Abstract
Background
Nasopharyngeal carcinoma (NPC) is a poorly differentiated malignant tumor, and 5-fluorouracil (5-FU) is one of the most effective chemotherapeutic drugs used for the treatment of NPC. Abnormal expression of RGS17 had been shown to improve the sensitivity of many cancers to chemotherapy; however, the effects of RGS17 on NPC remain unclear.
Methods
We cultured NPC cell lines and altered the RGS17 expression with vector. Subsequently colony formation assays and CCK8 cell viability assay was used to test the proliferation of NPC cells, flow cytometry was used to determine the percentage of apoptotic cells, MMP kit and flow cytometry was used to measure the mitochondrial membrane potential, and a xenograft tumour model was attached to investigate the effects of RGS17 on the growth of NPC cells in vivo. Additionally, RT-PCR and western blot was induced to examine the expression of RGS17 and the mechanism.
Results
Here, we report for the first time that RGS17 is downregulated in NPC cell lines and that RGS17 overexpression significantly reduces cell proliferation, decreases the mitochondrial membrane potential, and induces cell apoptosis in NPC cells. In vivo, RGS17 also inhibits the tumorigenicity of NPC. In addition, RGS17 could significantly improve the sensitivity of NPC cells to 5-FU. Furthermore, investigation into the underlying mechanisms showed that RGS17 upregulated the levels of IRE1α, p53, and active caspase-3 and cleaved PARP.
Conclusion
These results indicate that RGS17 could play important roles in the proliferation, apoptosis, and chemotherapeutic sensitivity of NPC cells.
Keywords: nasopharyngeal carcinoma, RGS17, cell apoptosis, mitochondrial membrane potential, fluorouracil chemotherapy
Introduction
Nasopharyngeal carcinoma (NPC) is associated with infection caused by a ubiquitous human herpesvirus, the Epstein–Barr virus (EBV),1 and it is more prevalent in Southern China, South Asia, North Africa, and the Arctic than in other parts of the world.2 Generally, traditional radiation therapy has been the most common treatment for NPC;3 however, the 5-year survival rate of patients with NPC is only approximately 60% in both adults and children.4 The molecular pathogenesis of NPC is a complicated process including the inactivation of tumor suppressor genes (TSGs). Thus, it is critical to identify novel TSGs and their related molecular mechanisms to search for more effective therapeutic strategies for the treatment of NPC.
G protein-coupled receptors (GPCRs) are the largest class of proteins in the human genome, and they are signal transducers. G proteins are heterotrimers consisting of α and βγ subunits. The regulators of G protein signaling (RGS) proteins bind directly to the Gα subunits to regulate the signaling functions of Gα.5 The mammalian family of RGS proteins is characterized by the RGS homology (RH) domain and contains more than 30 members that are divided into eight subfamilies according to the shared sequence identities found outside the RH domain.6 RGS17, an RGS member, could regulate Gα functioning and accelerate the rate of GTPase activity by the Gα subunits.7
Previous studies have shown that RGS17 expression is downregulated in patients with acute myelogenous lymphoma (AML) and ovarian cancer, in whom RGS17 acts as a TSG and improves the sensitivity of the cancer to chemotherapy agents. However, the function of RGS17 in patients with NPC is unclear. In this study, we used NPC cells and a mouse model to investigate the roles played by RGS17 in NPC, and we preliminarily explored the underlying mechanism.
Materials and methods
Cell lines and culture conditions
Human immortalized nasopharyngeal NP69 cells and three NPC cell lines (CNE2, CNE1, and HNE1) were obtained from Sun Yat-sen University (Guangzhou, China). The use of the cell lines has been approved by the Qingdao University Ethics Committee. The cell lines were maintained at 37°C in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS.
RNA extraction and semiquantitative RT-PCR
Total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and dissolved in diethyl pyrocarbonate (DEPC)-treated water according to the manufacturer’s instructions. Reverse transcription was performed with 1 µg total RNA. The mRNA expression levels of RGS17 were determined by semiquantitative RT-PCR with the GoTaq polymerase (Promega Corporation, Fitchburg, WI, USA). The transcription of the principal gene β-actin (forward primer, CATCCTCACCCTGAAGTACCC; reverse primer, AGCCTGGATGCAACGTACATG) was used as the internal control. Specific primers were designed according to the RGS17 sequence (RGS17 forward primer, AAGGCAGCAGTCCCAAAATG; RGS17 reverse primer, CCCGCATTTTCCCCT CTTTC).
Protein extraction and Western blot
Cells were harvested from the culture dishes and lysed in RIPA buffer (Beyotime, Hangzhou, Zhejiang, China) supplemented with protease inhibitors. The transfected cell proteins were collected and stored at −80°C after being centrifuged at 12,000× g for 15 minutes. Protein content was determined using bicinchoninic acid assay (BCA; Thermo Fisher Scientific). Cell lysates (40 µg protein/line) were separated on a 15% Tris-Tricine Ready Gel SDS-PAGE and transferred on to a nitrocellulose membrane (Bio-Rad Laboratories Inc., Hercules, CA, USA). After being blocked in 5% skim milk for 1 hour, the blotted membranes were incubated overnight at 4°C with the appropriate primary antibodies and then incubated with horseradish peroxidase – labeled secondary antibody (1:2,000; Cell Signaling Technology, Danvers, MA, USA) for 1 hour. The proteins were detected with chemiluminescent autoradiography (ECL Kit; Thermo Fisher Scientific). The band densities of the Western blots were quantified using Quantity One V4.62 software (Bio-Rad Laboratories Inc.).
Colony formation assays
For the colony formation assays, 1,000 cells were planted in a 10 cm diameter dish and allowed to grow for 2 weeks at 37°C in 5% CO2. The surviving colonies (≥50 cells/colony) were counted under a microscope after Giemsa staining. The experiments were performed in triplicate.
Cell viability assay
The cell viability assay was performed using the CCK8 Detection Kit (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. Cells were seeded in a 96-well plate at a density of 4,000 cells/well. The absorbance was measured on a microplate reader (Synergy H4 Hybrid Reader; BioTek Instruments, Inc., Winooski, VT, USA) at a wavelength of 450 nm. The experiments were performed in triplicate.
Cell apoptosis analysis
Flow cytometry was used to determine the percentage of apoptotic cells using the Pharmingen Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit I (BD Pharmingen, San Jose, CA, USA) according to the manufacturer’s instructions. Briefly, 5×105 cells were seeded into a six-well plate. After attachment overnight, 2.5 mg/L 5-fluorouracil (5-FU) was added at the concentrations indicated, and the cells were incubated for 24 hours. All cells, including the cells floating in the culture medium, were harvested. The cells were incubated with FITC Annexin V and propidium iodide (PI) for 15 minutes and then analyzed with a FACSCalibur Flow Cytometer (BD Biosystems, Heidelberg, Germany).
Measurement of the mitochondrial membrane potential (MMP)
An MMP Assay Kit (BestBio Co., Jiangsu, China) with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine iodide (JC-1) was used to determine the MMP. First, 5×105 cells were seeded into a six-well plate. After attachment overnight, 2.5 mg/L 5-FU was added, and the cells were incubated for 24 hours. The cells were collected and then incubated with 0.5 mL of JC-1 working solution for 20 minutes at 37°C before being washed twice, suspended in JC-1 buffer solution, and analyzed by flow cytometry (BD Biosystems). The experiments were performed in triplicate.
In vivo subcutaneous tumor model
All of the in vivo experimental protocols were approved by the animal care committee of Qingdao University. The guideline of the experiment for the welfare of the animals was followed by Institutional Animal Care and Use Committee (IACUC) of Qingdao University. First, 5×106 cells were injected subcutaneously into the flank of 5-week-old female BALB/c nude mice (six mice per group). Tumor volume was calculated by the following formula:
Tumors that formed in vivo were collected and embedded in paraffin for evaluation by immunohistochemistry (IHC). Proliferating cell nuclear antigen (PCNA) staining and RGS17 staining were performed by the pathology department.
Statistical analyses
Statistical analyses were performed using GraphPad Prism7 software (GraphPad Software, Inc., La Jolla, CA, USA). The data were analyzed using Student’s t-tests. For all experiments, P-values <0.05 were considered statistically significant.
Results
RGS17 was downregulated in NPC cell lines
First, we characterized the expression levels of RGS17 in human immortalized nasopharyngeal NP69 cells and three NPC cell lines (CNE2, CNE1, and HNE1). The Western blot results showed that NP69 cells exhibited a higher expression level of RGS17 (Figure S1A) than the levels observed in the three NPC cell lines, indicating that RGS17 might be downregulated in most NPC cell lines. To further investigate the biofunction of RGS17, we transfected CNE1 and HNE cells with the RGS17 expression plasmid. All the transfected cell lines were confirmed by semiquantitative RT-PCR (Figure S1B) and Western blotting (Figure S1C).
RGS17 inhibited colony formation and proliferation of NPC cells in vitro
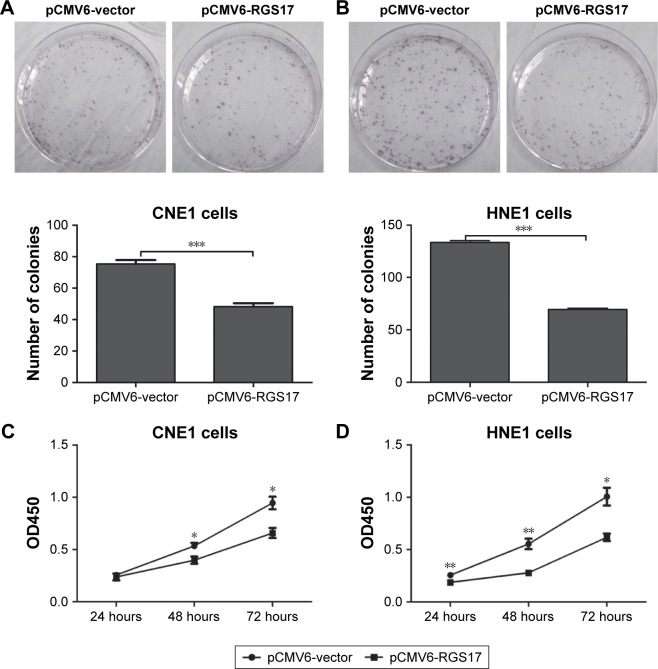
First, we investigated the effects of RGS17 expression on the proliferation of CNE1 and HNE cells in vitro by colony formation assays and cell viability assays, respectively. Colony formation assays showed that compared to control cells, cells overexpressing RGS17 formed significantly fewer colonies (Figure 1A and B). Similarly, the cell viability assays revealed that RGS17 significantly suppressed the proliferation of NPC cells (Figure 1C and D). These data indicate that RGS17 could suppress the colony formation and proliferation of NPC cells.
Figure 1.
RGS17 inhibited the proliferation of NPC cells.
Notes: (A and B) Colony formation assays were performed in NPC cells transfected with pCMV6-vector or pCMV6-RSG17 plasmids. Values were calculated and compared as shown in the graphs. (C and D) The cell viability of transfected CNE1 and HNE1 cells was measured at different time points. The data are presented as the mean ± SD. *P<0.05; **P<0.01; ***P<0.001.
Abbreviation: NPC, nasopharyngeal carcinoma.
RGS17 affected the apoptosis and MMP of NPC cells in vitro
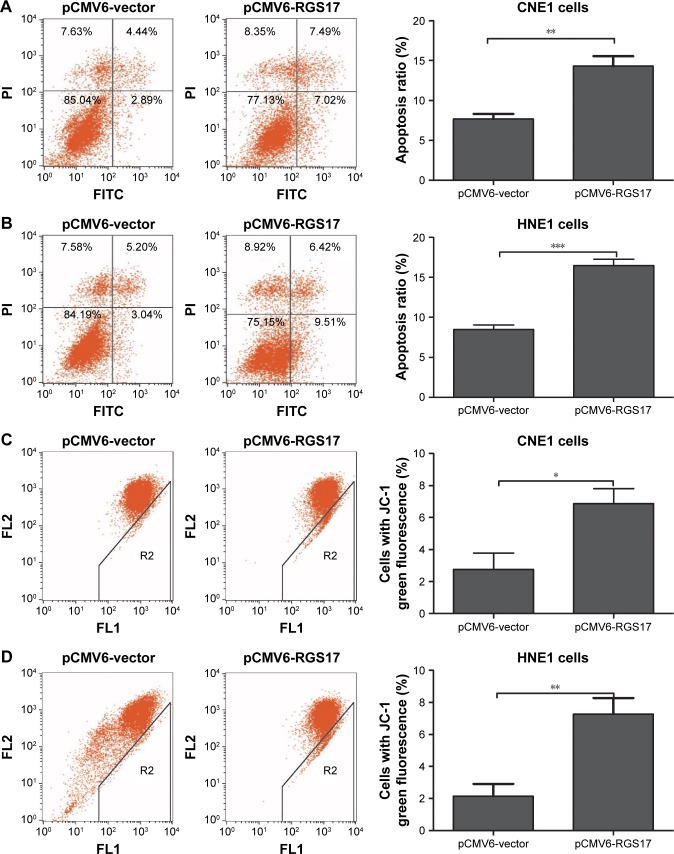
Then, we evaluated the role of RGS17 expression on the apoptosis of NPC cells. The results revealed that the rate of spontaneous apoptosis of cells overexpressing RSG17 were significantly higher than that of cells transfected with the pCMV6-vector (Figure 2A and B).
Figure 2.
RGS17 affected the apoptosis and mitochondrial membrane potential of NPC cells.
Notes: (A and B) Representative results of annexin V-FITC/PI staining and quantitative analyses of CNE1 and HNE1 cells. (C and D) Representative results of mitochondrial membrane potential and quantitative analyses of CNE1 and HNE1 cells. The values are represented as mean ± SD of three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: FITC, fluorescein isothiocyanate; NPC, nasopharyngeal carcinoma; PI, propidium iodide.
RGS17 enhanced the apoptosis of NPC cells. A decrease in the MMP is one of the establishing events of early apoptosis. We then tested the proportion of cells in early apoptosis by determining the proportion of cells with a lower MMP. Compared with cells transfected with the pCMV6-vector, there was a significantly higher percentage of pCMV6-RGS17 cells in early apoptosis (Figure 2C and D), indicating that RGS17 could enhance the proportion of cells in early apoptosis by decreasing the MMP.
RGS17 inhibited tumor growth in vivo
To strengthen our conclusion, we investigated the effects of RGS17 on the growth of NPC cells in vivo in a xenograft tumor model. Approximately 1×107 cells were injected subcutaneously into BALB/c nude mice (six mice per group). The average tumor size of pCMV6-RGS17 group was significantly smaller than that of the pCMV6-vector group (Figure 3A and B).
Figure 3.
RGS17 inhibited the tumorigenicity of NPC cells in vivo.
Notes: (A) Representative images of the in vivo subcutaneous tumor model. (B) Tumor volumes were compared in nude mice injected with cells transfected with CNE1-RGS17 and CNE1-vector plasmids. (C) Tumors formed in vivo were excised and embedded in paraffin for evaluation by IHC. The proportions of PCNA- and RSG-17-positive cells were measured in the pCMV6-vector and pCMV6-RGS17 groups. ***P<0.0016.
Abbreviations: IHC, immunohistochemistry; NPC, nasopharyngeal carcinoma; PCNA, proliferating cell nuclear antigen.
Tumors formed in vivo were acquired and embedded in paraffin for evaluation by IHC. The proportion of cells positive for PCNA, a cell proliferation marker, was higher in the pCMV6-vector group than in the pCMV6-RGS17 group (Figure 3C).
RGS17 increased the sensitivity of NPC cells to 5-FU
Since 5-FU is one of the most common chemotherapeutic drugs used in the clinical treatment of NPC patients, we further investigated the influence of RGS17 on the proliferation, apoptosis, and MMP of NPC cells treated with 5-FU.
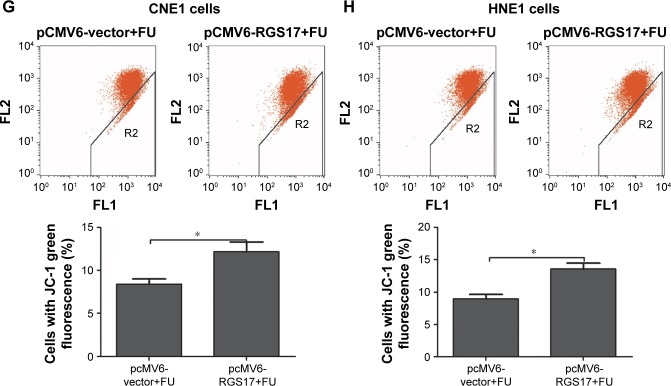
Our results showed that RGS17 overexpression significantly inhibited the proliferation, promoted the apoptosis, and increased the MMP of NPC cells treated with 5-FU (Figure 4A–H). These data suggest that RGS17 overexpression could increase the sensitivity of NPC cells to 5-FU.
Figure 4.
RGS17 affected the sensitivity of NPC cells to 5-FU.
Notes: (A and B) Colony formation assays were performed in transfected NPC cells treated with 2.5 mg/mL 5-FU. Values were calculated and compared (graphs on the right-hand side). (C and D) The cell viability of the transfected NPC cells treated with 2.5 mg/mL 5-FU was measured at different time points. (E and F) Apoptosis assays were performed with transfected NPC cells treated with 2.5 mg/mL 5-FU. Values were calculated and compared (graphs below). (G and H) Mitochondrial membrane potential was measured in transfected NPC cells treated with 2.5 mg/mL 5-FU. Values were calculated and compared (graphs on the right-hand side). The values are represented as mean ± SD of three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: NPC, nasopharyngeal carcinoma; 5-FU, 5-fluorouracil.
The potential mechanism of the inhibitory effects of RGS17 on NPC cells
As indicated above, RGS17 could act as a TSG in NPC cells by inhibiting proliferation and inducing apoptosis. Therefore, we investigated the potential mechanisms underlying the inhibitory effects of RGS17 on NPC cells.
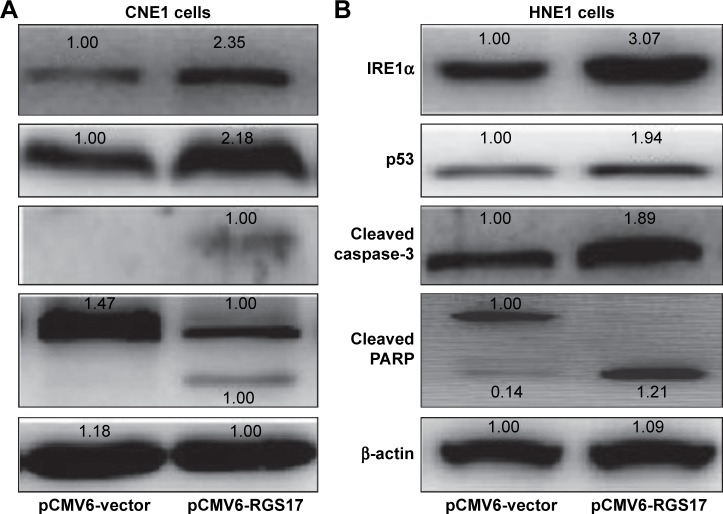
The results showed that RGS17 upregulated the expression of the endoplasmic reticulum (ER) stress response sensor IRE1α (Figure 5A and B). Since the IRE1α pathway plays important roles in the regulation of cell apoptosis,8 we then examined the expression levels of key regulators of the apop-totic pathway. Some proapoptotic proteins, including p53, cleaved PARP, and cleaved caspase-3, were all upregulated in cells transfected with RGS17 (Figure 5A and B).
Figure 5.
RGS17 affected the expression levels of ER stress response genes and cell apoptosis genes.
Note: The expression levels of IRE1α, p53, cleaved PARP, and cleaved caspase-3 were measured in CNE1 cells (A) and HNE1 cells (B) by Western blot.
Abbreviations: ER, endoplasmic reticulum; PARP, poly ADP-ribose polymerase.
Discussion
In this study, our results showed that RGS17 was downregulated in NPC cells. RGS17 expression could be precisely regulated by various factors such as DNA copy number alterations, DNA hypermethylation, miRNA inhibition, and protein degradation. Different patterns of regulation of RGS17 expression have been reported in various cancers. For instance, a significant reduction in the number of DNA copies and hypermethylation of the regulatory region of the RGS17 gene were observed in hepatocellular carcinoma (HCC) cells.9 Furthermore, in breast cancer cells, RGS17 was identified as a direct and functional target of miR-32.10
An increasing number of studies have revealed that RGS proteins could play important roles in the progression of various cancers such as RGS1 in melanoma,11 RGS2 in breast cancer,12 RGS3 in lung cancer,13 RGS5 in acute myeloid leukemia,14 RGS8 in breast cancer,15 RGS19 in ovarian cancer,16 and RGS20 in melanoma.17 These studies identified unique RGS-related targets that have the potential to lead to advances in cancer treatment, including new avenues to explore to identify novel anticancer therapies and drug options for patients with cancer.
In this study, we revealed that RGS17 may act as a tumor suppressor in NPC cells. Overexpression of RGS17 in CNE1 or HNE1 cells significantly attenuated cell viability and the colony-forming ability and induced cell apoptosis. It has been reported that cell proliferation is affected by the crosstalk among apoptosis, autophagy, and necroptosis.18 For instance, Feng et al19 reported that the knockdown of YY1 expression resulted in cell viability inhibition by blocking autophagy rather than influencing apoptosis. These results indicated that RGS17 might also inhibit cell viability and the colony-forming ability through other pathways such as autophagy or necroptosis, which is a suggestion that needs to be further investigated. The mechanisms underlying NPC tumorigenesis are complex, involving aberrations in a large variety of signaling pathways that are related to multiple abnormalities including the upregulation of cellular proliferation, abnormal cell adhesion, and aberrations in cell apoptosis.20 Further in vitro studies need to be conducted to determine the mechanisms by which RGS17 acts in NPC tumorigenesis. In ovarian cancer, knockdown of RGS17 is sufficient to increase cell survival and decrease cell sensitivity to drug treatment, while the overexpression of RGS17 leads to increased sensitivity to chemotherapeutic agents.21 In our study, RSG17 increased the sensitivity of NPC cells to the chemotherapeutic agent 5-FU.
The ER is a dynamic tubular network involved in metabolic processes. When cells encounter difficult conditions such as ER stress, they can activate a series of complementary adaptive mechanisms. If ER stress is not mitigated and homeostasis is not restored, cell apoptosis occurs.22 Interestingly, one of the three major ER stress sensors, IRE1α, was upregulated in NPC cells overexpressing RGS17. IRE1α could degrade certain mRNAs and activate the “alarm stress pathways” driven by JNK that play important roles in cell apoptosis.22 Furthermore, p53 is a key regulator of apoptosis, controlling the transcription of many apoptosis-related genes, and our research revealed that RGS17 also upregulates the expression of p53. All these results suggest that RGS17 acts as a tumor suppressor and might have the potential to become a new therapeutic target in the treatment of NPC. However, further study is still needed to reveal the precise mechanisms by which RGS17 acts in NPC tumorigenesis.
As the expression levels of RGS17 at the gene and protein levels have not been fully investigated, contradictory findings may be reported. Conversely, the aforementioned databases have demonstrated a different correlation between RGS17 mRNA/protein expression and prognoses in HCC,23 non-small cell lung cancer,24 acute myeloid leukemia,25 and breast cancer.10 Therefore, further investigation is required to understand these differing expression profiles.
Supplementary material
Analysis of RGS17 expression levels in the human immortalized nasopharyngeal cell line NP69 and three NPC cell lines. (A) The expression levels of RGS17 in the NP69 cells and the three NPC cell lines (CNE2, CNE1, and HNE1) according to Western blotting. (B) The RGS17 expression level in transfected cells was evaluated by reverse transcription PCR in CNE1 and HNE1 cells. (C) The RGS17 expression levels in transfected cells were confirmed by Western blotting in CNE1 and HNE1 cells.
Abbreviation: NPC, nasopharyngeal carcinoma.
Acknowledgments
This work was supported by grants from the Funding for Excellent Departments (Qingdao University).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yan F, Wang M, Chen H, et al. Gambogenic acid mediated apoptosis through the mitochondrial oxidative stress and inactivation of Akt signaling pathway in human nasopharyngeal carcinoma CNE-1 cells. Eur J Pharmacol. 2011;652(1–3):23–32. doi: 10.1016/j.ejphar.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 2.Razak AR, Siu LL, Liu FF, et al. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46(11):1967–1978. doi: 10.1016/j.ejca.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Peng G, Wang T, Yang KY, et al. A prospective, randomized study comparing outcomes and toxicities of intensity-modulated radiotherapy vs. conventional two-dimensional radiotherapy for the treatment of nasopharyngeal carcinoma. Radiother Oncol. 2012;104(3):286–293. doi: 10.1016/j.radonc.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Marcus KJ, Tishler RB. Head and neck carcinomas across the age spectrum: epidemiology, therapy, and late effects. Semin Radiat Oncol. 2010;20(1):52–57. doi: 10.1016/j.semradonc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002;520(1–3):97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 6.Cho H, Kehrl JH. Regulation of immune function by G protein-coupled receptors, trimeric G proteins, and RGS proteins. Prog Mol Biol Transl Sci. 2009;86:249–298. doi: 10.1016/S1877-1173(09)86009-2. [DOI] [PubMed] [Google Scholar]
- 7.Mao H, Zhao Q, Daigle M, et al. RGS17/RGSZ2, a novel regulator of Gi/o, Gz, and Gq signaling. J Biol Chem. 2004;279(25):26314–26322. doi: 10.1074/jbc.M401800200. [DOI] [PubMed] [Google Scholar]
- 8.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Lefave C, Sirosh I, et al. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med Genomics. 2015;8:28. doi: 10.1186/s12920-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Li L, Lin J, et al. Deregulation of RGS17 expression promotes breast cancer progression. J Cancer. 2015;6(8):767–775. doi: 10.7150/jca.11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grünebach F, Erndt S, Häntschel M, Heine A, Brossart P. Generation of antigen-specific CTL responses using RGS1 mRNA transfected den-dritic cells. Cancer Immunol Immunother. 2008;57(10):1483–1491. doi: 10.1007/s00262-008-0486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyu JH, Park DW, Huang B, et al. RGS2 suppresses breast cancer cell growth via a MCPIP1-dependent pathway. J Cell Biochem. 2015;116(2):260–267. doi: 10.1002/jcb.24964. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Wu Y, Meng Q, Xia Z. Elevated microRNA-25 inhibits cell apoptosis in lung cancer by targeting RGS3. In Vitro Cell Dev Biol Anim. 2016;52(1):62–67. doi: 10.1007/s11626-015-9947-2. [DOI] [PubMed] [Google Scholar]
- 14.Boss CN, Grünebach F, Brauer K, et al. Identification and characterization of T-cell epitopes deduced from RGS5, a novel broadly expressed tumor antigen. Clin Cancer Res. 2007;13(11):3347–3355. doi: 10.1158/1078-0432.CCR-06-2156. [DOI] [PubMed] [Google Scholar]
- 15.Wiechec E, Wiuf C, Overgaard J, Hansen LL. High-resolution melting analysis for mutation screening of RGSL1, RGS16, and RGS8 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(2):397–407. doi: 10.1158/1055-9965.EPI-10-0514. [DOI] [PubMed] [Google Scholar]
- 16.Hurst JH, Mendpara N, Hooks SB. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell Mol Biol Lett. 2009;14(1):153–174. doi: 10.2478/s11658-008-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riker AI, Enkemann SA, Fodstad O, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Yang Z, Li Y, et al. Cell apoptosis, autophagy and necroptosis in osteosarcoma treatment. Oncotarget. 2016;7(28):44763–44778. doi: 10.18632/oncotarget.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng L, Ma Y, Sun J, et al. YY1-MIR372-SQSTM1 regulatory axis in autophagy. Autophagy. 2014;10(8):1442–1453. doi: 10.4161/auto.29486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma – review of the molecular mechanisms of tumorigenesis. Head Neck. 2008;30(7):946–963. doi: 10.1002/hed.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooks SB, Callihan P, Altman MK, et al. Regulators of G-protein sig-naling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells. Mol Cancer. 2010;9:289. doi: 10.1186/1476-4598-9-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 23.Sokolov E, Iannitti DA, Schrum LW, Mckillop IH. Altered expression and function of regulator of G-protein signaling-17 (RGS17) in hepatocellular carcinoma. Cell Signal. 2011;23(10):1603–1610. doi: 10.1016/j.cellsig.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi Y, Jin Q, Liu X, et al. miR-203 inhibits cell proliferation, invasion, and migration of non-small-cell lung cancer by downregulating RGS17. Cancer Sci. 2017;108(12):2366–2372. doi: 10.1111/cas.13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosakhani N, Räty R, Tyybäkinoja A, et al. MicroRNA profiling in chemoresistant and chemosensitive acute myeloid leukemia. Cytogenet Genome Res. 2013;141(4):272–276. doi: 10.1159/000351219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of RGS17 expression levels in the human immortalized nasopharyngeal cell line NP69 and three NPC cell lines. (A) The expression levels of RGS17 in the NP69 cells and the three NPC cell lines (CNE2, CNE1, and HNE1) according to Western blotting. (B) The RGS17 expression level in transfected cells was evaluated by reverse transcription PCR in CNE1 and HNE1 cells. (C) The RGS17 expression levels in transfected cells were confirmed by Western blotting in CNE1 and HNE1 cells.
Abbreviation: NPC, nasopharyngeal carcinoma.