Abstract
Purpose
The aim of the study is to use Raman spectroscopy to analyze the biochemical composition of medulloblastoma and normal tissues from the safety margin of the CNS and to find specific Raman biomarkers capable of differentiating between tumorous and normal tissues.
Methods
The tissue samples consisted of medulloblastoma (grade IV) (n = 11). The tissues from the negative margins were used as normal controls. Raman images were generated by a confocal Raman microscope—WITec alpha 300 RSA.
Results
Raman vibrational signatures can predict which tissue has tumorous biochemistry and can identify medulloblastoma. The Raman technique makes use of the fact that tumors contain large amounts of protein and far less lipids (fatty compounds), while healthy tissue is rich in both.
Conclusion
The ability of Raman spectroscopy and imaging to detect medulloblastoma tumors fills the niche in diagnostics. These powerful analytical techniques are capable of monitoring tissue morphology and biochemistry. Our results demonstrate that RS can be used to discriminate between normal and medulloblastoma tissues.
Keywords: Raman, Spectroscopy, Medulloblastoma, Embryonal tumor
Medulloblastoma (MB) is the most common embryonal tumor of the central nervous system (CNS) (70% of all). It is located only in the posterior fossa. In the group of patients < 18 years of age, it constitutes 18% of all brain tumors and 30% concerning the posterior fossa. The median of age is 9 years with two occurrence peaks, 2–4 and 6–8 years of age. The frequency of occurrence revolves around 0.5/100,000 children. A more frequent occurrence in the male sex is suggested (♂:♀, 1.8:1).
The basic feature of MB biology is its huge local malice and the ability to spread through cerebrospinal fluid (CSF). The contemporary comprehensive treatment of tumors consists of surgical removal followed by pharmacological chemotherapy and megavoltage CNS therapy. The principle is the use of surgical treatment at the first stage. Radiotherapy is not used in patients under 3 years of age who are treated exclusively with chemotherapy and palliative surgical procedure in primary disseminated disease (M2–M4 Chang classification).
The recurrence of the neoplastic process occurs most frequently during the first 2 years after the end of treatment. As a rule, it occurs at the place of the original location.
The results of the last years’ research quite unequivocally indicate the possibility of improving the effects of MB treatment with the application and appropriate selection of conventional methods. However, the maximum 5-year survival rates achieved at 75–85% for the standard-risk groups and 60–69% in the high-risk groups seem to be the limit.
Although precise delineation of the tumor excision border is a crucial step in patient treatment and survival, there are currently no methods able to differentiate normal tissue from tumor during operation. Traditional methods such as radiography, ultrasonography, computed tomography, and magnetic resonance imaging are insufficient in spatial resolution and have limited intraoperative availability [14, 16]. This could be counteracted by Raman spectroscopy, a promising simple, quick, and non-invasive method.
Raman spectroscopy (RS) and imaging (RI) are methods that measure inelastic scattering of light, providing information about vibrations of tissue components in samples. As a result, Raman spectroscopy can provide biochemical information of tissues without using any contrast agents [1, 3].
Recent years brought a great number of papers indicating usefulness of Raman spectroscopy in brain research on animals [11, 17, 23] as well as preliminary research on human brain [10, 11, 13, 15, 20]. However, the studies included individual cases. Here, we studied 11 cases of medulloblastoma and 3 samples from the safety margin as a control.
The aim of the study is to use Raman spectroscopy to analyze the biochemical composition of medulloblastoma and normal tissues from the safety margin of the CNS and to find specific Raman biomarkers capable of differentiating between tumorous and normal tissues.
Materials and methods
Study participants and tissue preparations
All experiments were performed in compliance with relevant laws and guidelines of the Bioethical Committee at the Polish Mother’s Memorial Hospital Research Institute in Lodz (53/216) and of the Ministry of Health of the Republic of Poland. Written informed consent was obtained from patients. The tissue samples consisted of medulloblastoma (grade IV) (n = 11). The tissues from the negative margins were used as normal controls.
Microtomed 16-μm-thick tissue sections were obtained from frozen blocks of the material removed during surgical operation at the Polish Mother’s Memorial Hospital (Lodz, Poland) and placed on CaF2 substrates (CRYSTAL GmbH, Germany) for Raman spectroscopy and Raman imaging measurements. Parallel 6-μm tissue sections were obtained and stained with H&E followed by histology examination for all the specimens by a certified neuropathologist from the Polish Mother’s Memorial Hospital Research Institute in Lodz. The tissue sections were examined by Raman spectroscopy and Raman imaging. MRI images were used for visualization and location of the tumor region for each patient.
Raman spectroscopy and imaging
Raman images were generated by a confocal Raman microscope—WITec alpha 300 RSA (Ulm, Germany)—consisting of an Olympus microscope coupled with a 300-mm Czerny–Turner monochromator (Princeton Instruments Acton SP23000; 300-mm imaging triple-grating monochromator/spectrograph) and a thermoelectrically cooled CCD camera (ANDOR Newton DU970N-UVB-353; EMCCD chip with a 1600 × 200 pixel format, 16 μm dimension each) operating in the standard mode at − 64 °C with full vertical binning. The excitation laser beam was a second harmonic of the Nd:YAG laser (532 nm) which was focused on the sample with a × 40-magnification objective (NIKON CFI Plan Fluor C ELWD 40×: NA 0.60, WD 3.6–2.8 mm; DIC-M, C.C.0-2) to the laser spot of 1 μm determined by the laser wavelength and microscope objective being used. The average laser excitation power was 10 mW, with a collection (integration) time of 0.5 s and a spectral step of 2 cm−1 in the fingerprint range of 200–1800 cm−1 and high-frequency region of 1800–3600 cm−1. A piezoelectric table was used to record Raman images. The spectra were collected at one acquisition per pixel and a 1200-line-mm−1 diffraction grating with the spectral bandpass varying from about 5.5 cm−1 per pixel at about 200 cm−1 to about 3.3 cm−1 per pixel at 3600 cm−1. Raman images (50 × 50 μm, 100 × 100 points per line) from the fingerprint spectral regions of the human brain tissue from the tumor mass and from the safety margin were constructed.
Detailed methodology on data pre-processing and multivariate data analysis used in the paper is available elsewhere [6, 8, 9].
The percentage of blue and red areas of images presented in the “Results and discussion” section was calculated with Color threshold and Measure option of the ImageJ software (US National Institutes of Health, Bethesda, MD, USA).
Results and discussion
Here we show results of the examination of 11 tissue samples of medulloblastoma compared with normal tissue from the safety margin.
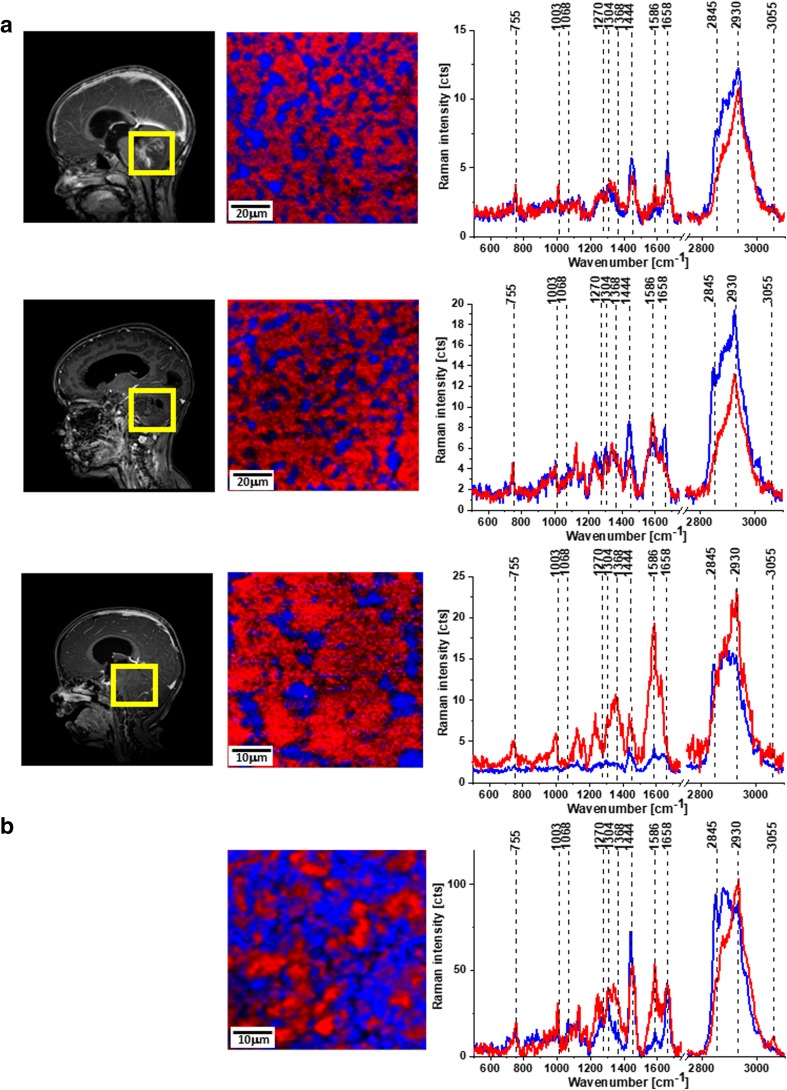
Figure 1 shows MRI, microscopy, and Raman images as well as Raman spectra for medulloblastoma (A) and normal (B) tissues.
Fig. 1.
MRI images, Raman images, and Raman spectra of medulloblastoma (P27, P34, P38) (A) and Raman image and Raman spectra of normal tissue (B). The line colors of the spectra correspond to the colors of the Raman maps. Integration time for images, 0.5 s; resolution step, 0.5 μm; laser excitation power, 10 mW
First of all, it is important to note that Raman mapping of human tissues can generate images as accurate as histology images with unique spatial resolution, sensitivity, and capabilities [1–4, 7].
A detailed insight into Fig. 1 shows spectral alterations in the chemical and morphological composition of the diagnosed medulloblastoma compared with normal tissue. The most important differences are in lipid and protein content marked on Raman images and spectra as blue (the band at 2845 cm−1 corresponds to CH2 sym. str. of lipids) and red (the band at 2930 cm−1 corresponds to CH3 sym. str. of lipids) areas, respectively.
The calculated areas of blue and red from Fig. 1 show 29, 24, and 20% of lipids for tumorous tissue and 58% area of lipids for normal tissue.
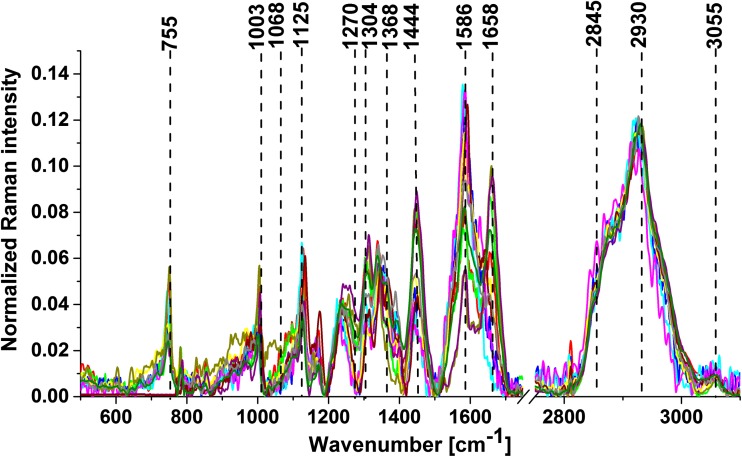
Figure 2 shows vector-normalized average spectra from Raman imaging for all analyzed samples of medulloblastoma. The results show that the spectra of all samples were highly reproducible.
Fig. 2.
Average spectra from Raman images for 11 cases of medulloblastoma
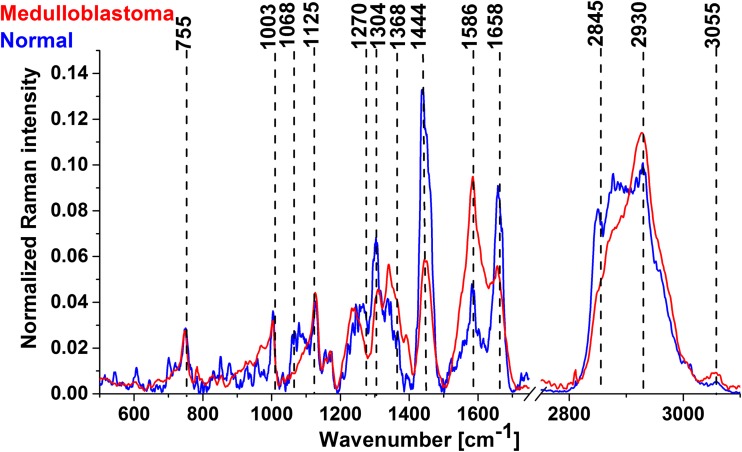
Figure 3 shows the vector-normalized average spectra for medulloblastoma and normal tissues.
Fig. 3.
Average Raman spectra for medulloblastoma and normal tissues
The results presented in Fig. 3 show that the Raman vibrational signatures can predict which tissue has tumorous biochemistry and can identify medulloblastoma. A detailed insight shows significant differences in lipid (1064, 1270, 1304, 1444, and 2845 cm−1) and protein (1240, 1368, 1586, 1658, and 2930 cm−1) content. Abramczyk et al. [1, 7, 26] showed that the Raman technique makes use of the fact that tumors contain large amounts of protein and far less lipids (fatty compounds), while healthy tissue is rich in both. Particularly interesting are the differences at 1586 cm−1, described as a marker of malignancy in tumors [21, 24]. Literature mostly merges this peak with the C=C bending mode of phenylalanine [5, 12, 19, 25], but our latest research shows that this also corresponds with phosphorylation of tyrosine. Also, the amide III band is shifted from 1270 to 1228 cm−1 as a result of phosphorylation. This is consistent with the latest research on phosphorylation inhibitors as a therapy for medulloblastoma [18].
For clinical application, the most important finding of the paper is the ratio of protein to lipid content presented in Table 1. This feature can be used to discriminate between normal and tumorous tissues. Table 1 shows the ratio for high frequencies and for the fingerprint region as well as the ratio calculated from the areas of proteins (Aproteins) and lipids (Alipids) from Raman images.
Table 1.
Raman intensity ratios at 2930/2845 cm−1 and 1586/1444 cm−1 for all analyzed medulloblastoma and normal samples
| Medulloblastoma patient number | I2930/I2845 | I1586/I1444 | Aproteins/Alipids |
|---|---|---|---|
| P3 | 2.74 | 1.30 | 1.85 |
| P4 | 2.76 | 0.81 | 8.00 |
| P9 | 3.2 | 3.81 | 3.34 |
| P18 | 2.97 | 7.89 | 1.77 |
| P27 | 1.90 | 4.64 | 4.00 |
| P34 | 2.56 | 2.21 | 3.16 |
| P38 | 3.25 | 0.56 | 2.44 |
| P43 | 3.42 | 0.60 | 4.00 |
| P44 | 2.86 | 2.90 | 2.70 |
| P48 | 2.89 | 2.10 | 3.55 |
| P49 | 3.17 | 1.13 | 6.69 |
| Average for medulloblastoma | 2.88 ± 0.26 | 2.54 ± 0.81 | 3.74 ± 1.24 |
| Average for normal | 1.6 ± 0.63 | 0.32 ± 0.18 | 0.77 ± 0.09 |
One can see that all studied cases of medulloblastoma tissue samples have the ratios I2930/I2845, I1586/I1444, and Aproteins/Alipids significantly higher than those for normal tissue, which demonstrates lower content of lipids in tumors. This is consistent with literature, where chromatography measurements show significantly reduced levels of polyunsaturated fatty acids (PUFA) and phospholipids in CNS tumors [22].
Both the ratios I2930/I2845 and I1586/I1444 can be used for diagnostic purposes. Although the results presented in the paper are highly reproducible, they are carried out on tissue slices, and for medical uses, more in vivo research should be conducted.
Conclusions
The ability of Raman spectroscopy and imaging to detect medulloblastoma tumors fills the niche in diagnostics. These powerful analytical techniques are capable of monitoring tissue morphology and biochemistry. Our results demonstrate that RS can be used to discriminate between normal and medulloblastoma tissues by monitoring alterations in lipid and protein content using I2930/I2845 and I1586/I1444 ratios.
Funding information
This work was supported by the National Science Center of Poland (grant UMO-2015/19/B/ST4/01878, Dz. St. 2018).
Compliance with ethical standards
All experiments were performed in compliance with relevant laws and guidelines of the Bioethical Committee at the Polish Mother’s Memorial Hospital Research Institute in Lodz (53/216) and of the Ministry of Health of the Republic of Poland.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Abramczyk H, Brozek-Pluska B. Raman imaging in biochemical and biomedical applications. Diagnosis and treatment of breast cancer. Chem Rev. 2013;113:5766–5781. doi: 10.1021/cr300147r. [DOI] [PubMed] [Google Scholar]
- 2.Abramczyk H, Brozek-Pluska B. New look inside human breast ducts with Raman imaging. Raman candidates as diagnostic markers for breast cancer prognosis: mammaglobin, palmitic acid and sphingomyelin. Anal Chim Acta. 2016;909:91–100. doi: 10.1016/j.aca.2015.12.038. [DOI] [PubMed] [Google Scholar]
- 3.Abramczyk H, Brozek-Pluska B, Surmacki J, Jablonska J, Kordek R. Raman ‘optical biopsy’ of human breast cancer. J Mol Liq. 2011;164:123–131. doi: 10.1016/j.molliq.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Abramczyk H, Surmacki J, Kopeć M, Olejnik AK, Lubecka-Pietruszewska K, Fabianowska-Majewska K. The role of lipid droplets and adipocytes in cancer. Raman imaging of cell cultures: MCF10A, MCF7, and MDA-MB-231 compared to adipocytes in cancerous human breast tissue. Analyst. 2015;140(7):2224–2235. doi: 10.1039/C4AN01875C. [DOI] [PubMed] [Google Scholar]
- 5.Austin LA, Osseiran S, Evans CL. Raman technologies in cancer diagnostics. Analyst. 2015;141:476–503. doi: 10.1039/C5AN01786F. [DOI] [PubMed] [Google Scholar]
- 6.Brozek-Pluska B, Musial J, Kordek R, Bailo E, Dieing T, Abramczyk H. Raman spectroscopy and imaging: applications in human breast cancer diagnosis. Analyst. 2012;137:3773–3780. doi: 10.1039/c2an16179f. [DOI] [PubMed] [Google Scholar]
- 7.Brozek-Pluska B, Kopec M, Abramczyk H. Development of a new diagnostic Raman method for monitoring epigenetic modifications in the cancer cells of human breast tissue. Anal Methods. 2016;8:8542–8553. doi: 10.1039/C6AY02559E. [DOI] [Google Scholar]
- 8.Butler HJ, Fogarty SW, Kerns JG, Martin-Hirsch PL, Fullwood NJ, Martin FL. Gold nanoparticles as a substrate in bio-analytical near-infrared surface-enhanced Raman spectroscopy. Analyst. 2015;140:3090–3097. doi: 10.1039/C4AN01899K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler HJ, Ashton L, Bird B, Cinque G, Curtis K, Dorney J, Esmonde-White K, Fullwood NJ, Gardner B, Martin-Hirsch PL, Walsh MJ, McAinsh MR, Stone N, Martin FL. Using Raman spectroscopy to characterize biological materials. Nat Protoc. 2016;11:664–687. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- 10.Desroches J, Jermyn M, Pinto M, Picot F, Tremblay MA, Obaid S, Marple E, Urmey K, Trudel D, Soulez G, Guiot MC, Wilson BC, Petrecca K, Leblond F. A new method using Raman spectroscopy for in vivo targeted brain cancer tissue biopsy. Sci Rep. 2018;8:1792. doi: 10.1038/s41598-018-20233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gajjar K, Heppenstall LD, Pang W, Ashton KM, Trevisan J, Patel II, Llabjani V, Stringfellow HF, Martin-Hirsch PL, Dawson T, Martin FL. Diagnostic segregation of human brain tumors using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis. Anal Methods. 2013;5:89–102. doi: 10.1039/C2AY25544H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghomi M (2012) Applications of Raman spectroscopy to biology: from basic studies to disease diagnosis. IOS press, ISBN 978-1-60750-999-8
- 13.Imiela A, Polis B, Polis L, Abramczyk H. Novel strategies of Raman imaging for brain tumor research. Oncotarget. 2017;8:85290–85310. doi: 10.18632/oncotarget.19668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeeves MA. Mind fields: reflections on the science of mind and brain. Grand Rapids: Baker Books; 1994. p. 21. [Google Scholar]
- 15.Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, Bernstein L, Guiot MC, Petrecca K, Leblond F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl med. 2015;7:274ra19. doi: 10.1126/scitranslmed.aaa2384. [DOI] [PubMed] [Google Scholar]
- 16.Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E, Pitter K, Huang R, Campos C, Habte F, Sinclair R, Brennan CW, Mellinghoff IK, Holland EC, Gambhir SS. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat Med. 2012;18:829–834. doi: 10.1038/nm.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirsch M, Schackert G, Salzer R, Krafft C. Raman spectroscopic imaging for in vivo detection of cerebral brain metastases. Anal Bioanal Chem. 2010;398:1707–1713. doi: 10.1007/s00216-010-4116-7. [DOI] [PubMed] [Google Scholar]
- 18.Klisch TJ, Vainshtein A, Patel AJ, Zoghbi HY. Jak2-mediated phosphorylation of Atoh1 is critical for medulloblastoma growth. eLife. 2017;6:e31181. doi: 10.7554/eLife.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin K, Zheng W, Lim CM, Huang Z. Real-time in vivo diagnosis of nasopharyngeal carcinoma using rapid fiber-optic Raman spectroscopy. Theranostics. 2017;7(14):3517–3526. doi: 10.7150/thno.16359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu CH, Das BB, Sha Glassman WL, Tang GC, Yoo KM, Zhu HR, Akins DL, Lubicz SS, Cleary J, Prudente R, Celmer E, Caron A, Alfano RR. Raman, fluorescence, and time-resolved light scattering as optical diagnostic techniques to separate diseased and normal biomedical media. J Photochem Photobiol B. 1992;16:187–209. doi: 10.1016/1011-1344(92)80008-J. [DOI] [PubMed] [Google Scholar]
- 21.Maier JS, Demuth JE, Cohen JK, Stewart S, MmcClelland LA (2010) Cytological analysis by Raman spectroscopic imaging. United States Patent. No. US 7,697,576 B2
- 22.Martin DD, Robbins ME, Spector AA, Wen BC, Hussey DH. The fatty acid composition of human glioma differs from that found in nonmalignant brain tissue. Lipids. 1996;31:1283–1288. doi: 10.1007/BF02587914. [DOI] [PubMed] [Google Scholar]
- 23.Meyer T, Bergner N, Bielecki C, Krafft C, Akimov D, Romeike BF, Reichart R, Kalff R, Dietzek B, Popp J. Nonlinear microscopy, infrared, and Raman microspectroscopy for brain tumor analysis. J Biomed Opt. 2011;16:021–113. doi: 10.1117/1.3533268. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro A, Gofrit ON, Pizov G, Cohen JK, Maier J. Raman molecular imaging: a novel spectroscopic technique for diagnosis of bladder cancer in urine specimens. Eur Urol. 2011;59:106–112. doi: 10.1016/j.eururo.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Stone N, Stavroulaki P, Kendall C, Birchall M, Barr H. Raman spectroscopy for early detection of laryngeal malignancy: preliminary results. Laryngoscope. 2000;110:1756–1763. doi: 10.1097/00005537-200010000-00037. [DOI] [PubMed] [Google Scholar]
- 26.Surmacki J, Brozek-Pluska B, Kordek R, Abramczyk H. The lipid-reactive oxygen species phenotype of breast cancer. Raman spectroscopy and mapping, PCA and PLSDA for invasive ductal carcinoma and invasive lobular carcinoma. Molecular tumorigenic mechanisms beyond Warburg effect. Analyst. 2015;140:2121–2133. doi: 10.1039/C4AN01876A. [DOI] [PubMed] [Google Scholar]