Abstract
Pompe disease, caused by deficiency of acid alpha-glucosidase (GAA), leads to widespread glycogen accumulation and profound neuromuscular impairments. There has been controversy, however, regarding the role of central nervous system pathology in Pompe motor dysfunction. We hypothesized that absence of GAA protein causes progressive activation of neuropathological signaling, including pathways associated with cell death. To test this hypothesis, genomic data (Affymetrix Mouse Gene Array 2.0ST) from the midcervical spinal cord in 6 and 16 mo old Pompe (Gaa−/−) mice were evaluated (Broad Institute Molecular Signature Database), along with spinal cord histology. The midcervical cord was selected because it contains phrenic motoneurons, and phrenic-diaphragm dysfunction is prominent in Pompe disease. Several clinically important themes for the neurologic etiology of Pompe disease emerged from this unbiased genomic assessment. First, pathways associated with cell death were strongly upregulated as Gaa−/− mice aged, and motoneuron apoptosis was histologically verified. Second, proinflammatory signaling was dramatically upregulated in the Gaa−/− spinal cord. Third, many signal transduction pathways in the Gaa−/− cervical cord were altered in a manner suggestive of impaired synaptic function. Notably, glutamatergic signaling pathways were downregulated, as were “synaptic plasticity pathways” including genes related to neuroplasticity. Fourth, many genes and pathways related to cellular metabolism are dysregulated. Collectively, the data unequivocally confirm that systemic absence of GAA induces a complex neuropathological cascade in the spinal cord. Most importantly, the results indicate that Pompe is a neurodegenerative condition, and this underscores the need for early therapeutic intervention capable of targeting the central nervous system.
Keywords: Pompe, neurodegeneration, neuroinflammation, plasticity, cervical
pompe disease is an autosomal recessive disorder caused by mutation in the gene for acid alpha-glucosidase (GAA), a hydrolase necessary for the degradation of lysosomal glycogen. Early-onset Pompe disease occurs when there is complete (or near complete) deficiency of functional GAA; late-onset patients have some residual GAA activity (20). GAA deficiency causes profound glycogen accumulation in many tissues including skeletal and cardiac muscle and throughout the central nervous system (CNS) (15). While somatic motor impairments in Pompe have historically been assumed to reflect skeletal muscle dysfunction, autopsy samples from Pompe patients (9, 30, 37, 51) and studies of Pompe animal models (9, 11, 45, 55) show neuropathology, and motoneurons appear to be particularly susceptible. To date, however, studies of CNS pathology in Pompe have been limited to histological assessments of brain and spinal cord (reviewed in Ref. 15). Accordingly, the molecular mechanisms of neuropathology triggered by GAA deficiency are unexplored.
Neuronal cell death and neuroinflammation are prominent in many lysosomal disorders (2), but these processes have not been evaluated in Pompe disease. There is also no information available regarding the impact of Pompe disease on the fundamental pathways associated with synaptic communication. These gaps in knowledge are particularly problematic considering the only Food and Drug Administration-approved therapy (enzyme replacement therapy or ERT) for Pompe is directed at correcting skeletal and cardiac muscle pathology (15, 27) and has little to no impact on the CNS. Intravenously infused GAA protein does not cross the blood-brain barrier to reach the CNS where it could impact neuropathology, and approximately half of patients with classical infantile disease treated with ERT do not survive ventilator-free beyond the age of 3 yr (28). In the absence of functional GAA, and without ERT, respiratory symptoms can present very early in infants (15), and have been detected by 6 mo of age in Gaa−/− mice (14). Pathology in respiratory motoneurons can be detected in Gaa−/− mice as young as 6 wk of age (55), and anatomical tracing studies indicate that adult Gaa−/− mice have fewer phrenic motoneurons compared with wild-type mice (9). Thus, an accumulation of evidence indicates that CNS pathology is relevant to both the progression and treatment of Pompe disease, but very little is known about the underlying mechanisms. Therefore, we used an established murine Pompe model (Gaa−/−, Ref. 44) to conduct an unbiased genomic screening (Affymetrix Mouse Gene Array 2.0ST) of CNS tissue mid- (6 mo old) and late stages (16 mo old) of the disease process, and complemented this with comparative histological assessments. We hypothesized that absence of GAA protein causes progressive activation of pathological signaling cascades within the spinal cord. In particular, we focused on the midcervical spinal cord because this region shows profound histopathology in Pompe mice (9, 42, 45) and patients (9, 37). The C3-5 cervical spinal cord also contains the phrenic motor nucleus, which innervates the diaphragm, and phrenic-diaphragm impairment is a prominent symptom in Pompe disease (15, 46). The data confirm the hypothesis and underscore the need for early therapeutic intervention in Pompe disease with specific targeting of the CNS. Furthermore, this genome-wide screening has provided new molecular targets for future mechanistic and therapeutic investigations in regard to neurologic dysfunction in Pompe disease. Aspects of these results have been previously reported in abstract form (53, 54).
METHODS
The University of Florida Institutional Animal Care and Use Committee approved all procedures. Mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited facility with a 12:12 light/dark cycle and ad libitum access to chow and water. The Gaa−/− mouse (44) (Taconic, Hudson, NY) was outbred to a 129SVE background (13) and studied at two ages, adult (6–8 mo, n = 8) and aged (12–16 mo, n = 8), and compared with age-matched mice from the syngeneic background strain (129SVE), henceforth referred to as wild type (WT).
Histology protocols.
These procedures were adapted from our recent reports (55). Mice (n = 4/group) were euthanized using Beuthanasia (260 mg/kg; Patterson Veterinary Supply, Alachua, FL); the CNS was removed and submerged into Gendre's fixative for 24–48 h, before transfer to 70% ethanol. The cervical spinal cord was cut at the C3 ventral root, and serial tissue sections were obtained throughout the C3-4 cord. Tissues underwent routine paraffin processing and embedding and were sectioned at 5 μm. Multiple tissue sections (3–4), separated by at least 30 μM, were slide-mounted (Fisher, Waltham, MA) for each stain described below. Sections were deparaffinized and rehydrated through a graded series of ethanol to water.
Periodic acid Schiff.
Glycogen accumulation was detected using the periodic acid Schiff (PAS) reaction, as described previously (13), with minor modifications. In brief, tissues were incubated in 0.5% periodic acid (Richard-Allan Scientific, Kalamazoo, MI) for 10-min at 60°C, rinsed with tap water, then stained with Schiff's reagent (Richard-Allan Scientific) for 5 min, and then rinsed again in tap water. Slides were counterstained using Select Tech hematoxylin 560 from Leica and cover-slipped in Vectamount (Vector Labs).
Terminal deoxynucleotidyl transferase dUTP nick end labeling.
Slides were blocked for endogenous peroxidase activity, placed in 0.1 M citrate buffer pH 6.0, and permeabilized by exposure to 4 min of microwave irradiation (600 W). A known positive sample and two negative control slides were included. Staining was performed using a commercially available apoptosis kit (ApopTag Peroxidase In Situ Apoptosis Detection Kit; Millipore, Billerica, MA) following the manufacturer's instructions. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) reaction mixture was incubated on the slides for 1 h at 37°C, with negative control slides receiving labeling mixture devoid of TdT enzyme. Positive signal was detected using ImmPACT DAB (Vector Labs). Slides were counterstained with hematoxylin before being coverslipped using Vectamount (Vector Labs).
Ionized calcium binding adaptor molecule 1.
The goal of these experiments was to provide a qualitative evaluation of the appearance and location of microglia. Slides were blocked in 3% horse serum for 1 h. Primary antibodies were then applied overnight at 4°C. The antibodies used were rabbit anti-Iba-1 (1:300; Wako Chemicals, Cape Charles, VA) and mouse anti-NeuN (1:1,000; Encor Biotechnology, Gainesville, FL). Antigen retrieval with Trilogy reagent (Cell Marque) at 95°C for 25 min was required for optimal staining. Immunoreactivity was detected using 1:500 dilutions of species appropriate Alexa Fluor antibodies raised in donkey anti-rabbit AlexaFluor 594 secondary and anti-mouse Alexa Fluor 488 secondary were used for ionized calcium binding adaptor molecule 1 (IBA-1) and NeuN stains, respectively. Sections were mounted in VectaShield with DAPI (Vector Labs) prior to imaging. Positive control tissues and concentration matched Ig controls were included with each immunoassay.
Microscopy and evaluation of sections.
Histological images were captured with an Olympus BX43 upright microscope and an Olympus DP80 camera with Olympus CellSens software. Using the software, we created images that were white or black balanced for bright field or fluorescence, respectively. All images were qualitatively evaluated by multiple investigators. Glycogen accumulation was visualized using the PAS stain, which imparts a deep pink color in neurons considered positive (31). Cells were considered to be TUNEL-positive if the nucleus had a clearly discernable dark rim of staining or if the entire soma was dark brown (29). IBA-1-positive microglia were characterized as “activated” based on a hypertrophied and/or “bushy” appearance or an amoeboid morphology (24).
The Gaa−/− and WT histological results were compared using a Fisher's exact test (Graphpad Software, http://www.graphpad.com) of the null hypothesis that there were no differences in the qualitative appearance of tissues between the two strains. Thus, data were evaluated in a tabular format in which the numbers of animals showing a particular histological characteristic were listed. For comparisons between the mutant (Gaa−/−) and WT strains, statistical significance was set at P < 0.05.
Genome-wide screening of mRNA expression.
Mice (n = 4/group) were injected intraperitoneally (ip) with Beuthanasia. After the loss of the hindlimb withdrawal reflex, cervical spinal tissues (C3–C5) were harvested, placed into RNA Later (Life Technologies, Carlsbad, CA), and stored at −80°C. RNA extraction was performed using TRIzol and isolated total RNA was purified using an RNeasy Mini kit (Qiagen, Valencia, CA), according to manufacturer's instructions. The resulting quantity and purity of total RNA were tested through absorbance spectrophotometry at 230, 260, and 280 nm. For microarray analysis, RNA samples from 6 mo and 16 mo Gaa−/− and WT mice (4 samples/group) were sent to the Boston University Medical Center Microarray Core Facility to measure the expression of well-annotated genes using the Affymetrix Mouse Gene Array 2.0ST. Raw Affymetrix CEL files were normalized to produce gene-level expression values using the robust multiarray average (RMA) (21) in the affy package (version 1.36.1; Ref. 17) included in the Bioconductor software suite (version 2.12; Ref. 19) and an Entrez Gene-specific probe-set mapping (17.0.0) from the Molecular and Behavioral Neuroscience Institute (Brainarray) at the University of Michigan (Ref. 8; http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF) that maps the probes on the array to unique Entrez Gene Identifiers. Expression values were log2-transformed by default. Array quality was assessed by computing relative log expression (RLE) and normalized unscaled standard error (NUSE) using the affyPLM Bioconductor package (version 1.34.0; Ref. 3). For each sample, median RLE values > 0.1 or NUSE values > 1.05 are considered out of the usual limits, although RLE is the quality metric most strongly associated with technical quality. All arrays had median RLE and NUSE values well within these limits. Principal component analysis (PCA) was performed using the prcomp R function with expression values that had been normalized across all samples to a mean of zero and a standard deviation of one. We assessed pairwise differential gene expression by performing Student's t-test on the coefficients of simple linear models created using the lmFit function in the limma package (version 3.14.4). Correction for multiple hypothesis testing was accomplished using the Benjamini-Hochberg false discovery rate (FDR; Ref. 1). Human homologs of mouse genes were identified using HomoloGene (version 68; Ref. 41a). All microarray analyses were performed using the R environment for statistical computing (version 2.15.1). Both .cel files and expression values were deposited into MIAME-compliant National Center for Biotechnology Information Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/query/.
Enriched signaling pathway analysis.
Genes significantly upregulated or downregulated in Gaa−/− mice were further analyzed with the National Institutes of Health-sponsored Broad Institute's Molecular Signatures Database (MSigDB; http://www.broadinstitute.org/gsea/msigdb/index.jsp) to identify enriched signaling pathways (32, 33). We created gene lists by using the differential gene expression analyses and filtering analyzed genes by P value (<0.05, t-test of the fold change value compared between two groups) and fold change (positive or negative) for a given comparison (e.g., 6 mo Gaa−/− vs. 6 mo WT). The four analyzed comparisons used to create gene lists included: 1) 6 mo Gaa−/− vs. 6 mo WT, 2) 6 mo Gaa−/− vs. 16 mo Gaa−/−, 3) 6 mo WT vs. 16 mo WT, 4) 16 mo Gaa−/− vs. 16 mo WT. Up- and downregulated genes were also analyzed for age-genotype interactions by a one-way ANOVA. The gene lists were analyzed using four or five distinct pathway databases within MSigDB: Canonical, Reactome, Kyoto Encyclopedia of Genes and Genomes (KEGG), Hallmark, and Biological processes for both up- and downregulated genes. These subdatabases emphasize distinct cellular processes; thus, this analytic approach provided a comprehensive overview of signaling changes. For example, the KEGG pathway database is a collection of pathway maps that represent molecular interaction networks for metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, human diseases, and drug development (22, 23). For pathway analysis, multiple comparisons were tested, thus we used the optimized false discovery rate q value (FDR-q) with statistical significance set to ≤ 0.05. In this setting, q values are more appropriate because a q value of 0.05 implies only 5% of significant tests will produce a false positive (rather than 5% of all tests as occurs with P values). Thus, all pathways meeting these significance criteria, up to the 100 most significant, were included in analysis. Specific outputs (e.g., canonical) were selected for the figures based on the most statistically robust findings and to illustrate the range of results.
RESULTS AND DISCUSSION
Data from Pompe animal models (9, 11, 45, 55) and patients (9, 16, 35, 37, 38, 50) support the hypothesis that neuropathology contributes to motor impairments in Pompe disease, but clinical treatments do not address this problem. Here we used unbiased genomic screening and histological evaluation of the Pompe (Gaa−/−) mouse to confirm progressive neuropathology and activation of neurodegenerative signaling cascades in cervical spinal cord. These data provide unequivocal support for the development of CNS-directed treatments in Pompe disease.
Histopathology and glycogen accumulation in the Gaa−/− cervical spinal cord.
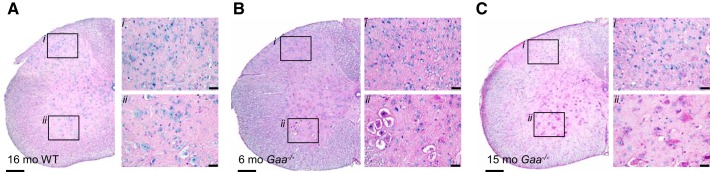
The histopathologic appearance of the cervical spinal cord in Gaa−/− mice was demonstrated with PAS staining (9, 55) (Table 1). The PAS stain imparts a pink color to glycogen and was negative in spinal cords from WT mice at any age (Fig. 1A). In contrast, all 6–8 mo Gaa−/− cervical spinal cords showed robust PAS staining in the ventral gray matter (lamina VIII), including the immediate vicinity of the phrenic motor nucleus (Refs. 43, 55; Table 1, P = 0.014). PAS staining extended dorsally to the intermediate gray (lamina V–VII; Fig. 1B, Table 1; P = 0.014) in all samples, but the cervical dorsal horn (lamina I–III) was positive in only 2/4 Gaa−/− cords at 6–8 mo (Fig. 1B, P = 0.214). By 12–15 mo, however, PAS staining extended from the ventral gray matter (lamina VIII) and dorsal horn (lamina II–III, Fig. 1C) in 4/4 Gaa−/− samples (P = 0.014 vs. WT). Dorsal horn PAS staining was mainly localized to lamina II–III and only extended to lamina I in 1/4 Gaa−/− spinal cords. Consistent with prior reports (9, 11, 12, 31), Gaa−/− motoneurons at both time points showed vacuolization throughout the soma (e.g., Fig. 1).
Table 1.
Summary of PAS-positive staining
| WT (6–8 mo) | Gaa−/− (6–8 mo) | WT (12–15 mo) | Gaa−/− (12–15 mo) | |
|---|---|---|---|---|
| Dorsal horn | 0/4 | 2/4 | 0/4 | 4/4* |
| Intermediate gray | 0/4 | 4/4* | 0/4 | 4/4* |
| Ventral horn | 0/4 | 4/4* | 0/4 | 4/4* |
The periodic acid Schiff (PAS) stain imparts a pink color in neurons considered positive (e.g., Fig. 1). The table depicts the proportion of Pompe (Gaa−/−) and wild-type (WT) mice showing positive PAS staining in specifically defined regions (e.g., 4/4 indicates that 4 of 4 mice examined showed PAS positive neurons). Proportions were examined by Fisher's exact test.
P < 0.05.
Fig. 1.
Periodic acid-Schiff (PAS) staining confirms glycogen accumulation in Pompe (Gaa−/−) but not wild-type (WT) mouse cervical spinal cords. Neuronal PAS staining is absent in the WT cervical spinal cord (A), but it is prominent in the ventral horn in 6–8 mo Gaa−/− cervical spinal tissues (B), and throughout the cervical spinal tissues from 12–15 mo Gaa−/− mice (C). In all panels, i indicates the dorsal horn region and ii indicates the ventral horn region. Calibration bars indicate 200 μM (lower-power images) and 20 μM (higher-power images).
Spinal cord transcriptome in Gaa−/− mice.
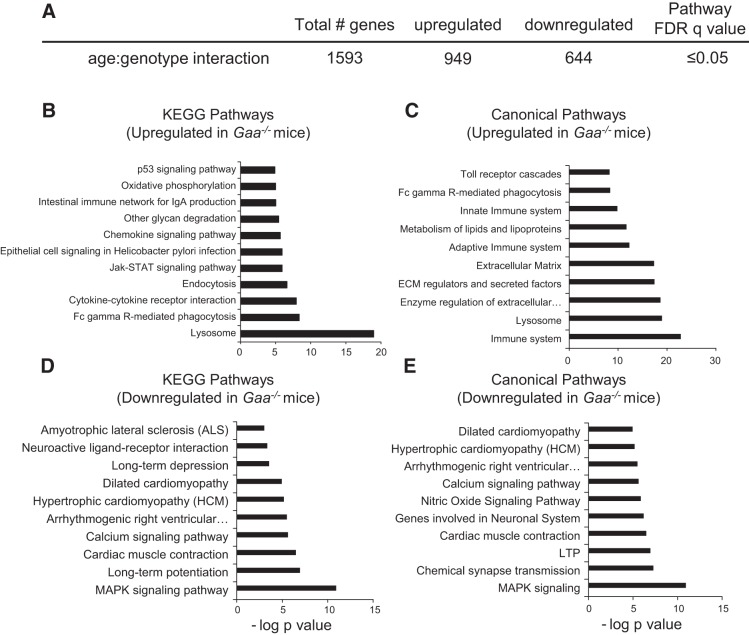
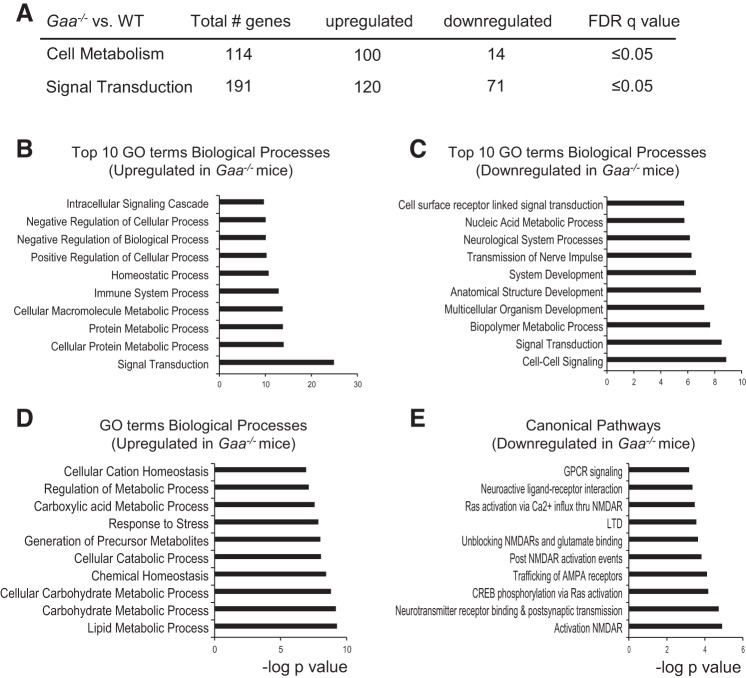
As summarized in Table 2, three different comparisons of pathway activation were performed: 1) 6 mo Gaa−/− vs. 6 mo WT, 2) 6 mo Gaa−/− vs. 16 mo Gaa−/−, and 3) 16 mo Gaa−/− vs. 16 mo WT. These initial analytical approaches confirm that highly significant changes in the spinal cord transcriptome were occurring as Gaa−/− mice were aging, and this is consistent with the progressive nature of Pompe disease (reviewed in Refs. 15, 34). We next examined age-genotype interactions within the top 100 enriched signaling pathways. The 10 most significantly enriched KEGG and canonical signaling pathways for up- and downregulated genes are detailed in Fig. 2. This analysis indicates the Gaa−/− spinal cord had an increase in neuroinflammatory signaling, an immune system response, and extracellular matrix alterations (Fig. 2, B and C). The proapoptotic p53 signaling pathway was upregulated in Gaa−/− mice, suggesting on-going cell death (Fig. 2B). Downregulated pathways included MAPK signaling (Fig. 2, D and E), long-term potentiation (LTP; Fig. 2, D and E), long-term depression (LTD, Fig. 2D), and synaptic transmission (Fig. 2, D and E). Thus, there were highly significant changes in mRNA expression related to 1) neurodegeneration and neuronal loss, 2) neuroinflammation, 3) signal transduction/synaptic plasticity, and 4) cell metabolism. These themes are relevant to the etiology, clinical progression, and treatment of Pompe disease, and each theme is considered separately.
Table 2.
Summary of differential gene expression between groups
| Comparison | Total Genes, n | Upregulated Genes | Top 5 Upregulated Canonical Pathways | Downregulated Genes | Top 5 Downregulated Canonical Pathways |
|---|---|---|---|---|---|
| 6 mo Gaa−/−vs. 6 mo WT | 829 | 449 | lysosome, immune system, extracellular matrix, metabolism of lipids & lipoproteins, glycan degradation | 380 | ErbB1 downstream signaling, neuroactive ligand-receptor interaction, Huntington's disease, PDGFR-beta signaling, respiratory electron transport & ATP synthesis |
| 6 mo Gaa−/−vs. 16 mo Gaa−/− | 2,342 | 1,414 | immune system, extracellular matrix, lysosome, metabolism of lipids & lipoproteins, hemostasis | 928 | neuronal system, chemical synapse transmission, calcium signaling pathway, long-term potentiation, neurotransmitter binding & postsynaptic transmission |
| 16 mo Gaa−/−vs. 16 mo WT | 2,522 | 1,487 | lysosome, immune system, extracellular matrix, metabolisms of lipids & lipoproteins, adaptive immune system | 1,035 | neuronal system, chemical synapse transmission, calcium signaling pathway, long-term potentiation, neurotransmitter binding & postsynaptic transmission |
The table summarizes the number of genes that changed when comparing specific groups (e.g., 6 mo Gaa−/−vs. 6 mo WT; t-test P value <0.05); and also lists the Top 5 Canonical signaling pathways that were up- or downregulated for each comparison. Genes were considered significantly different at P < 0.05. Enriched signaling pathways had q values < 0.005. The findings support the progressive nature of the disease as 2,342 genes changed between 6 mo Gaa−/−vs. 16 mo Gaa−/−mice.
Fig. 2.
Transcriptome assessment suggests altered signaling pathways in Gaa−/− cervical spinal tissues. Total gene changes based on age-genotype interaction analysis are summarized in A. The top 10 upregulated Kyoto Encyclopedia of Genes and Genomes (KEGG) (B) and Canonical (C) pathways show signaling changes related to the lysosomes, inflammation, and cell death. The top 10 downregulated KEGG (D) and Canonical (E) signaling pathways reveal decreases in neuronal signal transduction, long-term potentiation (LTP), and similarities to other diseases. Enriched signaling pathways are graphed as the negative log10 of their P value. Statistical significance was set to a false discovery rate (FDR) q value of <0.05.
Neurodegeneration and neuronal loss.
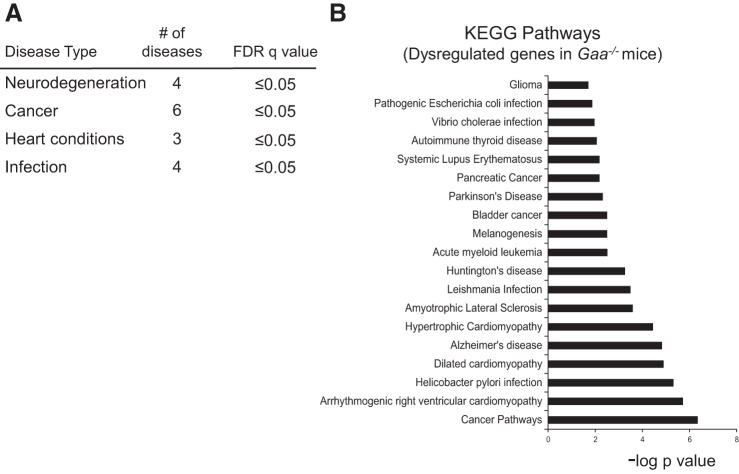
Pathway analyses indicated genetic similarities with other diseases including cancer (n = 6 types), heart conditions (n = 3), infections (n = 4), and neurodegenerative conditions (n = 4, Fig. 3A). Shifts in gene expression between Pompe and each disease category were specific and extended beyond the proinflammatory signaling also described in these conditions (see “Neuroinflammation” below). Of particular interest, there were several overlaps in the genetic signature of Pompe disease with other neurodegenerative conditions, as follows.
Fig. 3.
Transcriptome similarities between Pompe and other diseases. Analysis of the top 100 KEGG signaling pathways (FDR q value <0.05) revealed signaling changes in Gaa−/− tissues that were similar to pathways activated in other diseases. A: a summary of major disease categories. B: a list of specific diseases. Enriched signaling pathway data are expressed as the negative log10 of the P value.
Neurodegenerative diseases such as Parkinson's, Huntington's, and amyotrophic lateral sclerosis (ALS) are characterized by neuronal death from pathogenic cellular and molecular changes within specific CNS regions (10). These neurodegenerative diseases share several common features including abnormal protein deposition, dysfunctional cellular transport, mitochondrial deficits, glutamate excitotoxicity, iron accumulation, and inflammation (10). A recent mRNA microarray study of human brain tissue found that Alzheimer's, Huntington's, Parkinson's, and ALS had >60 dysregulated genes in common (10). This is consistent with the notion that neurodegenerative diseases have a “genetic signature” (5, 6, 10). The current study indicates that Pompe disease also has a neurodegenerative genetic signature. As shown in Fig. 3B, KEGG analyses revealed significant similarities between the Gaa−/− spinal cord and known genetic changes occurring in the CNS in Alzheimer's, Parkinson's, and Huntington's disease as well as ALS.
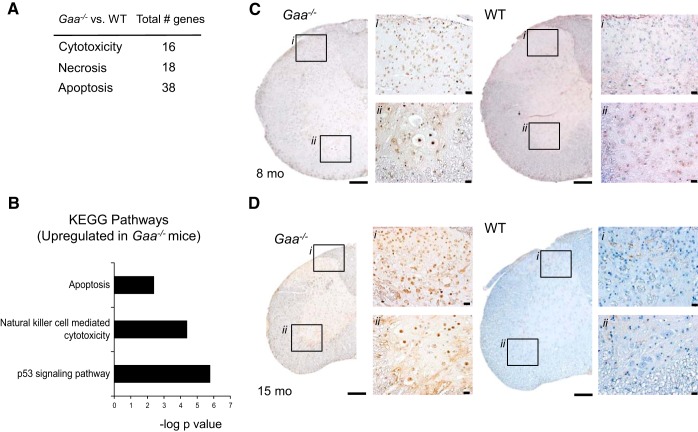
Neuronal loss is a hallmark of neurodegenerative processes, and there is suggestion from Gaa−/− mice (9) and clinical evaluations (46, 47) that neuronal loss may be occurring in Pompe disease. Therefore, we specifically evaluated pathways associated with cytotoxicity, necrosis, and apoptosis (Fig. 4A). Robust increases in p53, apoptotic, and natural killer cell cytotoxicity pathways were present in the Gaa−/− spinal cord (Fig. 4B). We therefore stained for DNA fragmentation using the TUNEL assay (18) to evaluate the possibility of cell death in spinal cords from 6–8 mo and 12–15 mo in Gaa−/− and WT mice. Clear TUNEL staining was noted in 6–8 mo Gaa−/− cords but was absent in WT tissues (Table 3). Spinal dorsal horn neurons (lamina I–III) had increased incidence of apoptotic cells (Fig. 4C), and the presence of positive cells spread through the intermediate gray (lamina V–VII) to motoneurons in the ventral gray (lamina VIII) in 4/4 Gaa−/− tissues (Table 3, P = 0.014). Importantly, Fig. 4C shows clear TUNEL labeling in putative phrenic motoneurons based on location and morphologic appearance (42). With respect to aged mice, positive TUNEL staining was present in 3/4 Gaa−/− cervical spinal cords and 0/4 WT cervical spinal cords (Table 3, P = 0.071). In the single cord with negligible TUNEL staining, very few neurons were present and we suspect that the greatest period of cell death had already occurred. In aged Gaa−/− sections, the densest TUNEL-positive staining was found in the intermediate gray (lamina V–VII, Fig. 4D), with a smaller proportion of ventral horn neurons (lamina VIII) labeled (Fig. 4D). Collectively, we noted TUNEL-positive cells in the dorsal horn, intermediate gray, and ventral horn regions in 7/8 Gaa−/− cervical spinal cords compared with 0/8 WT cords (P = 0.0007). Thus, collectively the current data confirm that the absence of functional GAA leads to neurodegenerative processes in the cervical spinal cord.
Fig. 4.
Evidence for cell death in Gaa−/− cervical spinal cord. Upregulation of mRNA expression of genes for cytotoxicity, necrosis, and apoptosis occurred in Gaa−/− tissues (A). Analysis of the top 100 upregulated KEGG signaling pathways (FDR q value <0.05) revealed enriched signaling pathways related to cell death (B). Enriched signaling pathway data are expressed as the negative log10 of the P value. Histological examination showed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells throughout cervical spinal tissue from Gaa−/− but not WT tissues at 6–8 mo (C) and 12–15 mo (D), indicating on-going neuronal cell loss. In C and D, i indicates the dorsal horn region, and ii indicates the ventral horn region. Calibration bars indicated 200 μM (lower-power images) and 20 μM (higher-power images).
Table 3.
Summary of TUNEL-positive labeling
| WT (6–8 mo) |
Gaa−/− (6–8 mo) |
WT (12–15 mo) |
Gaa−/− (12–15 mo) |
|
|---|---|---|---|---|
| Dorsal horn | 0/4 | 4/4* | 0/4 | 3/4 |
| Intermediate gray | 0/4 | 4/4* | 0/4 | 3/4 |
| Ventral horn | 0/4 | 4/4* | 0/4 | 3/4 |
The table depicts the proportion of Gaa−/−and WT mice showing positive staining in the region examined (e.g., 0/4 indicates that 0 of 4 mice examined showed positive staining). There were no terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells detected in WT tissues at either age. Gaa−/−tissues showed profound TUNEL-positive labeling throughout the cervical spinal cord, including in the region of phrenic motoneurons. The period of greatest cell loss may have already occurred in the 1 Gaa−/−animal that did not have positive labeling at 15 mo, as very few cells were visible with the hematoxylin counterstain. The proportion of animals showing positive staining in Gaa−/−vs. WT mice was examined by a Fisher's exact test.
P < 0.05.
Neuroinflammation.
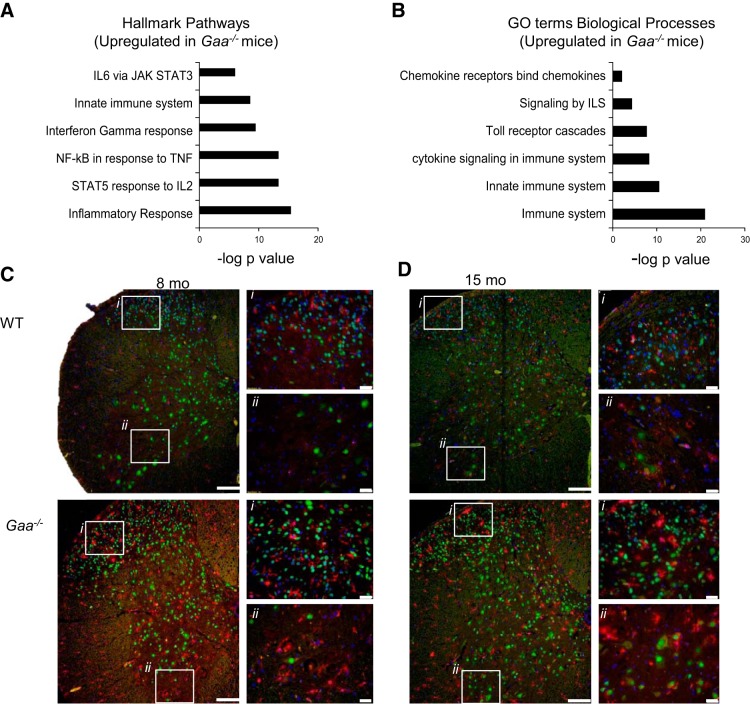
The pathogenesis of neurodegeneration is not restricted to neuronal processes but can also be at least partially attributed to abnormal neuronal-glial interactions (10). Indeed, neuroinflammation is well established in lysosomal storage diseases, and can lead to neuronal loss, astrocytosis and microglial proliferation (25, 40, 57). In Krabbe disease, for example, neuronal-glial metabolic disturbances correlate with the progression of glial activation and neurodegeneration (40). Activation of glial cells in the vicinity of degenerating neurons is also commonly observed in neurodegenerative conditions such as Alzheimer's (49) and ALS (6). A recent gene microarray study of human brain tissue found that the > 60 common genetic changes across a range of neurodegenerative diseases were primarily associated with the innate immune response (10). Moreover, isolated spinal microglia from a murine ALS model have a neurodegeneration-specific gene expression signature (6), and this supports the idea that neuronal-glial disturbances are an important feature underlying neurodegeneration. With respect to Pompe disease, little is known regarding the potential roles of neuroinflammation and neuronal loss to the overall disease progression. The transcriptome assessment indicates that neuroinflammatory and immune system processes are upregulated by 6 mo in Gaa−/− spinal cords, and these processes progress with age (Fig. 5, A and B). For example, Hallmark pathway analysis showed an enriched inflammatory response with FDR q value = 9.55e−15.
Fig. 5.
Proinflammatory signaling and ionized calcium binding adaptor molecule 1 (IBA-1) staining indicate neuroinflammation in Gaa−/− cervical spinal cord. Analysis of upregulated Hallmark signaling pathways (FDR q value <0.05; A) and Gene Ontology (GO) terms biological processes (FDR q value <0.05; B) showed clear increases in proinflammatory signaling in Gaa−/− compared with WT tissues. Enriched signaling pathway data are expressed as the negative log10 of the P value. Histological examination showed no signs of “activated” microglia in WT tissues. However, in Gaa−/− tissues, there were multiple examples of IBA-1-positive microglia with a “bushy” appearance, an amoeboid morphology, or colocalization with neurons at 6–8 mo (C) and 12–15 mo age points (D). In C and D, i indicates the dorsal horn region and ii indicates the ventral horn region. Calibration bars indicate 100 μM (lower-power images) and 20 μM (higher-power images). IBA-1 staining is shown in red, NeuN is shown in green, and DAPI is in blue.
Microglia in the cervical spinal cord were histologically evaluated with antibodies against IBA-1 (Table 4). There was no evidence of “activated” microglial morphology in any region of WT cervical spinal cords (Fig. 5, C and D). In contrast, IBA-1-positive cells in Gaa−/− tissues often displayed a hypertrophied or “activated” morphology, with examples of amoeboid phenotypes in the dorsal horn (lamina I–IV), the intermediate gray (lamina V–VII), and the ventral horn (Fig. 5, C and D). The IBA-1 staining was apparent in 6–8 mo Gaa−/− tissues with evidence of activated microglia in 3/3 samples in the dorsal horn (Fig. 5C), and 2/3 samples in the intermediate gray and ventral horn (P = 0.14, Fig. 5C). In Gaa−/− tissues from 12–15 mo old mice, 4/4 cords showed evidence of activated microglia in the dorsal horn, intermediate gray and ventral horn regions (P = 0.014, Fig. 5D). Of the total spinal cords evaluated, activated microglial phenotypes were seen in 0/8 WT cords but in comparison were seen in 7/8 Gaa−/− intermediate gray and ventral horn regions and 8/8 Gaa−/− dorsal horn regions, respectively. (P ≤ 0.001). Overall, these data confirm a strong immune and inflammatory component to Pompe neuropathology, and in turn this raises the possibility exploring anti-inflammatory treatments.
Table 4.
Summary of IBA-1 staining
| WT (6–8 mo) |
Gaa−/− (6–8 mo) |
WT (12–15 mo) |
Gaa−/− (12–15 mo) |
|
|---|---|---|---|---|
| Dorsal horn | 0/4 | 3/3* | 0/4 | 4/4* |
| Intermediate gray | 0/4 | 2/3 | 0/4 | 4/4* |
| Ventral jorn | 0/4 | 2/3 | 0/4 | 4/4* |
The table depicts the proportion of Gaa−/−and WT mice showing positive staining in the region examined (e.g., 0/4 indicates that 0 of 4 mice examined showed positive staining). Robust cervical spinal ionized calcium binding adaptor molecule (IBA-1) staining was observed in more Gaa−/− than WT tissues. The proportion of animals showing positive staining in Gaa−/−vs. WT mice was examined bya Fischer's exact test;
indicates P < 0.05.
Signal transduction.
Dysregulated synaptic transmission and reduced capacity for synaptic plasticity are common in neurodegenerative diseases. For example, attenuated synaptic plasticity and/or “metaplasticity” have been documented in Alzheimer's (39), Parkinson's (26), and Huntington's (41) disease. The transcriptome assessment of the Gaa−/− spinal cord indicates that similar processes are occurring in Pompe disease as evidenced by downregulation of pathways associated with glutamatergic synaptic transmission, LTP, and LTD (Figs. 2 and 6). Disrupted neural signal transmission was another theme that emerged at the 6 mo time point and progressed by 16 mo. Pathway analyses also showed that cell-cell signaling, signal transduction, cell surface receptor-linked signal transduction, and nerve impulse transmission were four of the 10 most significantly downregulated biological processes (Fig. 6C). Canonical pathway analysis (Top 100 pathways, FDR q value <0.05) revealed significant downregulation of glutamatergic receptor related processes including AMPA receptor trafficking, NMDA receptor unblocking, activation and glutamate binding, and attenuated G protein-coupled receptor signaling (Fig. 6E). Respiratory-related synaptic transmission relies heavily on glutamatergic signaling, and accordingly potential reductions in glutamatergic signaling could be part of a “molecular substrate” that contributes to decreases in respiratory nerve output in Gaa−/− mice (9) and/or Pompe patients (46). This possibility is supported by recent experiments showing that enhancement of AMPA-mediated glutamatergic currents can improve respiratory motor output in Gaa−/− mice. Specifically, systemic treatment with a pharmacologic compound that acts as a positive allosteric modulator of AMPA receptors (ampakine CX717) increases respiratory-related motor output by 300–400% in Gaa−/− mice, but the equivalent dose has no effect in WT mice (12). These findings are physiologically consistent with the current mRNA data indicating that spinal cord AMPA receptor subunits are significantly downregulated in 16-mo Gaa−/− mice and are unchanged in WT (Fig. 6). Collectively, these data suggest that spinal synaptic signaling may be impaired in Pompe disease and also serve to identify molecular targets for future evaluation of synaptic function in Gaa−/− mice.
Fig. 6.
mRNA changes indicate altered cell metabolism and signal transduction in Gaa−/− cervical spinal cord. Genes related to cell metabolism and signal transduction were dysregulated in Gaa−/− tissues (A). B and C: the top 10 up- and downregulated biological processes, respectively. D: further shifts in cell metabolism. There was also clear evidence for downregulation of genes related to excitatory neural transmission (E). Enriched signaling pathways are graphed as the negative log10 of the P value. Statistical significance was set to an FDR q value of <0.05.
Cell metabolism.
The accumulation of lysosomal glycogen can lead to dramatic shifts in cellular metabolic processes, and the current data confirm that this is happening in the cervical spinal cord. In 6 mo Gaa−/− mice, transcriptome changes were consistent with attenuated signaling in molecular pathways associated with the Krebs cycle, the respiratory transport chain, and ATP synthesis as well as large increases in other glycan degradation mechanisms. Overall, mRNA expression of 114 different cell metabolism-related genes were significantly different than WT (Fig. 6A), of which 100 were upregulated. Figure 6, B and C, shows the top 10 most significantly changed biological processes and illustrates a wide range of alterations in cellular metabolism, including lipid, carbohydrate, and protein metabolism (Fig. 6, B and D). Fewer metabolic processes were downregulated and were mainly limited to nucleic acid metabolism and biopolymer metabolism (Fig. 6C).
Conclusions and Significance
Overall, our study confirms that systemic absence of GAA, as occurs in the most severe cases of Pompe disease, is associated with activation of neuropathological signaling cascades in the cervical spinal cord. Analyses of Gaa−/− transcriptome age-genotype interactions indicated progressive activation of signaling pathways associated with cell death and inflammation, and alterations in pathways related to signal transduction and metabolism. Collectively, these pathways comprise a “neurodegenerative signature” related to Pompe disease progression, and coupled with the new histological evidence presented here regarding cell death (e.g., Fig. 4) and prior observations (9, 16, 35, 37, 38, 50), the data indicate that the current clinical paradigm of ERT for treating Pompe disease is likely to be insufficient. Indeed, muscle-directed ERT treatment can be expected to slow disease progression (56), but clinical results indicate that patients will eventually experience respiratory failure despite ERT (28). In addition, recent successes using implanted wires to electrically stimulate the diaphragm in Pompe (i.e., “diaphragm pacing”) are consistent with a neurologic contribution to respiratory insufficiency (47). Specifically, if skeletal muscle dysfunction alone were responsible for breathing impairments, then electrical activation of the diaphragm would not be capable of sustaining ventilation even for short periods. Another recent study documents reductions in evoked diaphragm compound muscle action potentials during bilateral phrenic magnetic stimulation in Pompe patients, a finding that is consistent with a reduction in the overall number of diaphragm motor units (i.e., as would be expected if neurodegenerative processes were ongoing) (46).
The current results support the hypothesis that neuropathology is prominent in the Pompe CNS and contributes to motor impairments (9, 15). In turn, this underscores the need for early therapeutic intervention capable of targeting the CNS (52). Gene therapy approaches show great promise in that regard since both intramuscular and intravenous delivery can effectively transduce neurons. Initial results from diaphragm gene transfer in Pompe patients are promising and show that direct diaphragm delivery of recombinant adeno-associated virus (serotype 1) encoding human GAA (rAAV1-hGAA) is safe and resulted in a >400% increase in the duration of unassisted breathing that could be tolerated with no ventilator support (48). Future work will focus on AAV serotypes capable of efficient retrograde movement and methods for widespread CNS transduction (4). A final consideration is that acceptance of Pompe as a neurodegenerative condition will necessitate additional metrics for evaluating therapeutic outcomes, particularly in preclinical studies using animal models. In addition to standard measures of GAA activity and glycogen clearance, assessments of neuronal cell death and neuroinflammation will be needed.
GRANTS
Funding sources: Clinical and Translational Science Award Grant U54-TR001012 (Boston University), National Institutes of Health Grants 2R01HD-052682-06A1 (D. D. Fuller and B. J. Byrne) and K01AR-066077, and Muscular Dystrophy Association Grant 216676 (D. J. Falk).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
S.M.F.T., D.J.F., B.J.B., and D.D.F. conception and design of research; S.M.F.T. performed experiments; S.M.F.T. analyzed data; S.M.F.T., D.J.F., B.J.B., and D.D.F. interpreted results of experiments; S.M.F.T. prepared figures; S.M.F.T. drafted manuscript; S.M.F.T., D.J.F., B.J.B., and D.D.F. edited and revised manuscript; S.M.F.T., D.J.F., B.J.B., and D.D.F. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Marda Jorgenson for expert help with histological and immunochemical procedures.
REFERENCES
- 1.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) : 289–300, 1995. [Google Scholar]
- 2.Bosch ME, Kielian T. Neuroinflammatory paradigms in lysosomal storage diseases. Front Neurosci : 417, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brettschneider J, Collin F, Bolstad BM, Speed TP. Quality assessment for short oligonucleotide microarray data. Technometrics : 241–264, 2008. [Google Scholar]
- 4.Byrne BJ, Falk DJ, Clement N, Mah CS. Gene therapy approaches for lysosomal storage disease: next-generation treatment. Hum Gene Ther : 808–815, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, Guan Q, Nie ZY, Jin LJ. Gene expression profile and functional analysis of Alzheimer's disease. Am J Alzheimers Dis Other Demen : 693–701, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O'Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, Myers RM, Maniatis T. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep : 385–401, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res : e175, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeRuisseau LR, Fuller DD, Qiu K, DeRuisseau KC, Donnelly WH Jr, Mah C, Reier PJ, Byrne BJ. Neural deficits contribute to respiratory insufficiency in Pompe disease. Proc Natl Acad Sci USA : 9419–9424, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durrenberger PF, Fernando FS, Kashefi SN, Bonnert TP, Seilhean D, Nait-Oumesmar B, Schmitt A, Gebicke-Haerter PJ, Falkai P, Grünblatt E, Palkovits M, Arzberger T, Kretzschmar H, Dexter DT, Reynolds R. Common mechanisms in neurodegeneration and neuroinflammation: a BrainNet Europe gene expression microarray study. J Neural Transm : 1055–1068, 2015. [DOI] [PubMed] [Google Scholar]
- 11.Elmallah MK, Falk DJ, Nayak S, Federico RA, Sandhu MS, Poirier A, Byrne BJ, Fuller DD. Sustained correction of motoneuron histopathology following intramuscular delivery of AAV in Pompe mice. Mol Ther : 702–712, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ElMallah MK, Pagliardini S, Turner SM, Cerreta AJ, Falk DJ, Byrne BJ, Greer JJ, Fuller DD. Stimulation of respiratory motor output and ventilation in a murine model of Pompe disease by ampakines. Am J Respir Cell Mol Biol : 326–335, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falk DJ, Mah CS, Soustek MS, Lee KZ, Elmallah MK, Cloutier DA, Fuller DD, Byrne BJ. Intrapleural administration of AAV9 improves neural and cardiorespiratory function in Pompe disease. Mol Ther : 1661–1667, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falk DJ, Soustek MS, Todd AG, Mah CS, Cloutier DA, Kelley JS, Clement N, Fuller DD, Byrne BJ. Comparative impact of AAV and enzyme replacement therapy on respiratory and cardiac function in adult Pompe mice. Mol Ther Methods Clin Dev : 15007, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller DD, ElMallah MK, Smith BK, Corti M, Lawson LA, Falk DJ, Byrne BJ. The respiratory neuromuscular system in Pompe disease. Resp Physiol Neurobiol : 241–249, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambetti P, DiMauro S, Baker L. Nervous system in Pompe's disease. Ultrastructure and biochemistry. J Neuropathol Exp Neurol : 412–430, 1971. [DOI] [PubMed] [Google Scholar]
- 17.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics : 307–315, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol : 493–501, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol : R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschhorn R, Reuser AJJ. Glycogen storage disease type II: acid α-glucosidase (acid maltase) deficiency. In: The Metabolic Basis of Inherited Disease, edited by Scriver CR, Beaudet AL, Sly WS, Valle D. New York: McGraw Hill, 2001, p. 3389–3420. [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics : 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res : 27–30, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res : D277–D280, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karperien A, Ahammer H, Jelinek HF. Quantitating the subtleties of microglial morphology with fractal analysis. Front Cell Neurosci : 3, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawashita E, Tsuji D, Toyoshima M, Kanno Y, Matsuno H, Itoh K. Prostaglandin E2 reverses aberrant production of an inflammatory chemokine by microglia from Sandhoff disease model mice through the cAMP-PKA pathway. PLoS One : e16269, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SJ, Udupa K, Ni Z, Moro E, Gunraj C, Mazzella F, Lozano AM, Hodaie M, Lang AE, Chen R. Effects of subthalamic nucleus stimulation on motor cortex plasticity in Parkinson disease. Neurology : 425–432, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishnani PS, Beckemeyer AA. New therapeutic approaches for Pompe disease: enzyme replacement therapy and beyond. Pediatr Endocrinol Rev , Suppl 1: 114–124, 2014. [PubMed] [Google Scholar]
- 28.Kishnani PS, Corzo D, Leslie ND, Gruskin D, Van der Ploeg A, Clancy JP, Parini R, Morin G, Beck M, Bauer MS, Jokic M, Tsai CE, Tsai BW, Morgan C, O'Meara T, Richards S, Tsao EC, Mandel H. Early treatment with alglucosidase alpha prolongs long-term survival of infants with Pompe disease. Pediatr Res : 329–335, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kressel M, Groscurth P. Distinction of apoptotic and necrotic cell death by in situ labelling of fragmented DNA. Cell Tissue Res : 549–556, 1994. [DOI] [PubMed] [Google Scholar]
- 30.Lee CC, Chen CY, Chou TY, Chen FH, Zimmerman RA. Cerebral MR manifestations of Pompe disease in an infant. AJNR Am J Neuroradiol : 321–322, 1996. [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KZ, Qiu K, Sandhu MS, Elmallah MK, Falk DJ, Lane MA, Reier PJ, Byrne BJ, Fuller DD. Hypoglossal neuropathology and respiratory activity in pompe mice. Front Physiol : 31, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol Biol : 153–160, 2014. [DOI] [PubMed] [Google Scholar]
- 33.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics : 1739–1740, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim JA, Li L, Raben N. Pompe disease: from pathophysiology to therapy and back again. Front Aging Neurosci : 177, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancall EL, Aponte GE, Berry RG. Pompe's disease (diffuse glycogenosis) with neuronal storage. J Neuropathol Exp Neurol : 85–96, 1965. [DOI] [PubMed] [Google Scholar]
- 37.Martin JJ, de Barsy T, van Hoof F, Palladini G. Pompe's disease: an inborn lysosomal disorder with storage of glycogen. A study of brain and striated muscle. Acta Neuropathol : 229–244, 1973. [DOI] [PubMed] [Google Scholar]
- 38.Martini C, Ciana G, Benettoni A, Katouzian F, Severini GM, Bussani R, Bembi B. Intractable fever and cortical neuronal glycogen storage in glycogenosis type 2. Neurology : 906–908, 2001. [DOI] [PubMed] [Google Scholar]
- 39.Megill A, Tran T, Eldred K, Lee NJ, Wong PC, Hoe HS, Kirkwood A, Lee HK. Defective age-dependent metaplasticity in a mouse model of Alzheimer's disease. J Neurosci : 11346–11357, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meisingset TW, Ricca A, Neri M, Sonnewald U, Gritti A. Region- and age-dependent alterations of glial-neuronal metabolic interactions correlate with CNS pathology in a mouse model of globoid cell leukodystrophy. J Cereb Blood Flow Metab : 1127–1137, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murmu RP, Li W, Szepesi Z, Li JY. Altered sensory experience exacerbates stable dendritic spine and synapse loss in a mouse model of Huntington's disease. J Neurosci : 287–298, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res : D8–D20, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu K, Falk DJ, Reier PJ, Byrne BJ, Fuller DD. Spinal delivery of AAV vector restores enzyme activity and increases ventilation in Pompe mice. Mol Ther : 21–27, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu K, Lane MA, Lee KZ, Reier PJ, Fuller DD. The phrenic motor nucleus in the adult mouse. Exp Neurol : 254–258, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raben N, Nagaraju K, Lee E, Kessler P, Byrne B, Lee L, LaMarca M, King C, Ward J, Sauer B, Plotz P. Targeted disruption of the acid alpha-glucosidase gene in mice causes an illness with critical features of both infantile and adult human glycogen storage disease type II. J Biol Chem : 19086–19092, 1998. [DOI] [PubMed] [Google Scholar]
- 45.Sidman RL, Taksir T, Fidler J, Zhao M, Dodge JC, Passini MA, Raben N, Thurberg BL, Cheng SH, Shihabuddin LS. Temporal neuropathologic and behavioral phenotype of 6neo/6neo Pompe disease mice. J Neuropathol Exp Neurol : 803–818, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith BK, Corti M, Martin AD, Fuller DD, Byrne BJ. Altered activation of the diaphragm in late-onset Pompe disease. Respir Physiol Neurobiol : 11–15, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith B, Fuller DD, Martin AD, Lottenberg L, Islam S, Lawson LA, Onders RP, Byrne BJ. Diaphragm pacing as a rehabilitative tool for patients with Pompe disease who are ventilator-dependent: case series. Physical Therapy : 696–703, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith BK, Collins S, Conlon T, Mah C, Lawson LA, Martin D, Fuller DD, Cleaver B, Clement N, Phillips D, Islam S, Dobjia N, Byrne B. Phase I/II trial of adeno-associated virus-mediated alpha-glucosidase gene therapy to the diaphragm for chronic respiratory failure in Pompe disease: initial safety and ventilatory outcomes. Hum Gene Ther : 630–640, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoeck K, Schmitz M, Ebert E, Schmidt C, Zerr I. Immune responses in rapidly progressive dementia: a comparative study of neuroinflammatory markers in Creutzfeldt-Jakob disease, Alzheimer inverted question marks disease and multiple sclerosis. J Neuroinflamm : 170, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teng YT, Su WJ, Hou JW, Huang SF. Infantile-onset glycogen storage disease type II (Pompe disease): report of a case with genetic diagnosis and pathological findings. Chang Gung Med J : 379–384, 2004. [PubMed] [Google Scholar]
- 51.Thurberg BL, Lynch Maloney C, Vaccaro C, Afonso K, Tsai AC, Bossen E, Kishnani PS, O'Callaghan M. Characterization of pre- and post-treatment pathology after enzyme replacement therapy for Pompe disease. Lab Invest : 1208–1220, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Todd AG, McElroy JA, Grange RW, Fuller DD, Walter GA, Byrne BJ, Falk DJ. Correcting neuromuscular deficits with gene therapy in Pompe disease. Ann Neurol : 222–234, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner S, ElMallah M, Falk D, Byrne B, Fuller D. Preferential neuropathology in respiratory-related motor nuclei in Pompe mice. FASEB J : 660 3, 2015. [Google Scholar]
- 54.Turner S, Hoyt A, Falk D, Byrne B, Fuller D. Genome-wide assessment of the Pompe (Gaa−/−) mouse cervical spinal cord confirms widespread neuropathology. FASEB J : 1285.–2., 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner SM, Hoyt AK, ElMallah MK, Falk DJ, Byrne BJ, Fuller DD. Neuropathology in respiratory-related motoneurons in young Pompe (Gaa(−/−)) mice. Respir Physiol Neurobiol : 48–55, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Ploeg AT, Clemens PR, Corzo D, Escolar DM, Florence J, Groeneveld GJ, Herson S, Kishnani PS, Laforet P, Lake SL, Lange DJ, Leshner RT, Mayhew JE, Morgan C, Nozaki K, Park DJ, Pestronk A, Rosenbloom B, Skrinar A, van Capelle CI, van der Beek NA, Wasserstein M, Zivkovic SA. A randomized study of alglucosidase alfa in late-onset Pompe's disease. N Engl J Med : 1396–1406, 2010. [DOI] [PubMed] [Google Scholar]
- 57.Vitner EB, Farfel-Becker T, Eilam R, Biton I, Futerman AH. Contribution of brain inflammation to neuronal cell death in neuronopathic forms of Gaucher's disease. Brain : 1724–1735, 2012. [DOI] [PubMed] [Google Scholar]