ABSTRACT
Circular RNA (circRNA) is shown to participate in various tumors, including lung cancer. Although a few circRNAs involved in lung cancer are reported, whether circRNA negatively regulates lung cancer development remains elusive. In this study, we showed hsa_circ_100395 expression was decreased in lung cancer tissues. Besides, hsa_circ_100395 level was inversely correlated with TNM stage and metastases in lung cancer and low hsa_circ_100395 expression in patients predicted poor prognosis. Overexpression of hsa_circ_100395 dramatically inhibited lung cancer cell proliferation, arrested cell-cycle progression and reduced cell migration and invasion in vitro. Xenograft experiments showed that hsa_circ_100395 overexpression delayed tumor growth in vivo. Mechanistically, we showed hsa_circ_100395 serves as a sponge for miR-1228 targeting TCF21 in lung cancer. Rescue assays indicated that hsa_circ_100395 regulates lung cancer cell proliferation, migration and invasion through modulating miR-1228/TCF21 pathway. Altogether, our study reveals a novel regulatory loop that hsa_circ_100395/miR-1228/TCF21 axis modulates lung cancer development.
Abbreviations: circRNA: circular RNA; miRNA: microRNA; RNA-FISH: RNA fluorescence in situy bridization; qRT-PCR: Reverse transcription and quantitative real-time PCR.
KEYWORDS: hsa_circ_100395, lung cancer, proliferation, migration, invasion
Introduction
As the most prevalent cancer, lung cancer has become a leading factor of tumor-associated death around the world [1]. Lung cancers consist of non-small cell lung cancer (NSCLC) including adenocarcinoma and squamous cell carcinoma, and small cell carcinoma (SCLC) [2]. Although advancement in lung cancer diagnosis and treatment, the five-year overall survival rate of lung cancer patients remains very low [3]. The outcomes of lung cancer patients with advanced stage or metastasis are quite poor [4]. Nowadays, lung cancer has become the global public health challenge. Thus, understanding the pathogenesis underlying lung cancer is crucial to develop novel therapeutic targets.
As the boom of sequencing technology and bioinformatics methods, increasing noncoding RNAs (ncRNAs) were identified [5,6]. CircRNA is featured with a covalently closed loop with neither a 5’ cap nor a 3’ polyadenylated tail [7]. Recent studies indicate circRNAs are very stable and transcribed in a tissue-specific manner [8]. Accumulating evidences imply that circRNA possesses important functions in tumorigenesis and might serve as biomarkers for disease [9]. For instance, Zhong et al. reported that circC3P1 inhibits liver cancer progression by miR-4641/PCK1 signaling [10]. Ma et al. showed that circMAN2B2 regulates miR-1275/FOXK1 pathway to promote lung cancer growth [11]. Zeng et al. indicated that circHIPK3 interacts with miR-7 to promote colorectal cancer progression [12]. A growing number of reports have illustrated the pivot roles of circRNAs in many biological processes, including cell survival and mobility [13]. Hence, exploring the functions of circRNAs in tumor may benefit for tumor therapy.
A few circRNAs are shown to participate in lung cancer, whether there are some circRNAs exerting tumor-suppressive roles in lung cancer remains elusive. In this study, we identified a novel circRNA termed hsa_circ_100395 (circBase ID: hsa_circ_0015278) that was downregulated in lung cancer tissues. Through inquiring from circBase (http://www.circbase.org), we found that hsa_circ_100395 was located at chromosome 1q25.1 and was composed of four exons of KLHL20 mRNA. We investigated the role and mechanism of hsa_circ_100395 in lung cancer. We demonstrated that hsa_circ_100395 suppressed lung cancer progression via modulating miR-1228/TCF21 signaling pathway, which suggested the hsa_circ_100395/miR-1228/TCF21 axis might be a promising therapeutic target. .
Results
hsa_circ_100395 was downregulated in lung cancer tissues
To screen out key circRNAs involved in lung cancer, we analyzed an online-available microarray dataset (GSE101586), including 5 pairs of lung cancer tissues and normal lung tissues. We found there are several circRNAs that are downregulated or upregulated in all five lung cancer tissues (Figure 1(a)). Among them, hsa_circ_100395 (Table 1) was the most downregulated in lung cancer tissues. To confirm it, we analyzed the levels of hsa_circ_100395 in 69 pairs of lung cancer tissues and non-tumor tissues. The results showed that hsa_circ_100395 was downregulated in lung cancer tissues (Figure 1(b)). We then grouped these sample tissues into different TNM stages and found that hsa_circ_100395 displayed lower expression in tissues of stage III/IV (n = 41) than that in tissues of stage I/II (n = 28) (Figure 1(c)). Besides, hsa_circ_100395 expression was negatively correlated with lung cancer metastases (Figure 1(d)). Moreover, Kaplan-Meier curve analysis indicated that patients with lower level of hsa_circ_100395 possesses poorer prognosis (Figure 1(e)). In sum, these results demonstrated that hsa_circ_100395 was downregulated in lung cancer and might participate in tumor progression.
Figure 1.
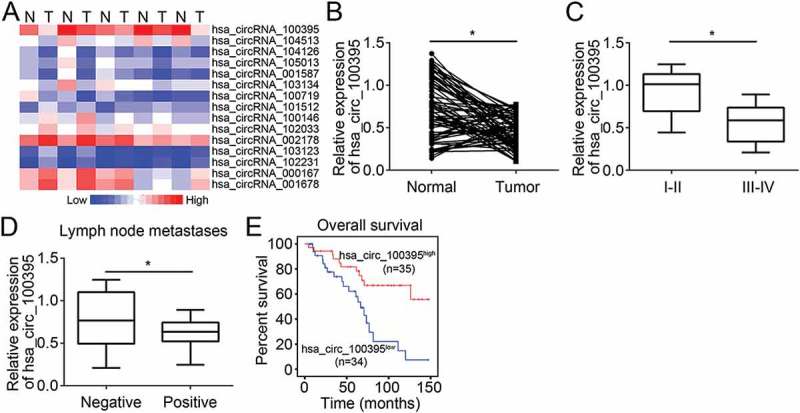
hsa_circ_100395 was downregulated in lung cancer tissues. (a) Heatmap of the most differentially expressed circRNAs in lung cancer tissues from five female lung adenocarcinoma patients with no smoking history compared to matched normal lung tissues according to circRNA microarray dataset (GSE101586). N, normal lung tissues; T; lung tumor tissues. (b) Relative expression of hsa_circ_100395 in 69 pairs of lung cancer tissues and adjacent normal tissues by qRT-PCR. (c) Relative expression of hsa_circ_100395 in different TNM stages of lung cancer tissues by qRT-PCR. (d) Relative expression of hsa_circ_100395 in lung cancer tissues with or without lymph node metastases. (e) Kaplan-Meier curve of overall survival of patients with lung cancers according to hsa_circ_100395 expression (mean value as cut-off). *P < 0.05.
Table 1.
Sequence of hsa_circ_100395.
| UUGGUGGUUGGUGCAGUGGAGAUGCCAUUUCCAGUGUUGAACGAUAUGAUCCACAGACCAAUGAAUGGAGAAUGGUGGCUUCAAUGAGCAAAAGGAGAUG |
| CGGAGUUGGGGUCAGUGUUCUUGAUGAUCUGUUAUAUGCAGUAGGAGGCCAUGAUGGAUC |
| CUCUUAUCUCAAUAGUGUUGAAAGGUAUGACCCCAAAACAAACCAGUGGAGCAGUGAUGUGGCCCCUACA |
| AGCACCUGCAGGACAAGUGUUGGUGUAGCAGUACUUGGAGGCUUUCUUUAUGCUGUGGGUGGCCAG |
| GAUGGUGUGUCUUGCCUCAACAUUGUUGAGAGGUAUGAUCCGAAGGAGAACAAGUGGACUCGGGUA |
| GCUUCUAUGAGUACCAGAAGACUAGGUGUGGCUGUGGCUGUGUUAGGAGGGUUCUUAUAUGCUGUAGG |
| UGGCUCUGACGGGACAUCUCCUCUCAACACAGUGGAACGUUACAAUCCUCAGGAAAACAGAUGGCACACUA |
| UAGCCCCUAUGGGGACCCGGAGGAAACACCUAGGCUGUGCAGUAUAUCAGGACAUGAUCUAUGCU |
| GUAGGAGGUAGAGAUGACACUACAGAGCUGAGCAGU |
| GCUGAGAGAUACAACCCCAGAACCAACCAGUGGUCUCCAGUGGUGGCCAUGACAUCACGCCGUAGUGGA |
Effects of hsa_circ_100395 overexpression on lung cancer cell proliferation, cell-cycle, migration and invasion
To investigate physiological roles of hsa_circ_100395, we first measured the expression of hsa_circ_100395 in lung cancer cell lines. Results showed that hsa_circ_100395 levels were lower in lung cancer cell lines than that in Beas-2B cells (Figure 2(a)). Among all lung cancer cell lines, hsa_circ_100395 expression was the lowest in A549 and H460 cells (Figure 2(a)). Thus, we chose these two cell lines for following experiments. We constructed two stable A549 and H460 cell lines with overexpressed hsa_circ_100395 (Figure 2(b)). Through MTT assay, we found that hsa_circ_100395 overexpression impaired the proliferation of A549 and H460 cells (Figure 2(c)). Decreased proliferation might be caused by aberrant cell-cycle. Therefore, we measured the effect of hsa_circ_100395 on cell-cycle progression. By FACS analysis, we found that hsa_circ_100395 overexpression led to more cells arrested in G0/G1 phase while less cells in S and G2/M phase (Figure 2(d)). We above have observed a negative association between hsa_circ_100395 expression and metastases. To confirm it, we performed transwell assays using A549 and H460 cells. The results showed that hsa_circ_100395 upregulation repressed the migration and invasion of A549 and H460 cells (Figure 2(e and f)).
Figure 2.
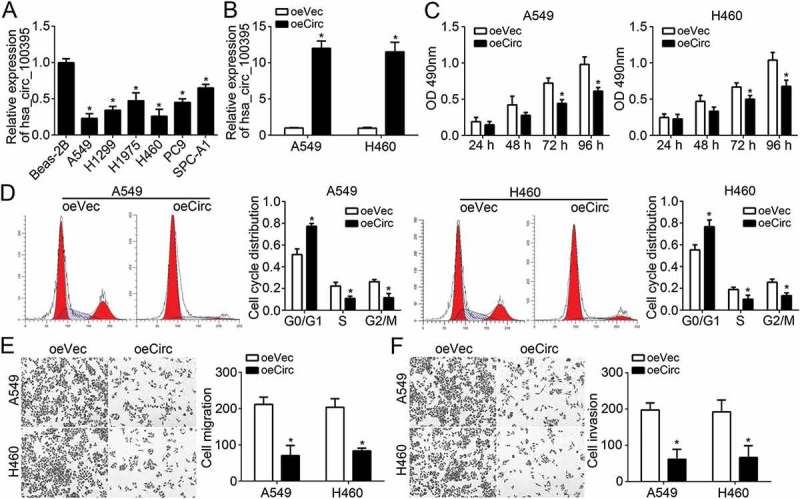
Effects of hsa_circ_100395 overexpression on lung cancer cell proliferation, cell-cycle, migration and invasion. (a) Relative expression of hsa_circ_100395 in different lung cancer cell lines by qRT-PCR. (b) Relative expression of hsa_circ_100395 in A549 and H460 cells transfected with hsa_circ_100395 ectopic expressing vector or empty vector. oeVec, overexpressing vector control, oeCirc, overexpressing hsa_circ_100395. (c) Overexpression of hsa_circ_100395 decreased the proliferation in both A549 and H460 cells. (d) Cell cycle distribution was measured in A549 and H460 cells transfected with hsa_circ_100395 ectopic expressing vector or empty vector. (E and F) Cell migration and invasion was determined by transwell assay in A549 and H460 cells transfected with hsa_circ_100395 ectopic expressing vector or empty vector. *P < 0.05.
hsa_circ_100395 served as a sponge of miR-1228
Circular RNAs often serve as sponges of miRNAs by interacting with RISC complex [14]. In order to search the mechanism of hsa_circ_100395, we analyzed the potential targets of hsa_circ_100395 using miRBase (http://www.mirbase.org/) and circinteractome (https://circinteractome.nia.nih.gov/). We identified miR-1228 as the most possible target of hsa_circ_100395. As shown, miR-1228 expression was increased in lung cancer tissues and negatively correlated with hsa_circ_100395 level (Figure 3(a and b)). To confirm the direct interaction between hsa_circ_100395 and miR-1228, we first conducted RNA pulldown assay with biotin-miR-1228 using A549 or H460 cell lysates. The results demonstrated that hsa_circ_100395 was significantly enriched by biotin-miR-1228 (Figure 3(c)). Then we constructed luciferase reporter plasmids (wild-type hsa_circ_100395 and mutant hsa_circ_100395) for luciferase reporter assays (Figure 3(d)). The results indicated that miR-1228 mimic transfection repressed the activity of WT-hsa_circ_100395 in A549 and H460 cells (Figure 3(e)). Moreover, RNA-FISH assay showed that hsa_circ_100395 was co-localized with miR-1228 in A549 cells (Figure 3(f)). And we also found that overexpression of hsa_circ_100395 significantly inhibited the levels of miR-1228 in A549 and H460 cells (Figure 3(g)). Taken together, these data demonstrated that hsa_circ_100395 acted as a sponge for miR-1228 and inhibited its availability.
Figure 3.
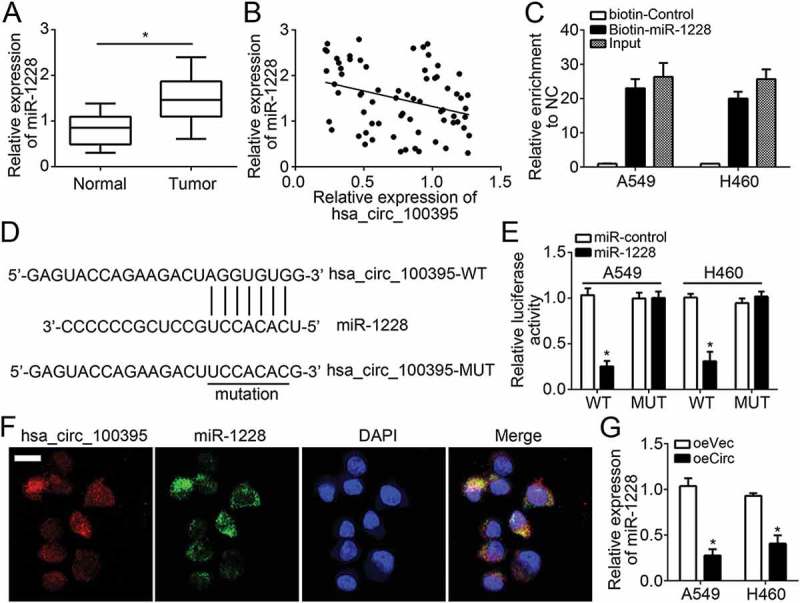
hsa_circ_100395 served as a sponge of miR-1228. (a) Relative expression of miR-1228 in lung cancer tissues and paired adjacent normal tissues. (b) Expression correlation between hsa_circ_100395 and miR-1228 in lung cancer tissues by qRT-PCR. (c) RNA pulldown assay was performed with biotin-miR-1228, control or 10% input using A549 or H460 cell extracts. RNA levels of hsa_circ_100395 in immunoprecipitates were detected by qRT-PCR. (d) The predicted wild-type or mutated miR-1228 binding site in hsa_circ_100395. (e) A549 and H460 cells were transfected with wide-type or mutant hsa_circ_100395 reporter plasmid, and the luciferase reporter was performed to confirm the direct target site. (f) RNA-FISH was utilized to examine the co-localization between hsa_circ_100395 and miR-1228 in A549 cells. (g) Overexpression of hsa_circ_100395 inhibited the levels of miR-1228 in A549 and H460 cells. *P < 0.05.
miR-1228 targeted TCF21 in lung cancer
fIncreasing reports demonstrate miRNAs exert roles via binding to 3ʹ-UTR of target mRNAs [15]. Thus, we looked for the potential target of miR-1228 by using TargetScan and miRanda. TCF21 was identified as a candidate. According to a microarray dataset (GSE19804) and qRT-PCR result, TCF21 expression was downregulated in lung cancer tissues (Figure 4(a and b)). In addition, we observed a negative association between miR-1228 and TCF21 levels in lung cancer tissues (Figure 4(c)). To validate the prediction, we performed RNA pulldown assay and found that biotin-miR-1228 could enrich TCF21 mRNA in A549 and H460 cells (Figure 4(d)), indicating miR-1228 interacts with TCF21 mRNA. To further confirm it, we performed luciferase reporter assay and found that miR-1228 mimics inhibited the activity of WT-TCF21 reporter (Figure 4(e and f)). Moreover, miR-1228 upregulation markedly suppressed TCF21 expression in A549 and H460 cells (Figure 4(g and h)). We have demonstrated hsa_circ_100395 inhibited miR-1228 and miR-1228 targeted TCF21 in lung cancer cells. Then we sought to explore whether hsa_circ_100395 regulates TCF21 expression via inhibiting miR-1228. As shown overexpression of WT hsa_circ_100395 significantly abolished the inhibitory effects of miR-1228 on TCF21 levels (Figure 4(i and j)). In a word, these data suggested that hsa_circ_100395 promoted TCF21 expression by sponging miR-1228.
Figure 4.
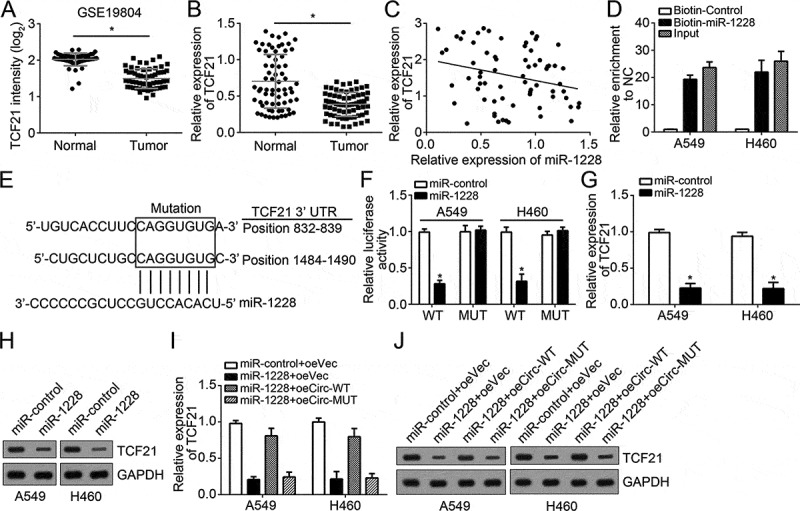
miR-1228 targeted TCF21 in lung cancer. (a) Relative expression of TCF21 in lung cancer tissues and lung normal tissues according to a microarray dataset (GSE19804). (b) Relative expression of TCF21 in pairs of lung cancer tissues and adjacent normal tissues by qRT-PCR. (c) Expression correlation between TCF21 and miR-1228 in lung cancer tissues by qRT-PCR. (d) RNA pulldown assay was performed with biotin-miR-1228, controls or 10% input using A549 or H460 cell extracts. RNA levels of TCF21 in immunoprecipitates were detected by qRT-PCR. (e) The predicted wild-type or mutated miR-1228 binding sites in the 3ʹ-UTR of TCF21 mRNA. (f) A549 and H460 cells were transfected with wide-type or mutant TCF21 reporter plasmid, and the luciferase reporter was performed to confirm the direct target sites. (g and h) Overexpression of miR-1228 inhibited the mRNA and protein levels of TCF21 in A549 and H460 cells by qRT-PCR and western blot. (I and J) Ectopic expression of wide-type hsa_circ_100395 abrogated the inhibitory effects of miR-1228 mimics on TCF21 mRNA and protein levels in A549 and H460 cells. *P < 0.05.
hsa_circ_100395 regulated lung cancer cell proliferation, migration and invasion by modulating miR-1228/TCF21 pathway
To further explore whether hsa_circ_100395 exerts its role by regulating miR-1228/TCF21 pathway, we conducted rescue assays by transfection with miR-1228 inhibitor or TCF21 siRNA in A549 and H460 cells. We first validated the transfection efficiency by analyzing TCF21 protein levels (Figure 5(a)), followed by MMT and transwell assays. As shown, hsa_circ_100395 knockdown increased the proliferation, migration and invasion of A549 and H460 cells (Figure 5(b–d)). However, miR-1228 inhibition suppressed lung cancer cell proliferation, migration and invasion while TCF21 knockdown promoted that of lung cancer cells (Figure 5(b–d)). These data demonstrated that hsa_circ_100395 and TCF21 exerted tumor suppressive roles while miR-1228 served as an oncogene in lung cancer. Moreover, hsa_circ_100395 played its roles by modulating miR-1228/TCF21 signaling pathway.
Figure 5.
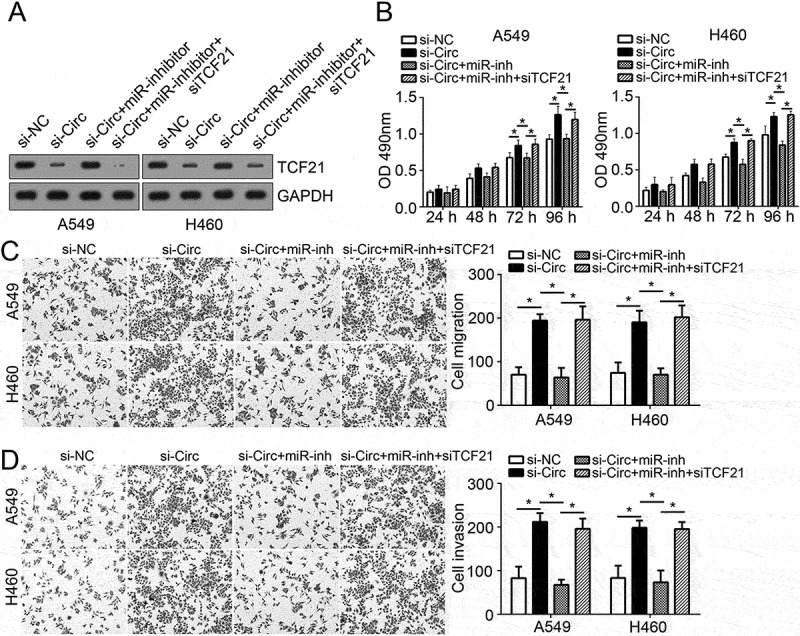
hsa_circ_100395 regulated lung cancer cell proliferation, migration and invasion by modulating miR-1228/TCF21 pathway. (a) Relative protein levels of TCF21 in A549 and H460 cells transfected with indicative plasmids. (b) MTT assay was used to determine cell proliferation in A549 and H460 cells transfected with indicative plasmids. (c and d) Transwell assay was utilized to examine cell migration and invasion in A549 and H460 cells transfected with indicative plasmids. *P < 0.05.
Effects of hsa_circ_100395 overexpression on tumor growth in vivo
We further assessed the effects of hsa_circ_100395 on tumor growth. We injected hsa_circ_100395-overexpressing or control A549 cells into nude recipient. At different time points, we calculated the tumor sizes. The results indicated that hsa_circ_100395 overexpression suppressed lung cancer cell growth in vivo (Figure 6(a)). At the end point, the tumor weights were determined. As shown, hsa_circ_100395 upregulation led to lighter weights (Figure 6(b)). Besides, hsa_circ_100395 levels were still elevated in oehsa_circ_100395-derived tumor tissues while miR-1228 was downregulated (Figure 6(c)). Moreover, hsa_circ_100395 overexpression reduced the expression of TCF21, Cyclin D1 and Ki67 in tumor cells (Figure 6(d)), indicating hsa_circ_100395 suppressed tumor cell proliferation via TCF21.
Figure 6.
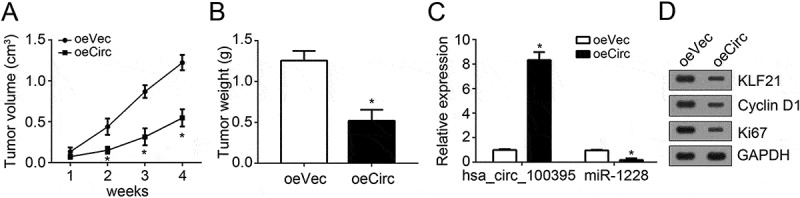
Effects of hsa_circ_100395 overexpression on tumor growth in vivo. (a) 2 × 106 conditional A549 cells (hsa_circ_100395 overexpression or control) were subcutaneously injected in rear flank of nude mice (5 per group). Then tumor volumes were calculated at indicative time points. (b) The expression of hsa_circ_100395 and miR-1228 in the resected tumors was examined by qRT-PCR. (c) Protein levels of TCF21, Cyclin D1 and Ki67 were examined in the resected tumors by western blot. (d) Tumors weights were measured after 4 weeks of injection. *P < 0.05.
Discussion
CircRNAs are critical regulators in various physiological and pathological processe [16]. Increasing evidences indicate that circRNA expression is closely correlated with clinical characteristics in patients and its aberrant expression often contributes to malignant behaviors, such as proliferation and metastasis [17]. Hence, it is crucial to determine the relationship between circRNA and tumor. In this study, we identified a new circRNA termed hsa_circ_100395 and demonstrate its tumor suppressor role in lung cancer.
dysregulation of circRNA expression is associated with the malignant behaviors in various cancers, including lung cancer [11], hepatocellular carcinoma [10], breast cancer [18], colorectal cancer [12], melanoma [19] and bladder cancer [20]. Many studies indicated circRNA is helpful markers in tumor diagnosis and prognosis [21]. Our results showed that hsa_circ_100395 expression is remarkably downregulated in lung cancer tissues and correlates with TNM stage and lymphoid node metastases. Our data also implied that higher expression of hsa_circ_100395 predicted better prognosis in lung cancers. Patients with higher TNM stages showed a worse prognosis [22]. Metastases is the most important cause of patients’ poor outcomes [23]. Thus, hsa_circ_100395 might be a potential biomarker for lung cancer diagnosis and prognosis.
CircRNAs have been shown to participate in various pathologic processes, including apoptosis, proliferation and invasion [24]. For instance, hsa_circ_0001649 knockdown modulates cholangiocarcinoma malignant behaviors [25]. Hsa_circ_0001564 is involved in tumor progression in osteosarcoma [26]. Additionally, circular RNA_LARP4 sponges miR-424-5p to suppress gastric cancer growth and metastasis [27]. In our study, we found that hsa_circ_100395 suppressed lung cancer cell proliferation, arrested cell-cycle progression, and reduced cell migration and invasion in vitro. Moreover, we showed that hsa_circ_100395 inhibited lung cancer growth in vivo. Our work revealed hsa_circ_100395 as a tumor suppressor for lung cancer. Notably, overexpression of hsa_circ_100395 did not affect the expression of its linear form mRNA (KLHL20) (Supplementary Fig. 1A and B). Thus, hsa_circ_100395 shows an independent effect on lung cancer. Additionally, we screened out hsa_circ_100395 according to the online dataset (GSE101586) which only contains five pairs of lung cancer tissues and normal tissues. The sample size is limited. Thus, we may omit other circRNAs that exert more important role in lung cancer.
Circular RNAs often serve as sponges of miRNAs [14]. To explore the functional mechanism of hsa_circ_100395, we search the potential target miRNA. We identified miR-1228. Through luciferase reporter assay, RNA pulldown and RNA-FISH, we validated the association between hsa_circ_100395 and miR-1228 in lung cancer cells. Previous studies indicated that miR-1228 promotes cancer development. For instance, Lin et al. reported that miR-1228 targets SCAI to facilitate breast cancer cell proliferation and invasion [28]. Zhang et al. showed that miR-1228 promotes hepatoma progression. However, the role of miR-1228 in lung cancer has not been illustrated [29]. We demonstrated that miR-1228 level was dramatically elevated in lung cancer tissues and also served an oncogene. Although we analyzed the association between hsa_circ_100395 and ten predicted target miRNAs (Data not shown), we only found that hsa_circ_100395 interacts with miR-1228 (Figure 3(e)). Whether other miRNAs are related to hsa_circ_100395 needs further investigation.
More and more studies demonstrate miRNAs exert roles via binding to 3ʹ-UTR of target mRNAs [15]. We showed miR-1228 might target TCF21 through bioinformatics analysis. Through a series of experiments, we also validated their direct interaction. Moreover, we showed hsa_circ_100395 promoted TCF21 expression via sponging miR-1228 in lung cancer. TCF21 has been acknowledged as a tumor suppressor in renal tumor [30], ovarian cancer [31], colorectal cancer [32] and lung cancer [33]. However, how TCF21 expression was regulated in lung cancer remains elusive. Our study indicated that TCF21 was underexpressed in lung cancer tissues and also played anti-tumor roles. Moreover, our results showed that TCF21 expression was regulated by hsa_circ_100395/miR-1228 axis in lung cancer.
In conclusion, our study for the first time demonstrated that hsa_circ_100395 inhibited lung cancer malignant behaviors via modulating miR-1228/TCF21 axis, which provided a novel potential mechanism involved in lung cancer progression.
Material and methods
Patient samples
69 pairs of lung cancer and non-tumor tissues were collected from Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University. All patients did not undergo radiotherapy or chemotherapy before surgery. Association of hsa_circ_100395 expression with clinicopathological features of lung cancer patients was in Table 2. Our study was approved by the Ethnic Committee of Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University. Informed consent was obtained from each patient.
Table 2.
Association of hsa_circ_100395 expression with clinicopathological features of lung cancer patients.
| Clinicopathological feature | Lower (n = 34) | Higher (n = 35) | P-value |
|---|---|---|---|
| Gender | 0.450 | ||
| Male | 21 | 25 | |
| Female | 13 | 10 | |
| Age(year) | 0.319 | ||
| ≤60 | 24 | 20 | |
| >60 | 10 | 15 | |
| Lymph node metastasis | 0.002* | ||
| Negative | 12 | 26 | |
| Positive | 22 | 9 | |
| Tumor size | 0.628 | ||
| ≤3 cm | 19 | 22 | |
| > 3 cm | 15 | 13 | |
| TNM stage | 0.027* | ||
| I/II | 9 | 19 | |
| III/IV | 25 | 16 |
*Indicated statistical significance (P < .05).
Cell culture and transfection
Lung cancer cell lines (A549, H460, PC9, H1299, H1975 and SPC-A1) were from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) containing 10% FBS (Gibco, USA), 100 mg/mL streptomycin and 100 IU/mL penicillin at 37 °C in a humidified 5% CO2 incubator. The immortalized human bronchial epithelium cells Beas-2B from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) were cultured according to the standard protocols.
sihsa_circ_100395 (5’-ATCACGCCGTAGTGGATTGGT-3’), siTCF21 (5’-GTGCTCTCTGTCTCTGCTT-3’), miR-1228 mimics, miR-1228 inhibitors and their respective controls were purchased from Gene-Pharma (Shanghai, China). Transfection (50 nM of miRNA mimics, inhibitors and siRNAs) were performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. To construct stable hsa_circ_100395 overexpression cell lines, hsa_circ_100395 coding sequence was constructed into pcD-ciR vector (Geenseed Biotech, Guangzhou, China). Then, A549 and H460 cells were transfected and screened using G418.
MTT assay
3 × 103 cells per well were seeded in 96-well plates. After incubation, 20 μl MTT solution (5 mg/ml; Sigma, St. Louis, Mo, USA) was added into cells and incubated for 4 hours. Then 200 μl DMSO (Sigma, St. Louis, MO, USA) was added and absorbance at 490 nm was measured using a microplate reader (Bio Tek Instruments, Inc., Winooski, VT, USA).
Transwell assay
Cell migration and invasion was measured using transwell chamber (Costar, Massachusetts, USA) according to the manufacturer’s protocol.
RNA pulldown
RNAs were labeled with biotin using Pierce RNA 3ʹ End Desthiobiotinylation Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols. Then biotin-labeled wild-type miR-1228 and negative control (mutant miR-1228) were incubated with cell lysates, as well as Magnetic beads. After incubation for 6 hours, the precipitated RNAs on beads were eluted and extracted, followed by qRT-PCR examination.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNAs were extracted using TRIzol solution (Invitrogen) from tumor cells according to manufacturer’s protocol. 500 ng RNAs were reversely transcribed into cDNAs using the Prime Script RT Master Mix. qRT-PCR analysis was conducted utilizing the SYBR Select Master Mix (Applied Biosystems) on ABI73900 system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocols. Expression values were normalized to GAPDH6 or U6, and calculated using 2−ΔΔCT method.
RNA fluorescence in situ hybridization (RNA-FISH)
RNA-FISH was performed according to a previous study [34].
Luciferase reporter assay
hsa_circ_100395 sequence and TCF21 3′UTR containing wide-type (WT) or mutant-type (Mut) binding site for miR-1228 were constructed into pGL3 luciferase vector (Promega, Madison, WI, USA) to obtain hsa_circ_100395-WT, hsa_circ_100395-Mut and TCF21-3′UTR-WT (WT), TCF21-3′UTR-Mut (Mut). Then reporter plasmid and miR-1228 mimics were transduced into lung cancer cells along with pRL-TK vector (Promega). Luciferase activity was measured according to the dual-luciferase Reporter assay system (Promega) 24 hours after transfection.
In vivo assay
Animal experiments were approved by the Animal Care and Use Committees at Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University. 2 × 106 A549 cells in 100 μl PBS were injected into the right flank of 5-weeks old female BALB/c nude mice. Tumor volumes and weights were measured as previously described [35].
Statistical analysis
SPSS 19.0 software and GraphPad Prism 6 were used for statistical analysis. All results were expressed as mean ± SD. Student’s t-test or one-way ANOVA analysis was utilized to calculate significant difference. Kaplan-Meier curve was used for overall survival analysis and log rank test was used for determining significant difference. Chi-square test was utilized to analyze association between hsa_circ_100395 expression and clinical features. Spearman’s rank analysis was used to identify the correlation between hsa_circ_100395 and miR-1228, or between TCF21 and miR-1228. P < 0.05 represented statistically significant.
Funding Statement
This work was supported by the 2nd Clinical Medicine College (Shenzhen People’s Hospital) of Jinan University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental Material
Supplemental data for this article can be accessed here.
References
- [1].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- [2].She KL, Yan H, Huang J, et al. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wu HY, Zhou CC.. Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem Biophys Res Commun. 2018;496:738–745. [DOI] [PubMed] [Google Scholar]
- [4].Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. [DOI] [PubMed] [Google Scholar]
- [5].Liu BY, Ye BQ, Yang LL, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. [DOI] [PubMed] [Google Scholar]
- [6].Chen BJ, Byrne FL, Takenaka K, et al. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2018;9:5786–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- [10].Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499(4):1044–1049. [DOI] [PubMed] [Google Scholar]
- [11].Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498:1009–1015. [DOI] [PubMed] [Google Scholar]
- [12].Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [13].Huang WJ, Wang Y, Liu S, et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70–80. [DOI] [PubMed] [Google Scholar]
- [14].Wang H, Xiao Y, Wu L, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kadmon CS, Landers CT, Li HYS, et al. MicroRNA-22 controls interferon alpha production and erythroid maturation in response to infectious stress in mice. Exp Hematol. 2017;56:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242:1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou J, Zhang WW, Peng F, et al. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang Q, Chen J, Wang A, et al. Differentially expressed circRNAs in melanocytes and melanoma cells and their effect on cell proliferation and invasion. Oncol Rep. 2018;39:1813–1824. [DOI] [PubMed] [Google Scholar]
- [20].Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li WH, Song YC, Zhang H, et al. Decreased expression of hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Luo YH, Zhu XZ, Huang KW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. [DOI] [PubMed] [Google Scholar]
- [23].Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. [DOI] [PubMed] [Google Scholar]
- [24].Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–461. [DOI] [PubMed] [Google Scholar]
- [26].Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369–2375. [DOI] [PubMed] [Google Scholar]
- [27].Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lin L, Liu D, Liang H, et al. MiR-1228 promotes breast cancer cell growth and metastasis through targeting SCAI protein. Int J Clin Exp Pathol. 2015;8:6646–6655. [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang Y, Dai J, Deng H, et al. miR-1228 promotes the proliferation and metastasis of hepatoma cells through a p53 forward feedback loop. Br J Cancer. 2015;112:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gooskens SL, Klasson TD, Gremmels H, et al. TCF21 hypermethylation regulates renal tumor cell clonogenic proliferation and migration. Mol Oncol. 2018;12:166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wei J, Zhang L, Li J, et al. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J Ovarian Res. 2017;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dai Y, Duan H, Duan C, et al. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration and invasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469:430–436. [DOI] [PubMed] [Google Scholar]
- [33].Wu H, Zhou J, Zeng C, et al. Curcumin increases exosomal TCF21 thus suppressing exosome-induced lung cancer. Oncotarget. 2016;7:87081–87090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li A, Peng R, Sun Y, et al. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1alpha signaling. Cell Death Dis. 2018;9:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang G, Jiang H, Lin Y, et al. lncAKHE enhances cell growth and migration in hepatocellular carcinoma via activation of NOTCH2 signaling. Cell Death Dis. 2018;9:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


