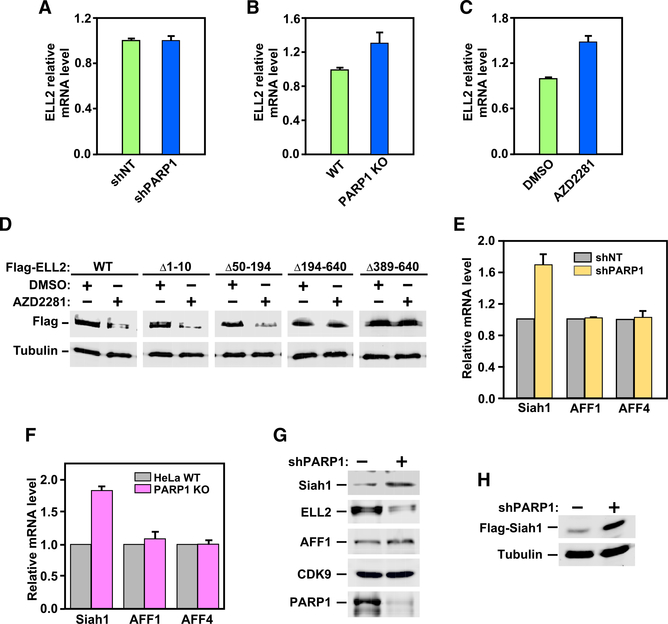
To investigate the mechanism by which PARP1 controls the levels of ELL2 and ELL2-SEC in cells, we found that the control was at the ELL2 protein but not mRNA level, because neither the depletion of PARP1 expression by KD or KO nor inhibition of PARP1 activity by AZD2281 decreased the ELL2 mRNA level, as revealed by qRT-PCR analysis (Figures 3A–3C). Furthermore, the C-terminal region of ELL2 was necessary for AZD2281 to decrease the ELL2 protein level. Although deletion of the N-terminal region up to position 194 still allowed ELL2 to be downregulated by AZD2281, the deletion of the C-terminal region beyond position 389 rendered ELL2 unresponsive to the drug (Figure 3D).
Figure 3. PARP1 Suppresses Expression of Siah1 at Both mRNA and Protein Levels.
(A–C) ELL2 mRNA isolated from HeLa cells expressing shNT or shPARP1 (A), from WT and PARP1 KO HeLa cells (B), and from HeLa cells treated with DMSO or AZD2281 (C) were measured using qRT-PCR, normalized to those of GAPDH, and displayed, with the value of the first column in each panel set to 1. Error bars represent mean ± SD from three separate measurements.
(D) WCE of HeLa cells expressing the indicated WT and mutant Flag-ELL2 and treated with DMSO or AZD2281 were examined using WB.
(E and F) The indicated mRNA isolated from HeLa cells expressing shNT or shPARP1 (E) or from WT and PARP1 KO HeLa cells (F) were measured using qRT-PCR, normalized to those of GAPDH, and displayed, with the values obtained in the shNT and WT cells adjusted to 1. Error bars represent mean ± SD from three separate measurements.
(G and H) Endogenous Siah1 (G), Flag-Siah1 expressed from a transfected plasmid (H), and other indicated proteins present in NE (G) or WCE (H) of control and PARP1 KD (shPARP1) cells were examined using WB.
PARP1 Inhibits Siah1 Expression at Both mRNA and Protein Levels
We have previously shown that Siah1 is an E3 ubiquitin ligase that targets the ELL2 C-terminal region for degradation (Liu et al., 2012). Given that the ELL2 protein but not mRNA level decreased upon PARP1 inhibition, we hypothesized that PARP1 affects ELL2 through controlling Siah1. Indeed, analysis using qRT-PCR indicates that both PARP1 KD and KO increased the Siah1 mRNA level by ~75%–80% but had no effect on control genes AFF1 and AFF4 (Figures 3E and 3F). In addition, KD also increased the protein levels of both endogenous Siah1 (Figure 3G) and the transiently transfected Flag-Siah1 (Figure 3H), which was expressed from the cytomegalovirus (CMV) promoter on a plasmid. These results indicate that PARP1 likely controls the Siah1 expression at both mRNA and protein levels.
PARP1 Associates with Siah1 Gene Promoter to Inhibit Transcription in a Process Likely Involving Transcriptional Co-repressor NCoR
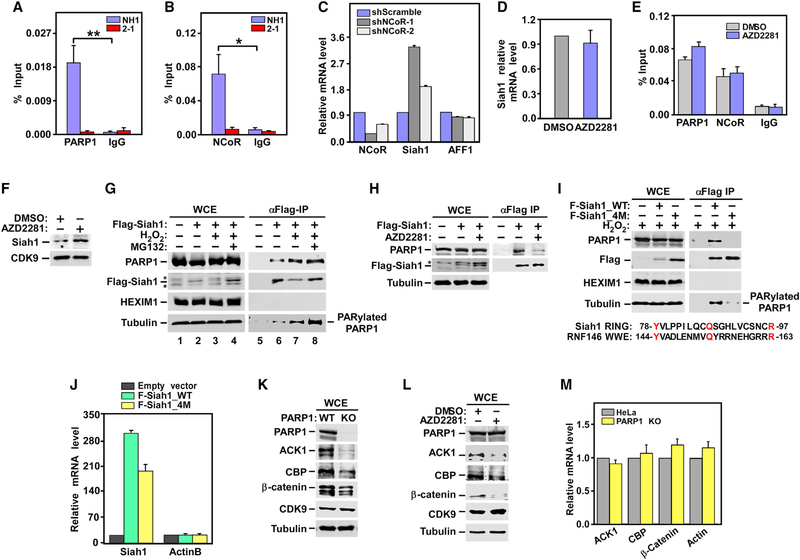
Consistent with a potential role of PARP1 in directly controlling Siah1 transcription, robust PARP1 signal was detected by chromatin immunoprecipitation (ChIP)-qPCR at the Siah1 promoter, and the signal dramatically decreased upon PARP1 KD (Figure 4A). At the same location, the well-known transcriptional co-repressor NCoR, which has been reported to form a suppressive complex with PARP1 at the estrogen-regulated TFF1 gene promoter prior to activation (Ju et al., 2006), was also detected in WT but not the PARP1 KD cell (Figure 4B). Furthermore, the presence of NCoR was required for transcriptional inhibition of Siah1, as the NCoR KD by two different shRNAs increased the Siah1 mRNA levels and the degree of the increases correlated with the extent of the KD (Figure 4C). Thus, PARP1 and NCoR likely act together to inhibit Siah1 transcription, a role that has been demonstrated previously on a number of other genes.
Figure 4. Mechanisms by which PARP1 Suppresses Siah1 Expression at Transcriptional and Protein Stability Levels: The Effect of PARP1 Depletion or Inhibition on Protein and mRNA Levels of Three Other Siah1 Substrates.
(A, B, and E) NH1 and the NH1-based PARP1 KD line 2–1 (A and B) or HeLa cells treated with DMSO or AZD2281 (E) were analyzed by ChIP-qPCR for the bindings of PARP1 and NCoR to the Siah1 gene promoter, with total mouse or rabbit IgG used as a negative control. The ChIP-qPCR signals were normalized to those of input DNA. Statistical significance was calculated from two-tailed Student’s t-test. *p < 0.05, **p < 0.005.
(C) Total RNAs isolated from HeLa cells expressing the indicated shRNAs were analyzed using qRT-PCR for the genes marked at the bottom. The signals were normalized to those of GAPDH and displayed.
(D and F) HeLa cells were treated with DMSO or AZD2281. Total RNAs were analyzed using qRT-PCR for the relative levels of Siah1 transcripts, which were normalized to those of GAPDH and shown (D). NEs were examined using WB for the indicated proteins (F).
(G and H) HeLa cells expressing WT Flag-Siah1 were treated with H2O2, pre-treated with MG132 and then followed by H2O2 (G), or treated with AZD2281 (H). WCE and anti-Flag immunoprecipitates (IPs) derived from WCE were examined using WB. Asterisks denote the positions of non-specific bands.
(I) HeLa cells were transfected with the same amount of plasmid expressing WT or the 4M mutant Flag-Siah1. WCE and anti-Flag IP derived from WCE wereexamined using WB. Also shown is an alignment between the RING domain of Siah1 and WWE domain of RNF146.
(J) Total RNAs isolated from HeLa cells either untransfected or transfected with the indicated expression constructs were analyzed using qRT-PCR as in (C).
(K and L) WCE from WT, the PARP1 KO (K), or the AZD2281-treated HeLa cells (L) were examined using WB for the indicated proteins.
(M) Total RNAs isolated from either WT or the PARP1 KO HeLa cells were analyzed using qRT-PCR as in (C)
The error bars in panels (A)–(E), (J), and (M) represent mean ± SD from three independent measurements. See also Figures S2–S4.
Furthermore, consistent with the reports that the PARP1 enzymatic activity is often dispensable for its transcriptional co-regulator function (Liu and Kraus, 2017), inhibition of PARP1 by AZD2281 did not elevate the Siah1 mRNA level (Figure 4D) and decrease the occupancy of PARP1 and NCoR at the Siah1 promoter (Figure 4E). This is different from our earlier results showing that the depletion of PARP1 by KD or KO increased the Siah1 mRNA level (Figures 3E and 3F). Thus, the physical presence but not catalytic activity of PARP1 at the Siah1 promoter was required for transcriptional inhibition. Nonetheless, given that the PARP1 enzymatic activity was required for maintaining the ELL2 protein level (Figure 2D), the above-described inhibition of Siah1 transcription by PARP1, although interesting and worth documenting, does not play a major role in controlling ELL2 level by the PARP1Siah1 axis.
PARP1 Promotes Siah1 Degradation Likely by Inducing PARdU of Siah1
As shown above, PARP1 also suppressed Siah1 expression at the protein level (Figures 3G and 3H). This effect depended on PARP1’s catalytic activity, because just like the KD of PARP1, inhibition by AZD2281 also elevated the Siah1 protein level in both HeLa (Figure 4F) and Jurkat 1G5 cells (Figure S2).
Siah1 is a RING-type E3 ubiquitin ligase that can catalyze selfubiquitination, leading to its own degradation (Liu et al., 2012). Recent studies have revealed how the RING-type E3 ubiquitin ligase RNF146 is activated by a PARylation signal (DaRosa et al., 2015; Wang et al., 2012). Iso-ADP-ribose (iso-ADPr), the smallest internal PAR structural unit, was shown to bind to the WWE and RING domains of RNF146 to function as an allosteric signal that switches the RING domain from an inactive to active state. In addition, RNF146 was found to bind directly to the PAR Pol tankyrase (TNKS).
To determine how PARP1 controlled the Siah1 protein level that depended on its enzymatic activity, we first tested whether Siah1 could bind to PARP1 and, if it could, whether the H2O2induced activation and auto-PARylation of PARP1 (Jungmichel et al., 2013) could enhance this binding. H2O2 treatment markedly decreased the Siah1 protein level in whole-cell extracts (WCE), which was blocked by pre-treating cells with the proteasome inhibitor MG132 (Figure 4G). This agrees with the idea that activated PARP1 decreases the Siah1 protein level by inducing degradation.
Although the overall Siah1 level decreased considerably in H2O2-treated cells, more total as well as the PARylated PARP1 became bound to the residual immunoprecipitated Siah1 (Figure 4G, lanes 6 and 7). Because the total PARP1 level in WCE was little affected by H2O2 (Figure 4G, lanes 2 and 3), this increased Siah1-PARP1 binding indicates that the binding was significantly enhanced by PARylation, which agrees with the model of PARdU-mediated Siah1 degradation. To confirm this from a different angle, we inhibited PARP1 with AZD2281 and found that even though the Flag-Siah1 level increased, the Siah1-PARP1 binding markedly decreased (Figure 4H), again indicating the dependence on PARylation for the binding.
Previous studies have identified amino acids Y144, Q153, and R163 in the RNF146 WWE domain as essential for RNF146’s binding to and activation by iso-ADPr (Wang et al., 2012; Zhang et al., 2011). In addition, K61 in the RING domain was also found to be key for PARdU (DaRosa et al., 2015). Although Siah1 does not have a canonical WWE domain, Y78, Q87, and R97 present in its RING domain are not only conserved but also display the same spacing as their counterparts in RNF146 (Figure 4I). These three residues, together with K99, the sole lysine in the entire Siah1 RING domain, were mutated to alanines to create a Siah1 mutant termed 4M. Compared with WT Siah1 expressed from the same amount of plasmid, 4M showed a significantly higher level of accumulation at the protein but not mRNA level (Figures 4I and 4J). More important, even though 4M was present ata higher level than WT Siah1 in WCE, it had drastically decreased binding to both total and the PARylated PARP1 (Figure 4I). These data are consistent with the model that PARP1 promotes Siah1 degradation likely through inducing PARdU of Siah1.
PARP1 Is Required for Stable Expression of Not Only ELL2 but Also Other Siah1 Substrates
Siah1 is an E3 ubiquitin ligase for not only ELL2 but also other proteins. Given that PARP1 controls the Siah1 expression, which in turn affects the ELL2 protein level, we asked whether three other known Siah1 substrates, ACK1 (activated Cdc42-associated kinase 1), CBP (CREB binding protein), and β-catenin (Qi et al., 2013) were similarly controlled by PARP1. Indeed, in both the PARP1 KO and AZD22 81-treated cells, the protein but not mRNA levels of the three substrates decreased to varying degrees, whereas the control proteins CDK9 and β-tubulin were unaffected (Figures 4–K4M). Notably, KD of Siah1 had no obvious effect on the expression of other known PARP1 target genes, such as NELL2 and Hsp70 (Figure S3). Collectively, these results have identified the PARP1-Siah1 axis as a key regulator of the expression of ELL2 and other Siah1 substrates.
DISCUSSION
Efficient HIV-1 replication in both human cell lines and primary monocyte-derived macrophages has been reported to rely on PARP1’s stimulation of HIV-1 transcription (Kameoka et al., 2004; Rom et al., 2015). Although the positive effect of PARP1 has been detected on the viral LTR, the precise mechanism by which PARP1 exerts this effect was unclear. In this study, we show that a PARP1-Siah1 axis promotes HIV-1 transcription, especially Tat transactivation, through elevating the cellular levels of ELL2 and ELL2-SEC. The latter is known to play an especially important role in mediating Tat transactivation (Li et al., 2016).
Notably, a recent study (Gibson et al., 2016) shows that PARP1 also stimulates transcription by modifying and inhibiting NELF, a negative elongation factor that pauses Pol II on genes under the elongation control. Thus, it is possible that both the PARP1-NELF pathway and the PARP1-Siah1 axis described here contribute to Pol II elongation on the HIV-1 proviral DNA. However, basal and PMA-activated HIV-1 transcription, both of which are Tat independent, was only mildly inhibited by the PARP1 KD (Figures 1A and 1B). In contrast, Tat transactivation, which strongly depends on ELL2-SEC, was more severely affected (Figure 1A). This key difference suggests that the PARP1-Siah1 axis that controls the ELL2-SEC formation plays a predominant role in controlling HIV-1 transcription, especially Tat transactivation, whereas the PARP1-NELF interplay may have only a minor role in this process.
When Jurkat 1G5 cells were exposed to PMA plus ionomycin, a combination that strongly activates T cells (Chatila et al., 1989), significantly elevated HIV-1 LTR activity was observed (Figure S4A). However, the treatment did not alter the PARP1 and PARP2 protein levels (Figure S4B). Thus, HIV-1 transactivation caused by T cell activation does not appear to involve an elevated PARP1/2 expression and the PARP1-Siah1 axis.
PARP1 has been shown to control transcription by binding to a sequence-specific motif at the promoter or other regions of its target genes in a few cases. In others, it uses alternative mechanisms such as forming co-regulatory complexes at the promoter and inducing chromatin remodeling, which can be dependent or independent of its catalytic activity (Kraus and Lis, 2003). In this study, we found that the transcriptional corepressor NCoR depends on PARP1 to bind to the Siah1 promoter region and also co-localizes with PARP1. Although NCoR was shown to be required for transcriptional repression of Siah1, the molecular details underlying its cooperation with PARP1 in this process remain to be elucidated.
PARylation has been shown to control the polyubiquitination and degradation of certain proteins through a mechanism called PARdU. So far, almost all known proteins regulated by PARdU are PARylated by the PAR polymerase (Pol) TNKS (Pellegrino and Altmeyer, 2016). Our data suggest that PARP1 could be another PAR Pol that can also use PARdU to activate Siah1. Consistently, a previous report shows that PAPR1 can bind and PARylate the E3 ligase UHRF1, which in turn facilitates UHRF1’s binding and polyubiquitination of its substrate DNMT1 (De Vos et al., 2014). In addition, PARP1 has also been reported to bind to the E3 ligase RNF146, which can target the PARylated PARP1 for proteasomal degradation (Kang et al., 2011). A major difference between our present finding and the well-studied TNKS-RNF146 case implicated in PARdU is that instead of PARylating an exogenous substrate, PARP1 can modify itself. The binding of Siah1 to this PAR moiety (probably through iso-ADPr) on PARP1 in turn activates the Siah1 RING domain, leading to the polyubiquitination and degradation Siah1’s exogenous substrates as well as Siah1 itself. Many details of this pathway are still unknown. Future studies will certainly shed more light on the intricate control and function of the PARP1-Siah1 axis especially during HIV-1 infection.
EXPERIMENTAL PROCEDURES
PARP1 KO
The HeLa-based PARP1 KO cell line was generated by using CRISPR-Cas9 with a single-guide RNA (sgRNA) (5’-CCACCTCAACGTCAGGGTGC-3’) that targets exon 2 of PARP1. The plasmid vector pSpCas9(BB)-2A-Puro (PX459), which expresses Cas9 and sgRNA, was from Addgene (plasmid 48139). Two days after transfection, cells were challenged with 2 μg/mL puromycin. The drug-resistant cells were diluted and allowed to grow into single colonies, which were subsequently examined for the loss of PARP1 expression by western blotting. The positive KO clones were verified by Sanger sequencing of the genomic amplicons obtained with the TA cloning kit (Life Technologies).
shRNA KD of PARP1
The shRNA constructs were annealed and ligated into the pSilencer vector (Life Technologies). Sequences for shPARP1–1-3 and shPARP1–2-1 are listed in Table S1. The procedures for shRNA-mediated gene KD have been described previously (He et al., 2008).
qRT-PCR Analyses
qRT-PCR reactions were performed as described previously (He et al., 2008). All reactions were run in triplicate. The primers used for the analyses are listed in Table S1.
Co-immunoprecipitation
Co-immunoprecipitation (coIP) was performed as described previously with minor changes. Briefly, 4 μg of CDK9 antibody was added to 700 mL nuclear extracts (NEs) from HeLa or PARP1 KD or KO cells and incubated overnight. Then, 25 μL protein A agarose beads (15918–014; Sigma-Aldrich) were added to the mixture for another 7 hr to collect the immunoprecipitated complex. After washing and elution, the immune complexes were analyzed using SDS-PAGE and western blotting with the indicated antibodies.
ChIP-qPCR Assay
The assay conditions were as previously described (Yang et al., 2005) with some modifications. Briefly, one 15 cm dish of NH1 WT and PARP1 KD cells at 100% confluency was harvested and subjected to the ChIP assay. Cells were lysed and sonicated by using a Covaris-S2 sonicator (Covaris) for a total processing time of 25 min (30 s on and 30 s off). For each immunoprecipitate (IP), 5 μg anti-PARP1 antibody (sc-74470X; Santa Cruz Biotechnology), 5 μg anti-NCoR antibody (A301–145A; Bethyl Laboratories), or 5 μg total rabbit IgG (sc-2025; Santa Cruz Biotechnology) was incubated with the diluted sheared chromatin. Twenty microliters of protein G Dyna beads (10003D; Life Technologies) or protein A Dyna beads (10002D; Life Technologies) were added to each tube and incubated at 4°C for 3 hr. After washing and elution, DNA was purified using the PCR purification kit and analyzed using qPCR.
Statistical Analysis
Statistical analysis was performed using two-tailed Student’s t-tests in Microsoft Excel 2010; p values < 0.05 were considered to indicate statistical significance in all analyses (*p < 0.05, **p < 0.005, ***p < 0.001).
Supplementary Material
Highlights.
PARP1 activity is required for optimal Tat activation of HIV-1 transcription
PARP1 increases levels of ELL2 protein and ELL2-SEC, key for Tat-transactivation
PARP1 and NCoR inhibit transcription of Siah1, an E3 ubiquitin ligase for ELL2
PARP1 induces proteosomal degradation of Siah1 likely through a PARdU mechanism
ACKNOWLEDGMENTS
This work is supported by grants from the National Key R&D Program of China (2018YFA0107303), the National Institutes of Health (R01A1041757), and the Tang Distinguished Scholarship.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.084.
REFERENCES
- Aguilar-Cordova E, Chinen J, Donehower L, Lewis DE, and Belmont JW (1994). A sensitive reporter cell line for HIV-1 tat activity, HIV-1 inhibitors, and T cell activation effects. AIDS Res. Hum. Retroviruses 10, 295–301. [DOI] [PubMed] [Google Scholar]
- Byun JS, Fufa TD, Wakano C, Fernandez A, Haggerty CM, Sung MH, and Gardner K (2012). ELL facilitates RNA polymerase II pause site entry and release. Nat. Commun 3, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila T, Silverman L, Miller R, and Geha R (1989). Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol 143, 1283–1289. [PubMed] [Google Scholar]
- DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, and Xu W (2015). Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADPribosyl)ation signal. Nature 517, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, El Ramy R, Qué net D, Wolf P, Spada F, Magroun N, Babbio F, Schreiber V, Leonhardt H, Bonapace IM, and Dantzer F (2014). Poly(ADP-ribose) polymerase 1 (PARP1) associates with E3 ubiquitin-protein ligase UHRF1 and modulates UHRF1 biological functions. J. Biol. Chem 289, 16223–16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, and Kraus WL (2016). Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Mé nissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JHJ, and de Murcia G (1990). The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl. Acad. Sci. USA 87, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, and Zhou Q (2008). A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 29, 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, and Zhou Q (2010). HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38, 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House CM, Mo¨ ller A, and Bowtell DDL (2009). Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 69, 8835–8838. [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, and Rosenfeld MG (2006). A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802. [DOI] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, and Nielsen ML (2013). Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 52, 272–285. [DOI] [PubMed] [Google Scholar]
- Kameoka M, Nukuzuma S, Itaya A, Tanaka Y, Ota K, Ikuta K, and Yoshihara K (2004). RNA interference directed against Poly(ADP-Ribose) polymerase 1 efficiently suppresses human immunodeficiency virus type 1 replication in human cells. J. Virol 78, 8931–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, et al. (2011). Iduna is a poly(ADP-ribose) (PAR)dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA 108, 14103–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL (2008). Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol 20, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, and Lis JT (2003). PARP goes transcription. Cell 113, 677–683. [DOI] [PubMed] [Google Scholar]
- Li Z, Lu H, and Zhou Q (2016). A minor subset of super elongation complexes plays a predominant role in reversing HIV-1 latency. Mol. Cell. Biol 36, 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, and Shilatifard A (2010). AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Kraus WL (2017). Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 65, 589–603.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Hsu J, Chan C, Li Z, and Zhou Q (2012). The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol. Cell 46, 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Li Z, Xue Y, and Zhou Q (2013). Viral-host interactions that control HIV-1 transcriptional elongation. Chem. Rev 113, 8567–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino S, and Altmeyer M (2016). Interplay between ubiquitin, SUMO, and poly(ADP-ribose) in the cellular response to genotoxic stress. Front. Genet 7, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Kim H, Scortegagna M, and Ronai ZA (2013). Regulators and effectors of Siah ubiquitin ligases. Cell Biochem. Biophys. 67, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli V, O’Farrell M, Me´ nissier-de Murcia J, and de Murcia G. (1997). Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry 36, 12147–12154. [DOI] [PubMed] [Google Scholar]
- Rom S, Reichenbach NL, Dykstra H, and Persidsky Y (2015). The dual action of poly(ADP-ribose) polymerase −1 (PARP-1) inhibition in HIV-1 infection: HIV-1 LTR inhibition and diminution in Rho GTPase activity. Front. Microbiol 6, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, and Conaway JW (1996). An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Duan DR, Haque D, Florence C, Schubach WH, Conaway JW, and Conaway RC (1997). ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc. Natl. Acad. Sci. USA 94, 3639–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, and Benkirane M (2010). HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 38, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, and Xu W (2012). Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Lowe AD, and Karn J (2001). Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J. Virol 75, 8524–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, and Zhou Q (2001). The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, and Zhou Q (2005). Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, et al. (2011). RNF146 is a poly(ADP-ribose)directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol 13, 623–629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.