Abstract
Background
Human transmembrane protein 176A (TMEM176A) is upregulated in several tumors. Growing evidence has suggested the high clinical value of TMEM176A as a biomarker for early tumor diagnosis. However, less is known about the function of TMEM176A in glioblastomas (GBMs).
Methods
In this study, we systematically analyzed the effect of TMEM176A knockdown and overexpression in GBM cells (U87, T98G and A172) on cell proliferation, cell cycle and cell apoptosis.
Results
Our results indicated that TMEM176A acted as a tumor-promoting factor in GBM cells. Moreover, a specific ERK1/2 inhibitor, U0126, suppressed the function of TMEM176A in GBM cells. Therefore, we proposed that TMEM176A may be involved in a pathway including ERK1/2 in the regulation of the cell cycle. Moreover, we also found that TMEM176A affected the expression of Bcl2 and played a central role in apoptosis of GBM cells.
Conclusion
Taken together, our results not only elucidated the multiple functions of TMEM176A in GBM cells but also provided a deep insight into the potential targets of TMEM176A in the growth of GBM cells.
Keywords: TMEM176A, cell cycle, cell apoptosis, ERK1/2, glioblastomas
Introduction
Glioblastomas (GBMs) are one of the most malignant brain tumors worldwide and are most commonly diagnosed in adults.1 More than half of the sufferers of GBM die within 1 year of the diagnosis. Much attention has been directed toward discovering effective therapies for GBM; however, the survival rate of GBM patients is still very low.2–4 Therefore, a better understanding of the key factors related to the mechanisms of GBM is urgently needed.
Human transmembrane protein 176A (TMEM176A) was mapped to human chromosome 7q36.1, which belongs to the TMEM family. Although the functions of TMEM176A are not well known in the context of cancers, growing reports indicated the potential value of TMEM176A as a useful biomarker for tumors. TMEM176A inhibits the growth of esophageal cancer cells in vivo and in vitro and acts as a diagnostic and prognostic biomarker in esophageal squamous cell cancer (ESCC).5 Moreover, reports have shown that dysregulation of TMEM176A is linked with cancer pathology, which also suggests the high potential value of TMEM176A in the treatment of certain cancers.6 Additionally, research focused on GBM has demonstrated that the knockdown of TMEM14A and TMEM45A suppresses the proliferation, migration, and invasion of glioma cells.7,8 Moreover, TMEM97 has been reported as a potential therapeutic target in GBM.9 However, the function of TMEM176A in GBM has scarcely been reported; therefore, it is meaningful to determine the functional characteristics of TMEM176A in GBM.
Cyclin D1 has been reported as an essential positive regulator of the cell cycle,10 and alteration of Cyclin D1 can influence cell cycle progression. The upregulation of Cyclin D1 promotes G1/S progression, which contributes to tumorigenesis.11 Moreover, high expression of Cyclin D1 is associated with an increased risk of mortality from breast cancer.12 Additionally, Cyclin D1 has been reported as a key target in treating cancer13 and has been regarded as a strong prognostic marker for cancers. Moreover, the expression of Cyclin D1 is upregulated in GBM cells compared with normal brain tissue and has been shown to be regulated by MiR-17 to affect cell viability and migration.14 In addition, it was previously reported that Cyclin D1 is targeted by MiR-15b in the regulation of GBM cell proliferation and apoptosis.3 Taken together, these findings indicate that Cyclin D1 is essential in the regulation of GBM cell development. Notably, in a previous study, the downregulation of Cyclin D1 was found to silence the expression of TMEM14A in human ovarian cancer cells.8 However, the homolog of TMEM14A remains unknown in GBM. Therefore, it is valuable to examine the relationship between TMEM176A and Cyclin D1 in GBM.
Previous reports have highlighted that the Cyclin D1/P21 signaling pathway plays a critical role in tumor growth and tumor cell invasion.15 Moreover, the Cyclin D1/P21 pathway is also important in the proliferation of GBM cells.16 P21, a universal inhibitor of Cyclin-dependent kinases (CDKs), negatively regulates the progression of the cell cycle. Research has suggested that P21 promotes the development of Proneural Glioma through tyrosine phosphorylation.17 Meanwhile, certain factors contribute to GBM by negatively regulating P21,18 and it is doubtful that those factors accelerate the phosphorylation of P21. However, whether TMEM176A affects the phosphorylation in GBM cells warrants further study.
The formation of a P21/Caspase-3 complex has been demonstrated to block the pro-Caspase-3 cleavage site and the subsequent activation of Caspase-3, resulting in the prevention of apoptosis.19 Caspase-3, which belongs to the cysteine protease family, plays a key role in apoptosis.20 Aberrant expression of Caspase-3 has often been reported in certain human cancers. For breast cancer, the expression level of Caspase-3 is negatively correlated with the overall survival rate.21,22 Moreover, it has been reported that Caspase-3 inhibits chemical-induced hepatocarcinogenesis.23 Additionally, a previous report indicated that Caspase-3 is involved in the metabolism of GBM cells.24
B-cell lymphoma protein 2 (Bcl2) acts as an antiapoptotic factor in the intrinsic apoptosis pathway.25 Reports have shown that Bcl2 is also essential for cell cycle progression.26 The dysregulation of Bcl2 not only disrupts normal cell development but also contributes to tumor progression.25 Some reports have indicated that Bcl2 is the primary target for microRNA-16 in the inhibition of glioma cell growth and invasion.27,28 BCL2L12 is a newly identified member of the Bcl2 family, which exhibits a substantially elevated expression in chronic lymphocytic leukemia patients.29 Moreover, BCL2L12 mediates the development of GBM through regulating Caspase-3 and Caspase-7.24 Hence, Bcl2 plays a central role in the metabolism of GBM. However, little is known about whether TMEM176A affects the expression of Bcl2. Extracellular signal-regulated kinase 1/2 (ERK1/2) belong to the mitogen-activated protein kinase (MAPK) group, which is related to the regulation of cell cycle progression, proliferation, and apoptosis.30 It has been reported that the apoptosis regulators Bcl2 and Caspase-3 can down-regulate the expression of ERK.31 The ERK1/2 signaling pathway has an important function related to the cell cycle checkpoint.32 Dysregulation of ERK1/2 through phospho-rylation (p-ERK1/2) contributes to cell development in gliomas.33 Moreover, a previous report has indicated that cell proliferation in gliomas is negatively regulated by ERK1/2 activity.34 Hence, the major function of ERK1/2 in gliomas is dependent upon the phosphorylation modification.
To examine the role of TMEM176A and its possible targets in GBM, RNA interference was used to knockdown the expression of TMEM176A in GBM cells. Meanwhile, a lentiviral vector was used to overexpress TMEM176A. Our data not only provide a deep understanding of the molecular mechanisms underlying TMEM176A but also provide the basis for determining the potential targets of TMEM176A in the growth of GBM cells.
Materials and methods
Tissue specimens and cell culture
GBM samples (n=84) and adjacent noncancerous tissues (n=12) were obtained from The Fourth People’s Hospital of Jinan, People’s Republic of China. Samples were snap-frozen in liquid nitrogen and stored at -80°C for further analysis. All patients provided informed and written consent. This study was approved by the independent ethics committee of The Fourth People’s Hospital of Jinan, People’s Republic of China.
The GBM cell lines used in this study (T98G, U87, U251, SHG44, U343, and A172) were obtained from the cell bank of the Shanghai Biology Institute (Shanghai, People’s Republic of China). Fetal bovine serum (10%) (GIBCO, Grand Island, NY, USA) was added to all culture media along with 2 mM L-glutamine and 1% penicillin/streptomycin (Solarbio, Shanghai, People’s Republic of China). Cells were grown in DMEM (Trueline, HyClone, Logan, UT, USA) and maintained in a 5% CO2 atmosphere at 37°C.
RNA isolation and real-time PCR
Total RNA from GBM samples was extracted using TRIzol Reagent (Invitrogen, Carlsbad, California, USA). Then, RNA was reverse transcribed into complementary DNA (cDNA) using the cDNA synthesis kit (Fermentas, Canada) according to the manufacturer’s instructions. The conditions for the Real-time PCR reaction were as follows: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 45 seconds. Gene expression was normalized to the expression of GAPDH, and relative gene expression was calculated using the 2-ΔΔCt method. All data represent the average of three replicates. The primers used are listed in Supplementary material S1.
Lentiviral-mediated RNA interference and overexpression of TMEM176A
Three plasmids containing siRNA targeting positions of human TMEM176A (NM_018487.2) were synthesized (Major, People’s Republic of China). The negative control (siNC) contained a nonspecific scrambled siRNA sequence. A lentiviral plasmid (pLVX-puro) containing the full-length human TMEM176A cDNA sequence and an empty plasmid acted as negative controls for overexpression (oeNC). All plasmids were transiently transfected into cells using Lipofectamine 2000 (Invitrogen, USA) according to the instructions of the manufacturer. Experiments were performed 48 hours after the transfection. Detailed information on the sequence of siTMEM176As is provided in Table S1.
Western blot
Whole protein lysates were extracted from the indicated cells (U87, T98G, and A172) using RIPA lysis buffer (JRDUN, Shanghai, People’s Republic of China) with an EDTA-free protease inhibitor cocktail (Roche, Germany). The protein concentration was estimated using an Enhanced BCA protein assay kit (Thermo Fisher, Shanghai, China). Equal amounts of total protein (25 µg) were fractionated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Millipore, MA, USA) overnight. Then, after being blocked with 5% nonfat dry milk for 1 hour at room temperature, the membranes were probed at 4°C overnight with the primary antibodies followed by incubation for 1 hour at 37°C with the secondary antibody (anti-mouse IgG) (1:1,000; Beyotime, Shanghai, People’s Republic of China). An enhanced chemiluminescence system (Tanon, People’s Republic of China) was used to detect the level of protein expression. Detailed information on the primary antibodies is provided in Table S2. Each sample was tested in triplicate, and GAPDH served as the internal reference.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) assay (SAB, MD, USA) was used to examine cell proliferation according to the protocol of the manufacturer. Briefly, cells transfected as indicated were seeded in 96-well plates and cultured for 0, 24, 48, and 72 hours, and then the CCK-8 solution (1:10) was mixed into each well and incubated for 1 hour. A microplate reader (Pulangxin, People’s Republic of China) was used to measure the optical density values (OD) at a wavelength of 450 nm. Each time point was tested in triplicate.
Cell cycle assay
DNA content was determined by propidium iodide (PI) staining. Cells with or without treatment were collected and resuspended in PBS. Then, 70% ethanol was added at -20°C for at least 2 hours to fix the cells. Then, all groups were treated with RNase A (Solarbio, People’s Republic of China) at 37°C for 15 minutes. After that, PI (7seabiotech, People’s Republic of China) was added to the cells, and then the cells were incubated at room temperature in darkness for 30 minutes. Flow cytometry (BD, NJ, USA) was used to determine the DNA content. The FlowJo cell cycle analysis program was used to analyze the ratio of cells at the G0/G1, S, and G2/M phases.
Cell apoptosis assay
In brief, cells (U87, T98G, and A172) were collected and stained using the Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Beyotime, People’s Republic of China) according to the instructions of the manufacturer at 48 hours after the viral infection. Then, flow cytometry (BD, USA) was used to assess the rate of apoptosis.
Xenograft model
The assay was carried out according to the Institute’s guidelines for animal experiments and was approved by the independent ethics committee of The Fourth People’s Hospital of Jinan, People’s Republic of China. The same number of cells for each cell line (n=2×106) was subcutaneously injected into the right flank of 4–6-week-old BALB/c nude mice (Shanghai Laboratory Animal Company). The length and width of the tumors were examined every 3 days for 33 days after the injection. The volume of the tumor was calculated as follows: length × (width2/2).
Statistical analysis
GraphPad Prism software Version 7.0 (CA, USA) was used for the statistical analyses. Data were displayed as the mean ± SD of at least three samples. Statistical significance was determined by one-way analysis of variance (ANOVA) for multiple comparisons. A P-value<0.05 was accepted to indicate statistical significance.
Results
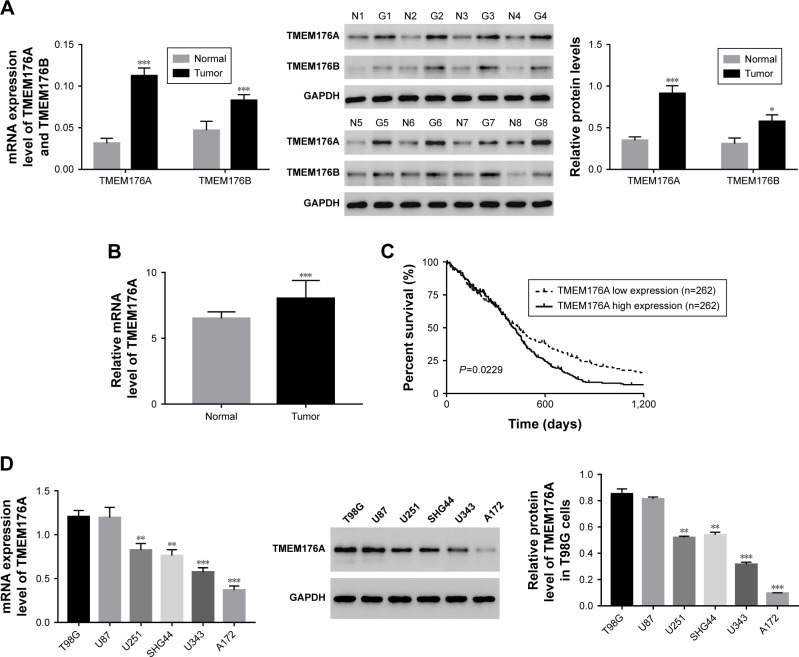
Expression of TMEM176A was upregulated in GBM samples and cell lines
The mRNA expression levels of TMEM176A and TMEM176B were examined using qRT-PCR performed on 84 GBM tissues and 12 normal brain samples. Meanwhile, eight pairs of GBM tissues and matched para-cancerous samples were used to determine the protein levels of TMEM176A and TMEM176B using Western blot. As shown in Figure 1A, both the mRNA and protein expression levels of TMEM176A and TMEM176B were upregulated in GBM compared with normal samples, which was in agreement with the data collected from The Cancer Genome Atlas (TCGA) GBM dataset (Figure 1B). Meanwhile, Kaplan–Meier survival analysis suggested that the survival time of patients with lower TMEM176A expression was clearly longer than that of patients with higher TMEM176A expression (Figure 1C, P<0.01). The analysis of GBM genomic profile was performed according to a previous report,35 and all relevant information can be found on the website https://cancergenome.nih.gov/search. Moreover, the mRNA and protein levels of TMEM176A were also detected in six GBM cell lines, including T98G, U87, U251, SHG44, U34, and A172. As shown in Figure 1D, the level of TMEM176A was significantly upregulated in T98G and U87 cells compared with the other cell lines. On the other hand, the A172 cells exhibited the lowest expression level of TMEM176A among all the GBM cell lines. Based on these results, T98G, U87, and A172 cells were chosen for further study.
Figure 1.
TMEM176A was upregulated in GBM tissues and cell lines.
Notes: (A) The mRNA and protein levels of TMEM176A and TMEM176B in GBM tissues *P<0.05 vs Normal ***P<0.001 vs Normal. (B) TMEM176A expression was significantly increased in GBM tissues (n=529) when compared with normal brain tissues (n=10) of patients from the TCGA GBM dataset (***P<0.001 vs Normal). (C) Survival analysis of patients from the TCGA GBM dataset. (D) The mRNA and protein levels of TMEM176A in different GBM cell lines. **P<0.01, ***P<0.001 vs T98G.
Abbreviations: GBM, glioblastoma; TCGA, The Cancer Genome Atlas.
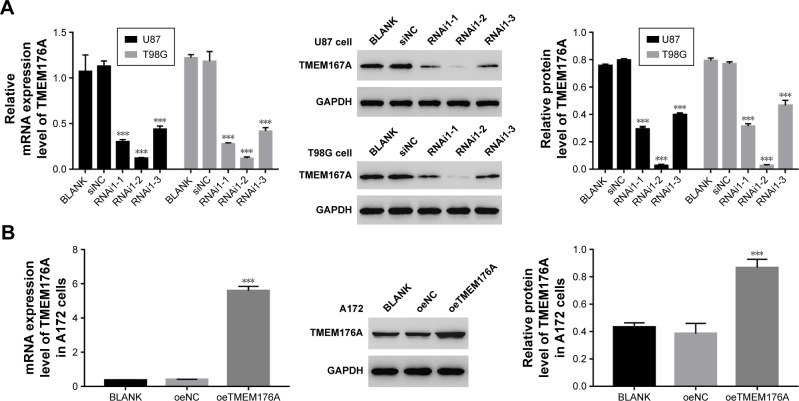
Establishing knockdown and overexpression of TMEM176Ain GBM cells
To silence the expression of TMEM176A, three short hairpin RNAs (shRNAs) targeting human TMEM176A (siTMEM176A; RNAi1-1, RNAi1-2, RNAi1-3) and a nonspecific scrambled siRNA (siNC) were synthesized and transfected into T98G and U87 cell lines. The untreated cells acted as a blank control (Blank). As shown in Figure 2A, all three TMEM176A-siRNAs strongly reduced the levels of endogenous TMEM176A. Cells transfected with RNAi1-1 and RNAi1-2 exhibited lower TMEM176A levels compared with cells transfected with RNAi1-3. Therefore, RNAi1-1 and RNAi1-2 were chosen for further study. In addition, A172 cells were transfected with a plasmid overexpressing TMEM176A (oeTMEM176A) and a mock plasmid (oeNC). The blank control was indicated above. Based on the TMEM176A mRNA and protein levels shown in Figure 2B, a remarkable overexpression of TMEM176A was achieved in oeTMEM176A-transfected cells. Hence, the oeTMEM76A cell line was chosen for the following TMEM176A overex-pression analysis.
Figure 2.
Knockdown and overexpression of TMEM176A in GBM cells.
Notes: (A) The mRNA and protein levels of TMEM176A following a knockdown by siRNAs in T98G and U87 cells, ***P<0.001 vs BLANK. (B) The mRNA and protein levels of TMEM176A following overexpression by oeTMEM176A in A172 cells, ***P<0.001 vs BLANK.
Abbreviation: GBM, glioblastoma.
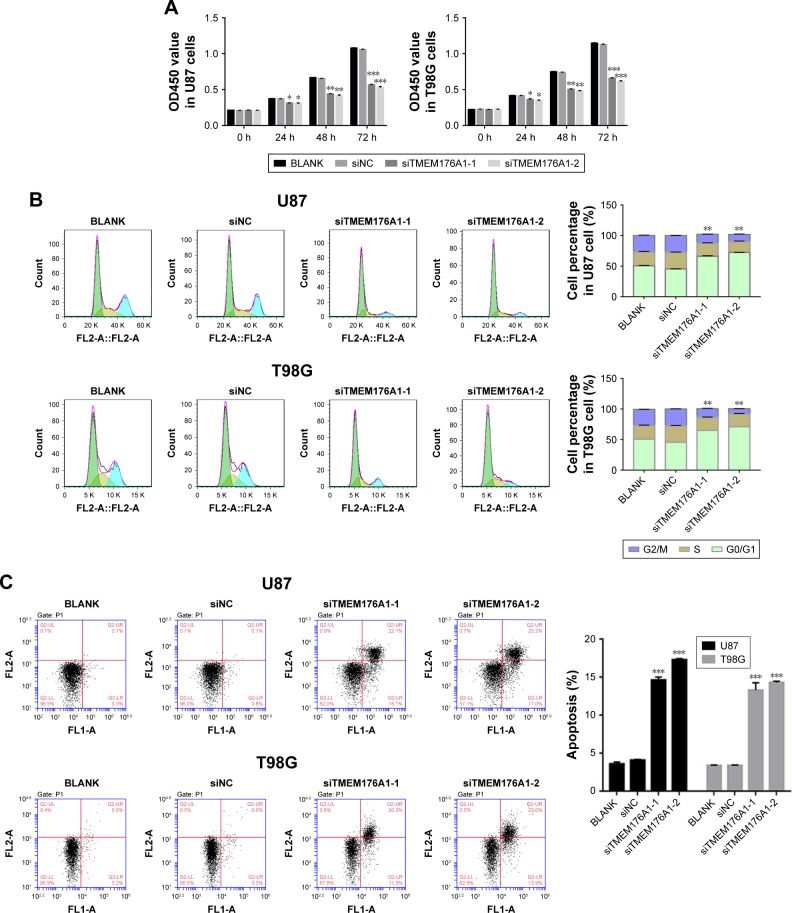
Knockdown of TMEM176A suppressed cell proliferation
The Cell Counting Kit-8 assay was used to examine the function of TMEM176A in the proliferation of GBM cells. BLANK, siNC, siTMEM176A1-1 and siTMEM176A1-2 plasmids were transfected into U87 and T98G cells. As shown in Figure 3A, there was no obvious difference in cell proliferation between the BLANK and NC groups. However, the cell growth rate of siTMEM176A cells was significantly decreased at 24, 48, and 72 hours compared with the NC group. Therefore, we concluded that TMEM176A was a positive regulator of cell proliferation in GBM.
Figure 3.
Knockdown of TMEM176A suppressed GBM cells growth.
Notes: (A) Cell proliferation was detected 0, 24, 48, and 72 hours after transfection with siNC, siTMEM176A1-1 and siTMEM176A1-2 in U87 and T98G cells *P<0.05 vs BLANK, **P<0.01 vs BLANK, ***P<0.001 vs BLANK. (B) Cell cycle profile of cells transfected with siNC, siTMEM176A1-1 and siTMEM176A1-2 in U87 and T98G cells **P<0.01 vs BLANK. (C) Cell apoptosis profile of cells transfected with siNC, siTMEM176A1-1, and siTMEM176A1-2 in U87 and T98G cells. (D) The protein levels of Cyclin D1, P21, cleaved-Caspase-3 and Bcl2 in various transfected cells. ***P<0.001 vs BLANK.
Abbreviation: GBM, glioblastoma.
Knockdown of TMEM176A suppressed G0/G1 phase arrest and cell apoptosis
Flow cytometry was utilized to assess the cell cycle in different cell groups as indicated above. As shown in Figure 3B, no obvious difference was found between BLANK- and siNC-transfected cells. However, the siTMEM176A-transfected cells exhibited an obvious abundance of G0/G1 phase cells. Meanwhile, TMEM176A siRNA caused a significant decrease in G2/M phase cells. In addition, an Annexin V-FITC/ PI staining assay was performed to explore the effect of TMEM176A on the rate of apoptosis of GBM cells. As shown in Figure 3C, the rate of apoptosis of cells transfected with TMEM176A siRNAs was significantly higher than that of cells transfected with siNC and BLANK. These results indicate an antiapoptotic function of TMEM176A in GBM.
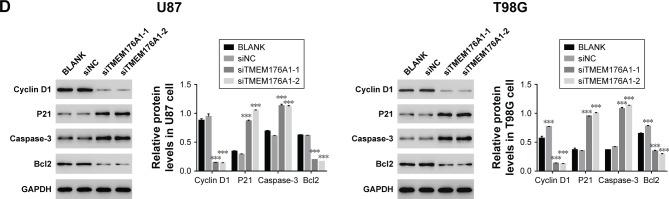
Depletion of TMEM176A influenced the expression levels of Cyclin D1, P21, Caspase-3 and Bcl2
The protein levels of Cyclin D1, P21, Caspase-3 and Bcl2 in different GBM cells, as indicated above, were identified by Western blot. As shown in Figure 3D, the protein level of Cyclin D1 was significantly downregulated in cells trans-fected with TMEM176A siRNAs. However, cells transfected with siTMEM176A exhibited remarkably elevated expression of P21, Caspase-3, and Bcl2. These results suggest the possible targets of TMEM176A in GBM cells.
Knockdown of TMEM176A influenced signal transduction in GBM cells
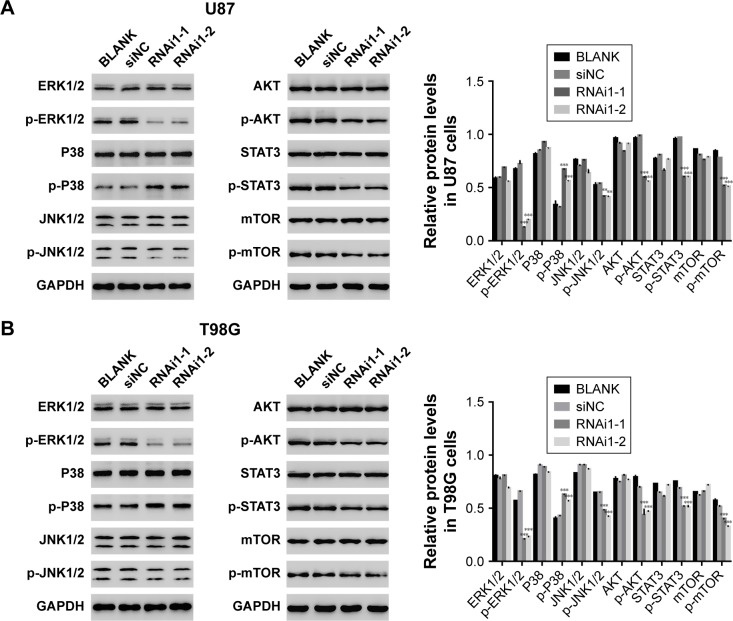
Many key proteins involved in signal transduction pathways are dependent upon phosphorylation modification, including ERK1/2, P38, c–Jun N–terminal kinases (JNKs), AKT, STAT3, and MTOR. Therefore, Western blot analysis was used to examine the phosphorylation level of several of these key proteins. As shown in Figure 4A and B, most of the signal transduction proteins exhibited a decrease in phosphorylation following the silencing of TMEM176A expression. These results indicate that TMEM176A is involved in the signal transduction in GBM cells. Interestingly, knockdown of TMEM176A strongly inhibited the expression of p-ERK1/2.
Figure 4.
Knockdown of TMEM176A affects the signal transduction in GBM cells.
Notes: (A) The protein level of signal transduction-related proteins in U87 cells transfected with siNC, siTMEM176A1-1, and siTMEM176A1-2, respectively, **P<0.01 vs BLANK, ***P<0.001 vs BLANK. (B) The protein level of signal transduction-related proteins in T98G cells transfected with siNC, siTMEM176A1-1 and siTMEM176A1-2, respectively, **P<0.01 vs BLANK, ***P<0.001 vs BLANK.
Abbreviation: GBM, glioblastoma.
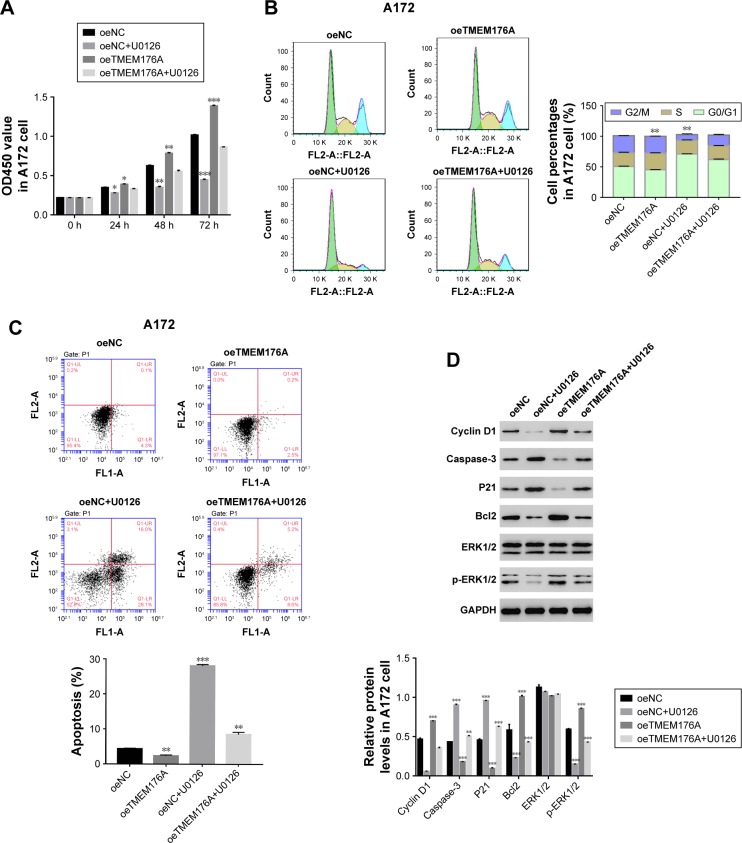
The function of TMEM176A was suppressed by a specific ERK1/2 inhibitor U0126
To determine the correlation between TMEM176A and ERK1/2, a specific ERK1/2 inhibitor, U0126, (10 µM) was added to oeNC- and oeTMEM176A-transfected cells. As shown in Figure 5A, the overexpression of TMEM176A strongly increased the cell proliferation rate of A172 cells compared with oeNC cells; however, the cell proliferation rate was obviously decreased following the addition of the inhibitor U0126. Moreover, the overexpression of TMEM176A promoted cell cycle progression in A172 cells; however, the G0/G1 cell phase was heavily arrested after treatment with U0126 (Figure 5B). As shown in Figure 5C, the cell apoptosis ratio was remarkably decreased in the oeTMEM176A cells; however, the cell apoptosis rate was highly significantly elevated in the U0126-treated groups, and oeTMEM176A also reduced cell apoptosis. Taken together, these results demonstrate a potential correlation between ERK1/2 and TMEM176A in GBM cells. Additionally, the protein expression level of p-ERK1/2 was also inhibited by U0126 in oeTMEM176A cells. On the other hand, the protein expression levels of Caspase-3 and P21 were recovered when U0126 was added to oeTMEM176A cells (Figure 5D). In addition, the possible positive correlation between Cyclin D1 and TMEM176A was weakened by the inhibitor U0126.
Figure 5.
ERK1/2 inhibitor U0126 inhibits the function of TMEM176A.
Notes: (A) Cell proliferation was detected 0, 24, 48, and 72 hours after transfection with oeNC, oeTMEM176A, oeNC+U0126 and oeTMEM176A+U0126 in A172 cells *P<0.05 vs oeNC, **P<0.01 vs oeNC, ***P<0.001 vs oeNC. (B) Cell cycle profile of cells transfected with oeNC, oeTMEM176A, oeNC+U0126 and oeTMEM176A+U0126 in A172 cells. **P<0.01 vs oeNC. (C) Cell apoptosis profile of cells transfected with oeNC, oeTMEM176A, oeNC+U0126 and oeTMEM176A+U0126 in A172 cells. **P<0.01 vs oeNC, ***P<0.001 vs oeNC. (D) The protein levels of Cyclin D1, P21, cleaved-Caspase-3, Bcl2, ERK1/2 and p-ERK1/2 in different transfected cells. ***P<0.001 vs oeNC.
Abbreviation: GBM, glioblastoma.
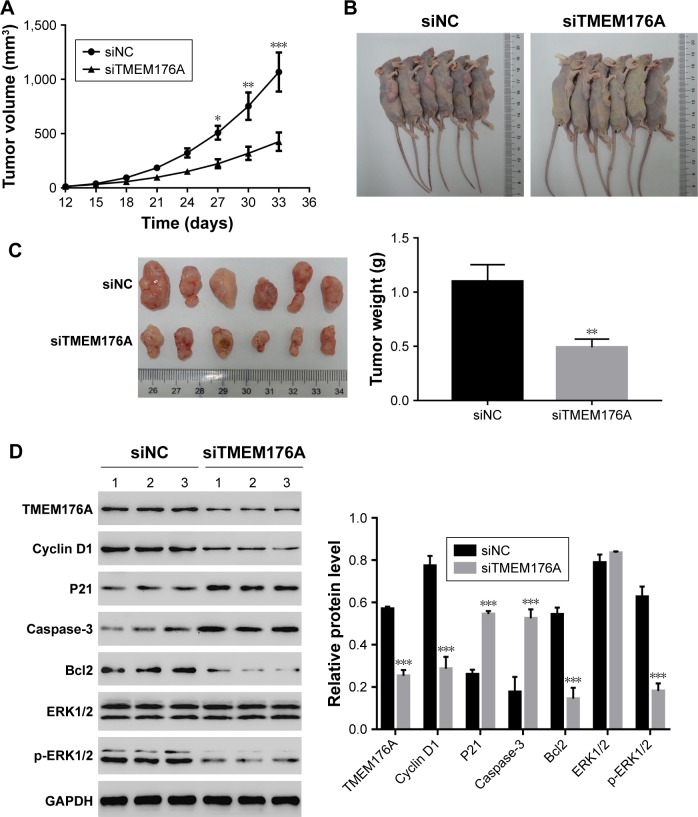
Knockdown of TMEM176A inhibited the tumorigenicity of GBM cells in nude mice
To investigate the effect of TMEM176A on tumorigenicity in vivo, an equal number of T98G cells transfected with scrambled siRNA or TMEM176A-siRNA were injected hypodermically into nude mice, and tumor formation was analyzed for 33 days. As shown in Figure 6A–C, although both of the injected cell types developed tumors, it was clear that the TMEM176A-siRNA cells significantly suppressed the tumor growth rate. Both the volume and weight of TME-M176A-siRNA tumors were highly significantly reduced compared with the siNC tumors. Moreover, the levels of cell cycle-related proteins (Cyclin D1, P21, and ERK1/2) were significantly decreased in the TMEM176A knockdown tumors. In contrast, the cell apoptosis-related proteins (Caspase-3 and Bcl2) were significantly increased in siTMEM176A tumors (Figure 6D). These results suggest an essential role of TMEM176A in the cell cycle and apoptosis.
Figure 6.
Knockdown of TMEM176A in GBM cells inhibits tumor growth in vivo.
Notes: T98G cells transfected with siRNA control (siNC) or siTMEM176A were subcutaneously injected into athymic nude mice. (A) The volume of the tumor was determined every 3 days for 33 days of each group. The growth of the tumor was clearly suppressed in nude mice injected with siTMEM176A cells. *P<0.05 vs siNC, **P<0.01 vs siNC, ***P<0.001 vs siNC. (B) Xenografts in nude mice at day 33 of each group (n=6). (C) Mice were sacrificed, and tumors were weighed at day 33. The six tumor tissues obtained from the siTMEM176A group were smaller and lighter than those of the siNC group, **P<0.01 vs siNC. (D) Protein levels of TMEM176A, Cyclin D1, P21, cleaved-Caspase-3, Bcl2, ERK1/2, p-ERK1/2 were analyzed by Western blot in xenografts from nude mice, ***P<0.001 vs siNC.
Abbreviation: GBM, glioblastoma.
Discussion
GBM is a common malignant brain tumor, for which the overall survival rates are limited to within 2 years. Although traditional therapy (tumor resection and radiotherapy) can help fight against the development of GBM, the prognosis is usually disappointing. Therefore, gaining a deeper insight into the molecular mechanisms underlying GBM may contribute to the development of a biomarker for diagnosing GBM at the early phase.
In this study, we systematically analyzed the function of a positive tumor regulator, TMEM176A, in GBM cells. RNA interference was employed to silence the expression of TMEM176A. Additionally, a lentiviral vector was used to mediate the overexpression of TMEM176A. The results from these two experimental phases were in agreement, and therefore, our results were more reliable.
From our CCK-8 assay, the cell proliferation rate was profoundly suppressed after silencing TMEM176A, which was upregulated in oeTMEM176A cells. Therefore, TMEM176A may serve to promote cell proliferation in GBM. Moreover, the ERK1/2 signaling pathway is essential in the cell cycle process. The activating of the ERK1/2 signaling pathway promotes tumorigenesis and progression of gastric cancer.36 A previous report demonstrated that dysregulation of ERK1/2 can trigger tumor cell development in GBM.33 Phosphorylation of ERK1/2 is necessary for cell proliferation and transformation,37 and in this study, the activity of ERK1/2 was heavily suppressed following the knockout of TMEM176A expression. Meanwhile, the level of p-ERK1/2 was significantly elevated in oeTMEM176A cells. Therefore, it appears that TMEM176A promotes the phosphorylation of ERK1/2. Interestingly, the effect of TMEM176A on cell proliferation, cell cycle, and cell apoptosis was remarkably inhibited by a specific ERK1/2 inhibitor, U0126. Our results suggest that ERK1/2 may be direct targets of TMEM176A in GBM cells.
Moreover, previous reports have demonstrated that Cyclin D1 is a central target in the regulation of cell proliferation and apoptosis in GBM cells.3,38 Downregulating the expression of Cyclin D1 contributes to the inhibition of tumor growth.39 In this study, the expression of Cyclin D1 was notably decreased once TMEM176A expression was silenced. Moreover, the expression of Cyclin D1 depends on the activation of the MEK/ERK pathway in oval cells.40 Therefore, our results not only demonstrated that TMEM176A may be involved in a pathway including ERK1/2 and Cyclin D1 but also indicated the important role of TMEM176A in the regulation of the cell cycle.
A previous report demonstrated that Bcl2 family members neutralize the maturation of Caspase-3, thereby increasing the tumor growth rate.24 In this study, we doubted that this neutralization effect had dropped off once the Bcl2 level was decreasing, which might promote the expression of Caspase-3 in siTMEM176A cells. Similar results were also obtained from the analysis of oeTMEM176A cells. Therefore, TMEM176A may regulate Bcl2 directly and may, therefore, be negatively correlated with Caspase-3. Taken together, these results suggest a central role of TMEM176A in the apoptosis of GBM cells. However, the ERK inhibitor U0126 promotes the expression of Caspase-3 in oeTMEM176A cells, which indicates that ERK may downregulate the expression of Caspase-3 in GBM cells. Therefore, further study is warranted to investigate the possible relationship between ERK and Caspase-3 in GBM cells.
In addition, it would be valuable to include the cell lines that exhibited moderate TMEM176A expression in our experiments. Moreover, TMEM14A and TMEM45A play a key role in cell migration and invasion;7,8 therefore, further study is warranted to investigate the function of TMEM176A in the migration and invasion of GBM cells.
Conclusion
In the present study, we examined the effect of TMEM176A knockdown and overexpression in GBM cells. Our results not only provide a better understanding of TMEM176A in GBM but also provide support for the clinical application of TMEM176A.
Supplementary materials
Supplementary material S1 Primer sequence information
1.1 Homo sapiens transmembrane protein 176A (TMEM176A), mRNA
NCBI reference sequence: NM_018487.2
Primer F 5′ AGTGCCTGCCGCATCTCC 3′
Primer R 5′ TCCAACATTTCCTTCTGGTCTC 3′
Pos: 445–692
Amplified product: Size: 248 bps
1.2 Homo sapiens transmembrane protein 176B (TMEM176B), transcript variant 5, mRNA
NCBI reference sequence: NM_001101314.1
Primer F 5′ TGTTGTCCTCTGCGTGAATAGC 3′
Primer R 5′ TTCCTCAGCATCTGCATGTAAG 3′
Pos: 315–491
Amplified product: Size: 177 bps
1.3 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase (GAPDH), transcript variant 1, mRNA
NCBI reference sequence: NM_001256799.2
Primer F 5′ AATCCCATCACCATCTTC 3′
Primer R 5′ AGGCTGTTGTCATACTTC 3′
Pos: 436–653
Amplified product: Size: 218 bps
Table S1.
Human gene TMEM176A (NM_018487.2) RNAi targeting locus information
| RNAi targeting locus
|
Sequence | |
|---|---|---|
| Name | Locus position | |
|
| ||
| RNAi1-1 | 61–79 | CAGACTGTGTCCCTGACAA |
| RNAi1-2 | 273–291 | GATCTTGAGTGCAGTCCTA |
| RNAi1-3 | 659–677 | TGCTCTTGGGTGTCTGGAT |
Table S2.
The primary antibodies information
| Antibody name | Source | Dilution factor |
|---|---|---|
|
| ||
| c-MET | Abcam, UK | 1:500 |
| Caspase-3 | Abcam, UK | 1:500 |
| AKT | CST, USA | 1:1,000 |
| p-AKT | CST, USA | 1:2,000 |
| GAPDH | CST, USA | 1:2,000 |
Abbreviation: CST, cell signaling technology.
Acknowledgments
The present study was supported by the China Postdoctoral Science Foundation (Grant no 2016M592196) and the special fund for postdoctoral innovation project of Shandong human resources and social security department (grant no 201502021).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sturm D, Bender S, Jones DT, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14(2):92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerber NK, Goenka A, Turcan S, et al. Transcriptional diversity of long-term glioblastoma survivors. Neuro Oncol. 2014;16(9):1186–1195. doi: 10.1093/neuonc/nou043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun G, Shi L, Yan S, et al. MiR-15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. Biomed Res Int. 2014;2014:687826. doi: 10.1155/2014/687826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi: 10.1186/s12943-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang Y, Herman JG, Linghu E, Guo M. Epigenetic silencing of TMEM176A promotes esophageal squamous cell cancer development. Oncotarget. 2017;8(41):70035–70048. doi: 10.18632/oncotarget.19550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuajungco MP, Podevin W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 2012;114(7):705–712. doi: 10.1016/j.acthis.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W, Qiu G, Zou Y, et al. Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int J Clin Exp Pathol. 2015;8(10):12657–12667. [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Chen X, Zhang X, Zhan J, Chen J. Knockdown of TMEM14A expression by RNAi inhibits the proliferation and invasion of human ovarian cancer cells. Biosci Rep. 2016;36(1):e00298. doi: 10.1042/BSR20150258. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Qiu G, Sun W, Zou Y, et al. RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells. Tumour Biol. 2015;36(10):8231–8238. doi: 10.1007/s13277-015-3552-6. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka T, Kubota M, Shinohara K, Yasuda K, Kato JY. In vivo Analysis of the Cyclin D1 Promoter during Early Embryogenesis in Xenopus. Cell Struct Funct. 2003;28:165–177. doi: 10.1247/csf.28.165. [DOI] [PubMed] [Google Scholar]
- 11.Liang S, Mu K, Wang Y, et al. CyclinD1, a prominent prognostic marker for endometrial diseases. Diagn Pathol. 2013;8:138. doi: 10.1186/1746-1596-8-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahlin C, Lundgren C, Embretsén-Varro E, Jirström K, Blomqvist C, Fjällskog M. High expression of cyclin D1 is associated to high proliferation rate and increased risk of mortality in women with ER-positive but not in ER-negative breast cancers. Breast Cancer Res Treat. 2017;164(3):667–678. doi: 10.1007/s10549-017-4294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Or CR, Chang Y, Lin WC, et al. Obatoclax, a Pan-BCL-2 Inhibitor, Targets Cyclin D1 for Degradation to Induce Antiproliferation in Human Colorectal Carcinoma Cells. Int J Mol Sci. 2016;18(1):E44. doi: 10.3390/ijms18010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun G, Sima G, Wu C, et al. Decreased MiR-17 in glioma cells increased cell viability and migration by increasing the expression of Cyclin D1, p-Akt and Akt. PLoS One. 2018;13(1):e0190515. doi: 10.1371/journal.pone.0190515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai M, Al-Odaini AA, Fils-Aimé N, et al. Cyclin D1 cooperates with p21 to regulate TGFβ-mediated breast cancer cell migration and tumor local invasion. Breast Cancer Res. 2013;15(3):R49. doi: 10.1186/bcr3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jun GJ, Zhong GG, Ming ZS. miR-218 inhibits the proliferation of glioma U87 cells through the inactivation of the CDK6/cyclin D1/ P21(Cip1/Waf1) pathway. Oncol Lett. 2015;9(6):2743–2749. doi: 10.3892/ol.2015.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hukkelhoven E, Liu Y, Yeh N, Ciznadija D, Blain SW, Koff A. Tyrosine phosphorylation of the p21 cyclin-dependent kinase inhibitor facilitates the development of proneural glioma. J Biol Chem. 2012;287(46):38523–38530. doi: 10.1074/jbc.M112.366542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JS, Xiao J, Patel P, et al. A novel tumor-promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro Oncol. 2014;16(2):191–203. doi: 10.1093/neuonc/not167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrillo García C, Riedt T, Li J, Dotten M, Brossart P, Janzen V. Simultaneous deletion of p21Cip1/Waf1 and caspase-3 accelerates proliferation and partially rescues the differentiation defects of caspase-3 deficient hematopoietic stem cells. PLoS One. 2014;9(10):e109266. doi: 10.1371/journal.pone.0109266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Q, Peng J, Liu W, et al. Elevated cleaved caspase-3 is associated with shortened overall survival in several cancer types. Int J Clin Exp Pathol. 2014;7(8):5057–5070. [PMC free article] [PubMed] [Google Scholar]
- 21.Pu X, Storr SJ, Zhang Y, et al. Caspase-3 and caspase-8 expression in breast cancer: caspase-3 is associated with survival. Apoptosis. 2017;22(3):357–368. doi: 10.1007/s10495-016-1323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Zhong DN, Qin H, et al. Caspase-3 over-expression is associated with poor overall survival and clinicopathological parameters in breast cancer: a meta-analysis of 3091 cases. Oncotarget. 2018;9(9):8629–8641. doi: 10.18632/oncotarget.23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang N, Bank T, Ding X, et al. Caspase-3 suppresses diethylnitrosamine-induced hepatocyte death, compensatory proliferation and hepatocarcinogenesis through inhibiting p38 activation. Cell Death Dis. 2018;9(5):558. doi: 10.1038/s41419-018-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegh AH, Kesari S, Mahoney JE, et al. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc Natl Acad Sci U S A. 2008;105(31):10703–10708. doi: 10.1073/pnas.0712034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui J, Placzek WJ. Post-Transcriptional Regulation of Anti-Apoptotic BCL2 Family Members. Int J Mol Sci. 2018;19(1):E308. doi: 10.3390/ijms19010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viant C, Guia S, Hennessy RJ, et al. Cell cycle progression dictates the requirement for BCL2 in natural killer cell survival. J Exp Med. 2017;214(2):491–510. doi: 10.1084/jem.20160869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang TQ, Lu XJ, Wu TF, et al. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci. 2014;105(3):265–271. doi: 10.1111/cas.12351. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Han J, Chen Q. MiR-16 modulate temozolomide resistance by regulating BCL-2 in human glioma cells. Int J Clin Exp Pathol. 2015;8(10):12698–12707. [PMC free article] [PubMed] [Google Scholar]
- 29.Papageorgiou SG, Kontos CK, Pappa V, et al. The novel member of the BCL2 gene family, BCL2L12, is substantially elevated in chronic lymphocytic leukemia patients, supporting its value as a significant biomarker. Oncologist. 2011;16(9):1280–1291. doi: 10.1634/theoncologist.2010-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey V, Bhaskara VK, Babu PP. Implications of mitogen-activated protein kinase signaling in glioma. J Neurosci Res. 2016;94(2):114–127. doi: 10.1002/jnr.23687. [DOI] [PubMed] [Google Scholar]
- 31.Moon DO, Park SY, Heo MS, et al. Key regulators in bee venom-induced apoptosis are Bcl-2 and caspase-3 in human leukemic U937 cells through downregulation of ERK and Akt. Int Immunopharmacol. 2006;6(12):1796–1807. doi: 10.1016/j.intimp.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Kolb RH, Greer PM, Cao PT, Cowan KH, Yan Y. ERK1/2 signaling plays an important role in topoisomerase II poison-induced G2/M checkpoint activation. PLoS One. 2012;7(11):e50281. doi: 10.1371/journal.pone.0050281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin L, Cao Y, Zhang T, et al. Effects of ERK1/2 S-nitrosylation on ERK1/2 phosphorylation and cell survival in glioma cells. Int J Mol Med. 2018;41(3):1339–1348. doi: 10.3892/ijmm.2017.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Zuo D, Luan C, et al. Glioma cell proliferation controlled by ERK activity-dependent surface expression of PDGFRA. PLoS One. 2014;9(1):e87281. doi: 10.1371/journal.pone.0087281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5(2):e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Yu Z, Zhou Q, et al. Tissue transglutaminase-2 promotes gastric cancer progression via the ERK1/2 pathway. Oncotarget. 2016;7(6):7066–7079. doi: 10.18632/oncotarget.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genome Res. 2002;12(1):47–56. doi: 10.1101/gad.962802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hui W, Yuntao L, Lun L, et al. MicroRNA-195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS One. 2013;8(1):e54932. doi: 10.1371/journal.pone.0054932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velpula KK, Dasari VR, Tsung AJ, et al. Regulation of glioblastoma progression by cord blood stem cells is mediated by downregulation of cyclin D1. PLoS One. 2011;6(3):e18017. doi: 10.1371/journal.pone.0018017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HY, Yang SL, Liang HF, Li CH, Ch L. HBx protein promotes oval cell proliferation by up-regulation of cyclin D1 via activation of the MEK/ERK and PI3K/Akt pathways. Int J Mol Sci. 2014;15(3):3507–3518. doi: 10.3390/ijms15033507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Human gene TMEM176A (NM_018487.2) RNAi targeting locus information
| RNAi targeting locus
|
Sequence | |
|---|---|---|
| Name | Locus position | |
|
| ||
| RNAi1-1 | 61–79 | CAGACTGTGTCCCTGACAA |
| RNAi1-2 | 273–291 | GATCTTGAGTGCAGTCCTA |
| RNAi1-3 | 659–677 | TGCTCTTGGGTGTCTGGAT |
Table S2.
The primary antibodies information
| Antibody name | Source | Dilution factor |
|---|---|---|
|
| ||
| c-MET | Abcam, UK | 1:500 |
| Caspase-3 | Abcam, UK | 1:500 |
| AKT | CST, USA | 1:1,000 |
| p-AKT | CST, USA | 1:2,000 |
| GAPDH | CST, USA | 1:2,000 |
Abbreviation: CST, cell signaling technology.