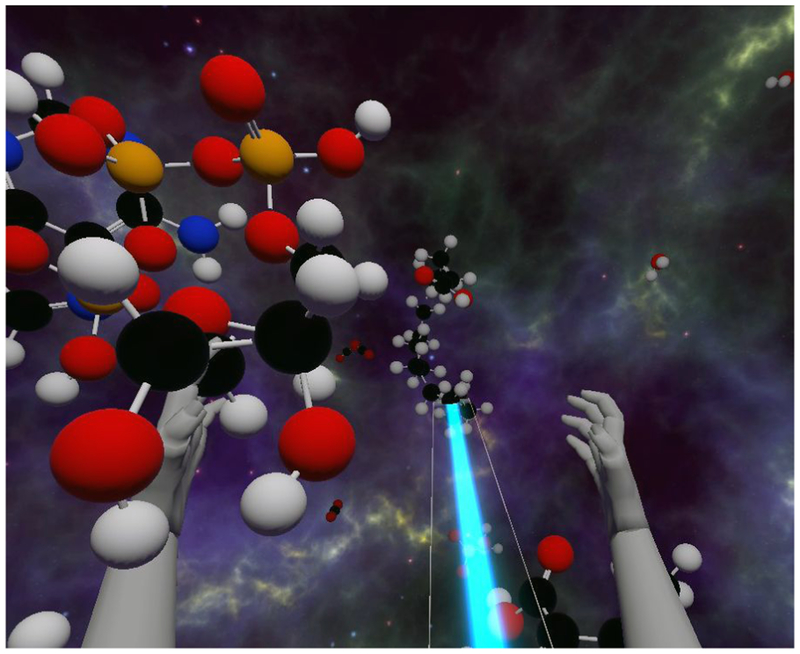
Abstract
Can virtual reality be useful for visualizing and analyzing molecular structures and 3-dimensional (3D) microscopy? Uses we are exploring include studies of drug binding to proteins and the effects of mutations, building accurate atomic models in electron microscopy and x-ray density maps, understanding how immune system cells move using 3D light microscopy, and teaching school children about biomolecules that are the machinery of life. What are the advantages and disadvantages of virtual reality for these applications? Virtual reality (VR) offers immersive display with a wide field of view and head tracking for better perception of molecular architectures and uses 6-degree-of-freedom hand controllers for simple manipulation of 3D data. Conventional computer displays with trackpad, mouse and keyboard excel at two-dimensional tasks such as writing and studying research literature, uses for which VR technology is at present far inferior. Adding VR to the conventional computing environment could improve 3D capabilities if new user-interface problems can be solved. We have developed three VR applications: ChimeraX for analyzing molecular structures and electron and light microscopy data, AltPDB for collaborative discussions around atomic models, and Molecular Zoo for teaching young students characteristics of biomolecules. We describe unique aspects of VR user interfaces, and the software and hardware advances needed for this technology to come into routine use for scientific data analysis.
Keywords: Virtual reality, ChimeraX, Molecular Zoo, AltPDB, Microscopy
Graphical Abstract
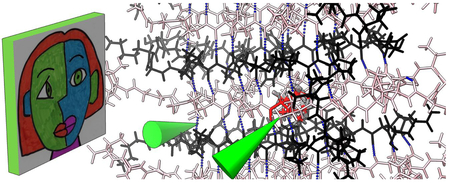
Introduction
A plethora of virtual, augmented and mixed reality display devices intended for video game entertainment and virtual experiences are now available. They have a display strapped to the user’s face providing images to the two eyes from two slightly different viewpoints to allow natural depth perception. They track the view direction and position in space and use this to update the display so that the user appears to occupy a computer-generated 3-dimensional (3D) hologram-like environment. Augmented and mixed reality devices use a see-through display superimposing the computer-generated scene on the actual physical surroundings, while virtual reality devices do not show the surroundings. Some devices include controllers held in each hand, with position and orientation tracked and shown in the scene. The controllers allow interaction with virtual objects using buttons, trackpads, and thumb sticks, and can vibrate for basic touch feedback. Many devices include stereo audio for spatially localized sounds and microphones. The combination of a visual 3D environment, use of hands, touch, and sound produce an immersive multi-modal experience that has more resemblance to the real world than to conventional computer screens, trackpads, mice and keyboards.
Immersive visualization of molecular and cellular structure has been employed for education, communicating results, and research data analysis. Examples include a virtual tour through a breast cancer cell based on serial block-face electron microscopy1, a tour through the bloodstream and into a cell nucleus called The Body VR [http://thebodyvr.com], a learning tool to build organic molecules called NanoOne from Nanome [http://nanome.ai], web tools such as Autodesk Molecule Viewer2 and RealityConvert3 that create virtual reality scenes with protein structures, a collaborative VR environment Arthea developed by Gwydion [https://gwydion.co] for viewing protein structures, and an augmented reality molecular analysis tool ChemPreview4. Molecular Rift5 and 3D-Lab6 facilitate drug design using virtual reality. Most of these applications are based on a video game engine Unity3D [https://unity3d.com] which does not have built-in support for molecules. An open source library, UnityMol7, allows molecular display in Unity3D. Older immersive computer systems called CAVEs (CAVE Automatic Virtual Environment8,9) project images on walls and the floor of a room and use 3D glasses and custom-built head and hand tracking devices; these have been used for quantum mechanical and classical molecular dynamics studies with Caffeine10, building and modeling nanotubes11, and teaching drug receptor interactions12. ConfocalVR™ for visualization of confocal microscope image stacks is described in a companion article in this issue [ref]. Ultimately, all of these systems can trace their lineage to ground breaking work by Ivan Sutherland in 1965 on what he called the “Ultimate Display”13.
We have developed three new virtual reality applications: ChimeraX VR for research data analysis and visualization, AltPDB for group discussions of molecular structures, and Molecular Zoo for teaching young students about familiar biomolecules. These applications focus on time-varying 3D data and multi-person interactions, two areas where immersive environments have substantial advantages over traditional computer displays. Two-dimensional (2D) computer screens can effectively convey the depth of molecular models by using multiple cues. Rotating the models gives a strong sense of depth (the “kinetic depth effect”), and occlusion of far atoms by near ones, dimming distant atoms, and shadows all work to improve 3D perception on a 2D screen. Time-varying 3D models can be thought of as 4-dimensional data, and such motions are difficult to clearly perceive on a 2D screen. Virtual environments with stereo depth perception excel at conveying this type of data. We illustrate this with molecular dynamics to study mutations in proteins and also with 3D light microscopy time series using ChimeraX VR. Our educational application Molecular Zoo presents familiar and simple molecules such as water, aspirin, a saturated fatty acid, and ATP as dynamic, fully flexible ball-and-stick models that can collide and have bonds broken and formed in qualitatively realistic ways. Our interpersonal communication application AltPDB currently does not support dynamics but offers the ability to have collaborating researchers meet and discuss via live audio molecular function using room-scale virtual molecular structures, or have teachers and students meet to discuss molecular mechanisms. Each participant can move and point at the structures, which can be scaled to permit the viewers (for example) to stand within a deeply buried drug binding site inside a protein.
There is a large difference in capabilities between the various virtual, augmented and mixed reality devices currently available. We developed our applications specifically for virtual reality headsets with tracked hand controllers that are driven by laptop and desktop computers. We use HTC Vive, Oculus Rift and Samsung Odyssey headsets which are tethered to a computer having a high performance graphics adapter by a 3-meter cable providing video, sound, tracking data, and power. In contrast, headsets that are driven by a smartphone such as Google Cardboard, GearVR, or Merge 360 are not currently suitable for our applications for several reasons: inadequate graphics performance, no hand controllers, and rotational but not positional tracking. The hand controller and positional tracking limitations are solved by standalone VR headsets such as Oculus Go, Vive Focus and Mirage Solo which do not require a separate computer, but inadequate graphics speed for interacting with molecular data remains a severe limitation of these devices for complex interactive scenes. Augmented and mixed reality devices such as Microsoft Hololens and Meta 2 are much less mature technologies currently offering only developer kit hardware. They have a limited field of view (30 – 50 degrees), and, unlike other application domains, there are few advantages to superimposing molecular data with human-scale physical surroundings. In the future, augmented reality devices may be a good choice for multi-person immersive environments allowing participants to see each other. In summary, consumer computer driven VR headsets are currently the only device type we feel are useful for the interactive visualizations described in this paper.
Results
The ChimeraX visualization program14 displays molecular structures and 3D microscopy data and has approximately one hundred diverse analysis capabilities. In 2014 we began adding the capability to display and manipulate this data using high-end VR headsets (https://www.rbvi.ucsf.edu/Outreach/Dissemination/OculusRift.pdf). In this section, we compare how a task is carried out using VR versus a conventional desktop interface, highlighting the strengths and weaknesses.
Replacing a residue
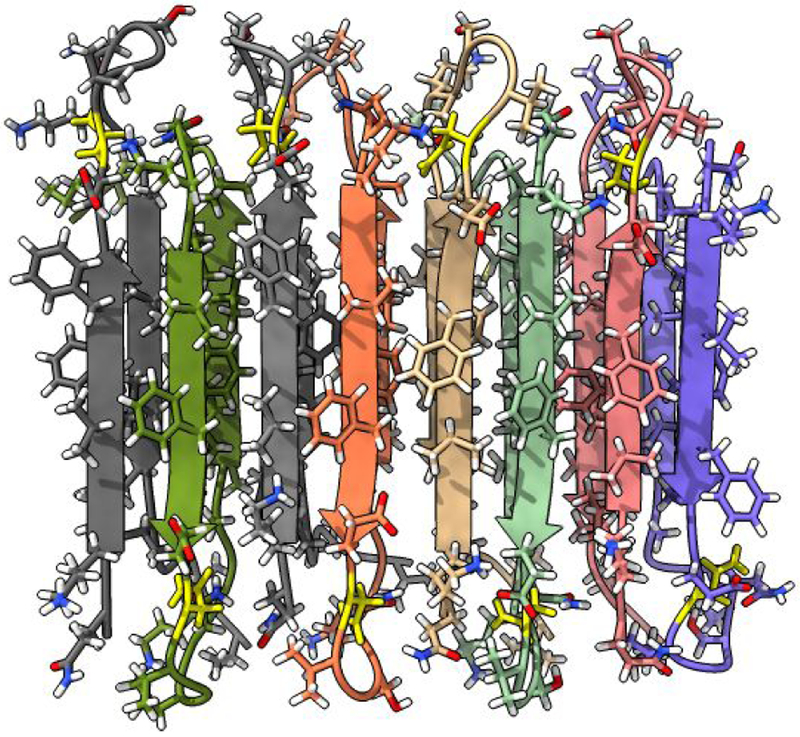
Our example task will be changing an amino acid, rotating a bond, running interactive molecular dynamics to repack nearby residues, and looking for changes in hydrogen bonds in a beta-amyloid fibril (PDB 2LNQ). A single amino acid mutation, aspartic acid to asparagine at residue position 23 of this beta-amyloid peptide is associated with early-onset neurodegeneration15. The mutation allows formation of anti-parallel beta-sheet fibrils (Figure 1). We start with a fibril structure deduced by solid state NMR and revert the mutation to look into why the wild-type peptide does not appear to form anti-parallel beta-sheets.
Figure 1:
Beta-amyloid fibril consisting of stacked anti-parallel beta strands. A single amino acid mutation favors anti-parallel fibril formation. The mutation can be manually reversed in VR and energy-minimized to explore how residue repacking may disrupt the fibril.
Performing these actions takes a few minutes using either conventional or virtual reality interfaces and entails essentially the same steps and achieves the same results. The structure is opened and the mutated residue is colored with commands, toolbar buttons are used to display the structure in stick style and show ribbons, and mouse modes allow swapping amino acids, rotating bonds and running molecular dynamics. In virtual reality the same toolbar buttons and mouse modes are activated using the desktop user-interface window that appears floating in the VR scene (Figure 2), with mouse clicks and drags replaced by hand controller clicks and motions. Although the steps are the same, the experience is substantially different, with the VR method having advantages and disadvantages we now elaborate.
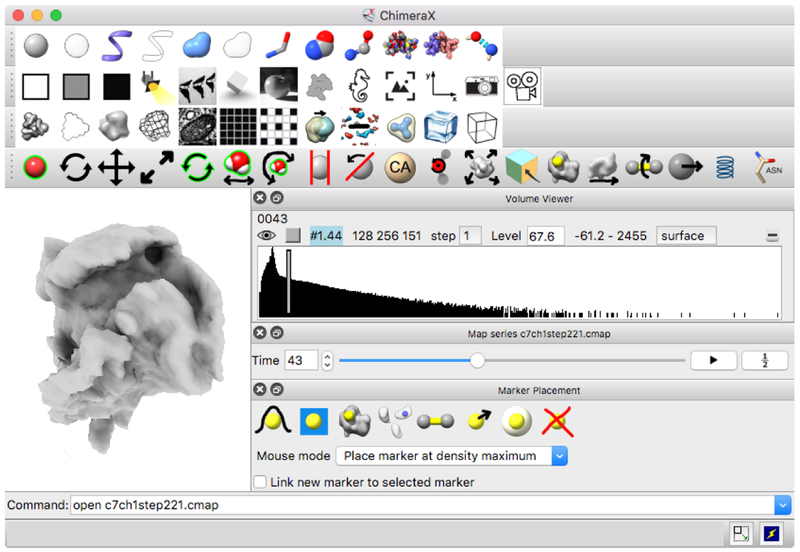
Figure 2:
ChimeraX user interface. Mouse modes, toolbar buttons and other controls can be used with hand controllers on this floating panel in the VR scene. A 3D optical microscopy time series of a crawling neutrophil is shown along with interface elements specific to this type of data in the lower right.
The first steps of opening and coloring involve typing commands, and are done prior to starting the VR which is also done with a command (vr on). This skirts the difficult problem that typing while wearing the VR headset is too cumbersome to be practical. The headset completely blocks the view of a physical keyboard, and the hands are holding controllers. Poking at a virtual keyboard floating in the VR scene is feasible but tedious. The reason these initial steps use typed commands is because they have parameters, the PDB code 2LNQ to be opened and the sequence number of the mutated residue. While there is no drawback of typing initial commands, analysis tasks during a VR session will frequently involve entering parameters such as numbers and names of data sets. A possible solution is to use speech recognition to enter commands while wearing the VR headset, and we describe our tests of speech input below. VR headsets have built-in microphones so no extra hardware is needed.
The steps of changing display style to sticks and ribbon can be done with toolbar buttons since no parameters are needed. In VR a button press on either hand controller shows the ChimeraX desktop window floating in the VR scene, it can be dragged to any position and is initially a meter wide. (When immersed in VR space you perceive distance in true physical dimensions, so one tends to describe scenes in real units such as meters.) The hand controllers are depictedas cones in the scene for accurate pointing, and clicks on any desktop user-interface control act like equivalent mouse clicks. This VR user interface is a poor experience compared to the conventional desktop window for several reasons. Extra effort is required to position a 3D input device on a floating 2D panel compared to using a trackpad or mouse, pointing accuracy is lower, and the panel can be inconveniently out of reach or view when moving or rotating the head to obtain different vantage points. The resolution of the VR display is also much inferior to conventional displays, making text in the user interface panel hard to read. Each eye in a VR device uses a screen with about half the number of pixels (1080 by 1200) of desktop screens (1920 by 1080) and covers 3 times the field of view (100 degrees versus 30 degrees) giving a factor of 6 reduction in pixels per angular degree. Increasing this resolution is the active focus of VR technology development companies, as can be seen by the recently announced HTC Vive Pro (1440 × 1600).
The next step in our task is to obtain a clear view of the mutated residue. In VR either the head can be moved (e.g. while seated in a rolling chair or walking) or either hand can grab the structure (with a button held down) and rotate and move it smoothly matching the hand motion. Bringing the two hands together or apart scales the structure. A typical viewpoint would have the mutated residue half a meter in size and one meter away with orientation chosen to avoid intervening atoms blocking the view. This process is simple and feels natural compared to the equivalent operations without VR.
The process for obtaining a useful 3D view with a conventional 2D user interface is difficult to describe, with several factors contributing to the complexity of the task. Because a mouse has only two degrees of freedom, ChimeraX uses two mouse buttons for translation and rotation. Rotation and translation each have three independent axes, so other techniques enable rotation and translation perpendicular to the screen: z-rotation is implied by initiating the mouse pointer near the edge of the window16, and z-translation is done by holding down the control key while moving the mouse. Scaling is done with the third mouse button or scroll wheel. Laptop computers with a trackpad require emulating three mouse buttons by holding additional keyboard keys (e.g., alt, option, command).
The desirable view will also differ substantially on a conventional display. Because the field of view is much narrower, typically 30 degrees versus 100 degrees in VR, it is undesirable to move in close to the residue of interest because the surrounding residues will then be out of view. But if the viewpoint is not close then other atoms are likely to block the view, a problem solved by use of clipping planes to hide nearby atoms, controlled with yet another mouse mode. Our example system is a fortuitous case where the residue of interest is on the surface and clipping is not needed. An alternative to clipping is to artificially increase the graphics field of view to allow a close vantage-point, but the high degree of distortion from a wide-angle perspective projection on a flat screen makes it hard to perceive spatial relationships and is rarely useful. Researchers become adept at the baroque process of using a 2D interface for 3D visualization, although only a minority discover the advanced techniques such as how to perform rotations and translations perpendicular to the screen. Aside from the difficulty in achieving useful structure views, the vantage achieved in VR gives a dramatically clearer perception of spatial relations due to the close-up view where stereoscopic depth perception is most effective.
The next steps are to compute and display hydrogen bonds as dashed lines, swap amino acid 23, replacing asparagine with aspartic acid, rotate bonds of the new amino acid if needed to avoid clashes with neighbor residues, and run molecular dynamics to equilibrate the new residue and its neighbors. These actions are all performed with the mouse or hand controllers. Hydrogen bonds are displayed by clicking a toolbar button. The swap amino acid mode is enabled by pressing a button on the mouse modes panel; then, a click and drag on the target residue flips through the 20 standard amino acids, replacing the atoms and displaying the new residue 3-letter code as a label, with button release choosing the new amino acid.
Currently a replaced residue is added in a single orientation by alignment of a template to the backbone. ChimeraX’s predecessor program, UCSF Chimera, can choose an appropriate rotamer from a library to reduce clashes and optimize hydrogen bonds, but that capability is not yet available in ChimeraX. So we instead can resolve serious clashes with the bond rotation mouse mode, which rotates atoms on one side of a bond by a click and mouse drag or hand controller twist on the bond. In VR, observing suitable bond rotations is easier than on a 2D screen because the clashes are perceived directly in stereo. Equally important is the ability to move the head to get different viewpoints while simultaneously performing the rotation with one hand. Alternatively, one hand can move the structure while the other hand rotates the bond. On a conventional screen with a mouse, the user instead must switch between rotating the bond and rotating the molecule with different mouse buttons to see the degree of clashes.
Finally, a mouse or VR mode that runs molecular dynamics on any clicked residue and its contacting neighbor residues is used to repack the residues into a lower energy state. Insight into residue motions is much clearer in VR due to the close-up direct 3D perception. The dynamics is calculated using OpenMM17, which requires hydrogen atoms that can be added with a ChimeraX command, and currently uses force field parameterization limited to standard amino acids.
The operations to perform the residue replacement are nearly identical with and without VR. In VR, the same user interface icons that are clicked with a mouse when using a conventional display are instead clicked with either of the VR hand controllers. For assigning VR modes, any of the three buttons on either hand controller can be clicked on the user-interface icons to assign that button to the desired operation. This allows up to six different operations using the two hand controllers. With one button assigned to rotate and move structures there are five additional operations that can be assigned to buttons for immediate use. Using a three-button mouse with scroll-wheel for zooming, typically only one button is reassigned over and over to new operations because the others are needed for rotation, translation and zooming.
In summary, the benefits of using VR in this residue mutation example are improved perception of the spatial arrangement of residues in combination with easier access to many hand controller operations. A recurring aspect in the VR uses that we find most compelling is structure dynamics. When parts of a molecule or cell are moving, understanding this four-dimensional data (space and time variation) on a conventional 2D screen is challenging. The direct stereo depth perception of VR combined with the ability to move one’s head to alter the viewpoint while hands are controlling the structure dynamics provides a clear view of the dynamics not possible with a conventional 2D display.
Refining atomic models in density maps
A second example where VR has substantial advantages is refining and correcting atomic models in X-ray or electron cryo-microscopy density maps. Initial atomic models are “built” into density with automated software such as Phenix18, which can correctly build more than 90% of most structures. Subsequently, interactive refinement is used, where a researcher fixes remaining problems such as incorrect residue positions or register shifts of strands and helices, and may incorporate additional residues. Maps of large systems at lower resolutions (~3 Angstroms) from cryoEM can require extensive interactive refinement because more errors are present. The ChimeraX tool ISOLDE19, developed by Tristan Croll, enables corrections and makes use of interactive molecular dynamics to maintain correct bonded geometry, minimize clashes, and move atoms into density. It also computes and displays 3D indicators for poor backbone geometry and bond angles. Interactive refinement requires excellent perception of the fit of atomic models in density map meshes combined with tugging residues to corrected positions, which we believe would benefit from a VR user interface. The complexity of the task when done with conventional displays is seen in an ISOLDE tutorial video [https://www.youtube.com/watch?v=limaUsNAVL8].
ChimeraX has a mouse and VR mode that allows tugging atoms while running molecular dynamics, but parallel computation of the graphics and dynamics is not yet implemented despite being needed to avoid flickering in VR, as discussed further below. Additional current capabilities assist visually inspecting the fit of residues in density maps. Map contour level can be changed using a hand controller mode dragging up and down. Maps can be cropped to regions of interest with another mode in which a map outline box is shown and faces of the box can be dragged. The main purpose of cropping is to reduce the scene complexity so it can render sufficiently fast for VR. Another mode allows clip planes to be moved and rotated, although clipping is usually not necessary because VR allows close-up views with no obstructing density between the viewer and region of interest. Manual fitting by rigidly moving and rotating component atomic models into density is supported by another hand controller mode, and positions can be optimized to maximize correlation between atomic models and maps. To identify residues, yet another mode allows clicking atoms to display a floating residue name and number.
We have found that the conventional mesh surface display of density maps used in refinement does not display well in VR. The limited VR display resolution results in pixelated (stair-stepped) mesh lines where the steps drift up and down the lines with even small head motions, creating an overwhelming shimmering appearance. Common recommendations in VR video game development call for minimizing use of lines due to these distracting low-resolution artifacts. We find that transparent solid surfaces perform well for atomic model and map comparison. This is initially surprising because such surfaces are not used with 2D displays because of poor perception of depth, but stereoscopic VR remedies that weakness.
Tracking features in 3D light microscopy
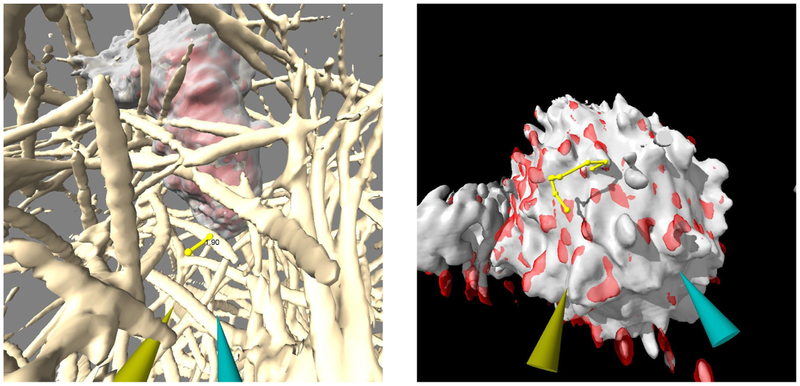
A third ChimeraX VR use is in tracking features in 3D light microscopy time series of live cells. An example is lightsheet microscopy of neutrophils crawling through collagen filaments20 imaged once per second for 3 minutes, with and without motility-affecting drugs. To examine whether the cells propel themselves by pushing or pulling on filaments, we can measure displacements of collagen in contact with cells (Figure 3). This is well-suited to VR as it benefits from an immersive view from within the collagen filament network. A VR mode allows placing markers on the filaments, while a second mode allows switching time points by dragging. Dragging up or down advances or regresses one time step for each 5 centimeters of hand motion. Quickly going back and forth through 20 frames allows moving filaments to be identified and marked. Markers placed on the same filament can be connected and distances shown on the connections using VR modes. Markers can be placed on a surface or on maximum density along a ray extending from the hand controller. Each marker can be moved to improve placement accuracy, resized, or deleted, all by direct action with hand controller buttons.
Figure 3:
Measuring and tracking features in 3D lightsheet microscopy. Measurement of deflection of a collagen filament over a 5-second interval as a crawling neutrophil passes it (left). Manually marked path of motion of a T-cell protrusion over a 15-second interval (right).
Another 3D lightsheet microscopy example involves tracking dozens of T-cell surface protrusions used to interrogate antigen presenting cells21. Again, VR is well suited to the task by combining stereo perception and hand-controlled time series playback to manually track paths of individual protrusions, determine their lifetimes, and assess localization of fluorescently labeled T-cell receptors seen in a second channel (Figure 3). The protrusions move large distances between time points, making automated tracking challenging. Both the neutrophil and T-cell examples would best be done with automated tracking to provide reproducible measurements. Similar to the refinement of atomic models, it is likely that automated tracking will need interactive correction to remove spurious results.
Handling large data
A critical requirement of VR displays is that graphics be rendered at the full 90 frames per second used by current devices. The headset displays we use are 1080 by 1200 pixels for each eye, but warping of the images to correct for optics distortions in the device involve rendering 1344 by 1600 pixels, a size comparable to rendering on conventional desktop displays. Two eye images are rendered for each frame. The rendering speed requirement is thus approximately 180 eye images per second. If the rendering rate is not maintained, the display flickers and the objects do not remain at fixed positions in space as the view direction changes, but instead jitter. Slow rendering causes motion sickness with symptoms of nausea persisting as long as 24 hours. This poses a serious and unfamiliar problem for software developers: applications can make users sick. The rapid performance improvements of Graphics Processing Units (GPUs) is one of the key technology developments that have enabled VR to move from what Fred Brooks, Jr. described in 1999 as “barely working”22 to the successful consumer-level systems of today; we anticipate continued GPU improvements in the future.
Conventional 2D computer displays typically refresh at a rate of 60 frames per second, but large science data sets that render at only 10 frames per second are common, and that frame update rate does not greatly interfere with visualization or analysis. VR requires nearly 20 times that rendering rate because the user’s head position can be constantly changing and it is imperative that the view tracks this head motion in order to avoid inducing nausea. The only way to achieve those speeds for large data sets with today’s GPUs is to reduce the level of detail shown.
As an example, we take an E. coli ribosome structure of 280,000 atoms with a bound antibiotic (PDB 4V7S) and a 130 Mbyte X-ray density map (grid size 253 × 363 × 372) used to determine this structure at 3.3 Angstroms resolution (Figure 4). We simplify the depiction by displaying the ribosomal RNA as ribbons with rungs for each base; proteins are shown with all-atom spheres, the antibiotic is shown in ball-and-stick and the density map is displayed only within a range of 10 Angstroms from the antibiotic. Those settings allow full-speed rendering in VR with a high-end Nvidia GTX 1080 graphics card. The atom spheres appear somewhat faceted due to automatic level-of-detail control which represents each sphere with only 90 triangles. The limited map range and ribbon style choices took some exploration to find settings that allow adequate VR rendering speed, and for many data sets the process of finding suitable settings can be challenging.
Figure 4:
Antibiotic telithromycin (purple) bound at the catalytic center of E. coli ribosome with antibiotic resistance methylation site on ribosomal RNA shown in yellow and X-ray density shown as a transparent surface. Level-of-detail tuning allows viewing complex scenes such as this in VR rendered at 90 frames per second.
In the case of neutrophils crawling through collagen filaments, the 3D image data was reduced by a factor of 2 along x and y axes (original grid size 256 × 512 × 151, 110 time steps, 32-bit floating point numbers) for VR to allow playing the time series while maintaining full frame update rate. The subsampling is done with a command to use every other grid point along the x and y axes on the fly so it does not require preprocessing, although preprocessing to produce a reduced file size makes the data load into memory faster. This reduction was necessary because the full-resolution data of 20 Gbytes did not fit in the graphics card memory, which is necessary for maintaining the fast rendering rate. (Most current high-performance graphics cards have 8 Gbytes of memory, although this continues to increase.)
Finding suitable display styles and settings to obtain the rendering speed needed for VR is burdensome and few researchers will have the detailed understanding of how to adjust inter-related graphics parameters to allow maximum detail to be shown. For example, a standard ChimeraX lighting method called ambient occlusion casts shadows from 64 different directions and even small models cannot render fast enough for VR with this setting. ChimeraX automatically turns off ambient occlusion in VR mode. More sophisticated automated level-of-detail control will be needed to allow researchers to routinely use VR for large data sets.
Handling slow calculations
A second performance issue critical for successful use of VR is that many calculations require longer than a single graphics frame update (1/90th second) to compute. In order for the graphics to render at full speed it must not wait for these calculations to complete. Examples of such calculations are hydrogen bond determination, molecular dynamics, adding hydrogen atoms to existing structures, molecular surface calculation, electrostatic potential calculation, computing new density map contour surfaces, optimizing rigid fits of atomic models into density maps, and dozens more. Currently ChimeraX performs the calculations and graphics rendering sequentially, and each calculation causes a pause in the VR rendering. Calculations that take a half a second or more cause the headset to gray out and show a busy icon. While these pauses have a good visual payoff for the wait, the effective use of VR requires a more sophisticated software design where the graphics renders in parallel to all other calculations. All objects in the VR scene continue to render at full speed, and the objects themselves are only updated to reflect the results of a calculation when it completes.
Speech recognition
Typed commands are common in ChimeraX usage. They are needed when specifying parameters, such as a residue number that cannot be easily chosen from a menu because the range of possible values is large. Other examples are fetching a known database entry by specifying its accession code, specifying a distance range for displaying atoms or density maps within a zone around a ligand, and typing a filename for saving a modified model. One approach is to type commands on a virtual keyboard floating in the VR scene by pressing each key with a hand controller. This is a general mechanism provided by the SteamVR [http://store.steampowered.com] environment but it is slow and tedious.
A potentially more effective solution is to use speech recognition, for example saying “color chain A residue 23 yellow” to perform the coloring operation discussed in the example above. Using the Windows Speech Recognition toolkit, one of us (JTD) developed a set of about 50 speech input macros for performing tasks within ChimeraX, and demonstrated their use in a video [https://www.youtube.com/watch?v=NHiQpNInEjM]. The spoken commands are not the literal typed-text command required by ChimeraX. For instance, the coloring command needed is color/A:23 yellow so the macros perform a translation, in this case changing the words “chain” and “residue” to the corresponding syntax characters used by ChimeraX. Although the video shows perfect speech recognition, errors are common. Our experience suggests that an essential element of speech recognition in VR will be display of words as the speech engine recognizes them, with the ability to fix mistakes, for instance by saying “back up” or “start over”. Speech input error rates could also be reduced by restricting to a limited vocabulary using open-source speech recognition toolkits such as Mozilla DeepSpeech [https://github.com/mozilla/DeepSpeech], Kaldi [http://kaldi-asr.org, http://publications.idiap.ch/index.php/publications/show/2265], or CMUSphinx [https://cmusphinx.github.io].
Beyond the cases of entering parameters, speech recognition could reduce tedious interactions with the VR user interface panel. Simple commands like “show hydrogen bonds” or changing a hand controller mode, e.g. “swap amino acid” with the desired hand controller button held could reduce interactions with the 2D user interface. VR headsets include built-in microphones intended for high-quality speech input, so no additional hardware is needed, and ChimeraX has the ability to create command aliases, so adding new voice commands should be straight forward.
Multi-person virtual reality
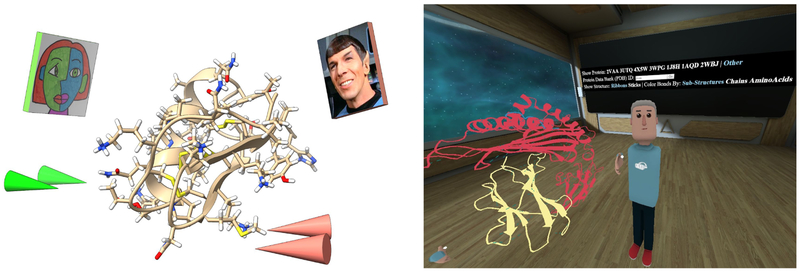
ChimeraX has basic capabilities for two or more people to join the same VR session. Each participant is shown with cones for hands and a postage stamp head (with image optionally specified by the user) that track the hand and head motions (Figure 5). Any participant can move or scale the models and point at features of interest as part of collaborative discussions. A session is started by one participant who loads data and initiates the meeting by invoking the ChimeraX command “meeting start” while others connect to the host’s machine with command “meeting <hostname>“, automatically receiving a copy of the scene from the host. Participants can be in the same room or at remote locations. This capability currently has many limitations, some of which are described below.
Figure 5:
ChimeraX VR meeting command shows each party as a postage-stamp head with customizable image and cone hands for pointing (left). AltPDB allows VR participants to discuss Protein Data Bank structures with custom human avatars, control molecule styles, and browse web pages within a popup pane (right).
The SteamVR toolkit that ChimeraX uses to communicate with VR headsets requires a separate computer (or virtual machine) running each headset. Any changes one participant makes to the VR scene must be communicated to the others over a network connection. Currently the only changes propagated to other participants are moving, rotating and scaling the displayed structures. Some additional synchronization is simple to add, such as for colors, display styles, and atom coordinates, while other changes such as swapping an amino acid (creating new atoms), or loading new data sets will likely require an approach that is slower to update, in which the new full ChimeraX session is sent to all participants. Large datasets such as 3D microscopy may take minutes or more to transfer because of their size.
While it is possible for participants to connect from remote sites, most academic institution firewalls block incoming network connections except for a select few and require administrative requests to have network ports opened up. A more usable design would have all participants connect to a hub machine set up to accept connections. To allow this to scale to hundreds of sessions running simultaneously by different groups of researchers, ChimeraX could start a cloud-based server on demand using a commercial service such as Amazon Web Services that accepts the connections and forwards changes to all participants. This would require accounts and payment for commercial services and is a complex undertaking. A further burden of remote connections in our existing implementation is that a third-party voice chat service is required.
We have developed another approach to allow multi-person molecular visualization called AltPDB using a social VR site called AltspaceVR. The technical complexity of initiating multi-person virtual reality sessions, maintaining synchronized views of data, audio connections, and customizable avatars for each participant is provided by AltSpaceVR [https://altvr.com/]. Two of us (TLS and SG) have added molecular display capabilities to this environment: ball-and-stick, ribbon, hydrogen bond, and surface depictions. Ribbon, surface and hydrogen bond depictions are computed by ChimeraX and passed to the AltspaceVR client as GLTF format [https://www.khronos.org/gltfy] geometric models. A user interface panel allows selecting entries from the Protein Data Bank by either typing an accession code or choosing one of several example structures from a list. Structures can be moved, rotated, scaled. The overall feel is richer with the meeting taking place in a room with web-browser and other controls available (Figure 6), contrasting with ChimeraX VR where only the structures in empty space are shown.
Figure 6:
Molecular Zoo educational application depicts fully flexible models in a large space. A tractor beam (blue) activated with hand controllers brings out-of-reach molecules to the VR user’s hand.
AltPDB is a powerful tool for education in which an expert explains the details of molecular structures to students. We have used it in presentations with up to 15 participants, explaining human T-cell receptor interactions with antigen-presenting cells. The ability to pass a human-sized molecular structures around, point at key features like an Ebola virus antigen, and explore inside molecules is by far the most effective method we have seen to make biology research accessible to a lay audience. Another important use is to allow researchers to collaborate to understand new structures.
While many of the multi-person VR difficulties are solved in AltPDB, the interaction is not directly with ChimeraX, which limits the analysis capabilities. Additional interfaces could allow participants to invoke any ChimeraX calculation or change in display, which would then run ChimeraX on a server and return a new scene (ChimeraX has a option for running on a server without a graphics display attached). Hand manipulation of the structure beyond moving and scaling will not allow real-time feedback using this relatively slow server-calculation approach. For example, rotating a bond would instead need new code added to AltPDB. Another current limitation is that the AltspaceVR platform only provides flat lighting. We adjust coloring of ribbons to include shadow effects to compensate for this.
Molecular Zoo: learning to love chemistry
We have developed a VR application called Molecular Zoo intended to allow young students to handle biomolecules they have heard of such as water, carbon dioxide, aspirin, caffeine, a saturated fatty acid, sulfuric acid, DNA, ATP, and methane. The aim is to familiarize students who have little or no knowledge of chemistry with the molecular world. Molecules and chemistry as presented in textbooks are abstract, and we hope that a concrete experience of fully flexible molecules, breaking their bonds, and assembling new molecules as if they were human-sized objects can contribute substantially to encouraging more students to become scientists.
The application starts with 100 molecules of many types flying in random directions, with sizes ranging from 30 centimeters for water to a few meters for a long fatty acid, shown in ball-and-stick style with standard coloring by element type (oxygen red, nitrogen blue, carbon black, hydrogen white, phosphorous orange, …) (Figure 6). The molecules vibrate, flex and collide with each other in qualitatively realistic ways, the dynamics simulated as springs and masses, with torsions and rotating joints implemented using the Nvidia PhysX engine [https://en.wikipedia.org/wiki/PhysX]. While this does not provide physically accurate molecular mechanics, the purpose is to capture the qualitative level of flexibility of small molecules and is visually highly engaging. Grabbing and shaking a molecule causes bonds to break. Breaking bonds make a snapping sounds, and fragments fly off with stub half-bonds able to form new compounds either when combined by hand or through collisions.
The virtual space is a 10-meter cube so that almost all of the molecules are flying out of easy reach. Virtual hands and arms allow interacting with the molecules using a variety of tools. The most basic is a tractor beam that produces a blue ribbon-like laser beam that pulls a molecule to your hand. Grasped molecules are identified in the headphones, for instance announcing “aspirin,” encouraging students to grab all the molecules to hear their names. In our experience showing this to hundreds of students, for example, at a local video game expo, many did not recognize even the simplest molecules, water and carbon dioxide, and such commonplace notions as the colors representing different elements were not obvious to them.
A basic aspect of the game is breaking molecules apart, and two hand tools allow launching hydrogen and oxygen atoms at other molecules. The oxygen atoms can break molecules, and both hydrogen and oxygen can bind to molecular fragments when valence electrons are available. Molecules can also be grabbed, held in two hands and pulled apart. Likewise, creating molecules is a central activity. We found that it takes considerable resolve to break apart existing molecules to produce ingredients for new molecules, so we added atoms of carbon, nitrogen, oxygen and hydrogen that sprout from the virtual arms and can be plucked off for an unlimited supply of at-hand atoms.
An important principle of our design was to favor user engagement over physical correctness, so the environment contains a rich, diverse and inconsistent set of items and behaviors including the planet Earth (2 meters in diameter and very massive, such that throwing it causes significant molecular damage on collisions), a user has four arms and hands (natural size), hydrogens are launched from a heme group held in the hand, oxygens are launched from an ATP synthetase complex (scaled down in size), a green cube floats as a remnant from the game’s early beginnings, an astronomy view of the universe is in the distance in all directions and a floor is at your feet. Launching atoms, tractor beaming, making and creating bonds have distinctive sound effects, and hitting Earth causes it to scream.
A common question from educators and other adults who have tried Molecular Zoo is “What are students expected to learn?”. We have not attempted to incorporate specific lessons, as we feel that our goal of producing future scientists will be better served by a playful rather than pedantic experience. But we believe much intuition about molecules and chemistry is conveyed, from the frequency of element types in biomolecules, the number of bonds each element can form, and the degree of flexibility of molecules. We consider this only an initial version of the application and plan to add other ingredients that will enhance student intuition about the molecular world. Some examples are: add water in ice, liquid and gas forms where the user heats with a flame to add energy and qualitatively realistic hydrogen-bonding orientations are favored; add duplex DNA that can be pulled apart and zipped back together -- again using qualitative hydrogen-bonding forces imposed by weak springs; allow the user to ignite natural gas (primarily methane) to observe the combustion cascade through dozens of intermediates ending in water, carbon dioxide and other compounds. We would like to provide a box of clinical drugs to familiarize students with the typical chemical characteristics needed to reach targets in cells. All of our current molecules are fetched from the PubChem database23, and we would like to add speech input that permits saying the name of a molecule and making it magically appear via a search and fetch from PubChem.
Molecular Zoo uses the Unity game engine [https://unity3d.com] which offers built-in support for spheres, cylinders, custom shapes, spring-and-mass physics, and virtual reality headsets. It enables rapid software development where we only need to implement molecules and molecular interaction behaviors. Our current scenes include about 1000 atoms and scenes with more objects or denser packing resulting in more collisions per second will require optimizations to achieve the high-speed needed for VR graphics. This has shaped development towards smaller simulations that demonstrate the beauty of a few unique molecules and how the player can interact with them.
A troublesome aspect of this application is that the ideal target audience is younger than the minimum recommended age for using VR of 12 or 13 years. These current recommended minimum ages from the headset manufacturers are based on uncertainty about adverse effects on the developing visual systems of younger children. We believe many children over the age of 12 years can benefit from Molecular Zoo, but the issue of how to minimize exposure to younger children needs careful consideration.
Discussion
The applications we describe have been demonstrated to hundreds of researchers, often visualizing their own data. In many cases they have claimed to observe features in their data they had not seen before. We believe the ability to perceive spatial relations especially in time-varying 3D molecular and cellular data is greatly enhanced using virtual reality compared to conventional displays. We describe the VR advantages as immersive visualization that includes the wide field of view, close-in vantage points, stereoscopic depth perception, 6-degree-of-freedom head and hand tracking with plentiful button inputs, and audio headphones and microphone. The combination of features rather than any single capability is the unique strength of the technology. While the novel combination of VR characteristics of immersive and multi-modal interaction on paper seem like small advances over prior 3D technologies, the user experience of VR headsets is qualitatively dramatically different and difficult to appreciate except by experiencing it.
The drawbacks of the VR headsets are numerous. The powerful devices we have used are bulky, heavy (typically 500 grams), tethered by a cable to a computer, require an expensive high-end graphics card, have poor resolution, interact poorly with 2D user interfaces, and completely block the user’s view of the real world. Future improvements in hardware and software can ameliorate these disadvantages to some extent.
The VR applications we have described are all at a proof-of-concept or demonstration stage with significant technical limitations and ease-of-use problems. The development of these tools has revealed many hurdles that will need to be overcome to make production-quality VR analysis tools for molecular and cellular structure researchers and for education. The problems for research analysis applications include input of parameters that would normally be typed, level-of-detail control for large data sets, and parallel processing for slow computations. These are challenging technical issues that will take months-to-years to adequately solve, but the potential solutions are well understood and likely to be successful. Conventional mouse, trackpad, keyboard and flat screen interfaces are clearly superior for essential 2D aspects of structure research such as web browsing, reading literature, and analyzing graphical data. Successful VR applications will need to minimize interleaving of 2D tasks which are difficult in current low-resolution headsets. Whether the advantages of VR analysis will outweigh the poorer interplay between 2D and 3D tasks will only be determined by reducing technical limitations and gauging adoption of VR in labs for routine research use.
Our efforts on multi-person VR reveal difficult technical challenges. Currently, few researchers have VR headsets, so the chance of collaborating researchers having the needed equipment is small and thus the potential audience for multi-person VR may be more limited than single-person applications. This suggests that near-term effort should focus more on single-person applications.
Educational applications such as Molecular Zoo avoid most of the technical difficulties encountered with research analysis: typed input, excessively large data sets, and slow computations. They have perhaps the greatest potential for making production-quality VR applications in the near term.
The poor resolution and more cumbersome interactions to perform 2D tasks (e.g. web browsing) in an immersive environment suggests that VR headsets should be used as an accessory to conventional screen, trackpad, mouse and keyboard, rather than trying to replace those interfaces. For analysis-intensive work done by researchers in ChimeraX, we can envision a more effective immersive environment combining 2D and 3D tasks using the older stereoscopic technologies of liquid-crystal display or polarized glasses combined with currently available ultrawide 4K or 5K resolution screens for a wide field of view, with added head tracking, and a tracked 6-degree-of-freedom hand controller. This setup would offer lightweight glasses, providing an augmented reality where both 2D user interfaces and 3D graphics can be mixed, and incorporates most of the advantages of current VR technology. While such a design is attractive for scientific data analysis, it seems less effective for entertainment applications than current VR approaches that allow 360-degree immersion. The success of any immersive technology will likely be controlled by mass-market entertainment and other consumer applications rather than the relatively small scientific data analysis and visualization market, so it will be necessary to focus effort developing science analysis on devices that are commercially the most successful.
Methods and Materials
ChimeraX is a program for the visualization and analysis of biomolecular structures, especially proteins and nucleic acids, integrative hybrid models of large molecular complexes, multiple sequence alignments, and 3D electron and light microscopy image data. It is free for academic use, runs on Windows, Mac and Linux operating systems, and can be obtained from https://www.rbvi.ucsf.edu/chimerax/. The use of a virtual reality headset is enabled with the ChimeraX command vr on, collaborative VR sessions with the command meeting, and 3D microscopy time-series viewing with command vseries. Hand-controller interaction modes are 3D versions of mouse modes associated with the mousemode command. Each of these commands is extensively documented on the ChimeraX web site. All Python source code is included with the ChimeraX distribution. Features described here are part of version 0.6 and earlier daily builds, and are continually being refined. ChimeraX VR requires the SteamVR runtime system to access connected headsets available with a free Steam account [https://steamcommunity.com]. We have only tested ChimeraX VR on the Windows 10 operating system.
The AltPDB collaborative viewing environment allows display of any models from the Protein Data Bank on Windows or Mac operating systems and can be used through a free AltspaceVR account [https://account.altvr.com/spaces/altpdb] with source code available at Github [https://github.com/AltPDB]. From the AltspaceVR web site select the “Molecule Viewer Activity” and to enter a protein VR viewing room.
The Molecular Zoo application and source code can be obtained from GitHub [https://github.com/alanbrilliant/MolecularZoo]. It is written in the C# programming language and uses the Unity3D video game engine. Molecular Zoo releases for the Windows 10 operating system are available and version 7 is described in this article. For Mac and Linux operating systems, the Unity project can be run with a free copy of the Unity3D development environment [https://unity3d.com]. We have only tested Molecular Zoo on Windows 10.
Highlights.
Are there compelling uses for virtual reality in molecular visualization?
VR allows clear views and analysis of time-varying atomic models and 3D microscopy.
Collaborative discussions benefit from room-scale virtual protein models.
Handling dynamic biomolecules can inspire young students to study science.
We give examples and identify key problems in using VR for scientific visualization.
Acknowledgements
We thank Lillian Fritz-Laylin and Dyche Mullins for neutrophil light microscopy data, En Cai for T-cell receptor light microscopy data, and Tristan Croll for help with OpenMM molecular dynamics and the ISOLDE ChimeraX plugin. Support for this work was provided by the UCSF School of Pharmacy, 2018 Mary Anne Koda-Kimble Seed Award for Innovation and NIH grant P41-GM103311.
Abbreviations
- VR
virtual reality
- 3D
3-dimensional
- 2D
2-dimensional
- cryoEM
cryo-electron microscopy
- GPU
graphics processing unit
References
- 1.Johnston APR, Rae J, Ariotti N, Bailey B, Lilja A, Webb R, Ferguson C, Maher S, Davis TP, Webb RI, McGhee J, Parton RG. Journey to the centre of the cell: Virtual reality immersion into scientific data. Traffic. 2018. February 01; 19(2): 105–110. [PubMed: 29159991] doi: 10.1111/tra.12538 [doi]. [DOI] [PubMed] [Google Scholar]
- 2.Balo AR, Wang M, Ernst OP. Accessible virtual reality of biomolecular structural models using the autodesk molecule viewer. Nat Methods. 2017. November 30; 14(12): 1122–1123. [PubMed: 29190274] doi: 10.1038/nmeth.4506 [doi]. [DOI] [PubMed] [Google Scholar]
- 3.Borrel A, Fourches D. RealityConvert: A tool for preparing 3D models of biochemical structures for augmented and virtual reality. Bioinformatics. 2017. December 01; 33(23): 3816–3818. [PubMed: 29036294] doi: 10.1093/bioinformatics/btx485 [doi]. [DOI] [PubMed] [Google Scholar]
- 4.Zheng M, Waller MP. ChemPreview: An augmented reality-based molecular interface. J Mol Graph Model. 2017. May 01; 73: 18–23. [PubMed: 28214437] doi: 10.1016/j.jmgm.2017.01.019 [pii]. [DOI] [PubMed] [Google Scholar]
- 5.Norrby M, Grebner C, Eriksson J, Bostrom J. Molecular rift: Virtual reality for drug designers. J Chem Inf Model. 2015. November 23; 55(11): 2475–2484. [PubMed: 26558887] doi: 10.1021/acs.jcim.5b00544 [doi]. [DOI] [PubMed] [Google Scholar]
- 6.Grebner C, Norrby M, Enstrom J, Nilsson I, Hogner A, Henriksson J, Westin J, Faramarzi F, Werner P, Bostrom J. 3D-lab: A collaborative web-based platform for molecular modeling. Future Med Chem. 2016. September 01; 8(14): 1739–1752. [PubMed: 27577860] doi: 10.4155/fmc-2016-0081 [doi]. [DOI] [PubMed] [Google Scholar]
- 7.Lv Z, Tek A, Da Silva F, Empereur-mot C, Chavent M, Baaden M. Game on, science - how video game technology may help biologists tackle visualization challenges. PLoS One. 2013; 8(3): e57990 [PubMed: 23483961] PMCID: PMC3590297 doi: 10.1371/journal.pone.0057990 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Neira C, Sandin DJ, DeFanti TA, Kenyon RV, Hart JC. The CAVE: Audio visual experience automatic virtual environment. Commun ACM. 1992; 35(6): 64–72. [Google Scholar]
- 9.Febretti A, Nishimoto A, Thigpen T, Talandis J, Long L, Pirtle J,D., Peterka T, Verlo A, Brown M, Plepys D, Sandin D, Renambot L, Johnson A, Leigh J. CAVE2: A hybrid reality environment for immersive simulation and information analysis. ; 2013. [Google Scholar]
- 10.Salvadori A, Del Frate G, Pagliai M, Mancini G, Barone V. Immersive virtual reality in computational chemistry: Applications to the analysis of QM and MM data. Int J Quantum Chem. 2016. November 15; 116(22): 1731–1746. [PubMed: 27867214] PMCID: PMC5101850 doi: 10.1002/qua.25207 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doblack BN, Allis T, Davila LP. Novel 3D/VR interactive environment for MD simulations, visualization and analysis. J Vis Exp. 2014. December 18; (94). doi(94): 10.3791/51384. [PubMed: 25549300] PMCID: PMC4396941 doi:10.3791/51384 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson A, Bracegirdle L, McLachlan SI, Chapman SR. Use of a three-dimensional virtual environment to teach drug-receptor interactions. Am J Pharm Educ. 2013. February 12; 77(1): 11 [PubMed: 23459131] PMCID: PMC3578324 doi: 10.5688/ajpe77111 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland IE. In: The ultimate display Proceedings of the IFIP congress; ; 1965. p. 506–508. [Google Scholar]
- 14.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2017. July 14 [PubMed: 28710774] PMCID: in-progress doi: 10.1002/pro.3235 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel beta-sheet architecture in iowa-mutant beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2012. March 20; 109(12): 4443–4448. [PubMed: 22403062] PMCID: PMC3311365 doi: 10.1073/pnas.1111305109 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Mountford SJ, Sellen A. A study in interactive 3-D rotation using 2-D control devices. SIGGRAPH Comput. Graph 1988. June; 22(4): 121–129. doi: 10.1145/378456.378497. [DOI] [Google Scholar]
- 17.Eastman P, Swails J, Chodera JD, McGibbon RT, Zhao Y, Beauchamp KA, Wang LP, Simmonett AC, Harrigan MP, Stern CD, Wiewiora RP, Brooks BR, Pande VS. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017. July 26; 13(7): e1005659 [PubMed: 28746339] PMCID: PMC5549999 doi: 10.1371/journal.pcbi.1005659 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010. February 01; 66(Pt 2): 213–221. [PubMed: 20124702] PMCID: PMC2815670 doi: 10.1107/S0907444909052925 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croll TI. ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Biol Crystallogr. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz-Laylin LK, Riel-Mehan M, Chen BC, Lord SJ, Goddard TD, Ferrin TE, Nicholson-Dykstra SM, Higgs H, Johnson GT, Betzig E, Mullins RD. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. Elife. 2017. September 26; 6: 10.7554/eLife.26990. [PubMed: 28948912] PMCID: PMC5614560 doi: 10.7554/eLife.26990 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, Gerard A, Liu TL, Chen BC, Betzig E, Bartumeus F, Krummel MF. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. 2017. May 12; 356(6338): 10.1126/science.aal3118. [PubMed: 28495700] doi:eaal3118 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks FP. What’s real about virtual reality? IEEE Computer Graphics and Applications. 1999; 19(6): 16–27. doi: 10.1109/38.799723. [DOI] [Google Scholar]
- 23.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem substance and compound databases. Nucleic Acids Res. 2016. January 04; 44(D1): 1202 [PubMed: 26400175] PMCID: PMC4702940 doi: 10.1093/nar/gkv951 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]