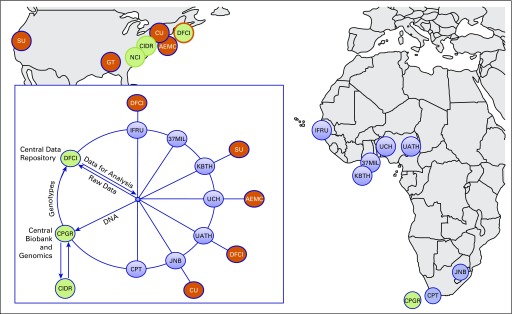
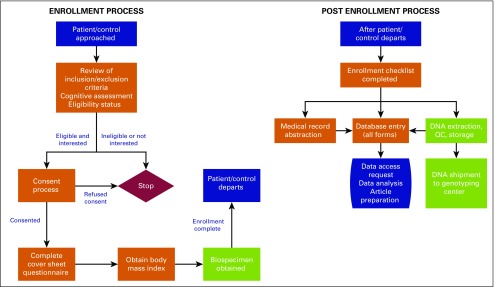
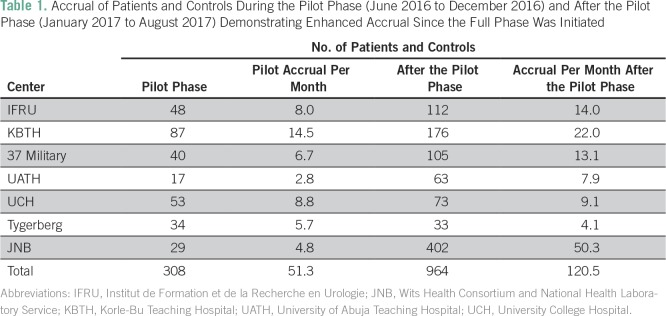
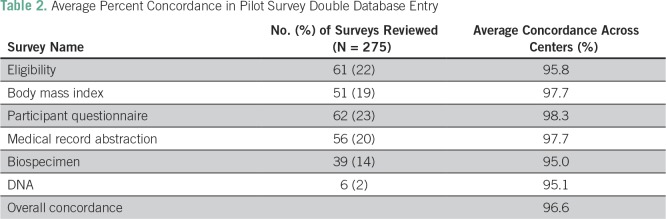
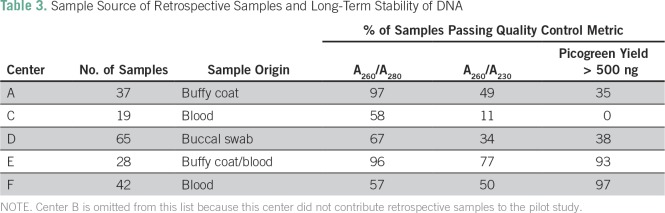
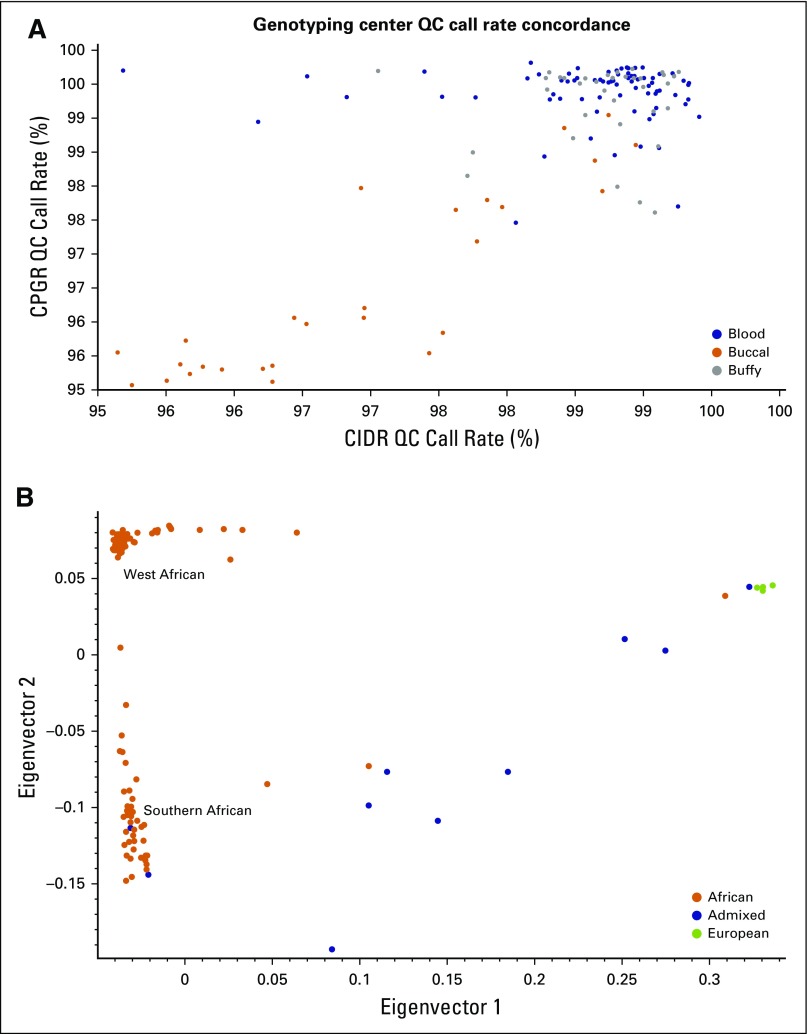
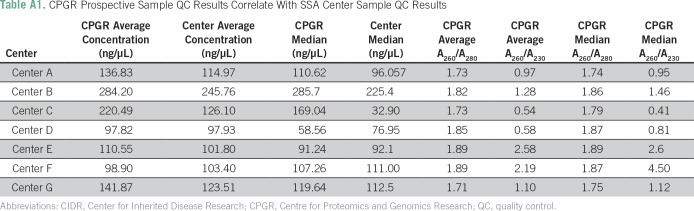
Caroline Andrews
Caroline Andrews
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Brian Fortier
Brian Fortier
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Amy Hayward
Amy Hayward
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ruth Lederman
Ruth Lederman
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Lindsay Petersen
Lindsay Petersen
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Jo McBride
Jo McBride
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Desiree C Petersen
Desiree C Petersen
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olabode Ajayi
Olabode Ajayi
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Paidamoyo Kachambwa
Paidamoyo Kachambwa
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Moleboheng Seutloali
Moleboheng Seutloali
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Aubrey Shoko
Aubrey Shoko
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Mamokhosana Mokhosi
Mamokhosana Mokhosi
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Reinhard Hiller
Reinhard Hiller
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Marcia Adams
Marcia Adams
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Chrissie Ongaco
Chrissie Ongaco
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Elizabeth Pugh
Elizabeth Pugh
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Jane Romm
Jane Romm
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Tameka Shelford
Tameka Shelford
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Frank Chinegwundoh
Frank Chinegwundoh
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ben Adusei
Ben Adusei
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Sunny Mante
Sunny Mante
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Nana Yaa Snyper
Nana Yaa Snyper
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ilir Agalliu
Ilir Agalliu
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
David W Lounsbury
David W Lounsbury
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Thomas Rohan
Thomas Rohan
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Alex Orfanos
Alex Orfanos
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Yuri Quintana
Yuri Quintana
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Judith S Jacobson
Judith S Jacobson
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Alfred I Neugut
Alfred I Neugut
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Edward Gelmann
Edward Gelmann
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Joseph Lachance
Joseph Lachance
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Cherif Dial
Cherif Dial
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Thierno Amadou Diallo
Thierno Amadou Diallo
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Mohamed Jalloh
Mohamed Jalloh
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Serigne Magueye Gueye
Serigne Magueye Gueye
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Papa Moussa Sène Kane
Papa Moussa Sène Kane
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Halimatou Diop
Halimatou Diop
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Anna Julienne Ndiaye
Anna Julienne Ndiaye
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Amina Sow Sall
Amina Sow Sall
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ndeye Coumba Toure-Kane
Ndeye Coumba Toure-Kane
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ezenwa Onyemata
Ezenwa Onyemata
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Alash’le Abimiku
Alash’le Abimiku
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Andrew A Adjei
Andrew A Adjei
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Richard Biritwum
Richard Biritwum
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Richard Gyasi
Richard Gyasi
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Mathew Kyei
Mathew Kyei
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
James E Mensah
James E Mensah
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Julian Okine
Julian Okine
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Vicky Okyne
Vicky Okyne
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Isabella Rockson
Isabella Rockson
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Evelyn Tay
Evelyn Tay
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Yao Tettey
Yao Tettey
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Edward Yeboah
Edward Yeboah
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Wenlong C Chen
Wenlong C Chen
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Elvira Singh
Elvira Singh
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Michael B Cook
Michael B Cook
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Christine N Duffy
Christine N Duffy
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ann Hsing
Ann Hsing
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Cassandra Claire Soo
Cassandra Claire Soo
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Pedro Fernandez
Pedro Fernandez
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Hayley Irusen
Hayley Irusen
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Oseremen Aisuodionoe-Shadrach
Oseremen Aisuodionoe-Shadrach
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Abubakar Mustapha Jamda
Abubakar Mustapha Jamda
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Peter Oluwole Olabode
Peter Oluwole Olabode
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Maxwell Madueke Nwegbu
Maxwell Madueke Nwegbu
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olalekan Hafees Ajibola
Olalekan Hafees Ajibola
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olushola Jeremiah Ajamu
Olushola Jeremiah Ajamu
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Yakubu Garba Ambuwa
Yakubu Garba Ambuwa
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Akindele Olupelumi Adebiyi
Akindele Olupelumi Adebiyi
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Michael Asuzu
Michael Asuzu
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olufemi Ogunbiyi
Olufemi Ogunbiyi
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olufemi Popoola
Olufemi Popoola
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olayiwola Shittu
Olayiwola Shittu
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Olukemi Amodu
Olukemi Amodu
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Emeka Odiaka
Emeka Odiaka
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Ifeoluwa Makinde
Ifeoluwa Makinde
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Maureen Joffe
Maureen Joffe
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Audrey Pentz
Audrey Pentz
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,
Timothy R Rebbeck
Timothy R Rebbeck
1Caroline Andrews, Brian Fortier, Amy Hayward, Ruth Lederman, and Timothy R. Rebbeck; Dana-Farber Cancer Institute; Alex Orfanos and Yuri Quintana, Beth Israel Deaconess Medical Center; Timothy R. Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA; Lindsay Petersen, Jo McBride, Desiree C. Petersen, Olabode Ajayi, Paidamoyo Kachambwa, Moleboheng Seutloali, Aubrey Shoko, Mamokhosana Mokhosi, and Reinhard Hiller, Centre for Proteomic and Genomic Research; Pedro Fernandez and Hayley Irusen, Stellenbosch University and Tygerberg Hospital, Cape Town; Wenlong C. Chen and Elvira Singh, National Cancer Registry, National Health Laboratory Service; Wenlong C. Chen, Elvira Singh, Maureen Joffe, Audrey Pentz, and Cassandra Claire Soo, University of Witwatersrand, Johannesburg, South Africa; Marcia Adams, Chrissie Ongaco, Elizabeth Pugh, Jane Romm, and Tameka Shelford, Center for Inherited Disease Research, Baltimore; Michael B. Cook, National Cancer Institute, National Institutes of Health, Bethesda, MD; Frank Chinegwundoh, Bart’s Health National Health Services Trust, London, United Kingdom; Ben Adusei, Sunny Mante, and Nana Yaa Snyper, 37 Military Hospital; Andrew A. Adjei, Richard Biritwum, Richard Gyasi, Mathew Kyei, James E. Mensah, Julian Okine, Vicky Okyne, Isabella Rockson, Evelyn Tay, Yao Tettey, and Edward Yeboah, Korle-Bu Teaching Hospital, Accra, Ghana; Ilir Agalliu, David W. Lounsbury, and Thomas Rohan, Albert Einstein College of Medicine, Bronx; Judith S. Jacobson, Alfred I. Neugut, and Edward Gelmann, Columbia University, New York, NY; Joseph Lachance, Georgia Institute of Technology, Atlanta, GA; Cristine N. Duffy and Ann Hsing, Stanford University, Stanford Cancer Institute, Stanford, CA; Cherif Dial, Thierno Amadou Diallo, Mohamed Jalloh, Serigne Magueye Gueye, and Papa Moussa Sène Kane, Hôpital Général de Grand Yoff, Institute de Formation et de la Recherche en Urologie et de la Santé de la Famillie; Halimatou Diop, Anna Julienne Ndiaye, Amina Sow Sall, and Ndeye Coumba Toure-Kane, Hôpital Aristide Le Dantec, Dakar, Senegal; Ezenwa Onyemata and Alash’le Abimiku, Institute of Human Virology, H3 African Biorepository Initiative; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, Maxwell Madueke Nwegbu, and Olalekan Hafees Ajibola, University of Abuja; Oseremen Aisuodionoe-Shadrach, Abubakar Mustapha Jamda, Peter Oluwole Olabode, and Maxwell Madueke Nwegbu, University of Abuja Teaching Hospital, Abuja; Olushola Jeremiah Ajamu and Yakubu Garba Ambuwa, Federal Medical Center, Keffi; Akindele Olupelumi Adebiyi, Michael Asuzu, Olufemi Ogunbiyi, Olufemi Popoola, Olayiwola Shittu, Olukemi Amodu, Emeka Odiaka, and Ifeoluwa Makinde, University College Hospital, Ibadan, Nigeria.
1,✉