ABSTRACT
Two of the most prevalent ovarian diseases affecting women’s fertility and health are Primary Ovarian Insufficiency (POI) and Polycystic Ovarian Syndrome (PCOS). Previous studies have shown that exposure to a number of environmental toxicants can promote the epigenetic transgenerational inheritance of ovarian disease. In the current study, transgenerational changes to the transcriptome and epigenome of ovarian granulosa cells are characterized in F3 generation rats after ancestral vinclozolin or DDT exposures. In purified granulosa cells from 20-day-old F3 generation females, 164 differentially methylated regions (DMRs) (P < 1 x 10−6) were found in the F3 generation vinclozolin lineage and 293 DMRs (P < 1 x 10−6) in the DDT lineage, compared to controls. Long noncoding RNAs (lncRNAs) and small noncoding RNAs (sncRNAs) were found to be differentially expressed in both the vinclozolin and DDT lineage granulosa cells. There were 492 sncRNAs (P < 1 x 10−4) in the vinclozolin lineage and 1,085 sncRNAs (P < 1 x 10−4) in the DDT lineage. There were 123 lncRNAs and 51 lncRNAs in the vinclozolin and DDT lineages, respectively (P < 1 x 10−4). Differentially expressed mRNAs were also found in the vinclozolin lineage (174 mRNAs at P < 1 x 10−4) and the DDT lineage (212 mRNAs at P < 1 x 10−4) granulosa cells. Comparisons with known ovarian disease associated genes were made. These transgenerational epigenetic changes appear to contribute to the dysregulation of the ovary and disease susceptibility that can occur in later life. Observations suggest that ancestral exposure to toxicants is a risk factor that must be considered in the molecular etiology of ovarian disease.
KEYWORDS: Ovary, granulosa, epigenetic, transgenerational, sperm, DNA methylation, ncRNA, transcriptome, PCO disease, POI
Introduction
Two of the most prevalent ovarian diseases affecting women’s fertility and health are Primary Ovarian Insufficiency (POI) and Polycystic Ovarian Syndrome (PCOS). POI is characterized by a marked reduction in the primordial follicle pool of oocytes and the induction of menopause prior to age 40 [1]. POI currently affects approximately 1% of female population [2]. While genetic causes can be ascribed to a minority of patients, around 90% of POI cases are considered idiopathic, with no apparent genetic link nor known cause [3].
PCOS is a multi-faceted disease that affects 6–18% of women [4,5]. It is characterized by infrequent ovulation or anovulation, high androgen levels in the blood, and the presence of multiple persistent ovarian cysts [6,7]. PCOS patients often show insulin resistance and a heightened risk for diabetes [8,9]. Both genetic and environmental factors have been linked to the development of PCOS, although these do not explain all cases [10,11]. For both PCOS and POI other underlying causes such as epigenetic transgenerational inheritance of disease susceptibility have seldom been considered.
Epigenetics refers to ‘molecular factors and processes around the DNA that regulate genome activity independent of DNA sequence, and that are mitotically stable’ [12]. Epigenetic factors include DNA methylation, histone modifications, expression of noncoding RNA (ncRNA), RNA methylation, and alterations in chromatin structure [13]. Epigenetic transgenerational inheritance is defined as ‘the germline transmission of epigenetic information and phenotypic change across generations in the absence of any continued direct environmental exposure or genetic manipulation’ [12]. Epigenetic changes can be induced by environmental factors such as nutrition or toxicant exposure and are an important mechanism by which organisms alter gene expression in response to their environment. Although transgenerational epigenetic changes must be inherited via germ cells (i.e., sperm or eggs), these germ cell epigenetic changes subsequently promote in the early embryo and stem cells alterations in epigenetics and gene expression that impacts all somatic cells and organs of the individual. This can lead to increased disease susceptibility later in life. Therefore, disease development in organs such as ovaries can in part be due to ancestral exposures and epigenetic inheritance [14].
Previous studies have shown that exposure to a number of environmental toxicants can promote the epigenetic transgenerational inheritance of ovarian disease. Exposure of gestating female rats (F0 generation) to the agricultural fungicide vinclozolin resulted in a transgenerational increase in ovarian abnormalities in great-grand-offspring (F3 generation) [15]. These abnormalities included a decrease in the primordial follicle pool of oocytes that was similar to what is seen in POI, and an increase in ovarian cysts that was similar to what is seen in PCOS. In addition, the ovarian granulosa cells from the antral follicles of 6-month-old F3 generation vinclozolin lineage rats showed transgenerational changes in gene expression and alterations in the pattern of DNA methylation compared to F3 generation control lineage rats [15]. Similarly, exposure of F0 generation pregnant rats to the insecticide DDT (dichlorodiphenyltrichloroethane) induced an epigenetic transgenerational increase in ovarian diseases in the F3 generation, which was comprized of primordial follicle loss and increased rates of ovarian cysts [16]. Ancestral exposure to DDT also induced transgenerational changes in F3 generation sperm DNA methylation patterns [16], but epigenetic changes in ovarian somatic cells such as granulosa cell have not been investigated.
The environmentally induced epigenetic transgenerational inheritance requires the germline transmission of epigenetic alterations (epimutations) by either the sperm or egg. The majority of transgenerational studies have examined sperm transmission of epigenetic changes due to limitations in oocyte numbers for efficient analysis. Although DNA methylation has been investigated more extensively, noncoding RNA (ncRNA) expression has also been shown to be involved in epigenetic transgenerational inheritance [17,18]. Differentially expressed ncRNAs have been shown to correlate with increased disease susceptibility originating from the ancestrally exposed male germline [19]. Both long (> 200 nt) and small (< 200 nt) ncRNAs have been implicated as contributing to epigenetic transgenerational inheritance [19,20]. Long noncoding RNAs are hypothesized to maintain epigenetic memory by posttranscriptional regulation and to assist in regulating DNA methylation, chromatin remodeling, and histone modifications [19]. Small noncoding RNAs are known to regulate gene expression by affecting transcript stability and have been shown to be abundant in sperm with a single spermatozoon containing above 20,000 long and short noncoding RNAs [21,22]. Recently, concurrent alterations of DNA methylation, ncRNA, and histone alterations have been identified in sperm mediating the epigenetic transgenerational inheritance of pathology [23,24]. Therefore, the current study investigates alterations in both DNA methylation, ncRNA expression and gene expression.
This study was designed to investigate transgenerational changes to the epigenome of ovarian granulosa cells isolated from F3 generation rats after ancestral vinclozolin or DDT exposure compared to controls. Elucidation of the epigenetic and gene expression changes that occur in the ovary after ancestral exposure to an environmental toxicant provide novel insights into the molecular etiology of the epigenetic transgenerational inheritance of ovarian disease. In addition, this improves our understanding of the risk factors that must be considered when investigating the underlying etiology of ovarian disease in the human population.
Results
Ovarian pathology analysis
Pregnant F0 generation female rats were transiently exposed to vinclozolin, DDT or control vehicle (dimethyl sulfoxide, DMSO) from days 8–14 of gestation, as described in Methods [25]. The in utero exposed offspring (F1 generation rats) were bred to produce the F2 generation, and similarly the F2 generation animals were bred to produce the transgenerational F3 generation. No sibling or cousin crosses were used to avoid inbreeding artifacts. Only the F0 generation rats received the experimental treatments. Granulosa cells were harvested from the ovaries of super-ovulated F3 generation females at 20–22 d of age. Granulosa cells were isolated and analyzed so as to characterize DNA methylation, mRNA gene expression and ncRNA expression as described in Methods. Additional F3 generation vinclozolin, DDT, and control lineage rats were aged to one year and their ovaries subjected to histopathological evaluation to detect signs of ovarian disease.
Ovaries were defined as diseased if there was a decrease in the number of primordial follicles at two standard deviations below those found in controls, and/or if there was an increase in the number of ovarian cysts at two standard deviations above those found in controls (see Methods), Supplemental Figure S1. There was a significant increase in ovarian disease in transgenerational F3 generation DDT and vinclozolin lineage rats at one year of age compared to F3 generation controls (Figure 1). Previous studies have shown that transgenerational increases in ovarian disease were detected following exposures to plastic derived compounds bisphenol A (BPA) and phthalates (DBT & DEHP) [26], dioxin (TCCD) [25], pesticides permethrin and DEET [27], jet fuel hydrocarbons [28], and methoxychlor [29], with nearly 100% disease frequency. Therefore, the transgenerational inheritance of increased ovarian disease can occur after exposure to a variety of environmental toxicants. There was no increase in ovarian disease in direct fetal exposed F1 or germline exposed F2 generation vinclozolin or DDT lineage rats compared to controls [30,31]. Therefore, as previously observed with most exposures, negligible ovarian disease is present following direct exposure [25,27–29], with the exception of BPA and phthalates [26]. This indicates that there was an epigenetic transgenerational increase in susceptibility to ovarian disease in rats ancestrally exposed to DDT or vinclozolin (Figure 1).
Figure 1.
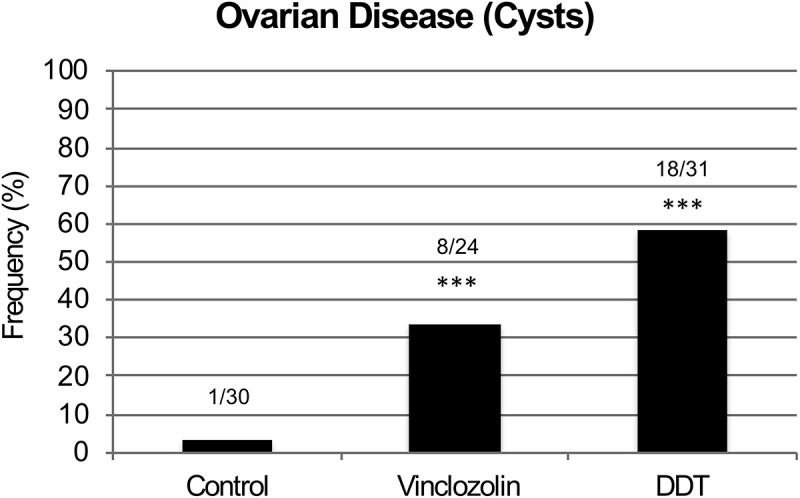
Ovarian pathology frequency. Transgenerational ovarian disease in F3 generation control, vinclozolin and DDT lineage rats at 1 y of age. Numbers for diseased individuals versus the total number of individuals analyzed is shown and (***) indicates statistical significance of P < 7 x 10−3 for vinclozolin and P < 1 x 10−5 for DDT by Fisher’s Exact Test. Transgenerational ovarian disease frequency (i.e., presence of ovarian cysts) from control, vinclozolin, and DDT lineage rats at 1 y of age.
DNA methylation analysis
Differences in sites of DNA methylation (i.e., differential DNA methylation regions, DMRs) between F3 generation control, vinclozolin, and DDT lineage rats were characterized for ovarian granulosa cells using an MeDIP-Seq procedure comprized of methylated DNA immunoprecipitation (MeDIP) followed by next-generation sequencing and bioinformatics techniques as described in Methods. A number of P value thresholds are assessed. In vinclozolin lineage granulosa cells compared to controls, there are 164 DMRs at a P value < 1 x 10−6, of which 33 DMRs are comprized of multiple neighboring genomic windows (Figure 2(A)). A list of these DMRs is presented in Supplemental Table S1. In DDT lineage cells compared to controls there are 293 DMRs at a P value < 1 x 10−6, of which 57 DMRs are comprized of multiple genomic windows (Figure 2(B)). A list of these DMRs is presented in Supplemental Table S2. Twenty-one DMRs overlapped between the vinclozolin and DDT lineages (Figure 2(C) and Supplemental Table S3). Chromosomal locations of the DMRs were examined. For vinclozolin lineage cells the DMRs are present on all chromosomes, while for DDT lineage cells the DMRs are present on all chromosomes except the small Y chromosome (Figure 3(A,B)). DMRs are not detected on the mitochondrial genome. The red arrowheads indicate the locations of the DMR and black boxes indicate clusters of DMRs.
Figure 2.
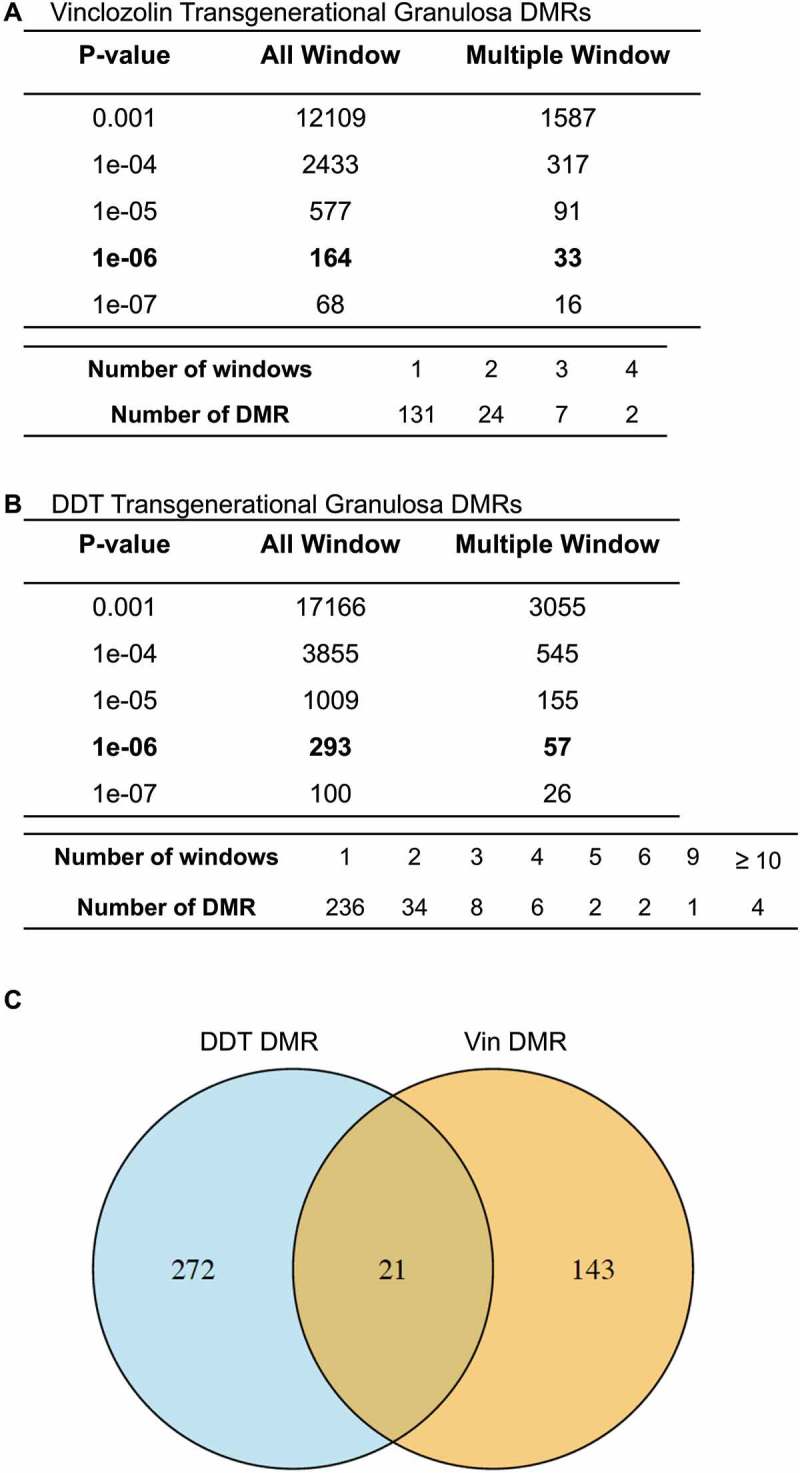
DMR identification. The number of DMRs found using different P value cutoff thresholds. The all window column shows all DMRs. The multiple window column shows the number of DMRs containing at least two significant windows. Lower table of each set shows the number of DMR having each specific number of significant windows at a P < 1x10−6. (a) Granulosa cell vinclozolin F3 generation DMRs P < 1x10−6. (b) Granulosa cell DDT F3 generation DMRs P < 1x10−6.
Figure 3.
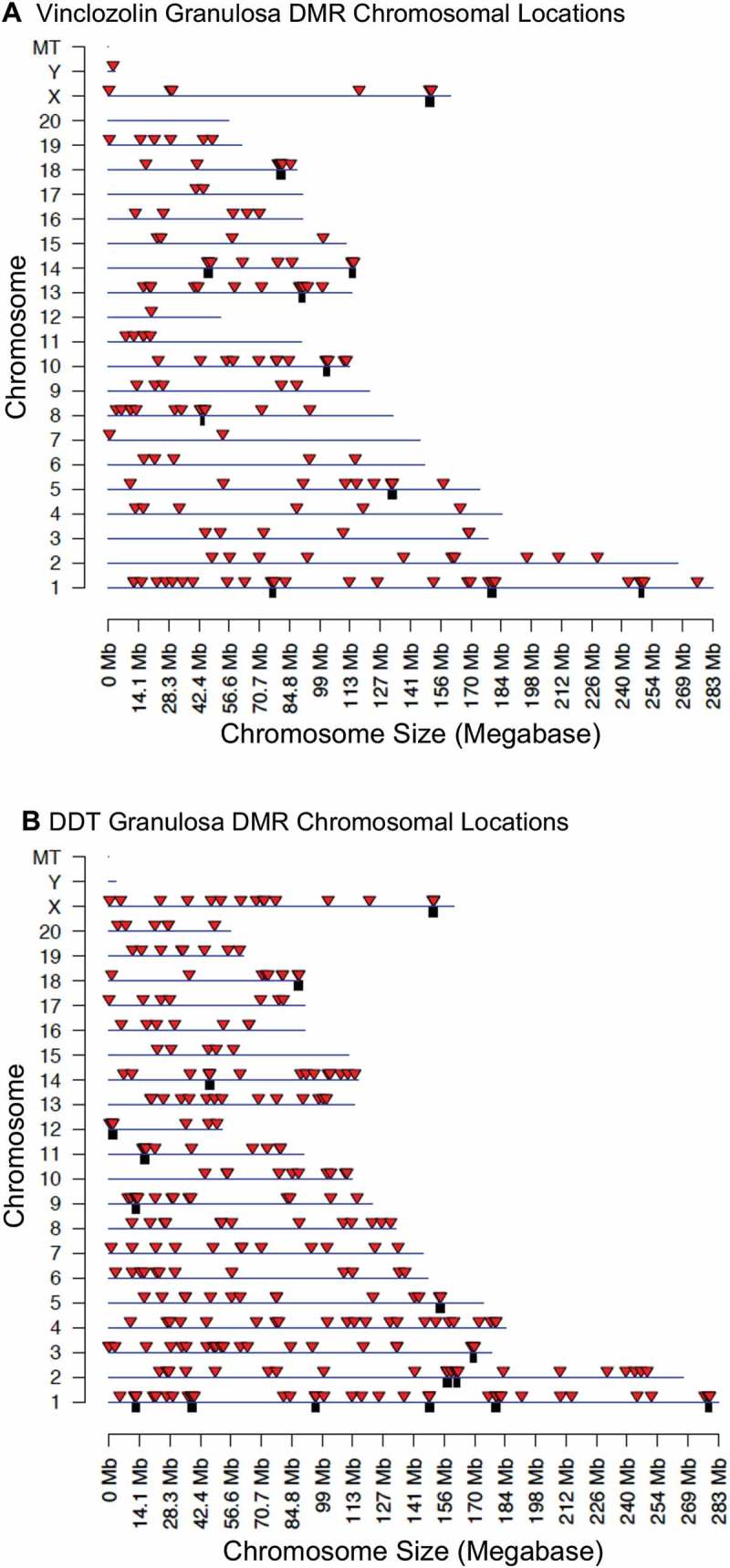
DMR chromosomal locations. The DMR locations on the individual chromosomes for all DMRs at a P value threshold of < 1x10−6. (a) Vinclozolin F3 generation. (b) DDT F3 generation. Red arrowheads indicate positions of DMRs and black boxes indicate clusters of DMRs.
Examination of the characteristics of the genomic sites where DMRs reside shows that for F3 generation vinclozolin lineage granulosa most DMRs are present in areas having on average of 1 or 2 CpG sites per 100 base pairs (Figure 4(A)). A CpG is a cytosine adjacent to a guanine on the DNA and it is primarily these cytosine bases that are methylated. For DDT lineage granulosa cells most DMRs are present in areas having on average of 1 to 3 CpG sites per 100 base pairs (Figure 4(C)). This indicates that most of the DMRs identified occur in areas of low CpG density, termed CpG deserts [32]. Most DMRs for both vinclozolin lineage and DDT lineage granulosa cells are shown to be one kilobase (kb) in length (Figure 4(B,D)). Within these 1 kb DMRs small clusters of CpG sites are anticipated to be regulatory as previously described [32].
Figure 4.
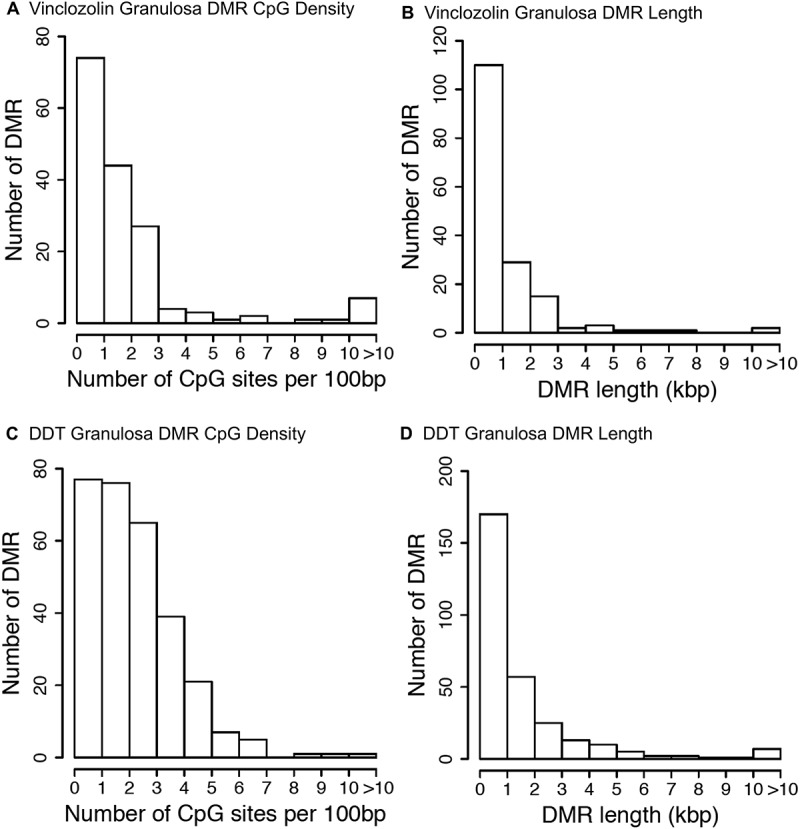
DMR genomic features. (a & c) The number of DMRs at different CpG densities for all DMRs at a p-value threshold of P < 1x10−6. (b & d) The DMR lengths for all DMRs are at a P value threshold of < 1x10−6. (a & b) Vinclozolin F3 generation. (c & d) DDT F3 generation.
Granulosa mRNA and noncoding RNA analysis
Differential gene expression and noncoding RNA expression between the granulosa control, DDT, and vinclozolin lineages were determined using RNA-seq as described in the Methods section. Differentially expressed RNAs were reported at a variety of different P value thresholds and a P < 1 x 10−4 was selected for subsequent analysis (Figure 5). Both DDT and vinclozolin lineage granulosa cells contained a similar number of differentially expressed mRNAs, while the vinclozolin lineage (Figure 5(A)) had more than twice the number of long noncoding RNAs (lncRNAs) than the DDT lineage (Figure 5(B)) at 123 vs. 51, respectively. In contrast, the DDT lineage contained twice the number of differentially expressed small noncoding RNAs (sncRNAs) at 1,085 compared to the vinclozolin lineage’s 492. The classes of differentially expressed RNAs were compared between the two lineages. The 492 sncRNAs from the vinclozolin lineage had a very high degree of overlap with the 483 sncRNAs from the DDT lineage (Figure 5(C)). Eight lncRNAs were similar between the two lineages (Figure 5(D)), while 21 mRNAs were common between DDT and vinclozolin lineage granulosa cells (Figure 5(E)). In addition, the differentially expressed sncRNAs were categorized by class (Figure 6). Notably, piRNAs accounted for nearly all affected sncRNA in both lineages. The high number of common affected sncRNAs between the two lineages (Figure 5(C)) were observed.
Figure 5.
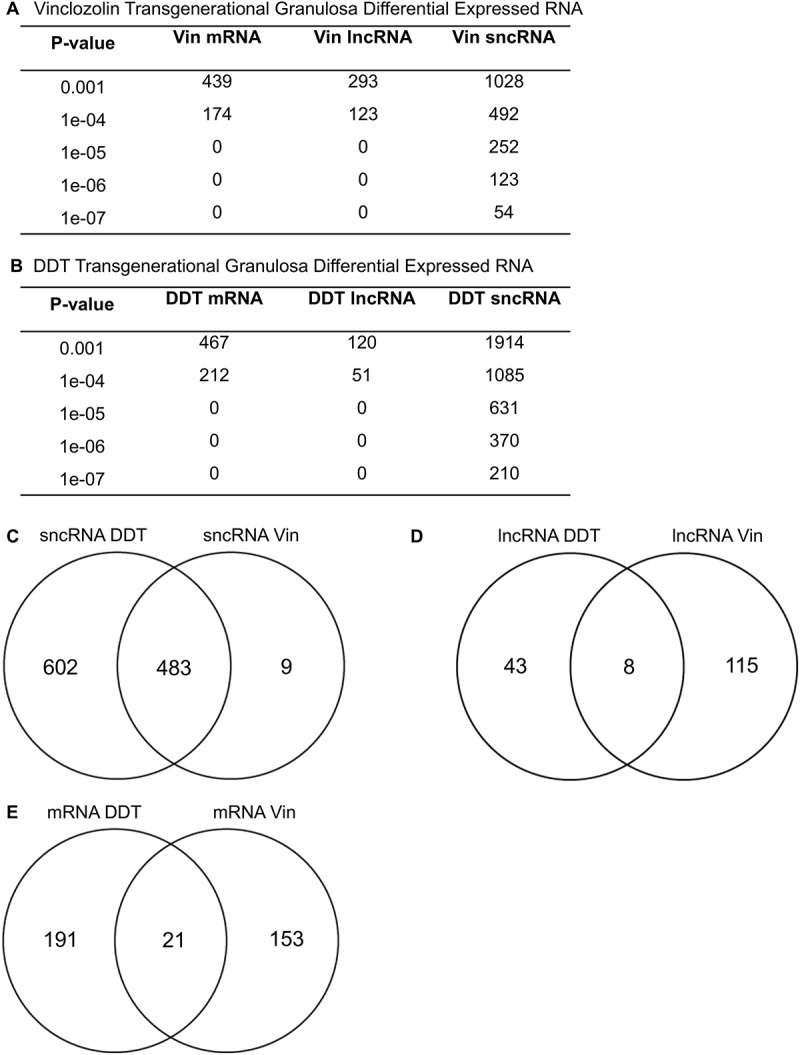
Differential RNA expression at different p-value thresholds for vinclozolin (a) and DDT (b). P value < 1x10−4 was used for subsequent analysis. Venn diagrams show overlap of RNA categories between the two lineages for (c) sncRNA, (d) lncRNA, and (e) mRNA.
Figure 6.

Differentially expressed small, noncoding RNA broken down by class and lineage. P value < 1 x 10−4 was used for analysis.
The chromosomal locations of the differentially expressed RNAs are presented in Figures 7 and 8 for each RNA type. The vinclozolin lineage’s sncRNAs showed a wide chromosomal distribution (Figure 7(A)). Both the differentially expressed lncRNAs (Figure 7(B)) and the mRNAs (Figure 7(C)) for vinclozolin lineage granulosa are present on all chromosomes except for the Y chromosome and the mitochondrial chromosomes. There was no overlap of vinclozolin lineage DMR with any differentially expressed RNA, and no overlap of the sites of the different classes of differentially expressed RNAs with each other (Figure 7(D)). DDT lineage differentially expressed RNAs of all classes were also widely distributed across chromosomes excepting the Y and the mitochondrial chromosomes (Figure 8(A-C)). In addition, similar to what was seen in the vinclozolin lineage, the DDT lineage DMRs and differentially expressed RNAs had very few overlaps with each other (Figure 8(D)).
Figure 7.
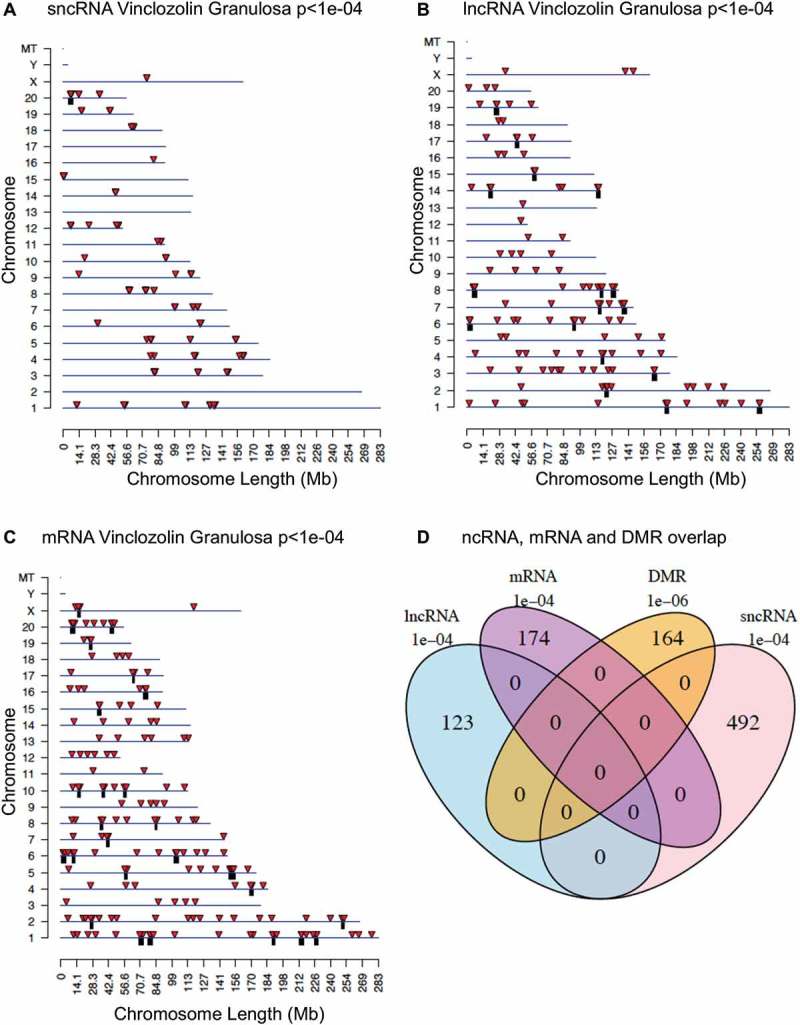
Differentially expressed RNAs from the vinclozolin lineage. Chromosomal locations of differentially expressed sncRNA (a), lncRNA (b), or mRNA (c). Individual RNAs are shown as red arrows and clusters are shown as black boxes. RNAs with unknown locations are not shown. (d) Venn diagram showing overlap of all differentially expressed epigenetic modifications from the vinclozolin lineage.
Figure 8.

Differentially expressed RNAs from the DDT lineage. Chromosomal locations of differentially expressed sncRNA (a), lncRNA (b), or mRNA (c). Individual RNAs are shown as red arrows and clusters are shown as black boxes. RNAs with unknown locations are not shown. (d) Venn diagram showing overlap of all differentially expressed epigenetic modifications from the DDT lineage.
Genes and pathway associations
The genes associated with differentially expressed lncRNAs (Supplemental Tables S7 and S8) and mRNAs (Supplemental Tables S10 and S11) were functionally categorized as described in Methods. The most predominant functional categories for differentially expressed mRNA associated genes are presented in Figure 9(A,B). The top functional category for mRNA associated genes for both the vinclozolin and DDT lineages was transcription (Figure 9(A,B), respectively). Both lineages also had high numbers of differentially expressed genes associated with signaling. Some DMRs occurred in the vicinity (within 10 kb) of known genes, Supplemental Tables S1, S2, S3. These DMR associated genes were categorized and evaluated for potential function. The DDT lineage DMR associated genes were most often involved in signaling and receptor functions (Figure 9(C)) while the vinclozolin lineage DMR associated genes were highest in receptor, metabolism, and transcription functions.
Figure 9.
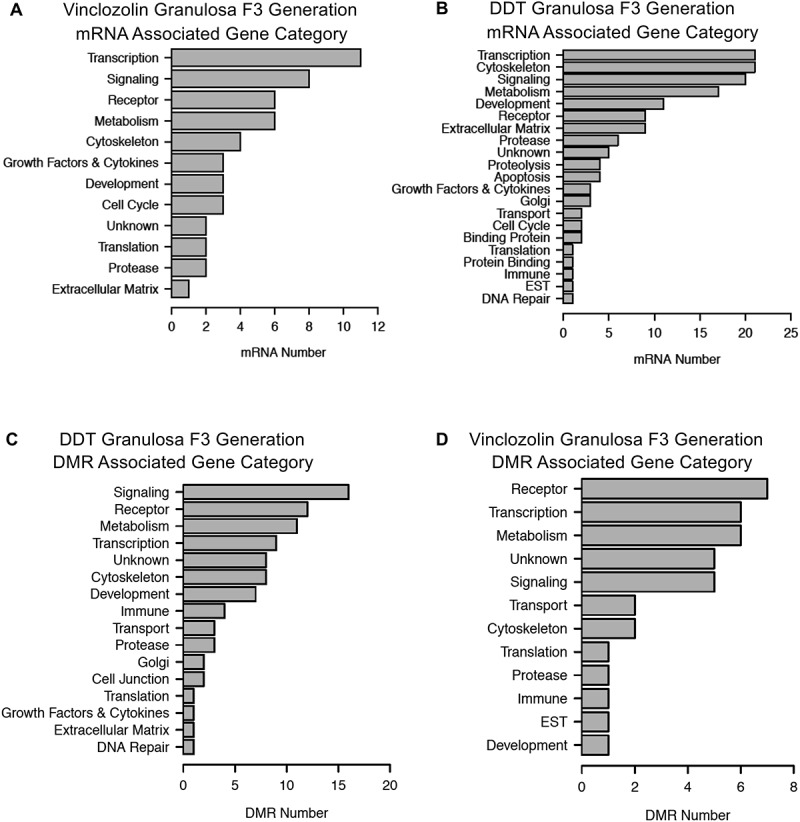
Differentially expressed epigenetic modifications were broken down by predicted associated gene category. Top row: mRNAs from vinclozolin (a) and DDT (b) lineages. Bottom row: DMRs from DDT (c) and vinclozolin (d). Genes or DMRs that could not be assigned a category are not shown.
The lists of differentially expressed DMRs and mRNAs are also compared to well-characterized physiological pathways in the KEGG database (http://www.kegg.jp/kegg/kegg2.html). Those pathways having the most DMR associated genes and differentially expressed mRNAs are presented in Figure 10(A,B). Metabolic pathways featured prominently, but since the KEGG metabolic pathway contains hundreds of genes the significance of this is unclear. The DDT lineage DMR associated genes occurred in cell adhesion, axon guidance, focal adhesion, specific signaling pathways and several disease-associated pathways. Examination of the genes involved revealed a high proportion of somewhat general-purpose signaling molecules. The vinclozolin lineage DMR associated genes included three olfactory receptors present in the olfactory transduction pathway. The differentially expressed mRNAs were present primarily in specific signaling pathways and disease-associated pathways (Figure 10(C,D)). Differentially expressed vinclozolin lineage mRNAs included the growth factors kit ligand (Kitlg), bone morphogenetic protein 15 (Bmp15), growth differentiation factor 9 (Gdf9), and zona pellucida proteins 1–4 (Zp1, Zp2, Zp3, Zp4). Differentially expressed DDT lineage mRNAs included insulin-like growth factor 1 (Igf1), the receptors platelet derived growth factor receptor a (Pdgfra), and growth hormone receptor (Ghr), and a selection of collagen and extracellular matrix genes (Col1a1, Col3a1, Col4a1, Col4a5, Col6a1, Col6a2, Col11a1, Itga9, Spp1). The majority of these genes have previously been shown to have functions in the ovary [33].
Figure 10.

Associated gene pathways for DMRs (a) DDT and (b) vinclozolin lineages and for mRNA (c) DDT and (d) vinclozolin lineages.
A final analysis correlated the transgenerational granulosa cell vinclozolin and DDT lineage gene associations with previously identified ovarian disease associated genes. Extensive reviews previously published have summarized the genes that have been associated with ovarian disease [34–41]. These published ovarian disease-associated genes were compiled into a list of 416 genes that are listed in Supplemental Table S13. An overlap of this published ovarian disease gene list with the transgenerational granulosa cell vinclozolin and DDT DMR associated genes (Figure 11(A)) and mRNA (Figure 11(B)) demonstrates several DMR associated genes and 20 mRNA genes overlapping. The specific overlapped genes are presented in Figure 11(C) and Table S14. Therefore, a number of ovarian disease-associated genes previously identified [34–41] were in common with the transgenerational granulosa cell DMRs and mRNA identified.
Figure 11.
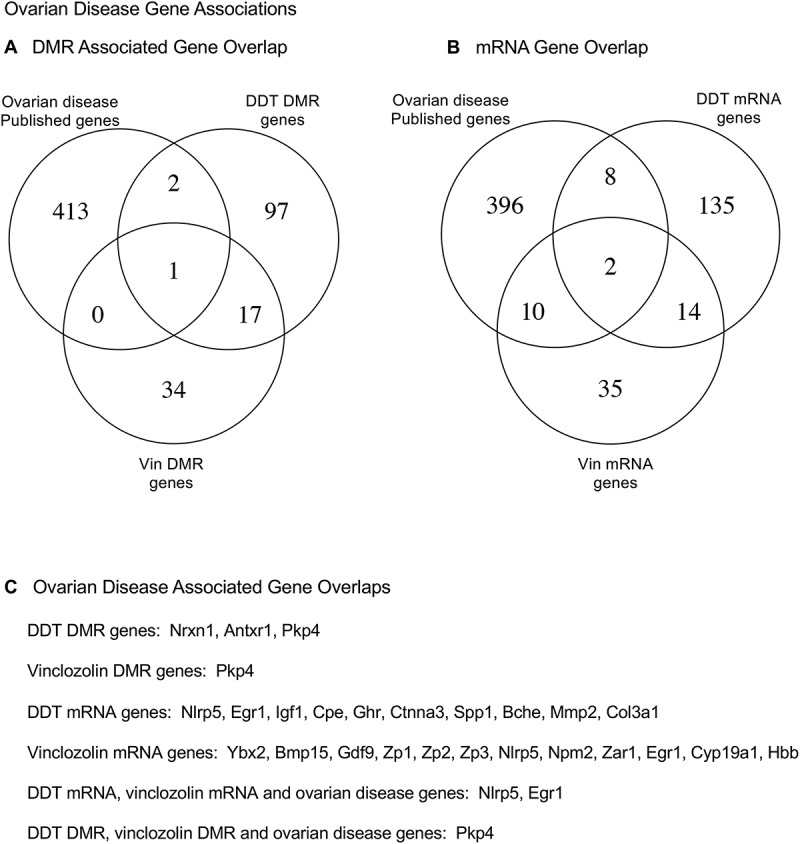
Ovarian disease gene associations. (a) DMR associated gene overlap with published ovarian disease genes. (b) mRNA gene overlaps with published ovarian disease genes. (c) Specific ovarian disease associated gene overlap with DMR associated genes and mRNA genes.
Discussion
Observations indicate that ancestral exposure to the environmental toxicants vinclozolin or DDT induced an epigenetic transgenerational increase in ovarian disease susceptibility in F3 generation rats. These results are in agreement with previous studies which also found transgenerational increases in susceptibility to ovarian diseases after exposure of F0 generation pregnant rats to vinclozolin [15] or DDT [16]. Experimental exposure of pregnant rats to other environmental toxicants such as jet fuel hydrocarbons, the plastics compounds bisphenol A (BPA) and phthalates, the pesticides permethrin and methoxychlor, and the industrial pollutant dioxin have also been shown to promote a transgenerational increase in ovarian disease [15,42]. This suggests that the ovary may be particularly sensitive to transgenerational epigenetic perturbations that disrupt somatic cell gene expression. Interestingly, these earlier studies demonstrated that the F1 generation direct fetal exposure did not induce ovarian disease later in life (1 y of age), but did promote ovarian disease in the transgenerational F3 generation at 1 y of age [15,42]. The exception was BPA and phthalate exposure that did promote ovarian diseases in both the F1 and F3 generations. In the current study, we also found negligible ovarian disease in the F1 generation, but significant ovarian disease in the F3 generation, Figure 1. When a gestating female is exposed the F0 generation female, the F1 generation fetus, and the germline within the F1 generation fetus that will generate the F2 generation are directly exposed to the environmental factor. Therefore, the first transgenerational generation is the F3 generation which has no direct exposure [43]. The direct exposure toxicology involves a signal transduction event and cellular response. The transgenerational molecular mechanism is distinct and involves the germline (sperm or egg) having an altered epigenome that following fertilization may modify the embryonic stem cells epigenome and transcriptome. This subsequently impacts the epigenetics and transcriptome of all somatic cell types derived from these stem cells [43]. Therefore, all somatic cells in the transgenerational animal have altered epigenomes and transcriptomes and those sensitive to this alteration will be susceptible to develop disease. Therefore, the F3 generation can have disease while the F1 and F2 generations do not, due to this difference in the molecular mechanisms involved. When disease is observed in the F1, F2 and F3 generations then the direct exposure actions and transgenerational actions have similar physiological responses, shown with BPA and phthalate induced ovarian disease [42].
Changes in DNA methylation were observed in F3 generation vinclozolin and DDT lineage granulosa cells compared to the control lineage. The sites of these DMRs were in genomic regions of relatively low CpG density ‘CpG deserts’ [32]. This finding is consistent with previous work in which transgenerational DMR in sperm were most often found in regions of low CpG density after ancestral toxicant exposure [25–29]. A previous study has also examined changes in DNA methylation in granulosa cells after ancestral exposure to vinclozolin [15]. This investigation used granulosa cells from 5–6-month-old F3 generation rats from vinclozolin and control lineages analyzed using a methylated DNA immunoprecipitation procedure (MeDIP) followed by a gene promoter microarray chip analysis. Similar to the current genome-wide analysis, there were DMRs identified in the vinclozolin lineage granulosa cells compared to controls [15]. In the current investigation the granulosa cells were isolated from 20-day-old rats which is prior to the onset of any clinical signs of ovarian disease. The current study used next-generation sequencing analysis which allows for a genome-wide investigation of the F3 generation vinclozolin and DDT lineage granulosa cells.
Changes in DNA methylation can affect genome activity and gene expression in concert with other epigenetic factors. DMRs were found in granulosa cells that were associated (within 10 kb) with genes, raising the possibility that these genes might be epigenetically regulated. An investigation of the putative functions of DMR associated genes revealed signaling, transcription, receptor and cytoskeleton genes to be predominant. These classes of genes are important for the interactions between granulosa cells and either oocytes or theca cells that are necessary for normal ovary function. Dysregulation of these functions may promote ovarian disease. In the current study there was limited correspondence between DMR associated genes and differential mRNA expression. However, the differentially expressed mRNAs were evaluated in granulosa cells collected from the healthy ovaries of young animals. The epigenetic changes observed may as the animals age activate associated gene expression to promote the dysregulation and increase disease susceptibility later in life. Alternatively, the DMR epimutations can influence distal gene expression through ncRNA.
Examination of the noncoding RNAs showed that F3 generation vinclozolin and DDT lineage granulosa cells differed in their classes of differentially expressed ncRNAs altered. The vinclozolin lineage had fewer differentially expressed lncRNAs and more sncRNAs when compared to the DDT lineage. Surprisingly, there was a significant overlap between the differentially expressed sncRNAs of each lineage accounting for 98% of the sncRNAs of the vinclozolin lineage and 45% of the DDT lineage. The differentially expressed sncRNAs of the vinclozolin lineage are a subset of those of the DDT lineage. The significance of this is unclear and warrants further investigation. However, looking into the functions of differentially expressed ncRNAs and mRNAs that are in common between the vinclozolin and DDT lineage granulosa cells may shed light on the underlying causes of the disease phenotypes.
Several growth factor and receptor mRNAs that were differentially expressed in F3 generation vinclozolin or DDT lineage granulosa cells have been previously implicated in normal ovarian function (Kitlg, Bmp15, GDF9, Pdgfra) [44–47]. In addition, abnormalities in Gdf9 and BMP15 expression are associated with polycystic ovarian syndrome (PCOS) and primary ovarian insufficiency (POI) in humans [48–50]. The receptor Scarb1 was differentially expressed in DDT lineage granulosa and has been associated with POI [51]. The growth factor IGF1 and the receptors Pdgfra and Ghr were also differentially expressed in the DDT lineage and have been implicated in PCOS [52–54]. Therefore, differentially expressed genes observed in the F3 generation of vinclozolin and DDT lineage granulosa cells have been correlated with PCOS and POI.
Correlations of genes previously identified to be associated with ovarian disease [34–41] (Supplemental Table S13) with the transgenerational granulosa cell genes of this study identified a number of genes associated with ovarian disease, Figure 11 and Supplemental Table S14. A gene that was present in the DDT and vinclozolin DMR associated genes and ovarian disease associated genes was Pkp4, plakophilins 4 [34,55]. The mRNA genes that were present in the DDT and vinclozolin mRNA genes and ovarian disease associated genes were Nlrp5 and Egr1 [34,56,57]. The Nlrp5 is associated with mitochondrial function in oocytes and embryo [56]. The Egr1 is associated with granulosa cell apoptosis during atresia through the NF-KB pathway [57]. The majority of transgenerational granulosa cell DMR associated genes and differentially expressed mRNA were not in common with these previously identified ovarian disease associated genes [34–41]. A more complete list of ovarian disease genes would likely have greater overlap. Observations demonstrate some genes previously shown to be involved in ovary disease are similar to the transgenerational granulosa cell associated genes identified.
The chromosomal locations for differentially expressed RNAs and DMRs for both vinclozolin and DDT lineages are generally genome-wide. There was a marked lack of overlap between the different epimutations in either lineage. It will become important to determine the gene targets of these epimutations to establish the mechanism behind the granulosa associated transgenerational disease. Interestingly, the epimutations and gene expression differences observed are present in granulosa cells in the late pubertal female rats at 22–24 d of age, which is long before any visible signs of ovarian disease are detectable. This indicates that the underlying factors that can contribute to adult-onset diseases like PCOS and POI appear to be present early in life. This helps explain the molecular mechanisms behind the developmental origins of ovarian disease.
In summary, these studies show that exposure to the environmental toxicants vinclozolin and DDT can promote the epigenetic transgenerational inheritance of ovarian disease susceptibility. Granulosa cells from young F3 generation vinclozolin and DDT lineage animals had epigenetic changes in DNA methylation and ncRNA expression, as well as in mRNA gene expression. These changes appear to contribute to the dysregulation of the ovary that can promote later life disease susceptibility. Future studies will need to translate these observations to investigate similar mechanisms in human females with POI or PCOS. Ancestral exposure to toxicants is now a risk factor that must be considered when investigating the underlying causes of ovarian disease in the human population.
Methods
Animal studies and breeding
Female and male rats of an outbred strain Hsd:Sprague Dawley®™SD®™ (Harlan) at about 70 to 100 d of age were fed ad lib with a standard rat diet and ad lib tap water for drinking. To obtain time-pregnant females, the female rats in proestrus were pair-mated with male rats. The sperm-positive (day 0) rats were monitored for diestrus and changes in body weight. If pregnant, then on days 8 through 14 of gestation [58], the females were administered daily intraperitoneal injections of vinclozolin (100 mg/kg BW/day, Chem Services, Westchester, PA), DDT (dichloro-diphenyl-trichloroethane) (25 mg/kg BW/day, Chem Services), or dimethyl sulfoxide (vehicle) as previously described [42]. Treatment groups were designated ‘vinclozolin’, ‘DDT’ and ‘control’ lineages. The gestating female rats treated were considered to be the F0 generation. The offspring of the F0 generation rats were the F1 generation. Non-littermate females and males aged 70–90 days from F1 generation lineages were bred to obtain F2 generation offspring. The F2 generation rats were bred to obtain F3 generation offspring. Only the pregnant F0 generation rats were treated directly with vinclozolin or DDT. All experimental protocols for the procedures with rats were pre-approved by the Washington State University Animal Care and Use Committee (IACUC approval # 06252).
Histopathology and ovarian disease classification
Rats at 12 months of age were euthanized by CO2 inhalation and cervical dislocation for tissue harvest. Ovaries were removed and fixed in Bouin’s solution (Sigma) followed by 70% ethanol, then processed for paraffin embedding by standard procedures for histopathological examination. Tissue sections (5 µm) were cut and every 30th section was collected and hematoxylin/eosin stained.
The three stained sections (150 µm apart) through the central portion of the ovary with the largest cross-section were evaluated microscopically for number of primordial follicles, small cystic structures and large cysts, as previously described [15]. The mean number of each evaluated structure per section was calculated across the three sections. Follicles had to be non-atretic and have the oocyte nucleus visible in the section in order to be counted. Primordial follicles are in an arrested state and have an oocyte surrounded by a single layer of either squamous or both squamous and cuboidal granulosa cells [59]. Cysts were defined as fluid-filled structures of a specified size that were not filled with red blood cells and which were not follicular antra. A single layer of cells may line cysts. Small cysts were 50–250 µm in diameter measured from the inner cellular boundary across the longest axis. Large cysts were greater than 250 µm in diameter. A cut-off was established to declare a tissue ‘diseased’ based on the mean number of histopathological abnormalities plus two standard deviations from the mean of control tissues as assessed by each of three individual observers blinded to the treatment groups. This number was used to classify rats into those with and without ovarian disease in each lineage. A rat tissue section was finally declared ‘diseased’ only when at least two of three observers marked the same tissue section ‘diseased’ for the same type of abnormality. Results were expressed as the proportion of affected animals and were analyzed using Fisher’s exact test.
Granulosa cell isolation
F3 generation rats from vinclozolin, DDT and control lineages were treated with Pregnant Mare Serum Gonadotropin (Sigma cat, St. Louis, MO) (10 IU PMSG injected IP) at 20–22 d of age. Two days later animals were sacrificed and ovaries removed. The ovarian bursa and its adherent fat were removed from each ovary and the ovaries processed for granulosa cell collection [60]. The ovaries were suspended in Ham’s F-12 base medium (Thermo Scientific, Waltham, MA). Following sequential 30-minute incubations at 37 °C in 6 mM EGTA in F-12 (to decrease Ca2+ – mediated cell adhesion) and then 0.5 M sucrose in F-12 (to increase osmotic pressure within follicles), ovaries were returned to F-12. Granulosa cells were released into the medium from antral follicles using 30-gauge needles and gentle pressure. Oocytes were removed by aspiration under a dissecting microscope. Granulosa cells from 4–9 rats from the same treatment group were pooled and collected into 1.5 ml tubes, allowed to settle for 10 minutes and the supernatant discarded. Three pools of granulosa cells were prepared from different animals and ovaries for each treatment group. Samples were stored at −70° until the time of RNA and DNA isolation.
DNA isolation
The granulosa cell pellet was resuspended in 100 μl PBS and then mixed with 820 μl DNA extraction buffer. Then 80 μl proteinase K (20 mg/ml) was added and the sample was incubated at 55°C for 2 hours under constant rotation. Then 300 μl of protein precipitation solution (Promega, Madison, WI, Genomic DNA Purification Kit, A795A) were added, the sample mixed thoroughly and incubated for 15 min on ice. The sample was centrifuged at 17,000xg for 20 minutes at 4°C. One ml of the supernatant was transferred to a 2 ml tube and 2 μl of Glycoblue (Thermo-Fisher AM9515) and 1 ml of cold 100% isopropanol were added. The sample was mixed well by inverting the tube several times then left in −20°C freezer for at least one hour. After precipitation, the sample was centrifuged at 17,000 x g for 20 min at 4°C. The supernatant was taken off and discarded without disturbing the (blue) pellet. The pellet was washed with 70% cold ethanol then centrifuged for 10 min at 4°C at 17,000 x g and the supernatant discarded. The pellet was air-dried at room temperature (about 5 minutes). The pellet was then resuspended in 100 μl of nuclease free water and DNA concentration determined on a NanoDrop.
Methylated DNA Immunoprecipitation (MeDIP)
Methylated DNA Immunoprecipitation (MeDIP) with genomic DNA was performed. The genomic DNA was sonicated to fragment using the Covaris M220. Granulosa cell genomic DNA was diluted to 130 μl with TE buffer (10 mM Tris HCl, pH7.5; 1 mM EDTA) and put into a Covaris tube. The Covaris was set to the 300 bp program and 10 μl of each sonicated DNA was run on 1.5% agarose gel to verify fragment size. The remaining DNA was diluted with TE buffer to 400 μl, heat-denatured for 10min at 95°C, then immediately cooled on ice for 10 min. Then 100 μl of 5X IP buffer and 5μg of antibody (monoclonal mouse anti 5-methyl cytidine; Diagenode #C15200006) were added, and the DNA-antibody mixture was incubated overnight with rotation at 4°C.
The following day 50μl of pre-washed anti-mouse magnetic beads (Dynabeads M-280 Sheep anti-Mouse IgG; Life Technologies 11201D) were added to the DNA-antibody mixture, then incubated for 2 h on a rotator at 4°C. The DNA-antibody-bead mixture was placed into a magnetic rack for 1–2 minutes and the supernatant discarded, then the pellet washed with 1x IP buffer 3 times. The washed bead mixture was then resuspended in 250 μl digestion buffer (5 mM Tris PH8, 10 mM EDT4, 0.5% SDS) with 3.5 μl Proteinase K (20 mg/ml) added. The sample was then incubated for 2–3 hours on a rotator at 55°. Buffered Phenol-Chloroform-Isoamyl alcohol solution was added (250 μl) to the sample and the tube, vortexed for 30 sec, then centrifuged at 17,000 x g for 5 min at room temperature. The aqueous supernatant was carefully removed and transferred to a fresh microfuge tube. Then, 250 μl chloroform were added to the supernatant from the previous step, vortexed for 30 sec and centrifuged at 17,000 x g for 5 min at room temperature. The aqueous supernatant was removed and transferred to a fresh microfuge tube. To the supernatant 2 μl of Glycoblue (20 mg/ml) (Invitrogen AM9516), 20 μl of 5 M NaCl and 500 μl 100% ethanol were added and mixed well, then precipitated at −20°C for > 1 hour.
The DNA precipitate was centrifuged at 17,000 x g for 20 min at 4°C and the supernatant removed. The pellet was washed with 500 μl cold 70% ethanol and incubated at −20°C for 15 min. then centrifuged again at 17,000 x g for 5 min at 4°C and the supernatant discarded. The pellet was air-dried at room temperature (about 5 min), then resuspended in 20 μl H2O or TE. DNA concentration was measured using a Qubit (Life Technologies) with ssDNA kit (Molecular Probes Q10212).
MeDIP-seq analysis
The MeDIP DNA was used to create libraries for next generation sequencing (NGS) using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB #E7530S) (San Diego, CA) starting at step 1.4 of the manufacturer’s protocol to generate double stranded DNA. After this step the manufacturer’s protocol was followed. Each pool or individual sample received a separate index primer. NGS was performed at WSU Spokane Genomics Core using the Illumina HiSeq 2500 with a PE50 application, with a read size of approximately 50 bp and approximately 45 million reads per pool. Five to six libraries were run in one lane.
RNA isolation and sequencing
Granulosa cell pellets were stored in 1.2 ml of Trizol reagent (Thermo Fisher) at – 80°C until use. Total RNA was extracted using Trizol reagent following the manufacturer’s protocol with one exception: during RNA precipitation, 1 ml of isopropanol was added to recover small RNAs. RNA was eluted in 50 μL of water and 0.5 μl murine RNase inhibitor (NEB) was added. The final RNA concentration was determined using the Qubit RNA High Sensitivity Assay Kit (Thermo Fisher), and quality control analysis was performed using an RNA 6000 Pico chip on the Agilent 2100 Bioanalyzer.
Large RNA libraries (noncoding and messenger RNA) were constructed using the KAPA RNA HyperPrep kit with RiboErase according to the manufacturer’s instructions with some modifications. NEBNext Multiplex Oligos for Illumina was used for the adaptor and barcodes. Libraries were incubated at 37°C for 15 minutes with the USER enzyme (NEB) before the final amplification. qPCR was used to determine cycle number with the KAPA RealTime Library Amplification Kit. Size selection (200–700 bp) was done using KAPA Pure beads. Quality control analysis was done with the Agilent DNA High Sensitivity chip and final concentration was determined with the Qubit dsDNA high sensitivity assay. Pooled libraries+ were sequenced with paired-end 100 bp sequencing on the Illumina HiSeq 4000 sequencer.
Small RNA libraries were constructed with the NEBNext Multiplex Small RNA Library Prep Set for Illumina and were barcoded with NEBNext Multiplex Oligos for Illumina. Purification and size selection were done with the KAPA Pure beads following the protocol. An additional size selection (115–160 bp) was performed using the Pippin Prep 3% gel with marker P (Sage Science). Concentration was determined using the Qubit dsDNA high sensitivity assay (Thermo Fisher) and quality control was done with Agilent DNA High Sensitivity Chip. Libraries were pooled and concentrated using KAPA Pure beads (2.2X), and sequenced with a custom sequencing primer: 5ʹ-ACA CGT TCA GAG TTC TAC AGT CCG A-3ʹ on the Illumina HiSeq 4000 sequencer (single-end 50 bp).
DMR statistics and bioinformatics
The basic read quality was verified using summaries produced by the FastQC program. The new data was cleaned and filtered to remove adapters and low-quality bases using Trimmomatic [61]. The reads for each MeDIP sample were mapped to the Rnor 6.0 rat genome using Bowtie2 [62] with default parameter options. The mapped read files were then converted to sorted BAM files using SAMtools [63]. To identify DMRs, the reference genome was broken into 100 bp windows. The MEDIPS R package [64] was used to calculate differential coverage between control and exposure sample groups. The edgeR P value [65] was used to determine the relative difference between the two groups for each genomic window. Windows with an edgeR P value less than an arbitrarily selected threshold were considered DMRs. The DMR edges were extended until no genomic window with an edgeR p-value less than 0.1 remained within 1000 bp of the DMR. CpG density and other information was then calculated for the DMR based on the reference genome. DMR clusters were identified as previously described [66].
DMRs were annotated using the biomaRt R package [67] to access the Ensembl database [68]. The genes that overlapped with DMR were then input into the KEGG pathway search [69,70] to identify associated pathways. The DMR associated genes were then sorted into functional groups by consulting information provided by the DAVID [71], Panther [72], and Uniprot databases incorporated into an internal curated database (www.skinner.wsu.edu under genomic data).
All molecular data has been deposited into the public database at NCBI (GEO # GSE118381 and SRA # PRJNA472849) and R code computational tools available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.
Funding Statement
This work was supported by the NIH NIEHS under Grant ES012974.
Acknowledgments
We acknowledge Ms. Jayleana Barton, Ms. Hannah Kimbel and Mr. Hayden McSwiggin for technical assistance and Ms. Heather Johnson for assistance in preparation of the manuscript. We thank the Genomics Core laboratory at WSU Spokane.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementry material
Supplemental data for this article can be accessed here.
References
- 1.Maclaran K, Panay N.. Premature ovarian failure. J Fam Plann Reprod Health Care. 2011;37(1):35–42. [DOI] [PubMed] [Google Scholar]
- 2.Luborsky JL, Meyer P, Sowers MF, et al. Premature menopause in a multi-ethnic population study of the menopause transition. Hum Reprod. 2003;18(1):199–206. [DOI] [PubMed] [Google Scholar]
- 3.Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med. 2009;360(6):606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azziz R, Woods KS, Reyna R, et al. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89(6):2745–2749. [DOI] [PubMed] [Google Scholar]
- 5.March WA, Moore VM, Willson KJ, et al. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum Reprod. 2010;25(2):544–551. [DOI] [PubMed] [Google Scholar]
- 6.Bozdag G, Mumusoglu S, Zengin D, et al. The prevalence and phenotypic features of polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod. 2016;31(12):2841–2855. [DOI] [PubMed] [Google Scholar]
- 7.Rotterdam group Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group, Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod. 2004;19(1):41–47. [DOI] [PubMed] [Google Scholar]
- 8.Lizneva D, Suturina L, Walker W, et al. Criteria, prevalence, and phenotypes of polycystic ovary syndrome. Fertil Steril. 2016;106(1):6–15. [DOI] [PubMed] [Google Scholar]
- 9.Glintborg D, Andersen M. An update on the pathogenesis, inflammation, and metabolism in hirsutism and polycystic ovary syndrome. Gynecol Endocrinol. 2010;26(4):281–296. [DOI] [PubMed] [Google Scholar]
- 10.De Leo V, Musacchio MC, Cappelli V, et al. Genetic, hormonal and metabolic aspects of PCOS: an update. Reprod Biol Endocrinol. 2016;14(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welt CK, Duran JM. Genetics of polycystic ovary syndrome. Semin Reprod Med. 2014;32(3):177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol. 2014;398(1–2):4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anway MD, Skinner MK. Epigenetic programming of the germ line: effects of endocrine disruptors on the development of transgenerational disease. Reprod Biomed Online. 2008;16(1):23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson E, Larsen G, Manikkam M, et al. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS One. 2012;7(5):1–18, e36129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skinner MK, Manikkam M, Tracey R, et al. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med. 2013;11:228, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rechavi O, Houri-Ze’evi L, Anava S, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158(2):277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gapp K, Jawaid A, Sarkies P, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan W. Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol Cell Endocrinol. 2014;398(1–2):24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. [DOI] [PubMed] [Google Scholar]
- 21.Krawetz SA, Kruger A, Lalancette C, et al. A survey of small RNAs in human sperm. Hum Reprod. 2011;26(12):3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Gao F, Fu J, et al. Systematic identification and characterization of long non-coding RNAs in mouse mature sperm. PLoS One. 2017;12(3):e0173402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skinner MK, Ben Maamar M, Sadler-Riggleman I, et al. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin. 2018;11(1):8, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben Maamar M, Sadler-Riggleman I, Beck D, et al. Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate Vinclozolin induced epigenetic transgenerational inheritance of disease. Environ Epigenetics. 2018;4(2):1–19, dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manikkam M, Tracey R, Guerrero-Bosagna C, et al. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One. 2012;7(9):1–15, e46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manikkam M, Tracey R, Guerrero-Bosagna C, et al. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 2013;8(1):1–18, e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manikkam M, Tracey R, Guerrero-Bosagna C, et al. Pesticide and insect repellent mixture (Permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod Toxicol. 2012;34(4):708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tracey R, Manikkam M, Guerrero-Bosagna C, et al. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod Toxicol. 2013;36:104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manikkam M, Haque MM, Guerrero-Bosagna C, et al. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult onset disease through the female germline. PLoS One. 2014;9(7):1–19, e102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King SE, McBirney M, Beck D, et al. Sperm epimutation biomarkers of obesity and pathologies following DDT induced epigenetic transgenerational inheritance of disease. Environmental Health. In review 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson E, King SE, McBirney M, et al. Vinclozolin induced epigenetic transgenerational inheritance of pathologies and sperm epimutation biomarkers for specific diseases. PLoS One. 2018;13(8):e0202662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skinner MK, Guerrero-Bosagna C. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics. 2014;15(1):692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skinner MK. Regulation of primordial follicle assembly and development. Hum Reprod Update. 2005;11(5):461–471. [DOI] [PubMed] [Google Scholar]
- 34.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortuno C, Labarta E. Genetics of primary ovarian insufficiency: a review. J Assist Reprod Genet. 2014;31(12):1573–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greene AD, Patounakis G, Segars JH. Genetic associations with diminished ovarian reserve: a systematic review of the literature. J Assist Reprod Genet. 2014;31(8):935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossetti R, Ferrari I, Bonomi M, et al. Genetics of primary ovarian insufficiency. Clin Genet. 2017;91(2):183–198. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen FC, van Overeem Hansen T, Sorensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16(9):599–612. [DOI] [PubMed] [Google Scholar]
- 39.Vichinsartvichai P. Primary ovarian insufficiency associated with autosomal abnormalities: from chromosome to genome-wide and beyond. Menopause. 2016;23(7):806–815. [DOI] [PubMed] [Google Scholar]
- 40.Guo Y, Sun J, Lai D. Role of microRNAs in premature ovarian insufficiency. Reprod Biol Endocrinol. 2017;15(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Lu Z, Unruh AK, et al. Clinically relevant microRNAs in ovarian cancer. Mol Cancer Res. 2015;13(3):393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manikkam M, Guerrero-Bosagna C, Tracey R, et al. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One. 2012;7(2):1–12, e31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson E, Sadler-Riggleman I, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet. 2018;4(2):1–13, dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilsson EE, Skinner MK. Kit ligand and basic fibroblast growth factor interactions in the induction of ovarian primordial to primary follicle transition. Mol Cell Endocrinol. 2004;214(1–2):19–25. [DOI] [PubMed] [Google Scholar]
- 45.Nilsson EE, Skinner MK. Growth and differentiation factor-9 stimulates progression of early primary but not primordial rat ovarian follicle development. Biol Reprod. 2002;67(3):1018–1024. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson EE, Detzel C, Skinner MK. Platelet-derived growth factor modulates the primordial to primary follicle transition. Reproduction. 2006;131(6):1007–1015. [DOI] [PubMed] [Google Scholar]
- 47.Paulini F, Melo EO. The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis. Reprod Domest Anim. 2011;46(2):354–361. [DOI] [PubMed] [Google Scholar]
- 48.Patino LC, Walton KL, Mueller TD, et al. BMP15 mutations associated with primary ovarian insufficiency reduce expression, activity, or synergy with GDF9. J Clin Endocrinol Metab. 2017;102(3):1009–1019. [DOI] [PubMed] [Google Scholar]
- 49.Wei LN, Huang R, Li LL, et al. Reduced and delayed expression of GDF9 and BMP15 in ovarian tissues from women with polycystic ovary syndrome. J Assist Reprod Genet. 2014;31(11):1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Y, Jiao X, Simpson JL, et al. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21(6):787–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsuiko O, Noukas M, Zilina O, et al. Copy number variation analysis detects novel candidate genes involved in follicular growth and oocyte maturation in a cohort of premature ovarian failure cases. Hum Reprod. 2016;31(8):1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong Z, Li F, Li Y, et al. Inhibition of microRNA-19b promotes ovarian granulosa cell proliferation by targeting IGF-1 in polycystic ovary syndrome. Mol Med Rep. 2018;17(4):4889–4898. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Liu Q, Li Y, Feng Y, et al. Single-cell analysis of differences in transcriptomic profiles of oocytes and cumulus cells at GV, MI, MII stages from PCOS patients. Sci Rep. 2016;6:39638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Wang L, Zhao Y, et al. Evaluation of the association between GHR exon 3 polymorphism and polycystic ovary syndrome among Han Chinese women. Growth Horm IGF Res. 2011;21(5):248–251. [DOI] [PubMed] [Google Scholar]
- 55.Papagerakis S, Shabana AH, Depondt J, et al. Immunohistochemical localization of plakophilins (PKP1, PKP2, PKP3, and p0071) in primary oropharyngeal tumors: correlation with clinical parameters. Hum Pathol. 2003;34(6):565–572. [DOI] [PubMed] [Google Scholar]
- 56.Fernandes R, Tsuda C, Perumalsamy AL, et al. NLRP5 mediates mitochondrial function in mouse oocytes and embryos. Biol Reprod. 2012;86(5):138, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan S, Wen J, Cheng J, et al. Age-associated up-regulation of EGR1 promotes granulosa cell apoptosis during follicle atresia in mice through the NF-kappaB pathway. Cell Cycle. 2016;15(21):2895–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nilsson EE, Anway MD, Stanfield J, et al. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction. 2008;135(5):713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meredith S, Dudenhoeffer G, Jackson K. Classification of small type B/C follicles as primordial follicles in mature rats. J Reprod Fertil. 2000;119(1):43–48. [DOI] [PubMed] [Google Scholar]
- 60.Peters CA, Maizels ET, Hunzicker-Dunn M. Activation of PKC delta in the rat corpus luteum during pregnancy. Potential role of prolactin signaling. J Biol Chem. 1999;274(52):37499–37505. [DOI] [PubMed] [Google Scholar]
- 61.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Handsaker B, Wysoker A, et al. Genome project data processing, the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lienhard M, Grimm C, Morkel M, et al. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30(2):284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haque MM, Nilsson EE, Holder LB, et al. Genomic Clustering of differential DNA methylated regions (epimutations) associated with the epigenetic transgenerational inheritance of disease and phenotypic variation. BMC Genomics. 2016;17:418, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durinck S, Spellman PT, Birney E, et al. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cunningham F, Amode MR, Barrell D, et al. Ensembl 2015. Nucleic Acids Res. 2015;43(Database issue):D662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 72.Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


