Abstract
Chinese wolfberry (Lycium spp.) is an important edible and medicinal plant, with a long cultivation history. The genetic relationships among wild Lycium species and landraces have been unclear for a number of reasons, which has hindered the breeding of modern Chinese wolfberry cultivars. In this study, we collected 19 accessions of Chinese wolfberry germplasm, and constructed the genetic relationship based on RAD-seq markers. We obtained 30.32 Gb of clean data, with the average value of each sample being 1.596 Gb. The average mapping rate was 85.7%, and the average coverage depth was 6.76 X. The phylogeny results distinguished all accessions clearly. All the studied landraces shared their most recent common ancestor with L. barbarum, which indicated that L. barbarum may be involved in cultivation of these landraces. The relationship of some landraces, namely the ‘Ningqi’ series, ‘Qingqi-1’ and ‘Mengqi-1,’ has been supported by the phylogeny results, while the triploid wolfberry was shown to be based on a hybrid between ‘Ningqi-1’ and a tetraploid wolfberry. This study uncovered the genetic background of Chinese wolfberry, and developed the foundation for species classification, accession identification and protection, and the production of hybrid cultivars of wolfberry.
Electronic supplementary material
The online version of this article (10.1007/s10528-018-9861-x) contains supplementary material, which is available to authorized users.
Keywords: Wolfberry, Goji, RAD-seq, Genetic background
Introduction
The wolfberry (Goji) genus Lycium belongs to the Solanaceae family, and comprises about 80 species, which are mainly distributed in South America (30 species), southwestern North America (21 species), South Africa (17 species), and temperate Europe and Asia (Hitchcock 1932; Fukuda et al. 2001; Miller 2002). In China, there are seven species and three taxonomic varieties, mostly distributed in Gansu, Qinghai Provinces, Xinjiang, and Ningxia Autonomous regions (Kuang and Lu. 1978).
The Chinese wolfberry has been widely used as traditional medicine and healthcare products for centuries. The classic Chinese herbal texts, “Shen Nong’s Herbal Classic” and “Ben Cao Gang Mu,” have recorded its medicinal value to liver, kidney, eyes, and other organs. The latest phytochemical analyses have demonstrated that there are many valuable components in wolfberry fruits and leaves, including zeaxanthin (Inbaraj et al. 2008) and β-carotene (Peng et al. 2005), polysaccharides (Gao et al. 2008), and low molecular-weight chemicals such as betaine, cerebroside, β-sitosterol, p-coumaric acid, and various vitamins (Dong et al. 2009; Duan et al. 2010; Inbaraj et al. 2010).
The cultivation of Chinese wolfberry can be traced back to 1400 years ago, where selection for the edible young leaves and leaf buds was the main breeding direction, and the main propagation route was by seed. By 600 years ago, the main cultivation areas for Chinese wolfberry were in the northwest region of China, especially in Zhongning County of the Ningxia Autonomous region, where propagation was mainly based on cuttings from the excellent plants with high-quality fruits. During the long history of improvement of Chinese wolfberry, by natural selection and artificial selection, more than ten traditional landraces have been developed in the Ningxia Autonomous region, with names such as ‘Damaye,’ ‘Xiaomaye,’ ‘Black leaf,’ ‘White stick,’ and ‘Leaf medlar.’ In the past 30 years, some earlier landraces have been further improved, to exhibit superior economic characters, such as hundred grain weight, fruit size, and total sugar. Improved landraces, such as ‘Ningqi-1,’ ‘Ningqi-2,’ and ‘Ningqi-4,’ were bred from the traditional landrace ‘Damaye’ by selection from natural superior mutants, and were quickly distributed to other cultivation regions by cutting propagation. Other improved landraces include ‘Mengqi-1’ (Wang et al. 2007), developed by selection, and ‘Ningcaiqi-1,’ which was a product of hybridization and selection for superior leaf traits, while triploid wolfberry was bred by hybridizing ‘Ningqi-1’ with a tetraploid. Modern breeding methods (induced mutation breeding, space mutation breeding, and plant biotechnology) have also contributed to the development of wolfberry breeding.
However, some factors limited the modern breeding of Chinese wolfberry. Firstly, the long artificial breeding and hybridization history has had a negative influence on the conservation of superior lines, and has led to the rapid deterioration of germplasm resources (Shao et al. 2015). Secondly, there was a shortage of germplasm resources; although more than 10 landraces have been identified, only four landraces were widely planted in northwestern China, namely ‘Ningqi-1,’ ‘Ningqi-4,’ ‘Mengqi-1,’ and ‘Ningcaiqi-1’ (An et al. 2009), with the other landraces being cultivated only in restricted regions. Furthermore, most landraces were concentrated in the Ningxia Autonomous region (Hu et al. 2016). The limited genetic background of Chinese wolfberry could increase the risk of disease problems, while the relative lack of germplasm resources will not only restrict the development of wolfberry breeding, but will also slow the development of lines exhibiting high yield and high quality as demanded by consumers. Finally, the identity of individual landraces is in question, because of the absence of scientific verification. Some landraces with similar morphology have been identified as the same landrace, or the same varieties cultivated in different regions were assumed to be different landraces (An et al. 2009).
The restriction site-associated DNA (RAD-seq) method can identify polymorphic variants in genomic regions (Miller et al. 2007; Zhou et al. 2015), and has been proven to be particularly suitable for species without reference genome (Wang et al. 2017; Verdu et al. 2016). It can provide thousands of genome-scale SNP sites (Pavinato et al. 2017; Wang et al. 2017) that have successfully revealed valuable information for phylogeny (Razkin et al. 2016; Wagner and Wagner. 2017), phylogeography (Jeffries et al. 2016), population genetics (Ellegren 2014; Shih et al. 2018) and species identification (Herrera and Shank. 2016). Also, the genome-wide SNPs have provided a fundamental data resource for investigating the genetic germplasm in tea (Hua et al. 2016), grapevines (Marrano et al. 2017), peanut (Hong et al. 2015), cassava (Wosula et al. 2017), and loquat (Wang et al. 2017).
In order to detect the genetic divergence of germplasm between wolfberry landraces and their wild relatives, the third-generation sequencing technology was used, by developing RAD-seq markers, in order to investigate the genetic background of the Chinese wolfberry, and to lay down a strong foundation for the development of comprehensive Chinese wolfberry breeding project. Based on the high resolution of the RAD-seq, combined with the breeding background of Chinese wolfberry, we speculated that L. barbarum was the main participant during the breeding process in the studied landraces. And some of the classic cultivated landraces, like Ningqi-1 -2, -3, were involved in breeding of the other studied landraces.
Materials and Methods
A total of 19 accessions were used in this study (Table 1), including four wild species (L. ruthenicum Murr., L. barbarum L., L. chinense Mill. var. potaninii (Pojark.) A. M. Lu, L. yunnanense Kuang) and a mutant with white fruits found in wild; these four species are genetically close, cross-compatible with cultivated landraces and, thus, could contribute to the improvement of Chinese wolfberry. In addition, fourteen landraces (‘Ningqi-1’ to ‘-8,’ ‘Ningcaiqi-1,’ ‘Qingqi-1,’ ‘Ningqi-v3,’ ‘Mengqi-1,’ ‘Zhongkelvchuan-1,’ and a cultivated triploid wolfberry; Table 1) have all been registered officially and are commercially planted in China. All the accessions were conserved with cutting propagation in the Germplasm Nursery of Qinghai Gouqi (Nuomuhong Co., Qinghai province), from which mature plant material was supplied.
Table 1.
Lycium accessions used in this research
| No. | Accession name | Origin |
|---|---|---|
| 1 | ‘Ningqi-1’ | Landrace ‘Damaye’ |
| 2 | ‘Ningqi-2’ | Landrace ‘Damaye’ |
| 3 | ‘Ningqi-3’ | The elite clone selected from L. barbarum |
| 4 | ‘Ningqi-4’ | Landrace ‘Damaye’ |
| 5 | ‘Ningqi-5’ | Landrace ‘Ningqi-1’ |
| 6 | ‘Ningqi-6’ | The descendant from natural hybrid of L. barbarum |
| 7 | ‘Ningqi-7’ | The elite clone selected from L. barbarum |
| 8 | ‘Ningqi-8’ | The elite clone selected from L. barbarum |
| 9 | ‘Ningqi-v3’ | – |
| 10 | ‘Mengqi-1’ | Plantation in Inner Mongolia |
| 11 | ‘Ningcaiqi-1’ | Hybrid between wild and cultivated wolfberry |
| 12 | ‘Qingqi-1’ | Landrace from Ningqi-1 seeds treated with mutagenesis |
| 13 | ‘Zhongkelvchuan-1’ | Landrace from the descendant of Ningqi-1 |
| 14 | cultivated triploid wolfberry | Hybrid between Ningqi-1 and tetraploid wolfberry |
| 15 | Wild white fruit wolfberry | |
| 16 | L. yunnanense Kuang | |
| 17 | L. barbarum L. | |
| 18 | L. chinense Mill. var. potaninii | |
| 19 | L. ruthenicum Murr. |
Fresh leaves were collected in August 2014. All tissue samples were stored at −80 °C prior to genomic DNA isolation. Genomic DNA was extracted with the plant genomic DNA extraction kit DP305 (Tiangen, Beijing, China). DNA purity and integrity were monitored by agarose gel electrophoresis and the use of a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), the latter mainly focusing on the OD260/OD280 ratio.
Based on the method of Baird et al. (2008), genomic DNA was digested with EcoRI (G∣AATTC) restriction enzyme, and the fragments were ligated to the Solexa P1 Adapter, which is complementary to the sticky ends. Subsequently, the adapter-ligated fragments were pooled, randomly sheared, and the 300–700-bp-sized fragments were selected to ligate the second adapter (P2, a Y adapter with divergent ends). Only the fragments with both P1 and P2 adapters could be amplified successfully, with amplified fragments ranging in length from 200 to 400 bp and 400–600-bp-sized fragments were collected for library construction.
To analyze the quality of the libraries, the Qubit 2.0 kit (Life Technologies, Grand Island, NY, USA) was used. Agilent 2100 (Agilent Technologies, Palo Alto, CA, USA) was used to check the insert size of the libraries after dilution of the library to 1 ng DNA/μl. Q-PCR was performed to detect the effective concentration of libraries (the effective concentration of library > 2 nM) after the fragments were appropriately inserted.
The libraries were sequenced by Illumina HiSeq 2500 platform (San Diego, CA, USA) and 125-bp paired-end reads were generated. The raw sequencing reads were firstly processed through In-House scripts to remove the bad reads that included adapter-or ploy-N containing reads, or low-quality or repetitive reads. The reads with RAD sites below a default threshold values (sequencing error rate > 1%, the depth of cover < 4 X) were also removed, because these reads could lack sufficient coverage for calling SNPs or lead to the formation of sequence error-created artifacts.
The paired-end reads of L. barbarum from each RAD site passing the above tests were sent to the Velvet Optimiser assembler (Zerbino and Birney. 2008). According to the record, most of the studied landraces are genetically close with L. barbarum (Table 1), which means that it had the largest genetic identity with the landraces, and could provide the highest sequencing contribution for sequence assemble. So the contigs of L. barbarum assembled by Velvet were selected as the reference genome for the next analysis.
Sequencing reads were aligned to the reference genome using BWA software (settings: mem -t 4 -k 32 –M) (Li and Durbin, 2009). Then the aligned reads were converted to BAM files using SAMtools software (Li et al. 2009). Variant allele calling was performed for each individual using SAMtools (settings: mpileup –q 1 –C 50 –S –D –m 2 –F 0.002), and SNP (Single Nucleotide Polymorphism) sites were initially filtered by vcfutils (settings: -Q 20 -d 2 -D 1000). In addition, the SNP sites with a max missing range higher than 10% were filtered out.
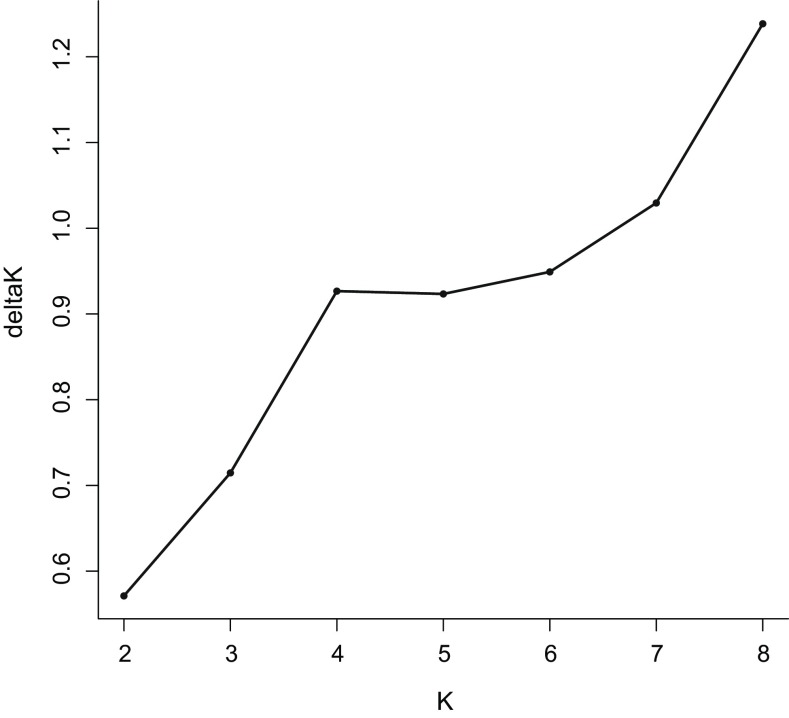
Based on the SNP sites, a neighbor-joining tree was generated by TreeBeST (http://treesoft.sourceforge.net/treebest.shtml). Genetic structure was inferred using the program frappe7c (http://med.stanford.edu/tanglab/software/frappe.html), which can implement an expectation maximization algorithm. To explore the convergence of individuals, the number of genetic clusters (K) was set from 2 to 8. The maximum iteration of the expectation–maximization algorithm was set to 100 in the frappe analysis. We computed the delta K statistics with the program admixture (http://www.genetics.ucla.edu/software/admixture/index.html) which can calculate rapidly using a fast numerical optimization algorithm.
Results
Using the RAD-tag sequencing approach, a total of 31.26 Gb raw paired-end reads were generated from the 19 accessions. After strict filtering, 30.32 Gb clean data were retained, with the average value of raw reads being 1.65 Gb, and that of clean reads being 1.596 Gb. The average effective rate was 97.0%, the average Q20 was 93.5%, and the average Q30 was 88.2% in the 19 accessions. All information on the sequencing data is shown in Supplementary File 1.
After removing all repetitive reads, single mismatch derivatives and reads with RAD sites below the threshold value and the high-quality RAD-tags were retained. The mean number of clean reads was 6,383,952.9, and L. barbarum had the highest number of reads (23,122,684 reads). The mean number of duplicated reads removed was 5,667,781.9, while the mean clean duplication rate was 12.59%. The mean number of digestion reads was 5,491,317.1, and the mean digestion ratio was 96.80% (Supplementary File 2).
The paired-end reads of L. barbarum were assembled by Velvet 1.2.10 as the reference genome for the next analysis. The number of contigs was 880,315, which were obtained from 160,163,757 bp reads, with an average length of 295 bp.
The mapping rates of accessions reflected the similarity of accession and reference genomes, with the coverage depth demonstrating the homology with the reference genome. Statistics of the mapping rate and coverage are shown in Supplementary File 3. The average mapping rate was 85.7%, and the average coverage depth was 6.76 X. At last, the number of SNP sites that reconstructed phylogeny and genetic subdivision is 721,813. The raw and clean sequencing data were deposited in Genbank (PRJNA351229).
The phylogeny tree was explored to determine the genetic relationships among the 19 accessions. Most accessions were separated from each other with high support (Bootstrap value = 100%). The 19 accessions were separated into two main branches: the first branch contained two clusters, one being L. chinense var potaninii and L. yunnanense, and the other being L. ruthenicum and the white fruit Chinese wolfberry. The other branch contained all the studied landraces, with L. barbarum located in the root of the phylogeny tree. The landraces formed three branches: cultivated triploid Chinese wolfberry clustered together with ‘Ningqi-4,’ ‘Ningcaiqi-1,’ and ‘Ningqi-3’; ‘Ningqi-1’ clustered with ‘Ningqi-2’ and ‘Qingqi-1’; and the third branch included ‘Zhongkelvchuan-1,’ ‘Mengqi-1,’ and ‘Ningqi-6’ (Fig. 1).
Fig. 1.
The phylogeny tree of the 19 samples based on the RAD data
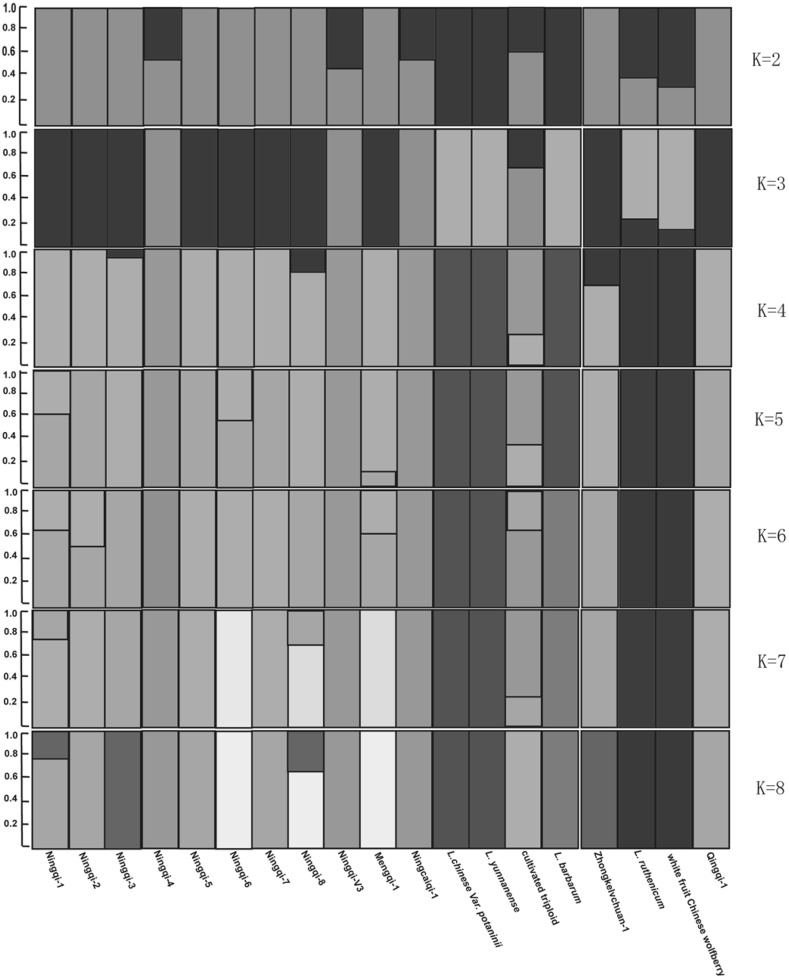
The genetic subdivision relationships of the landraces were uncovered by genetic structure analysis. The delta K value pointed to K = 4 (number of genetic clusters) as the strongest structure level in the data set (Fig. 2), the landraces Ningqi-1, -2, -5, -6, -7, Mengqi-1, and Qingqi-1 shared the same genetic background, while Ningqi-4, Ningqi-v3 and Ningcaiqi-1 shared the same genetic background. The same genetic subdivision was shared by L chinense var potaninii and L. yunnanense, and by L. ruthenicum and white fruit Chinese wolfberry when K was 4, 5, 6, 7, or 8. When K = 8, close relationships among the landraces were proved, such as ‘Ningqi-1,’ ‘Ningqi-2,’ and ‘Qingqi-1’; ‘Ningqi-5’ and ‘Ningqi-7’; ‘Ningqi-3’ and ‘Zhongkelvchuan-1’; ‘Ningqi-6’ and ‘Mengqi-1’; ‘Ningqi-4,’ ‘Ningqi-v3,’ and ‘Ningcaiqi-1.’ The cultivated triploid Chinese wolfberry contained two genetic components: one was from ‘Ningqi-4,’ ‘Ningqi-1,’ or ‘Ningqi-v3,’ while the other came from ‘Ningqi-3,’ which indicated the hybrid origin of triploid Chinese wolfberry (Fig. 3).
Fig. 2.
The plots for detecting the number of K groups that best fit the data
Fig. 3.
Plots of 19 individuals for K = 2 to 8, with colors representing proportion of genetic component
Discussion
RAD-seq is not limited by the reference genome, and has the advantage of simple operation, with the related sequences of specific enzyme-cutting sites being scanned quickly, and rapid identification of high-density SNP-based map. In this study, the 19 accessions were selected to include the landraces that were grown over the planting area and four wild species, which may be involved in the domestication of Chinese wolfberry. Based on RAD-seq markers, the average coverage of the accession genome reached 6.76 X. The phylogeny tree obtained greater resolution (Bootstrap ≥ 72%) than had previous research, which was based on RAPD and ITS markers (Sang et al. 2010; Li et al. 2014).
A close relationship was found between L. ruthenicum and the white fruit Chinese wolfberry. As only a few plants were found in wild, we speculated that the white fruit Chinese wolfberry may be a mutant of L. ruthenicum. L. barbarum located in the root of clade which includes all landraces, indicating that all the studied landraces shared their most recent common ancestor with L. barbarum. The main distribution range of L. barbarum covers Ningxia, Gansu, and Qinghai area. The Ningxia area was the first cultivated region of Chinese wolfberry, so it is possible that L. barbarum has involved in the cultivation of some original landraces, before some landraces were cultivated in other areas. According to the literature (Li et al. 2004), the classic landraces, ‘Damaye’ and ‘Xiaomaye,’ were selected from wild Chinese wolfberry in Ningxia area.
These results also provided insights into the parentage of certain landraces. Landraces ‘Ningqi-1,’ ‘-2,’ and ‘-4’ were bred from elite lines of the classic landrace ‘Damaye,’ which, in turn, was selected from L. barbarum, based on the optimization of economic character. While ‘Ningqi-1’ and ‘Ningqi-2’ had a closer genetic relationship (Li et al. 2011), Ningqi-4 appeared to be quite genetically distant from the other two landraces. Some landraces were bred from L. barbarum directly, such as ‘Ningqi-3’ (Hu and Zhou 2005), ‘Ningqi-6,’ ‘-7,’ and ‘-8’ (Nan et al. 2014), but the results of genetic structure analysis and phylogeny did not support their close genetic relationship. This may be caused by the different parentage of landraces. Based on our field investigation, self-pollination always took place in some plants of L. barbarum, which could result in increased genetic variation within populations. And the wide and different environments also could increase the genetic differences among populations L. barbarum. Therefore, the landraces bred from different plants of L. barbarum could exist larger genetic differentiation.
Some landraces were developed from L. barbarum indirectly; ‘Ningcaiqi-1,’ for example, is a hybrid between wild and cultivated wolfberry (Li et al. 2004). ‘Ningqi-1’ was involved in the formation of some landraces, such as ‘Ningqi-5,’ ‘Qingqi-1’ (an induced mutant selected from mutagenized ‘Ningqi-1’ seeds), and ‘Zhongkelvchuan-1.’ ‘Mengqi-1’ developed from the dominant strain of the ‘Ningqi’ series (Wang et al. 2007), and our results showed that ‘Mengqi-1’ shared the same genetic background as ‘Ningqi-6,’ indicating that ‘Mengqi-1’ was bred from ‘Ningqi-6.’ A previous study had demonstrated that the triploid wolfberry was bred as a hybrid of ‘Ningqi-1’ and tetraploid wolfberry (An et al. 1998). Our results found that the triploid wolfberry possessed a genetic subdivision of ‘Ningqi-1’ or ‘Ningqi-3,’ and one of ‘Ningqi-4,’ ‘Ningcaiqi-1,’ or ‘Ningqi-v3.’
All the landraces had the common ancestor with L. barbarum, which could have been the cause of the close genetic relationship between the landraces. The wild wolfberry had greater genetic diversity than did the landraces. The low diversity of the landraces may be a consequence of long-term artificial directional selection, eliminating the lines (and genes) unsuitable for crop production and resulting in similar morphology (“convergent evolution”) among landraces from different parentages. From the perspective of long-term development of Chinese wolfberry, in order to prevent quality deterioration caused by inbreeding, the Chinese wolfberry industry should introduce more wolfberry germplasm material into the breeding program to enrich the Chinese wolfberry gene pool.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the Qinghai Province Natural Science Foundation for Youths (2015-ZJ-926Q), Major Science and Technology of Qinghai Province (2015-NK-A2), and Science and Technology Plan Projects of Qinghai Province (2014-SF-142).
References
- An W, Li Y, Jiao E, Li R. Breeding of triploid seedless variety of wolfberry (Lycium barbarum L.) J Ningxia Agric Coll. 1998;3:41–44. [Google Scholar]
- An W, Zhang HX, Jun HE, LI XY, Fan YF. Progress and prospects of wolfberry breeding. North Hortic. 2009;5:125–128. [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3(10):e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JZ, Lu DY, Wang Y. Analysis of flavonoids from leaves of cultivated Lycium barbarum L. Plant Food Hum Nutr. 2009;643:199–204. doi: 10.1007/s11130-009-0128-x. [DOI] [PubMed] [Google Scholar]
- Duan H, Chen Y, Chen G. Far infrared-assisted extraction followed by capillary electrophoresis for the determination of bioactive constituents in the leaves of Lycium barbarum Linn. J Chromatogr A. 2010;1217(27):4511–4516. doi: 10.1016/j.chroma.2010.04.069. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 2014;29(1):51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yokoyama J, Ohashi H. Phylogeny and biogeography of the genus Lycium (Solanaceae): inferences from chloroplast DNA sequences. Mol Phylogenet Evol. 2001;19(2):246–258. doi: 10.1006/mpev.2001.0921. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ali Z, Khan IA. Glycerogalactolipids from the fruit of Lycium barbarum. Phytochemistry. 2008;69(16):2856–2861. doi: 10.1016/j.phytochem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Herrera S, Shank TM. RAD sequencing enables unprecedented phylogenetic resolution and objective species delimitation in recalcitrant divergent taxa. Mol Phylogenet Evol. 2016;100:70–79. doi: 10.1016/j.ympev.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Hitchcock CL. A monographic study of the genus Lycium of the Western Hemisphere. Ann Mo Bot Gard. 1932;19(2–3):179–374. doi: 10.2307/2394155. [DOI] [Google Scholar]
- Hong Y, Pandey MK, Ying L, Chen X, Hong L. Varshney RK (2015) Identification and evaluation of single-nucleotide polymorphisms in allotetraploid peanut (Arachis hypogaeal.) based on amplicon sequencing combined with high resolution melting (hrm) analysis. Front Plant Sci 6:1068 [DOI] [PMC free article] [PubMed]
- Hu ZQ, Zhou QL. Breeding of Ningqi-4. Ningxia J Agric Fore Sci Technol. 2005;4:11–13. [Google Scholar]
- Hu BF, Zhang B, Cai G, Wang S, Zhon L, Wu L. Genetic diversity by SSR markers in seventeen Chinese and American Lycium barbarum L. Northern Hortic. 2016;01:90–94. [Google Scholar]
- Hua Y, Wei CL, Liu HW, Wu JL, Li ZG, Liang Z. Genetic divergence between Camellia sinensis and its wild relatives revealed via genome-wide SNPs from RAD sequencing. PLoS ONE. 2016;11(3):e0151424. doi: 10.1371/journal.pone.0151424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbaraj BS, Lu H, Hung CF, Wu WB, Lin CL, Chen BH. Determination of carotenoids and their esters in fruits of Lycium barbarum Linnaeus by HPLC–DAD–APCI–MS. J Pharm Biomed. 2008;47(4):812–818. doi: 10.1016/j.jpba.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Inbaraj BS, Lu H, Kao TH, Chen BH. Simultaneous determination of phenolic acids and flavonoids in Lycium barbarum Linnaeus by HPLC–DAD–ESI-MS. J Pharm Biomed. 2010;51(3):549–556. doi: 10.1016/j.jpba.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Jeffries DL, Copp GH, Lawson Handley L, Olsén KH, Sayer CD, Hänfling B. Comparing RADseq and microsatellites to infer complex phylogeographic patterns, an empirical perspective in the crucian carp, carassius carassius, L. Mol Ecol. 2016;25(13):2997–3018. doi: 10.1111/mec.13613. [DOI] [PubMed] [Google Scholar]
- Kuang KR, Lu AM (1978) Flora of China (Solanaceae). Science Press, Beijing, pp 8–18
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RH, Shi ZG, An W, Li YX, Jiao EN. New Chinese wolfberry varieties Ningcaiqi-1 for vegetable purpose. China Veg. 2004;1(5):48–48. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YL, Fan YF, Dai GL, An W, Cao YL. Analysis of genetic diversity for wolfberry germplasms by AFLP technology. Chinese Tradit Herb Drugs. 2011;42(4):770–773. [Google Scholar]
- Li XG, Wang YK, Li J, Zang XN, Ge J, He LJ (2014) Study on10 species of Lycium L. Genetic relationship based on nrDNA-ITS sequence. Chinese Agric Sci Bull 30(25):128–135
- Marrano A, Birolo G, Prazzoli ML, Lorenzi S, Valle G, Grando MS. SNP-discovery by RAD-sequencing in a germplasm collection of wild and cultivated grapevines (V. vinifera L.) PLoS ONE. 2017;12(1):e0170655. doi: 10.1371/journal.pone.0170655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JS. Phylogenetic relationships and the evolution of gender dimorphism in Lycium (Solanaceae) Syst Bot. 2002;27(2):416–428. [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17:240–248. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan XX, Wang JX, Chang HY, Wang H, Li YH, Shen XD. A new variety of Lycium barbarum Ningqi 8. Scientia silvae sinicae. 2014;50(12):170–170. [Google Scholar]
- Pavinato VA, Margarido GR, Wijeratne AJ, Wijeratne S, Meulia T, Souza AP. Restriction site associated DNA (RAD) for de novo sequencing and marker discovery in sugarcane borer, Diatraea saccharalis fab. (lepidoptera: crambidae) Mol Ecol Res. 2017;17:454–465. doi: 10.1111/1755-0998.12583. [DOI] [PubMed] [Google Scholar]
- Peng Y, Ma C, Li Y, Leung KSY, Jiang ZH, Zhao Z. Quantification of Zeaxanthin dipalmitate and total carotenoids in Lycium fruits (Fructus Lycii) Plant Food Hum Nutr. 2005;60(4):161–164. doi: 10.1007/s11130-005-9550-5. [DOI] [PubMed] [Google Scholar]
- Razkin O, Sonet G, Breugelmans K, Madeira MJ, Gómez-Moliner BJ, Backeljau T. Species limits, interspecific hybridization and phylogeny in the cryptic land snail complex pyramidula: the power of RADseq data. Mol Phylogenet Evol. 2016;101:267–278. doi: 10.1016/j.ympev.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Sang J, Li S, Zhang KW (2010) Genetic diversity analysis of Lycium babarum L. by RAPD. Bull Bot Res 30(1):116–119
- Shao QS, Gao L, Nan XX, Xu ML, Li YH, Wang JX. Analysis of genetic diversity and construction of fingerprint of Lycium barbarum L.using SSR technology. Northern Hortic. 2015;12:91–95. [Google Scholar]
- Shih KM, Chang CT, Chung JD, Chiang YC, Hwang SY. Adaptive genetic divergence despite significant isolation-by-distance in populations of taiwan cow-tail fir (Keteleeria davidiana var.formosana) Front Plant Sci. 2018;9:92. doi: 10.3389/fpls.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu CF, Guichoux E, Quevauvillers S, De Thier O, Laizet Y, Delcamp A. Dealing with paralogy in RADseq data: in silico detection and single nucleotide polymorphism validation in Robinia pseudoacacia l. Ecol Evol. 2016;6(20):7323–7333. doi: 10.1002/ece3.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N, Wagner N (2017) Insights into reticulate evolution and hybridization in the phylogeny of willows (Salix L.) using RAD Sequencing. IX International botanical congress
- Wang JM, Wand JP, Gto XP, Lei ZR, Jiang QX, Zhong YP. Mengqi-1 wolfberry breeding of new varieties. Crop Res. 2007;21(3):415–417. [Google Scholar]
- Wang Y, Shahid MQ, Lin S, Chen C, Hu C. Footprints of domestication revealed by RAD-tag resequencing in loquat: SNP data reveals a non-significant domestication bottleneck and a single domestication event. Bmc Genom. 2017;18(1):354. doi: 10.1186/s12864-017-3738-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosula EN, Chen W, Fei Z, Legg JP. Unravelling the genetic diversity among cassava bemisia tabaci white flies using next RAD sequencing. Genom Biol Evol. 2017;9(11):2958–2973. doi: 10.1093/gbe/evx219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Zhang Q, Zhang XQ, Tan C, Li C. Construction of high-density genetic map in barley through restriction-site associated DNA sequencing. PLoS ONE. 2015;10(7):e0133161. doi: 10.1371/journal.pone.0133161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.